Abstract
Accumulating recent evidence identified the ribosome as binding target for numerous small and long non-protein-coding RNAs (ncRNAs) in various organisms of all 3 domains of life. Therefore it appears that ribosome-associated ncRNAs (rancRNAs) are a prevalent, yet poorly understood class of cellular transcripts. Since rancRNAs are associated with the arguable most central enzyme of the cell it seems plausible to propose a role in translation control. Indeed first experimental evidence on small rancRNAs has been presented, linking ribosome association with fine-tuning the rate of protein biosynthesis in a stress-dependent manner.
Translation Regulation by Non-coding RNAs
Gene expression is a complex and multistep cellular process, where transcription, mRNA export, mRNA degradation, translation, and protein turnover rates represent the major regulatory hubs.Citation1 Studies measuring the transcriptomes and proteomes of mammalian cells in parallel, demonstrated that for the vast majority of protein coding genes, the transcript levels do not reflect the actual protein levels. Although the correlation is higher as initially reported,Citation2 this new data highlight that mRNA levels do not represent protein levels and most of the differences could be explained by translation regulation control mechanisms.Citation3 The same is also true for prokaryal organisms, where no correlation between mRNA and protein copy numbers could be found using a single cell approach in Escherichia coli.Citation4 Regarding the observation that the transcriptome does not entirely correlate with the proteomeCitation1,5 the term ‘ribonome’ was proposed.Citation3 The ‘ribonome’ is defined by the total cellular RNA content and its regulatory factors, including ribosomes and their regulatory non-coding RNAs (ncRNAs).
Translation control utilizing structural features and regulatory sequences within the untranslated regions (UTRs) of messenger RNAs (mRNAs) on the one hand and protein-based targeting of translation initiation on the other hand, are reasonably well understood mechanisms.Citation6 Since Ambros and coworkers discovered the first micro RNA (miRNA) in 1993, ncRNAs came into focus of translational control.Citation7,8 Shortly thereafter miRNAs turned out to be a widespread family of endogenous ncRNAs, processed from larger hairpin structured precursors to a length of ∼22 nucleotides (nt). mRNAs are the target of miRNAs loaded on the RISC complex, to which they bind by imperfect base-pairing, leading to mRNA decay, translational repression, or sequestration of mRNAs to specific cellular compartments.Citation9,10 In addition to miRNAs, genome-encoded small interfering RNAs (endo-siRNAs) have been described in a variety of multicellular organisms.Citation11 While these siRNAs are biochemically indistinguishable from miRNAs, they differ by their origin and mode of action.Citation12 siRNAs are usually cis-encoded and processed from long double-stranded RNAs, also loaded to the RISC complex to be functional.Citation13 Generally, the 21–23 nt long siRNAs bind to mRNAs by perfect base-pairing and thereby trigger endonucleolytic mRNA cleavage and degradation.Citation14 Analogous to miRNAs and siRNAs in eukaryotes, bacterial antisense RNAs have been shown to be the main ncRNA regulators of translation. In general antisense RNAs can be clustered into 2 families, the cis- and trans- acting small regulatory RNAs (asRNAs).Citation15 In case of trans- encoded antisense RNAs, multiple mRNAs are targeted via imperfect base pairing. In contrast cis- encoded antisense RNAs, derived from the opposite strand of the same genomic region, accomplish translation repression of their target mRNAs by perfect complementarity.Citation15
All of the above mentioned ncRNA translation regulators (miRNAs, siRNAs, asRNAs) share one common feature: they all target mRNAs. This restricts regulation of protein synthesis typically to specific target messages and thus allows fine-tuning of gene expression in time and space of a defined subset of mRNA transcripts. However, is it also possible to regulate the ribosome, the central enzyme of the translation machinery, directly with ncRNAs? By targeting the ribosome, RNA molecules would allow a fast and direct regulation of protein production. Such a rapid response is important under sudden environmental changes and allows the required massive reprogramming of the gene expression pattern.Citation16 However, conventional signaling pathways, including the synthesis, degradation or modification of protein factors are comparably time and energy consuming.
