ABSTRACT
Mesenchymal stem cells (MSC) and induced pluripotent stem cells (iPSC) have been reported to be able to differentiate to hepatocyte in vitro with varying degree of hepatocyte maturation. A simple method to decellularize liver scaffold has been established by the Department of Histology, Faculty of Medicine, Universitas Indonesia, in SCTE IMERI lab.15 This study aims to evaluate hepatocyte differentiation from iPSCs compared to MSCs derived in our decellularized liver scaffold. The research stages started with iPSC culture, decellularization, seeding cell culture into the scaffold, and differentiation into hepatocytes for 21 days. Hepatocyte differentiation from iPSCs and MSCs in the scaffolds was characterized using hematoxylin–eosin, Masson Trichrome, and immunohistochemistry staining to determine the fraction of the differentiation area. RNA samples were isolated on days 7 and 21. Expression of albumin, CYP450, and CK-19 genes were analyzed using the qRT-PCR method. Electron microscopy images were obtained by SEM. Immunofluorescence examination was done using HNF4-α and CEBPA markers. The results of this study in hepatocyte-differentiated iPSCs compared with hepatocyte-differentiated MSCs in decellularized liver scaffold showed lower adhesion capacity, single-cell-formation and adhered less abundant, decreased trends of albumin, and lower CYP450 expression. Several factors contribute to this result: lower initial seeding number, which causes only a few iPSCs to attach to certain parts of decellularized liver scaffold, and manual syringe injection for recellularization, which abruptly and unevenly creates pattern of single-cell-formation by hepatocyte-differentiated iPSC in the scaffold. Hepatocyte-differentiated MSCs have the advantage of higher adhesion capacity to collagen fiber decellularized liver scaffold. This leads to positive result: increase trends of albumin and higher CYP450 expression. Hepatocyte maturation is shown by diminishing CK-19, which is more prominent in hepatocyte-differentiated iPSCs in decellularized liver scaffold. Confirmation of mature hepatocyte-differentiated iPSCs in decellularized liver scaffold maturation is positive for HNF4-a and CEBPA. The conclusion of this study is hepatocyte-differentiated iPSCs in decellularized liver scaffold is mature with lower cell–ECM adhesion, spatial cell distribution, albumin, and CYP450 expression than hepatocyte-differentiated MSCs in decellularized liver scaffold.
Introduction
Liver transplantation is the mainstream medical therapy for patients with end-stage liver disease.Citation1 Significant restriction in liver transplantation is the limited number of available and suitable living liver donors. Liver transplant failure could be due to acute or chronic rejection. Standard post-transplant regiment to overcome rejection is the long-term use of immunosuppressive drugs. This increases the risk of severe viral or fungal infection in addition to malignancy. In brief, hepatocyte replacement therapy uses isolated hepatocytes of liver resected from a relative. They are expanded in vitro before being infused to the patient. This procedure has been shown to reduce immunosuppressive drug use and related complication.Citation2
The problem with hepatocyte replacement therapy is the low hepatocytes yield due to cell proliferation arrest in culture. Stem cells have higher capacity to meet the expected number. Mesenchymal stem cells (MSC) and induced pluripotent stem cells (iPSC) are able to differentiate to hepatocyte in vitro with varying degree of hepatocyte maturation.Citation3–10
In a study conducted by Blackford et al.,Citation11 validation of current Good Manufacturing Practice (cGMP) in vitro hepatocytes differentiation from iPSCs using 2D monolayer culture or 3D culture with with differentiation induction medium. The medium comprises basal medium, cytokine, growth factors, and small molecules. It is switched periodically to recapitulate stages of in vivo hepatocyte development. The first differentiation stage starts from pluripotent or multipotent stem cells, which become definitive endodermal cells by using Activin A/transforming growth factor β (TGF-β). At this stage, increase in Wnt/β-catenin signaling pathway is important. The second stage occurs when the definitive endoderm differentiates into hepatoblast by using bone morphogenetic protein 4 (BMP4) and fibroblast growth factor (FGF) to induce the expression of specific liver genes. The third stage is differentiation from hepatoblasts to hepatocytes using hepatocyte growth factor (HGF), oncostatin M (OSM), dexamethasone, and/or 3D reaggregation with other cell types to specify their fate as hepatocytes.Citation12,Citation13
A simple method to decellularize liver scaffold has been established and characterized by the Department of Histology, Faculty of Medicine, Universitas Indonesia, in SCTE IMERI lab.Citation14,Citation15 This study aims to compare the profile of hepatocyte differentiation from iPSCs or MSCs in decellularized liver scaffold. The research was conducted from January 2020 to June 2021 at the Stem Cell and Tissue Engineering (SCTE) laboratory of IMERI FKUI, Histology laboratory, Center laboratory for Materials and Processing Failure Analysis (CMPFA) Metallurgical Engineering FTUI, Molecular Laboratory Biology and Proteomic Core Facilities (MBPCF) IMERI FKUI, and the Human Genetic Research Center (HGRC) laboratory of IMERI FKUI.
Materials and methods
Materials
MSC cell line from human umbilical cord Wharton jelly was developed in SCTE IMERI FKUI laboratory. Complete MSC medium included 10% PRP, 1% heparin (1000 U/ml), 1% amphotericin B (250ug/ml), 1% pen-strep (10.000 U/ml penicillin and 1000 ug/ml streptomycin), and αMEM.
iPSC cell line from human bone marrow mesenchymal stem cells was purchased from EBiSC, UK, catalog number UKKi006-A. Complete iPSC medium included Essential 8 medium and supplement 50X and plate coated with Vitronectin Recombinant Human Protein GibcoTM.S. Harvest reagents included Dulbecco’s phosphate-buffered saline (DPBS), ethylenediaminetetraacetic acid (EDTA), and rho-associated protein kinase inhibitor (ROCK inhibitor).
Liver scaffold decellularization included sodium dodecyl sulfate (SDS) 1st BASE, Triton X-100 10%, EGTA, aquabides, NaCl 0.9%, and NaOH.
Hepatocyte differentiation induction medium included RPMI-1640, human serum albumin (HSA) 10%, Abam 1%, Glutamax 1%, Hepatozyme, and fetal bovine serum (FBS) 10%. Small molecules and growth factor for hepatocyte differentiation included 1.5 μM CHIR9901, 5 ng/ml bone morphogenetic protein 4 (BMP4), 5 μM LY29004, 40 ng/ml fibroblast growth factor 2 (FGF2), 50 ng/ml Activin A dan 25 ng/ml, 5 ng/ml Oncostatin M (OSM), dan 25 ng/ml hepatocyte growth factor (HGF).
