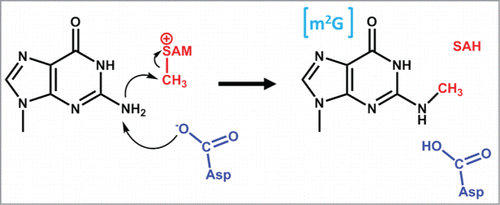
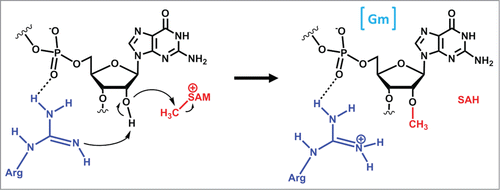
Abstract
tRNA molecules undergo extensive post-transcriptional processing to generate the mature functional tRNA species that are essential for translation in all organisms. These processing steps include the introduction of numerous specific chemical modifications to nucleotide bases and sugars; among these modifications, methylation reactions are by far the most abundant. The tRNA methyltransferases comprise a diverse enzyme superfamily, including members of multiple structural classes that appear to have arisen independently during evolution. Even among closely related family members, examples of unusual substrate specificity and chemistry have been observed. Here we review recent advances in tRNA methyltransferase mechanism and function with a particular emphasis on discoveries of alternative substrate specificities and chemistry associated with some methyltransferases. Although the molecular function for a specific tRNA methylation may not always be clear, mutations in tRNA methyltransferases have been increasingly associated with human disease. The impact of tRNA methylation on human biology is also discussed.
Introduction
RNA methyltransferases are a diverse group of post-transcriptional RNA modification enzymes responsible for the transfer of a methyl group from a methyl donor, most commonly S-adenosylmethionine (SAM or AdoMet), to any of several different locations on a target RNA nucleotide. Although all known major classes of cellular RNAs are subject to methylation, tRNA molecules remain the most heavily methylated molecules characterized to date and contain the most diversity in terms of types of methylation events that are observed.Citation1,2 While the 4 canonical RNA bases, adenosine (A), guanosine (G), cytosine (C) and uridine (U), are the most common substrates for tRNA methyltransferases, methyl groups are also added to modified nucleotides, such as pseudouridine (ψ), inosine (I) and many more complex species, as well as to the ribose 2′-hydroxyl.
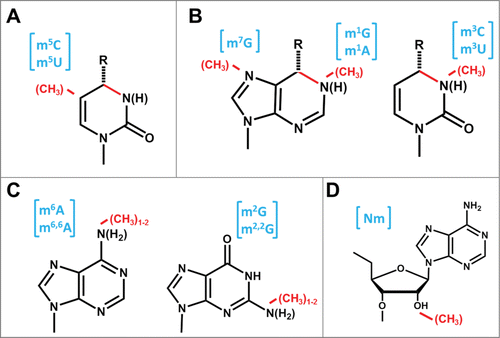
tRNA methylation is an apparently ancient process; out of 18 individual modified nucleotides that occur in tRNA from all 3 domains of life, 13 are methylated nucleotides (Cm, Gm, Um, m5C, m5U, m3U, m1G, m7G, m2G, m2,2G, m1A, m6A and m6,6A) ().Citation3,4 Consequently, characterization of the enzymes that carry out tRNA methylation can provide insight into the evolution and function of these essential macromolecules. The advent of genome sequencing and sophisticated biochemical methods facilitated an explosion in the identification of tRNA methyltransferase enzymes over the past few decades, such that the corresponding enzymes are now known for the majority of the most common tRNA methylations.Citation5,6 Yet, recent investigations into the molecular mechanisms and functions of these tRNA methyltransferases and their orthologs have revealed some new surprises, including unexpected diversity in chemistry and substrate specificity that suggests complex functions for these abundant tRNA modification enzymes.
Figure 1. Common methylated nucleotides in tRNA. Sites of methylation are indicated by the red methyl group; abbreviations for the resulting methylated species are shown in brackets in blue. (A) Methylation at C5 of pyrimidines. The bond shaded in red represents a single or double bond in U or C, respectively, and the resulting presence or absence of a proton at N3 is indicated by parentheses. The group at the 4 position (carbonyl in U and amino in C) is indicated by R. (B) Methylation at endocyclic nitrogens of purines and pyrimidines. The red colored bond on the nucleotide base represents a single or double bond, in G/U or A/C, respectively, and the resulting presence or absence of a proton at N3 or N1 is indicated by parentheses. The methylated nucleotides m7G, m1A and m3C are likely to exist as the positively charged species at physiological pH, which has also been experimentally verified for m7G and m1A in the context of an intact tRNA.Citation135 The group at the 6 position of purines (carbonyl in G and amino in A) or the 4 position of pyrimidines (as above) is indicated by R. (C) Methylation at exocyclic nitrogens of purines. Either mono- or di-methylation is observed for both purine nucleotides, as indicated, and the subsequent replacement of one or more protons on the exocyclic amino groups caused by addition of the methyl groups is indicated by parentheses. (D) Methylation at the 2′-oxygen of ribose. A 2′-O-methylated adenosine is shown (Am); each of the other 4 bases, as well as other modified nucleotides can also carry the 2′-O methylation.