A Hitchhiker's Guide to The Ribosome
With the notable exceptions of the bacterial transfer-messenger RNA (tmRNA) and the universally conserved signal recognition particle (SRP) RNA, all functionally characterized ncRNAs capable of regulating protein biosynthesis target the mRNA rather than the ribosome directly. This is unexpected given the central role the ribosome plays during cell metabolism and the assumption that the ribosome evolved in the ‘RNA world’, where it likely learned receiving regulatory input from non-proteinous co-factors. Thus it is conceivable that such ribosome-bound ncRNAs have survived the evolutionary transition from the ‘RNA world’ to contemporary biology but have so far escaped the detection in transcriptome screens. The kinetic and energetic advantage of ribosome-bound ncRNA translation regulators compared to protein sensors would be the immediate availability and biological functionality of the ncRNA upon changing environmental conditions without the need of prior production of a costly regulatory polypeptide. While initially ribosome-bound ncRNAs and ncRNA fragments were serendipitously found in mRNA-based RNA-seq approaches as ‘contaminants’,Citation17-Citation19 recently more focused studies on the ribosomal ncRNA interactome have been conducted (refs. 20–24 and our unpublished data). A plethora of small and long ncRNAs has been identified to be enriched in the polysomal and sub-polysomal fractions, thus emphasizing their putative roles in translation control. First experimental data support the view that these ncRNA entities do not represent passive hitchhikers of the translation machinery but appear what can be called an emerging class of non-coding ribo-regulators of protein biosynthesis.Citation22,24,25
Ribosome-bound small ncRNAs. In our lab we performed targeted transcriptome screens for ribosome-associated ncRNAs (rancRNAs) that potentially regulate protein biosynthesis. Therefore we have applied numerous environmental stress conditions to various model systems spanning all 3 domains of life followed by ribosome preparation, small RNA isolation, and finally RNA-seq analyses. By this approach we have picked up thousands of different small RNA molecules in the size range between 20 and ∼300 nt (refs. 21,22, and our unpublished data). The RNAs either originate from intergenic regions of the genomes, and thus represent so far unrecognized ncRNA genes, or they are processed out of functional precursor transcripts such as mRNAs, tRNAs, snoRNAs, SRP RNA, and rRNAs. Post-transcriptional RNA cleavage events have been demonstrated to further expand the spectrum and functionality of transcriptomes.Citation26,27 In-depth analyses on the fate of these processing products are largely lacking, or are restricted to investigations on RNAi-related trans-silencing activities.Citation26 The rancRNAs in our screens were not only processed from specific sites of the parental RNA, but also showed stress-specific expression or ribosome-association.Citation21,22,24 Some of these ncRNAs are able to inhibit protein production on the global scale,Citation22,24 others obviously have a stimulating effect on translation (our unpublished data) (). Two ribosome-bound ncRNAs that were investigated in more detail originate from the TRM10 open reading frame in S. cerevisiae,Citation24 or from the 5’ parts of valine and alanine tRNAs of the halophilic archaeon Haloferax volcanii (ref. 22 and unpublished data). These ncRNAs down regulate protein synthesis on a global level by interacting with the large or small ribosomal subunit, respectively. It is important to note that the mode of action of these 2 examples is different. Whereas the tRNA-derived fragment of H. volcanii competes with mRNA binding to the small ribosomal subunit, the ncRNA originating from the S. cerevisiae TRM10 mRNA interferes with P-tRNA occupancy (our unpublished data). It was shown that these regulatory events are stress-dependent and occur quickly in response to sudden environmental changes. This highlights the power of ribosome-bound ncRNAs for rapid global translation attenuation. In S. cerevisiae we could demonstrate that a ribosome-bound ncRNA is needed for rapid shutdown of global translation and efficient growth resumption under hyperosmotic conditions.Citation24 Obviously this fast and global attenuation of metabolic activity as a consequence of high salt stress is crucial to open a time window for stress-specific adaptation programs to be established. Both the mRNA-derived fragment in yeast, as well as the tRNA-derived fragments in H. volcanii seem to inhibit the translation initiation process. However, there is no reason to assume that translation initiation is the sole step that can be regulated by small rancRNAs. Indeed, certain ncRNA candidates appear to specifically interfere with the elongation phase of protein biosynthesis (our unpublished data) but in principal it is conceivable that every sub-step of the translation cycle could be affected by ncRNA-mediated regulation.