HE staining: formaline 10%, alcohol 70%, alcohol 80%, alcohol 96%, alcohol 100%, xylol, paraffin wax, Hematoxylin dye, Eosin dye, aquadest, entellan. Masson Trichrome staining: Bouin’s solution, Weigert Hematoxylin, biebrich scarlet, phosphomolybdic acid, aniline blue. Immunohistochemistry: hydrogen peroxidase 30%, methanol, Recombinant Anti-Albumin antibody [EPSISR1] (ab240109 Abcam, UK), Recombinant Anti-Cytochrome P450 3A4/CYP3A4 antibody [EPR6202] (ab245774 Abcam, UK), Rabbit specific HRP/DAB Detection IHC Detection Kit Micro-polymer (ab236469 Abcam, UK), paraffin embedded rat liver slide from the Department of Histology, Faculty of Medicine, Universitas Indonesia lab, frozen rabbit liver stored in −20° freezer from SCTE IMERI FKUI lab, PBS 1X, Triton X-100 10%, Hematoxylin, and lithium carbonate.
qRT-PCR: Quick-RNATM Miniprep Plus Kit, Toyobo ReverTraAce qPCR RT Master Mix with gDNA remover, and SensiFASTTM SYBR Lo-ROX One-Step Kit.
SEM: Silica gel.
IF: HNF4-alpha antibody [EPR3648] (ab92378 Abcam, UK), Recombinant anti-CEBP Alpha/CEBPA antibody [EP708Y] (ab40761 Abcam, UK), Goat F(ab’) 2 anti-rabbit IgG Fc (FITC) (ab6018 Abcam, UK).
Methods
iPSC culture
iPSC cell line was thawed and cultured with the E8 culture medium in vitronectin-coated 12-well plates. Medium change was done every 2 days by removing 50% medium and adding equal volume of fresh medium. Microscopic observation and documentation was performed daily to evaluate colony morphology and confluency. Passage was done when confluency reaches 25–50% in each well. EDTA/Versene solution was used to disintegrate iPSC colony and to separate the colony from vitronectin after 4–5 minutes incubation in 37°C, 5% CO2 incubator. Cell scraper was used to ensure complete removal of iPSC from the vitronectin-coated well. iPSC suspension was collected in 5 ml complete medium with 10 µM ROCK inhibitor to halt the dissociative reaction. Split ratio was between 1:2 and 1:4.
Making native liver scaffold
Native liver scaffold from New Zealand White Rabbit liver was made with decellularization methods by using multiple syringe injection. This method based on previous study that conducted.Citation14 Ten lobules of liver were cut to a size of 1.5 cm x 1.5 cm with a thickness of 0.7–1 cm. The liver cubes were immersed in 0.001 M EGTA for 30 min and placed in a petri dish. A 1- ml syringe was fixed with a fixation device, with red wire attached with a toothpick on top of a styrofoam. The injection using fixated syringe was started with aquadest, continued with graded concentrations of 0.1%, 0.25%, 0.5%, 0.75%, and 1% SDS, and distilled water 25 times at the same point. Injections were carried out at the same site until the liver cube became clear. Further injection was repetitively done on 4–7 sites in the cube. The scaffolds were stored in 0.9% NaCl solution and placed in a freezer at −20°C (Supplementary Figure 1).
Prior to use in the study, the liver biological scaffolds were removed from the freezer and thawed in BSC. Then the scaffold was cutted into three parts with sterile surgical scissors and placed in a 12-well plate. UV sterilization was done in BSC for 1 hour prior to recellularization.
Hepatocyte differentiation by using native liver scaffold
The recellularization stage was carried out by injecting 125,000 from total harvested iPSC with 1 ml syringe into 9 pieces of the liver biological scaffolds (Supplementary Figure 2). Since the number of total harvested MSCs were higher, each liver scaffold was recellularized with approximately 50,000 MSCs. After recellularization, the liver scaffold was cultured in a static 12-well plate culture with the medium change based on modified Blackford protocolCitation11 for 21 days.
During the differentiation process, microscopic observations were made on the scaffold with an inverted microscope. After the differentiation process was carried out, the scaffold samples were retrieved on day 7 and day 21 for further analysis.
Histology analysis for cell adhesion to ECM in scaffold and measurement of collagen area fraction
Hematoxylin eosin (HE) staining was carried out by means of preparations deparaffinized with xylol, rehydration by in decreasing graded alcohol (100%, 96%, 80%, and 70%). The preparations were then incubated in hematoxylin solution, then washed with running water for a short time, and continued with eosin incubation. The preparation was covered with entellan and cover slip.
MT staining was carried out by deparaffinizing preparations with xylol and rehydration by soaking in decreasing graded alcohol (100%, 96%, 80%, and 70%). The preparation was then put into Bouin’s fixative solution and soaked in Weigert hematoxylin solution and Biebrich scarlet acid fuchsin. The preparation is then immersed in phosphomolybdic acid and incubated in aniline blue. The results from the microphoto with Optilab were then analyzed with ImageJ software for quantification of the collagen area on MT staining.
Cellular spatial distribution observed with scanning electron microscopy
The steps in this study were iPSC culture samples with a comparison of MSCs that were differentiated in liver biological scaffolds; histological technique was used to convert them into paraffin blocks. Then the sample was put into a container containing silica gel. Furthermore, the sample was coated with a gold layer and examined with the SEM model FEI Inspect F50 carried out at CMPFA FTUI. Data were collected by taking SEM photos at 5000X magnification.
Immunohistochemical examination for hepatocyte marker albumin and CYP3A7
The preparation was immersed in 6% hydrogen peroxidase solution and incubated in protein block solution for 1 hour and washed with 0.1% PBST. Each preparation was then separately incubated with anti-albumin antibody (1:5000 dilution) and antiCYP3A4 (1:1000 dilution). Incubation was carried out at room temperature for 2 hours in a moist chamber. The preparations were then incubated with HRP-conjugated secondary antibody for 1 hour. The preparations were then added with 3-3’-diaminobenzidine (DAB) for 30 seconds and washed. The preparation was then counterstained with hematoxylin and dripped with lithium carbonate solution. The preparations were then observed with a light microscope and photographed with Optilab at 40x and 400x magnification, and then the data in the form of photographs were compared to the histological picture and the staining results between the treatment and control groups. The results of the photographs are then analyzed with ImageJ software for quantification of the area fraction in the IHC.
Molecular analysis of hepatocyte differentiation from iPSCs or MSCs on rabbit liver biological scaffolds: albumin and CYP450 gene expression
The steps of RNA isolation from samples were carried out using the Quick-RNATM Miniprep Plus Kit (Zymo Research, USA). The RNA concentration was calculated using a NanoDrop spectrophotometer.
Complementary DNA (cDNA) synthesis using the protocol from Toyobo ReverTraAce qPCR RT Master Mix. The qRT-PCR in this study used the Applied Biosystems 7500 Fast machine and the SensiFASTTM SYBR Lo-ROX One-Step Kit reagent. The primer used was designed using NCBI and IDT PrimerQuest, with housekeeping gene using 18SS primer. The following is a list of the primer nucleotide sequences used in this study ().
Table 1. Primers nucleotide sequence
Each sample was repeated three times. The samples were then put into a PCR machine and run according to polymerase activation at 95°C, denaturation at 95°C, annealing at 60°C, and extension at 72°C. The CT values obtained were then processed using the Livak formula to determine the relative expression of albumin and CYP450 to the housekeeping gene 18SS gene.
Determination of hepatocyte maturation from differentiated iPSCs or MSCs by qRT-PCR Cytokeratin-19 (CK-19) and immunofluorescence of hepatocyte transcription factors HNF4a and CEBPA
CK19 is an intermediate filament with a molecular weight of around 40 kDa. CK19 was detected in the primitive hepatic progenitor cells at the 4–10 weeks’ gestation. The expression of CK19 is diminished in mature hepatocytes.Citation12 Primer nucleotide sequences of CK-19 are as follows: forward primer: TCGACAACGCCCGTCTG and reverse primer: CCACGCTCATGCGCAG.
The steps of the IF technique on samples of iPSC differentiation in liver biologic scaffolds were the preparations deparaffinized with xylol, rehydrated, and then incubated with namely anti-HNF4-α primary antibody (1:100 dilution) and anti-CEBPA (1:250 dilution). Incubation was carried out at 4°C for 2 hours in a moist chamber. The preparations were then washed with 0.1% PBST, followed by incubation of 1:2500 diluted anti-rabbit IgG Fc secondary antibody for 1 hour and washed with 0.1% PBST. The results of the IF staining were examined with a fluorescence microscope at HGRC IMERI FKUI.
Data analysis
Data analysis in this study was carried out by statistical tests using Graph pad prism 9 software. Statistical analysis was performed on gene expression variables and histological quantitative analysis using ImageJ software (collagen area, albumin expression, CYP3A4). The normality test was carried out using the Shapiro test, and the homogeneity test was carried out using the Levene test. The p value <.05 indicates a significant difference between the two groups.
Results
Cell–ECM adhesion to scaffold
From , histology of normal liver showed tightly packed hepatocytes with eosinophilic cytoplasm and radial arrangement of hepatocytes with basophilic nuclei. In decellularized rabbit liver scaffolds with MSC differentiation d7 and d21, there was extracellular matrix with pores and hepatocyte-differentiated MSC cells attached to the scaffold with basophilic nuclei (see ).
Figure 1. Histological features of hepatocyte differentiation in decellularized liver scaffold (Mag. 400x). A: HE-stained Liver control, B: HE-stained decellularized liver scaffold, C: HE-stained hepatocyte-differentiated MSC d7 in decellularized liver scaffold, D: HE-stained hepatocyte- differentiated iPSC d7 in decellularized liver scaffold, E: HE-stained hepatocyte-differentiated MSC d21 in decellularized liver scaffold, F: HE-stained hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, G: Masson trichrome-stained liver control, H: Masson trichrome-stained decellularized liver scaffold, I: Masson trichrome-stained hepatocyte-differentiated MSC d7 in decellularized liver scaffold, J: Masson trichrome-stained hepatocyte-differentiated iPSC d7 in decellularized liver scaffold, K: Masson trichrome-stained hepatocyte-differentiated MSC d21 in decellularized liver scaffold, L: Masson trichrome-stained hepatocyte-differentiated iPSC d21 in decellularized liver scaffold cells suspected of being MSCs were clearly observed (marked with circles), M: Brightfield image from inverted microscope of hepatocyte-differentiated iPSC d7 in decellularized liver scaffold, N: Brightfield image from inverted microscope of hepatocyte-differentiated iPSC d7 in decellularized liver scaffold. Circles marked cell adherence to scaffold. O. Quantification collagen area using Image J measurement from hepatocyte differentiated MSCs (n = 3) or iPSCs (n = 3) in decellularized liver scaffold (%).
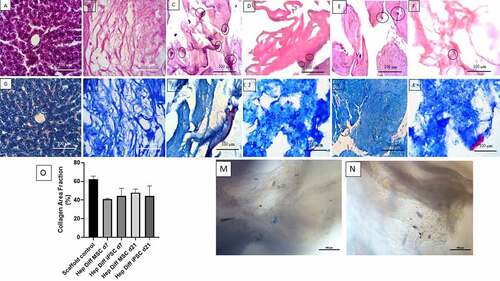
There was higher number of cellular adhesion to the scaffold with wider distribution in hepatocyte differentiated from MSCs than iPSCs (see vs ). Homogenous cell morphology was frequently found in hepatocyte differentiated from MSCs d7 and d21 as showed in HE result (see ). Diverse cell morphology ranging from flattened spindle shape to ovoid shape with heterogenous size was observed microscopically in hepatocyte derived from iPSCs on d7 and d21 (see ). On d21, the attached cells seen were larger in size with distinction between the nucleus and cytoplasm appearing clearly both in hepatocyte-differentiated MSCs or iPSCs in decellularized liver scaffold (see vs ). From the results of the examination with HE staining, cell adhesion to ECM in scaffold was observed both in d7 and d21 hepatocyte-differentiated MSCs or iPSCs in decellularized liver scaffold with different profile.
Based on observations made on MT staining from , it was found that in normal liver parenchyma tightly packed with eosinophilic hepatocyte cytoplasm and blue-colored connective tissue. In decellularized liver scaffolds (), the extracellular matrix was composed of blue-colored collagen fibers with interspersed pores (appeared as hollow white spaces between fibers). In , the hepatocyte-differentiated MSC d7 and d21 with eosinophilic cytoplasm and blue-black-colored nuclei. They were seen on the surface of collagen fibers or filled the scaffold pores. In , the hepatocyte-differentiated iPSC d7 and d21 also were seen on the surface of collagen fibers or filled the scaffold pores.
In the measurements with ImageJ in , it was found that the mean percentage of collagen area in the decellularized liver scaffold control was 62.56% with standard error of mean 1.12%. The mean percentage of collagen area in the hepatocyte-differentiated MSC d7 was 41.04% ± 0.42% and 47.88% ± 2.26% in hepatocyte-differentiated MSC d21. The mean percentage of collagen area in the hepatocyte-differentiated iPSC d7 was 44.43% ± 3.66% and 44.42% ± 4.77% in hepatocyte-differentiated iPSC d21. There was no statistical difference of collagen area result between groups (p > .5).
Spatial cell distribution profile comparison between hepatocyte differentiated from iPSCs with MSCs in scaffold
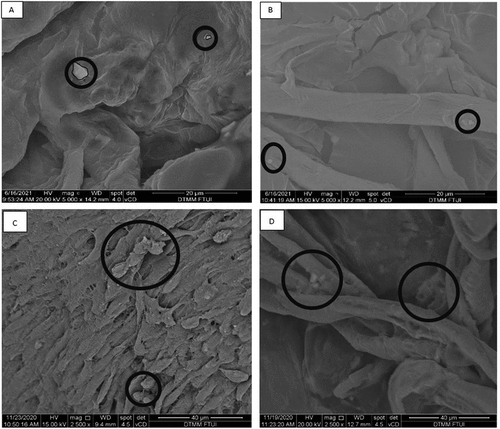
As a comparison in this study, the SEM images of hepatocyte differentiated from iPSCs and MSCs are showed in . The result showed a three-dimensional topographic image of the extracellular matrix collagen fibers with the presence of a few single-formation hepatocyte-differentiated iPSCs d7 attached to collagen fibers in decellularized liver scaffold (see ). The distance between each single cell was far apart. The results were also seen on hepatocyte-differentiated iPSCs d21 in decellularized liver scaffold (see ). In comparison, the results from showed that hepatocyte-differentiated MSC d7 and d21 distributed in different pattern than iPSC. The hepatocyte-differentiated MSCs d7 attached to the collagen fibers of the decellularized liver scaffold forming clusters of adherent cells in contact or in close proximity with each other (see ) and more abundant. Similar results were also seen in hepatocyte-differentiated MSC d21. Different cellular spatial distribution in the decellularized liver scaffold between iPSCs and MSCs hepatic differentiation was described as single-isolated cell for hepatocyte-differentiated iPSCs and clustered-adjacent cells for hepatocyte-differentiated MSCs.