Figure 1. Functional consequences of ncRNA-ribosome interactions. Short or long rancRNAs can target ribosomes (dark gray) either as naked molecules or as RNPs (light gray). As a consequence global (e.g., the yeast TRM10 mRNA-derived 18-mer ncRNA)Citation24 or mRNA-specific translation regulation can occur. Loading ribosomes on ncRNAs can also affect the cellular stability and/or localization of rancRNAs (ref. 19 and references therein). Size and line thickness of the arrows on the right correspond to experimentally supported (thick and solid), predicted (thin and solid), or in principle possible (dotted) rancRNA functions.
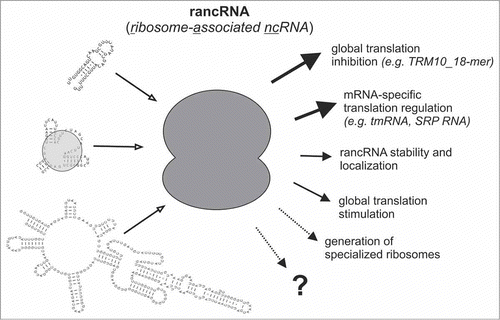
Besides regulating protein synthesis on a global level, rancRNAs have also the potential to target the translation of specific mRNAs (). Two prominent examples for the latter scenario are the bacterial tmRNA and the SRP RNA. tmRNA mediates a unique global quality control system that combines translational surveillance with the rescue of stalled ribosomes.Citation28 tmRNA specifically recognizes and binds to ribosomes that got stuck on open reading frames (ORFs) due to the absence of stop codons or due to other unproductive pausing events. This ribosome-targeted ncRNA functions as both, mRNA and tRNA, and thus enables ribosome recycling and simultaneously tagging of the incompletely translated protein for degradation.Citation29 The second example for a well-known mRNA-specific rancRNA is the SRP RNA, which is an integral part of the abundant, cytosolic, and universally conserved SRP ribonucleoprotein (RNP) complex. The SRP is involved in targeting of certain nascent polypeptides to protein-conducting membrane channels, enabling transportation of nascent polypeptide chains across membranes as well as their integration into the membrane itself.Citation30 Thereby the ncRNA component of the complex (7SL RNA in eukaryotes and 4.5S RNA in bacteria) is not only necessary for binding to the ribosome and recognition of the emerging peptide signal sequence,Citation31,32 but also for the whole complex assembly and thus represents the functional core of the SRP.Citation33 Recently an additional function for a 7SL-derived ncRNA has been proposed. The Alu RNA (a repetitive element originating from the 7SL RNA) has been suggested to deliver the protein dimer SRP9/14 to the small ribosomal subunit.Citation34 As a consequence reduced polysome levels were observed resulting in global translation inhibition.Citation35
The small rancRNAs are reminiscent of known low molecular weight effectors, such as antibiotics and other secondary metabolites, which have been shown to be capable of tuning the ribosome.Citation36 The 2 functionally studied small rancRNAs in yeast and H. volcanii have been demonstrated to target functional hotspots of the ribosomes and possess K d values in the low micromolar range, comparable to ribosome-targeted antibiotics. These examples of ribosome-bound small ncRNAs likely represent only the forefront of a so far largely elusive class of translation regulators and can pave the way for novel mechanisms to be uncovered.