Figure 2. Representative SEM images of hepatocyte differentiation in decellularized liver scaffold (Mag. 5000x). A: hepatocyte-differentiated iPSC d7 in decellularized liver scaffold (n = 3), B: hepatocyte-differentiated iPSC 21 in decellularized liver scaffold (n = 3), C: hepatocyte-differentiated MSC d7 in decellularized liver scaffold (n = 3), F: hepatocyte-differentiated MSC d21 in decellularized liver scaffold (n = 3). Circles marked cell adherence to scaffold.
Detection of albumin from hepatocyte-differentiated MSCs and iPSCs in decellularized liver scaffold
Based on observations made on IHC staining with albumin antibody from , positive immunoreactivity to albumin appeared as brown color in hepatocyte cytoplasm and less frequently in the nucleus or outside hepatocyte as shown in positive liver control (see ). Negative immunoreactivity to albumin is shown in .
Figure 3. Albumin detection from hepatocyte differentiation in decellularized liver scaffold. A: IHC albumin liver negative control (Mag. 400x), B: IHC albumin liver positive control (Mag. 400x), C: IHC albumin hepatocyte-differentiated MSC d7 in decellularized liver scaffold (Mag. 40x), D: IHC albumin hepatocyte-differentiated iPSC d7 in decellularized liver scaffold (Mag. 40x), E: IHC albumin hepatocyte-differentiated MSC d21 in decellularized liver scaffold (Mag. 40x), F: IHC albumin hepatocyte-differentiated iPSC d21 in decellularized liver scaffold (Mag. 40x), G: Percentage of albumin area fraction using Image J measurement from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), H: Percentage of albumin area fraction using Image J measurement from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3), I: qRT-PCR mean normalized albumin gene expression from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), J: qRT-PCR mean normalized albumin gene expression from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3). Comparison between groups is not significant (p > .5).
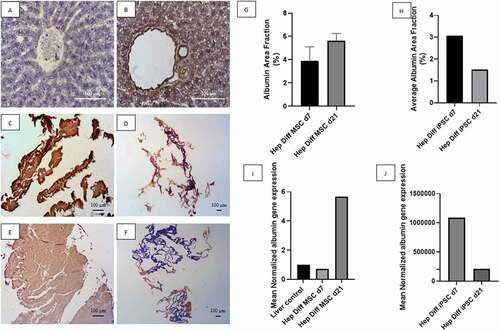
Low power field magnification captures the whole view of positive and negative area of albumin immunoreactivity against hepatocyte-differentiated MSCs and iPSCs in decellularized liver scaffold. Hepatocyte-differentiated MSCs in decellularized liver scaffold d7 () revealed albumin-positive area depicted in brown almost covering the entire structure. Increment of albumin-positive area of hepatocyte-differentiated MSCs in decellularized liver scaffold d21 () showed as bulkier brown stained structure. Hepatocyte-differentiated iPSCs in decellularized liver scaffold d7 () showed partial albumin-positive area with more restricted patch of albumin-positive area in d21 (). Quantification of albumin-positive area using image analysis is in line with the descriptive result. Mean albumin area fraction in hepatocyte-differentiated MSC in decellularized liver scaffold d7 was 3.88% ± 0.7% and 5.63% ± 0.3% in hepatocyte-differentiated MSC d21 (). The mean albumin area fraction is higher in hepatocyte-differentiated MSC in decellularized liver scaffold d21 than d7. There was no statistical difference of albumin area fraction result between d21 and d7 (p > .5). The mean of albumin area fraction in the hepatocyte-differentiated iPSC in decellularized liver scaffold d7 was 3.07% and 1.52% in hepatocyte-differentiated iPSC d21 (). There is a decrease in albumin area fraction d7 to d21 from hepatocyte-differentiated iPSC in decellularized liver scaffold.
qRT-PCR albumin result supports the IHC albumin result. Mean normalized albumin gene expression from hepatocyte-differentiated MSC in decellularized liver scaffold d7 was 0.73 and increased up to 5.68 in hepatocyte-differentiated MSC d21 (). This result confirms increment of albumin expression from hepatocyte-differentiated MSC in decellularized liver scaffold d7 to d21. Mean normalized albumin gene expression from hepatocyte-differentiated iPSC in decellularized liver scaffold d7 was 1085052.43 and decreased to 207919.66 in hepatocyte-differentiated iPSC d21 ().
Identification of CYP450 from hepatocyte-differentiated MSCs and iPSCs in decellularized liver scaffold
Based on observations made on IHC staining with CYP450 antibody from , positive immunoreactivity to CYP450 appeared as brown color in hepatocyte cytoplasm as shown in positive liver control (see ). Negative immunoreactivity to CYP450 is shown in as blue or purple stained nuclei with clear cytoplasm. High-power field magnification of positive and negative CYP450 immunoreactivity against hepatocyte-differentiated MSCs and iPSCs in decellularized liver scaffold showed larger details (see ). Hepatocyte-differentiated MSCs in decellularized liver scaffold d7 () revealed CYP450-positive cells in some area. Frequently found positive cells from hepatocyte-differentiated MSCs in decellularized liver scaffold d21 displayed as uniform light brown color covering almost the entire structure () with the exclusion of some peripheral cells with blue-stained nuclei. Hepatocyte-differentiated iPSCs in decellularized liver scaffold d7 () contained some CYP450-positive cells, which diminishes to a few CYP450-positive cells in d21 (). Quantification of CYP450-positive cells using image analysis is in line with the descriptive result. Mean CYP450 area fraction from hepatocyte-differentiated MSC in decellularized liver scaffold d7 was 1.63% ± 0.08% and 1.95% ± 0.6% in hepatocyte-differentiated MSC d21 (). There was no statistical difference of CYP450 area fraction result between d7 and d21 (p > .5). The mean of CYP450 area fraction in the hepatocyte-differentiated iPSC in decellularized liver scaffold d7 was 1.67% ± 0.53% and 0.75%± 0.2% in hepatocyte-differentiated iPSC d21 (). There is a decrease in CYP450 area fraction d7 to d21 from hepatocyte-differentiated iPSC in decellularized liver scaffold with no statistical significant difference.