Ribosome-bound long ncRNAs. Long ncRNAs (lncRNAs) have recently received considerable attention in the field. This class of ncRNA molecules is vaguely defined by the length range of >200 nt to several kilobases.Citation37 Initially lncRNAs have been connected to chromosome dosage compensation in mammals (Xist RNA) and regulation of imprinting (e.g. HOTAIR RNA). Recent years have witnessed a burst of lncRNA identification (primarily by bioinformatic means) and have expanded the scope of lncRNA functions to transcription enhancer, miRNA sponging, RNA turnover, or translation control roles (reviewed in refs. 37, 38). These capabilities of lncRNAs in turn affect the regulation of crucial cellular processes such as embryogenesis, cell cycle, maintenance of pluripotency, apoptosis, and differentiation. In general lncRNAs share common features with mRNAs, such as transcription by polymerase II, splicing, 5’-capping, and 3’-polyadenylation. What distinguishes them from genuine mRNAs is the lack of reasonably long and evolutionarily constrained ORFs, the predominant nuclear localization, and the lack of encoded peptide fragments detectable in mass spectrometry studies. Recent ribosome profiling, translating ribosome affinity purification (TRAP), as well as polysome profiling approaches, however, presented evidence that some lncRNAs are in fact cytoplasmic and associate with ribosomal and poly-ribosomal fractions.Citation17,20 , Citation23,39-41 This raises the possibility that lncRNAs are ribosome-bound to fine-tune the speed or specificity of the translation machinery (). It has been suggested that lncRNAs decorated with multiple ribosomes would be a means for titrating out and storing ribosomes for later use.Citation23 Alternatively it has been proposed that lncRNAs pair sequence specifically with certain mRNAs promotingCitation42 or inhibitingCitation43 their translation. Strictly speaking, the latter anti-sense lncRNAs are in fact not genuine rancRNAs since they associate with polysomes via their hybridization with mRNAs. On the other hand polysomal lncRNAs might in fact co-sediment with translating ribosomes because they actually encode proteins or short peptides. The currently available data fueled an intense discussion about the possible protein coding potential of several lncRNAsCitation19,44 and called into question the annotation of non-protein-coding transcripts.Citation37,38 A study in zebrafish has shown that up to 45% of previously proposed lncRNAs possess detectable ORFs and therefore might actually represent genuine mRNAs,Citation19 thus stressing the fact that the protein/peptide coding potential of lncRNAs has been underestimated. In support of this view, 2 very recent whole human proteome studies identified up to ∼450 peptides to be encoded in annotated lncRNAs, pseudogenes, and other transcripts of uncertain coding potential.Citation45,46 It therefore appears that conventional gene annotations over-estimated the number of lncRNAs in vertebrate genomes. Nevertheless the ribosome association of genuine lncRNAs implies an attractive possibility for translation control and awaits further investigations.
Conclusion & Outlook
Thousands of putative ribosome-associated ncRNAs (rancRNAs) have been recently identified, yet not all of them are expected to alter the performance of the ribosome. It is possible that some, or even the majority, of these RNA molecules are ribosome-bound by unspecific interactions and thus represent biological noise. Others might be ribosome-bound because the ribosome is in a state that can be referred to as ‘default translation initiation’ mode (). This might represent the basal program of the translation machinery attempting to spuriously bind initiation codons on all encountered cytoplasmatic RNAs.Citation23 On the other hand, first experimental evidence on some small rancRNA molecules has been presented that suggest indeed translation control functions.Citation22,24,25 These first examples demonstrated a very rapid global attenuation of protein production in a stress-dependent manner (). Furthermore, mRNA-specific effects on protein biosynthesis by rancRNAs have also been observed with the well characterized SRP RNA and tmRNA as role models for this subclass of ribo-regulators. Future work will need to clarify whether or not ribosome-bound ncRNAs are capable of eliciting additional regulatory cues to the translation machinery and thus further expand the known repertoire of translation regulation and ncRNA biology ().
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Miriam Koch for valuable comments on the manuscript and all lab members for stimulating discussions.
Funding
The research was partly supported by the NCCR ‘RNA & Disease’ funded by the Swiss National Science Foundation. Additional grant support derives from the Swiss National Science Foundation project 31003A_143388/1 to N.P.
References
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature 2011; 473:337-42; PMID:21593866; http://dx.doi.org/10.1038/nature10098
- Maier T, Guell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett 2009; 583:3966-73; PMID:19850042; http://dx.doi.org/10.1016/j.febslet.2009.10.036
- Mansfield KD, Keene JD. The ribonome: a dominant force in co-ordinating gene expression. Biol Cell 2009; 101:169-81.