Figure 4. CYP450 identification from hepatocyte differentiation in decellularized liver scaffold (Mag. 400x). A: IHC CYP450 liver negative control, B: IHC CYP450 liver positive control, C: IHC CYP450 hepatocyte-differentiated MSC d7 in decellularized liver scaffold, D: IHC CYP450 hepatocyte-differentiated iPSC d7 in decellularized liver scaffold, E: IHC CYP450 hepatocyte-differentiated MSC d21 in decellularized liver scaffold, F: IHC CYP450 hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, G: Percentage of CYP450 area fraction using Image J measurement from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), H: Percentage of CYP450 area fraction using Image J measurement from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3), I: qRT-PCR mean normalized CYP450 gene expression from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), J: qRT-PCR mean normalized CYP450 gene expression from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3). Comparison between groups is not significant (p > .5).
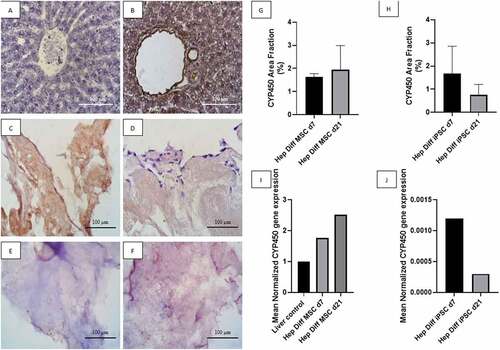
qRT-PCR CYP450 result supports the previous result. Mean normalized CYP450 gene expression from hepatocyte-differentiated MSC in decellularized liver scaffold d7 was 1.76 and increased up to 2.52 in hepatocyte-differentiated MSC d21 (). This result confirms increment of CYP450 expression from hepatocyte-differentiated MSC in decellularized liver scaffold d7 to d21. Mean normalized CYP450 gene expression from hepatocyte-differentiated iPSC in decellularized liver scaffold d7 was 0.0012 and decreased to 0.0003 in hepatocyte-differentiated iPSC d21 ().
Determination of hepatocyte maturation from differentiated iPSCs or MSCs in decellularized liver scaffold
qRT-PCR CK-19 result is important to determine hepatocyte maturation stage from differentiated iPSCs or MSCs in decellularized liver scaffold. Mean normalized CK-19 gene expression from hepatocyte-differentiated MSC in decellularized liver scaffold d7 was 0.84 and decreased to 0.57 in hepatocyte-differentiated MSC d21 (). Mean normalized CK-19 gene expression from hepatocyte-differentiated iPSC in decellularized liver scaffold d7 was 0.0030 and decreased to 0.0009 in hepatocyte-differentiated iPSC d21 (). The rate of diminishing CK-19 expression during hepatocyte differentiation in decellularized liver scaffold is more prominent in iPSC than MSC (d7/d21 ratio = 3.33 vs. 1.47).
Figure 5. Determination of hepatocyte maturation stage by qRT-PCR CK-19 and IF HNF4-a and CEBPA. A: qRT-PCR mean normalized CK-19 gene expression from hepatocyte-differentiated MSCs in decellularized liver scaffold (n = 3), B: qRT-PCR mean normalized CK-19 gene expression from hepatocyte-differentiated iPSCs in decellularized liver scaffold (n = 3), C: Brightfield image from fluorescence microscope liver control, D: IF HNF4a liver-positive control, E: Brightfield image from fluorescence microscope decellularized liver scaffold, F: IF HNF4a decellularized liver scaffold control, G: Brightfield image from fluorescence microscope hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, H: IF HNF4a hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, I: Brightfield image from fluorescence microscope hepatocyte-differentiated iPSC d21 in decellularized liver scaffold, J: IF CEBPA hepatocyte-differentiated iPSC d21 in decellularized liver scaffold.
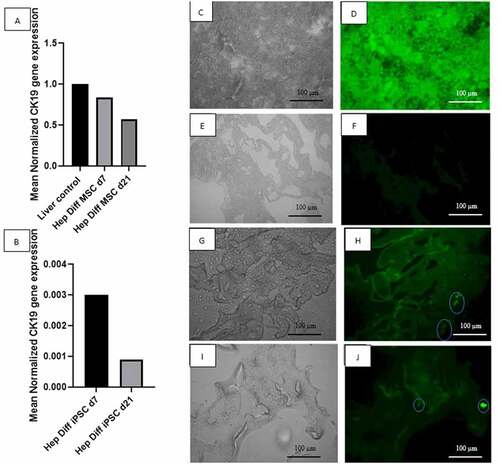
This result required further investigation with two hepatocyte transcription factors that regulate mature hepatocyte gene expression: HNF4a and CEBPA. HNF4a immunofluorescence result from hepatocyte-differentiated iPSC in decellularized liver scaffold d21 () showed the presence of HNF4a-positive cells attached to the scaffold surface. CEBPA immunofluorescence result from hepatocyte-differentiated iPSC in decellularized liver scaffold d21 () also showed the presence of CEBPA-positive cells attached to part of the scaffold.
Discussion
Reconstruction of liver tissue engineeringCitation18 is a challenge for Indonesia, which categorized among lower middle income country in world bank list. Currently, one of the liver transplant centers available in Indonesia is Cipto Mangunkusumo National General Hospital as adjacent academic hospital of Faculty of Medicine, Universitas Indonesia, as the core institution where this study took place. The need of bioartificial liver from liver tissue engineering construct as bridge therapy for transplant waiting list patients who lived far from the liver transplant centers is pressing. This study is the beginning of developing bioartificial liver from liver tissue engineering studies at Faculty of Medicine, Universitas Indonesia, which is the frontier for stem cell and tissue engineering research in Indonesia. This is the first report of hepatocyte differentiation from iPSCs or MSCs in decellularized liver scaffold established by Department of Histology, Faculty of Medicine, Universitas Indonesia, in SCTE IMERI lab.
HE staining for iPSC differentiation showed fewer cells compared to MSC differentiation, with the scaffolds remaining intact in both cultures. In addition, cells attached to the liver biological scaffold were seen in iPSC differentiation but fewer than MSC differentiation. The initial number of cells seeded into the scaffold differ with lower number of iPSC than MSC. Initial seeding number of iPSCs to scaffold in this study is lower than in other studies. Those studies harvested iPSCs from culture after more than 80% confluence.Citation19–21 In this study, prior to seeding, the iPSCs culture confluence nearly half from the reported value. Despite the theory of infinite proliferative capacity of iPSCs, harvesting lower than 80% confluence ultimately yields fewer iPSCs. This results in lower initial seeding number, which causes only a few iPSCs to attach to certain parts of the liver’s biological scaffold.
Collagen area that is occupied by hepatocyte-differentiated iPSCs or MSCs in this study represent cell–ECM interaction. Masson trichrome staining results indicated that the iPSCs differentiated in the scaffold interacts in different behavior than MSCs. The comparison shows that the hepatocyte-differentiated MSCs in scaffold d21 showed more reduction in collagen area than d7, while collagen area remains similar for hepatocyte-differentiated iPSC d21 and d7. Reduction in collagen area refers to the fact that more collagen area are occupied by growing number or size of cells. This could be due to proliferation and increase in cell size of the attached MSCs in the scaffold during in vitro hepatocyte differentiation. The similar collagen area percentage of hepatocyte-differentiated iPSC d21 and d7 indicates stable interaction between differentiating iPSCs and collagen fibers in the scaffold. This means the amount of collagen area whose spaces are occupied by the differentiating iPSCs remains constant during in vitro hepatocyte differentiation process. Collagen area difference could contribute to the different behavior of hepatocyte-differentiated iPSC compared with hepatocyte-differentiated MSCs. Heterogenous morphology and more restricted distribution of hepatocyte-differentiated iPSC in the scaffold in contrast to homogenous morphology and wider distribution of hepatocyte-differentiated MSC in the scaffold profile.
The results of SEM images further clarify the pattern of spatial cell distribution in the collagen fibers and scaffold pores. It shows that hepatocyte-differentiated MSC in the scaffold were cluster-adjacent cells and adhered more abundant to the collagen fibers than of hepatocyte-differentiated iPSC in the scaffold with single-cell-formation. In addition, the created scaffold retains a 3D structure that is useful as a microenvironment for the hepatocyte differentiation. The results of this study are unique compared with previous study SEM images, which showed more abundant iPSC adherence and denser iPSC filling the interconnected scaffold pores. Limitation of this study is the limited resource by which the recellularization technique was done manually by syringe injection of stem cell suspension to the decellularized liver scaffold. This causes recellularization to be abrupt and uneven as the pattern of single-cell-formation by hepatocyte-differentiated iPSC in the scaffold. Previous studies showed higher density of recellularization using perfusion pump with designated flow rate that facilitates balance spatial cell distribution in decellularized liver scaffold.Citation22–24 This study shows hepatocyte-differentiated MSCs in the scaffold were cluster-adjacent cells and adhered more abundant to the collagen fibers. Plausible explanation may relate to the higher adhesion capacity from hepatocyte-differentiated MSC to collagen fibers in decellularized liver scaffold compared with hepatocyte-differentiated iPSCs. This result supports that of previous studies, which showed higher amount of hepatocytes differentiated from MSC with collagen fiber-based scaffold.Citation25–27
The level of two hepatocytes markers albumin and CYP450 from hepatocyte-differentiated iPSC in decellularized liver scaffold is below that of hepatocyte-differentiated MSC. This was evidenced by a decreased trend in the area fraction analyzed with albumin and CYP3A4 IHC on hepatocyte-differentiated iPSC in decellularized liver scaffold d21. In contrast, hepatocyte-differentiated MSC in decellularized liver scaffold d21 showed an increase trend in the area fraction of albumin and CYP450. In addition, the results on albumin and CYP450 gene expression showed similar trends. These result are not in accordance with previous studies with comparable high level of albumin and CYP450 from hepatocyte-differentiated iPSC in decellularized liver scaffold.
Constant space occupying differentiating iPSCs with collagen fibers in the scaffold, abrupt, and uneven manual syringe injection recellularization are factors from this study which lead to low and decreased trends of albumin and CYP450 from hepatocyte-differentiated iPSC in decellularized liver scaffold. The distribution of hepatocyte-differentiated iPSC in decellularized liver scaffold that are far apart in this study have been identified by previous study to affect the communication between cells and the microenvironment, which causes the proliferation and differentiation processes to be less optimal. The multicellular mode of connection is a physical property of the interactions between cells. However, those physical properties have a significant impact on cell density, ligand receptor interactions, signal gradient processing, intracellular signal transduction, and the iPSC microenvironment.Citation28–30 The next factor is the different degrees of differentiation between cells, where the differentiation process occurs faster in some parts of the scaffold, but in other parts of the scaffold, the differentiation process is slower. This may occur because many parts of the scaffold are not filled by cells or undergo cell death during the differentiation process, which affects differentiation and maturation into hepatocytes.
This study in in vitro hepatocyte differentiation protocol is one continuous set of iPSC differentiation in decellularized liver scaffold for 21 days with periodic hepatocyte induction medium change without perfusion pump machine. Previous studies showed higher hepatocyte differentiation efficiency by dividing each differentiation step, thus providing differences of culture conditions (enriched microenvironment), which set the stage for developmental milestones.Citation31,Citation32 Further selections of iPSC colony size, separation of undifferentiated iPSCs at the time of differentiation step, monitor iPSCs density before and after endoderm induction or further differentiation step.Citation33–36 The microchip fluidic PMDS platform has been shown to recapitulate liver morphology and various liver functions for 4–6 weeks in vitro.Citation37,Citation38 In addition, previous study suggests arginylglycylaspartic acid (RGD) incorporated into scaffold designs to support cell attachment.Citation39 Other approach is to involve these various techniques, including 3D co-culture with non-parenchymal cells (endothelial cells and mesenchymal cells) in the form of organoid.Citation40,Citation41
The positive result from this study is further maturation of hepatocyte stage from hepatocyte-differentiated iPSC in decellularized liver scaffold than hepatocyte-differentiated MSCs at transcriptional level. Diminishing CK-19 expression during hepatocyte differentiation in decellularized liver scaffold is more prominent in iPSC than MSC. If result confirms that there is positive signals for HNF4-α and CEBPA on hepatocyte-differentiated iPSC in decellularized liver scaffold d21.
Previous study elaborates the role of HNF4-a and CEBPA in hepatocyte differentiation. HNF4-α is a transcription factor that plays a central role in the differentiation of mature hepatocytes and forms the basis for building a network of transcription factors that regulate hepatic mRNA expression. The dependence on HNF4-α caused the transcriptional regulation of gene expression to stabilize as the liver transcription factor network increases in complexity during hepatocyte maturation. CEBPA is a member of the liver-specific CEBP transcription factor family and is correlated with hepatocyte maturation. CEBPA regulates the expression of albumin and AFP genes.Citation42–44
CK-19, HNF4-A, and CEBPA expression from hepatocyte-differentiated iPSCs in decellularized liver scaffold appears to contradict the decreased trends of albumin and low CYP450 expression in this study.
Limitation of this study are fewer number of recellularized iPSCs than MSCs without prior optimization, lack of live/dead staining or proliferation marker Ki67 to evaluate cell survival in the scaffold, and absence of HNF4a and CEBPA immunofluorescence of the MSC-derived hepatocytes in the scaffold.
Conclusion
Hepatocyte-differentiated iPSCs in decellularized liver scaffold differentiation is mature with lower cell–ECM adhesion, spatial cell distribution, albumin, and CYP450 than hepatocyte-differentiated MSC in decellularized liver scaffold.
Statement of ethics
This research has been approved by the Medical Research Ethics Committee at the Faculty of Medicine, University of Indonesia, as stated in the Certificate of Passing Review Ethics (Ethical Clearance) with protocol number 19-10-1232 dated March 20, 2020.