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010; 329:533-8; PMID:20671182; http://dx.doi.org/10.1126/science.1188308
- Ghazalpour A, Bennett B, Petyuk VA, Orozco L, Hagopian R, Mungrue IN, Farber CR, Sinsheimer J, Kang HM, Furlotte N, et al. Comparative analysis of proteome and transcriptome variation in mouse. PLoS Gen 2011; 7:e1001393
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 2004; 5:827-35; PMID:15459663; http://dx.doi.org/10.1038/nrm1488
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009; 136:642-55; PMID:19239886; http://dx.doi.org/10.1016/j.cell.2009.01.035
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843-54; PMID:8252621; http://dx.doi.org/10.1016/0092-8674(93)90529-Y
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Gen 2010; 11:597-610.
- Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Critical Rev Biochem Mol Biol 2013; 49:16-32; http://dx.doi.org/10.3109/10409238.2013.844092
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol 2008; 9:673-8; PMID:18719707; http://dx.doi.org/10.1038/nrm2479
- Latronico MV, Condorelli G. RNA silencing: small RNA-mediated posttranscriptional regulation of mRNA and the implications for heart electropathophysiology. J Cardio Electrophys 2009; 20:230-7; http://dx.doi.org/10.1111/j.1540-8167.2008.01357.x
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev 2006; 20:515-24; PMID:16510870; http://dx.doi.org/10.1101/gad.1399806
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/10.1016/S0092-8674(04)00045-5
- Thomason MK, Storz G. Bacterial antisense RNAs: how many are there, and what are they doing? Ann Rev Gen 2010; 44:167-88; http://dx.doi.org/10.1146/annurev-genet-102209-163523
- Lintner NG, Cate JH. Regulating the ribosome: a spotlight on RNA dark matter. Mol Cell 2014; 54:1-2; PMID:24725592; http://dx.doi.org/10.1016/j.molcel.2014.03.042
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009; 324:218-23; PMID:19213877; http://dx.doi.org/10.1126/science.1168978
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011; 147:789-802; PMID:22056041; http://dx.doi.org/10.1016/j.cell.2011.10.002
- Chew GL, Pauli A, Rinn JL, Regev A, Schier AF, Valen E. Ribosome profiling reveals resemblance between long non-coding RNAs and 5' leaders of coding RNAs. Development 2013; 140:2828-34; PMID:23698349; http://dx.doi.org/10.1242/dev.098343
- Jiao Y, Meyerowitz EM. Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol Sys Biol 2010; 6:419.
- Zywicki M, Bakowska-Zywicka K, Polacek N. Revealing stable processing products from ribosome-associated small RNAs by deep-sequencing data analysis. Nucleic Acids Res 2012; 40:4013-24; PMID:22266655; http://dx.doi.org/10.1093/nar/gks020
- Gebetsberger J, Zywicki M, Kunzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012; 2012:260909; PMID:23326205; http://dx.doi.org/10.1155/2012/260909
- van Heesch S, van Iterson M, Jacobi J, Boymans S, Essers PB, de Bruijn E, Hao W, MacInnes AW, Cuppen E, Simonis M. Extensive localization of long noncoding RNAs to the cytosol and mono- and polyribosomal complexes. Genome Biol 2014; 15:R6; PMID:24393600; http://dx.doi.org/10.1186/gb-2014-15-1-r6
- Pircher A, Bakowska-Zywicka K, Schneider L, Zywicki M, Polacek N. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol Cell 2014; 54:147-55; PMID:24685157; http://dx.doi.org/10.1016/j.molcel.2014.02.024
- Sobala A, Hutvagner G. Small RNAs derived from the 5' end of tRNA can inhibit protein translation in human cells. RNA Biol 2013; 10:553-63; PMID:23563448; http://dx.doi.org/10.4161/rna.24285
- Tuck AC, Tollervey D. RNA in pieces. Trends Gen 2011; 27:422-32; http://dx.doi.org/10.1016/j.tig.2011.06.001
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 2013; 10:1798-1806; PMID:24351723; http://dx.doi.org/10.4161/rna.27177
- Janssen BD, Hayes CS. The tmRNA ribosome-rescue system. Adv Prot Chem Struct Biol 2012; 86:151-91; http://dx.doi.org/10.1016/B978-0-12-386497-0.00005-0
- Himeno H, Kurita D, Muto A. tmRNA-mediated trans-translation as the major ribosome rescue system in a bacterial cell. Front Gen 2014; 5:66.