Supplemental Material
Download Zip (346.1 KB)Acknowledgments
The authors would like to thank Prof Ismail Hadisoebroto Dilogo and Prof Jeanne Adiwinata Pawitan for insightful stem cell expertise, direction, and guidance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website
Additional information
Funding
References
- Kwong A, Kim W, Lake J, Smith J, Schladt D, Skeans M, Noreen SM, Foutz J, Booker SE, Cafarella M, et al. OPTN/SRTR 2019 annual data report: liver. Am J Transplant. 2021;21(S2):208–15. doi:10.1111/ajt.16494.
- Iansante V, Mitry R, Filippi C, Fitzpatrick E, Dhawan A. Human hepatocyte transplantation for liver disease: current status and future perspectives. Pediatr Res. 2017;83(1–2):232–40. doi:10.1038/pr.2017.284.
- Alizadeh E, Eslaminejad MB, Akbarzadeh A, Sadeghi Z, Abasi M, Herizchi R, Zarghami N. Upregulation of MiR-122 via trichostatin a treatments in hepatocyte-like cells derived from mesenchymal stem cells. Chem Biol Drug Des. 2016;87(2):296–305. doi:10.1111/cbdd.12664.
- Banas A, Teratani T, Yamamoto Y, Tokuhara M, Takeshita F, Osaki M, Kato T, Okochi H, Ochiya T. Rapid hepatic fate specification of adipose-derived stem cells and their therapeutic potential for liver failure. J Gastroenterol Hepatol. 2009;24(1):70–77. doi:10.1111/j.1440-1746.2008.05496.x.
- Li J, Xing F, Chen F, He L, So K, Liu Y. Functional 3D human liver bud assembled from MSC-derived multiple liver cell lineages. Cell Transplant. 2018;27(1):1–12. doi:10.1177/0963689717745332.
- Ma X, Duan Y, Tschudy-Seney B, Roll G, Behbahan IS, Ahuja TP, Tolstikov V, Wang C, McGee J, Khoobyari S, et al. Highly efficient differentiation of functional hepatocytes from human induced pluripotent stem cells. Stem Cells Transl Med. 2013;2(6):409–19. doi:10.5966/sctm.2012-0160.
- Banas A, Teratani T, Yamamoto Y, Tokuhara M, Takeshita F, Quinn G, Okochi H, Ochiya T. Adipose tissue-derived mesenchymal stem cells as a source of human hepatocytes. Hepatology. 2007;46(1):219–28. doi:10.1002/hep.21704.
- Szkolnicka D, Zhou W, Lucendo-Villarin B, Hay DC. Pluripotent stem cell– derived hepatocytes: potential and challenges in pharmacology. Annu Rev Pharmacol Toxicol. 2013;53(1):147–59. doi:10.1146/annurev-pharmtox-011112-140306.
- Ang L, Tan A, Autio M, Goh S, Choo S, Lee K, Tan J, Pan B, Lee JJH, Lum JJ. A roadmap for human liver differentiation from pluripotent stem cells. Cell Rep. 2018;22(8):2190–205. doi:10.1016/j.celrep.2018.01.087.
- Nakai S, Shibata I, Shitamichi T, Yamaguchi H, Takagi N, Inoue T, Nakagawa T, Kiyokawa J, Wakabayashi S, Miyoshi T, Higashi E, Ishida S, Shiraki N, Kume S, et al. Collagen vitrigel promotes hepatocytic differentiation of induced pluripotent stem cells into functional hepatocyte- like cells. Biol Open. 2019;8(7):1. doi:10.1242/bio.042192.
- Blackford SJI, Ng SS, Segal JM, King AJF, Austin AL, Kent D, Moore J, Sheldon M, Ilic D, Dhawan A, Mitry RR, Rashid ST, et al. Validation of current good manufacturing practice compliant human pluripotent stem cell-derived hepatocytes for cell-based therapy. Stem Cells Transl Med. 2018;1. doi:10.1002/sctm.18-0084.
- Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and development of the liver. Dev Cell. 2010;18(2):175–89. doi:10.1016/j.devcel.2010.01.011.
- Ober E, Lemaigre F. Development of the liver: insights into organ and tissue morphogenesis. J Hepatol. 2018;68(5):1049–62. doi:10.1016/j.jhep.2018.01.005.
- Antarianto RD, Dewi AA, Pragiwaksana A, Pawitan JA. Decellularization of liver cubes using multiple site syringe injection for generating native liver scaffold: preliminary report. AIP Conf Proc. 2019;2193:040005.
- Dewi AAAAP, Antarianto RD, Pawitan JA. Characterization of Indonesia decellularized liver cubes scaffold using scanning electron microscopy. JBBBE. 2021;52:38–46. 10.4028/www.scientific.net/JBBBE.52.38.
- Chen KG, Mallon BS, McKay RD, Robey PG. Human pluripotent stem cell culture: considerations for maintenance, expansion, and therapeutics. Cell Stem Cell. 2014;14(1):13–26. doi:10.1016/j.stem.2013.12.005.
- Induced Pluripotent Stem Cell Culture Protocols [Internet]. Sigmaaldrich.com. 2021 [cited 20 June 2021]. Available from: https://www.sigmaaldrich.com/ID/en/technical-documents/protocol/cell-culture-and-cell-culture-analysis/stem-cell-culture/human-ipsc-culture-protocols.
- Mirdamadi E, Kalhori D, Zakeri N, Azarpira N, Solati-Hashjin M. Liver tissue engineering as an emerging alternative for liver disease treatment. Tissue Eng Part B Rev. 2020;26(2):145–63. doi:10.1089/ten.teb.2019.0233.
- Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, Duris C, North PE, Dalton S, Duncan SA. Highly efficient generation of human hepatocyte–like cells from induced pluripotent stem cells. Hepatology. 2010;51(1):297–305. doi:10.1002/hep.23354.
- Minami T, Ishii T, Yasuchika K, Fukumitsu K, Ogiso S, Miyauchi Y, Kojima H, Kawai T, Yamaoka R, Oshima Y. Novel hybrid three-dimensional artificial liver using human induced pluripotent stem cells and a rat decellularized liver scaffold. Regen Ther. 2019;10:127–33. doi:10.1016/j.reth.2019.03.002.
- Takeishi K, de L’hortet AC, Wang Y, Handa K, Guzman-Lepe J, Matsubara K, Morita K, Jang S, Haep N, Florentino RM, et al. Assembly and function of a bioengineered human liver for transplantation generated solely from induced pluripotent stem cells. Cell Rep. 2020;31(9):107711. doi:10.1016/j.celrep.2020.107711.
- Debnath T, Shekar Mallarpu C, Kiran Chelluri L. Development of bioengineered organ using biological acellular rat liver scaffold and hepatocytes. Organogenesis. 2020;16(2):61–72. doi:10.1080/15476278.2020.1742534.
- Ogiso S, Yasuchika K, Fukumitsu K, Ishii T, Kojima H, Miyauchi Y, Yamaoka R, Komori J, Katayama H, Kawai T, et al. Efficient recellularisation of decellularised whole-liver grafts using biliary tree and foetal hepatocytes. Sci Rep. 2016 Oct 21;6(1):1–10. doi:10.1038/srep35887.
- Hillebrandt KH, Everwien H, Haep N, Keshi E, Pratschke J, Sauer IM. Strategies based on organ decellularization and recellularization. Transpl Int. 2019 Jun;32(6):571–85. doi:10.1111/tri.13462.
- Ong S-Y, Dai H, Leong KW. Inducing hepatic differentiation of human mesenchymal stem cells in pellet culture. Biomaterials. 2006;27(22):4087–97. doi:10.1016/j.biomaterials.2006.03.022.
- Aleahmad F, Ebrahimi S, Salmannezhad M, Azarnia M, Jaberipour M, Hoseini M, Talaei-Khozani T. Heparin/Collagen 3D scaffold accelerates hepatocyte differentiation of Wharton’s Jelly-derived mesenchymal stem cells. Tissue Eng Regen Med. 2017;14(4):443–52. doi:10.1007/s13770-017-0048-z.
- Khodabandeh Z, Vojdani Z, Talaei-Khozani T, Bahmanpour S. Hepatogenic differentiation capacity of human Wharton’s jelly mesenchymal stem cell in a co-culturing system with endothelial cells in matrigel/collagen scaffold in the presence of fetal liver extract. Int J Stem Cell. 2017;10(2):218. doi:10.15283/ijsc17003.
- Graffmann N, Ncube A, Wruck W, Adjaye J. Cell fate decisions of human iPSC-derived bipotential hepatoblasts depend on cell density. PLOS ONE. 2018;13(7):e0200416. doi:10.1371/journal.pone.0200416.
- Muncie JM, Weaver VM. The physical and biochemical properties of the extracellular matrix regulate cell fate. Curr Top Dev Biol. 2018;130:1–37.
- Wang B, Jakus A, Baptista P, Soker S, Soto-Gutierrez A, Abecassis M, Shah RN, Wertheim JA. Functional maturation of induced pluripotent stem cell hepatocytes in extracellular matrix—A comparative analysis of bioartificial liver microenvironments. Stem Cells Transl Med. 2016;5(9):1257–67. doi:10.5966/sctm.2015-0235.
- Jaramillo M, Yeh H, Yarmush ML, Uygun BE. Decellularized human liver extracellular matrix (hDLM)‐mediated hepatic differentiation of human induced pluripotent stem cells (hIPSCs). J Tissue Eng Regen Med. 2018;12(4):e1962–73. doi:10.1002/term.2627.
- Luo Y, Lou C, Zhang S, Zhu Z, Xing Q, Wang P, Liu T, Liu H, Li C, Shi W, et al. Three-dimensional hydrogel culture conditions promote the differentiation of human induced pluripotent stem cells into hepatocytes. Cytotherapy. 2018 Jan 1;20(1):95–107. doi:10.1016/j.jcyt.2017.08.008.
- Kehtari M, Beiki B, Zeynali B, Hosseini FS, Soleimanifar F, Kaabi M, Soleimani M, Enderami SE, Kabiri M, Mahboudi H. Decellularized Wharton’s jelly extracellular matrix as a promising scaffold for promoting hepatic differentiation of human induced pluripotent stem cells. J Cell Biochem. 2019;120(4):6683–97. doi:10.1002/jcb.27965.
- Sekine K, Tsuzuki S, Yasui R, Kobayashi T, Ikeda K, Hamada Y, Kanai E, Camp JG, Treutlein B, Ueno Y, et al. Robust detection of undifferentiated iPSC among differentiated cells. Sci Rep. 2020 Jun 24;10(1):1–9. doi:10.1038/s41598-020-66845-6.
- Schwartz RE, Fleming HE, Khetani SR, Bhatia SN. Pluripotent stem cell- derived hepatocyte-like cells. Biotechnol Adv. 2014;32(2):504–13. doi:10.1016/j.biotechadv.2014.01.003.
- Gieseck IIIRL, Hannan NR, Bort R, Hanley NA, Drake RA, Cameron GW, Wynn TA, Vallier L. Maturation of induced pluripotent stem cell derived hepatocytes by 3D-culture. PloS one. 2014 Jan 22;9(1):e86372. doi:10.1371/journal.pone.0086372.
- Starokozhko V, Hemmingsen M, Larsen L, Mohanty S, Merema M, Pimentel RC, Wolff A, Emnéus J, Aspegren A, Groothuis G, et al. Differentiation of human‐induced pluripotent stem cell under flow conditions to mature hepatocytes for liver tissue engineering. J Tissue Eng Regen Med. 2018 May;12(5):1273–84. doi:10.1002/term.2659.
- Park K, Hussein K, Hong S, Ahn C, Yang S, Park S, Kweon O-K, Kim B-M, Woo H-M. Decellularized liver extracellular matrix as promising tools for transplantable bioengineered liver promotes hepatic lineage commitments of induced pluripotent stem cells. Tissue Eng Part A. 2016;22(5–6):449–60. doi:10.1089/ten.tea.2015.0313.
- Bertucci T, Dai G. Biomaterial engineering for controlling pluripotent stem cell fate. Stem Cells Int. 2018;2018:1–12. doi:10.1155/2018/9068203.
- Cotovio J, Fernandes T. Production of human pluripotent stem cell-derived hepatic cell lineages and liver organoids: current status and potential applications. Bioengineering. 2020;7(2):36. doi:10.3390/bioengineering7020036.
- Abbey D, Elwyn S, Hand N, Musunuru K, Rader D. Self‐organizing human induced pluripotent stem cell hepatocyte 3D organoids inform the biology of the pleiotropic TRIB1 gene. Hepatol Commun. 2020;4(9):1316–31. doi:10.1002/hep4.1538.
- DeLaForest A, Di Furio F, Jing R, Ludwig-Kubinski A, Twaroski K, Urick A, Pulakanti K, Rao S, Duncan SA. HNF4A regulates the formation of hepatic progenitor cells from human iPSC-derived endoderm by facilitating efficient recruitment of RNA Pol II. Genes. 2019;10(1):21. doi:10.3390/genes10010021.
- Viiri LE, Rantapero T, Kiamehr M, Alexanova A, Oittinen M, Viiri K, Niskanen H, Nykter M, Kaikkonen MU, Aalto-Setälä K. Extensive reprogramming of the nascent transcriptome during iPSC to hepatocyte differentiation. Sci Rep. 2019;9(1):1–2. doi:10.1038/s41598-019-39215-0.
- Tauran Y, Poulain S, Lereau-Bernier M, Danoy M, Shinohara M, Segard BD, Kato S, Kido T, Miyajima A, Sakai Y, et al. Analysis of the transcription factors and their regulatory roles during a step-by-step differentiation of induced pluripotent stem cells into hepatocyte-like cells. Molecular Omics. 2019;15(6):383–98. doi:10.1039/C9MO00122K.