- Saraogi I, Shan SO. Co-translational protein targeting to the bacterial membrane. Biochim Biophys Acta 2014; 1843:1433-41; PMID:24513458; http://dx.doi.org/10.1016/j.bbamcr.2013.10.013
- Gu SQ, Jockel J, Beinker P, Warnecke J, Semenkov YP, Rodnina MV, Wintermeyer W. Conformation of 4.5S RNA in the signal recognition particle and on the 30S ribosomal subunit. RNA 2005; 11:1374-84; PMID:16043501; http://dx.doi.org/10.1261/rna.7219805
- Keenan RJ, Freymann DM, Walter P, Stroud RM. Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell 1998; 94:181-91; PMID:9695947; http://dx.doi.org/10.1016/S0092-8674(00)81418-X
- Akopian D, Shen K, Zhang X, Shan SO. Signal recognition particle: an essential protein-targeting machine. Ann Rev Biochem 2013; 82:693-721; http://dx.doi.org/10.1146/annurev-biochem-072711-164732
- Berger A, Ivanova E, Gareau C, Scherrer A, Mazroui R, Strub K. Direct binding of the Alu binding protein dimer SRP9/14 to 40S ribosomal subunits promotes stress granule formation and is regulated by Alu RNA. Nucleic Acids Res 2014; 42:11203-11217; PMID:25200073; http://dx.doi.org/10.1093/nar/gku822
- Häsler J, Strub K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res 2006; 34:2374-2385; http://dx.doi.org/10.1093/nar/gkl246
- Davies J. What are antibiotics? Archaic functions for modern activities. Mol Microbiol 1990; 4:1227-32; PMID:2280684; http://dx.doi.org/10.1111/j.1365-2958.1990.tb00701.x
- Cohen SM. Everything old is new again: (linc)RNAs make proteins! EMBO J 2014; 33:937-8; PMID:24719208; http://dx.doi.org/10.1002/embj.201488303
- Karapetyan AR, Buiting C, Kuiper RA, Coolen MW. Regulatory roles for long ncRNA and mRNA. Cancers 2013; 5:462-90; PMID:24216986; http://dx.doi.org/10.3390/cancers5020462
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA 2014; 111:E203-12; PMID:24367078; http://dx.doi.org/10.1073/pnas.1317811111
- Kratz A, Beguin P, Kaneko M, Chimura T, Suzuki AM, Matsunaga A, Kato S, Bertin N, Lassmann T, Vigot R, et al. Digital expression profiling of the compartmentalized translatome of Purkinje neurons. Genome Res 2014; 24:1396-410; PMID:24904046; http://dx.doi.org/10.1101/gr.164095.113
- Zhou P, Zhang Y, Ma Q, Gu F, Day DS, He A, Zhou B, Li J, Stevens SM, Romo D, et al. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc Natl Acad Sci USA 2013; 110:15395-400; PMID:24003143; http://dx.doi.org/10.1073/pnas.1304124110
- Carrieri C, Cimatti L, Biagioli M, Beugnet A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C, et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012; 491:454-7; PMID:23064229; http://dx.doi.org/10.1038/nature11508
- Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M. LincRNA-p21 suppresses target mRNA translation. Mol Cell 2012; 47:648-55; PMID:22841487; http://dx.doi.org/10.1016/j.molcel.2012.06.027
- Guttman M, Russell P, Ingolia NT, Weissman JS, Lander ES. Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 2013; 154:240-51; PMID:23810193; http://dx.doi.org/10.1016/j.cell.2013.06.009
- Wilhelm M, Schlegl J, Hahne H, Moghaddas Gholami A, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. Mass-spectrometry-based draft of the human proteome. Nature 2014; 509:582-7; PMID:24870543; http://dx.doi.org/10.1038/nature13319
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. A draft map of the human proteome. Nature 2014; 509:575-81; PMID:24870542; http://dx.doi.org/10.1038/nature13302