The importance of tRNA methylation has often been clouded by the observation that loss of most individual modifications has little or no observable effect on cell growth. For example, only 3 tRNA modification enzymes in S. cerevisiae are strictly essential for cell viability (the A37 deaminase TAD2/TAD3, m1A58 methyltransferase TRM6/TRM61 and the tRNAHis guanylyltransferase THG1), although S. cerevisiae strains with single deletions in many of the non-essential tRNA modification enzymes exhibit slow growth or other phenotypes, such as hypersensitivity to the presence of 5-fluorouracil.Citation7,8 However, the importance of some tRNA methylations (and by extension, the enzymes that catalyze them) is highlighted by several examples of convergent evolution among methyltransferase families. For example, Trm5, an archaeal and eukaryal tRNA methyltransferase, catalyzes N1 methylation at the G37 position of tRNA.Citation9 Likewise, TrmD catalyzes this same methyl group addition to G37-containing tRNAs in Bacteria.Citation10 While both enzymes are responsible for catalyzing essentially the same methylation reaction, the 2 are structurally unrelated and possess different recognition mechanisms, which are reminiscent of differences between the 2 classes of convergently-evolved aminoacyl tRNA synthetases.Citation11-16 Moreover, loss of tRNA methylation can lead to biologically significant effects on tRNA stability and function.Citation17-23 This review provides a current survey of the diverse landscape of known tRNA methyltransferases, with a particular emphasis on recent developments in terms of alternative functions and substrate specificities for these enzymes, and newly-revealed associations of these enzymes with human disease.
Functional classes and conserved mechanistic features of tRNA methyltransferases
Enzymes that catalyze SAM-dependent methylation comprise at least 5 independent structural classes, with tRNA methyltransferases found overwhelmingly in 2 of these: Class I and IV.Citation24 Class I, which also contains most DNA methyltransferases, is the largest class identified to date and is characterized by the presence of a Rossmann-fold domain that accommodates the SAM cofactor. Class IV methyltransferases, or so-called SPOUT (named for its members SpoU and TrmD) methytransferases, are characterized by 3 β strands folded into a deep trefoil knot.Citation25,26 Interestingly, the recent discovery of the m6A methyltransferase that catalyzes formation of m6t6A37 in E. coli (TrmO) reveals that enzymes belonging to additional structural classes are possible, since this "Class VIII" enzyme comprises a novel protein fold.Citation27 Indeed, even among the relatively well-studied Class I and Class IV enzymes, some aspects of the overall structure and mechanism of distantly related family members is not necessarily obvious from sequence comparison alone.Citation26 For example, SPOUT methyltransferases generally assemble as homodimers in which each monomer contributes to the binding of SAM, but the recent identification of a monomeric SPOUT family enzyme (Trm10) suggests that alternative types of interactions with the methyl donor are possible.Citation28,29 A comprehensive survey of currently available tRNA methyltransferase structures and their corresponding properties has been reviewed in detail.Citation6
In terms of overall catalytic features associated with SAM-dependent methyltransferases, some common themes have emerged. The chemistry of nearly all tRNA methylation reactions formally requires removal of at least one proton from the substrate nucleotide, which suggests that acid-base catalysis likely plays a role. The only exception to this theme is for methylation at any of several endocyclic nitrogen positions (including guanine N7, adenine N1 and cytosine N3), in which a nucleotide with an overall +1 charge results from addition of the methyl group (see ). Indeed, for the relatively few tRNA methyltransferases for which detailed mechanistic information is known, protein residues that participate as acid-base catalysts have been identified, although the relative impact of these residues on the overall rates of reaction varies considerably. Detailed information about specific stereochemical mechanisms employed by diverse methyltransferases has been thoroughly reviewed elsewhere,Citation30 and here we focus on some general catalytic themes that have emerged.
Kinetic and mutational analyses identified important protein residues for several of the major classes of SAM-dependent tRNA methyltransferases, including amino acids that act as general base residues to facilitate proton removal. Not coincidentally, some of the best-characterized systems in terms of molecular mechanism are the 2 systems (bacterial TrmA and archaeal Trm5) for which co-crystal structures of the methyltransferase with tRNA or tRNA fragments have been obtained.Citation31,32 Nonetheless, proposed mechanisms have been indicated for many more enzymes by the numerous methyltransferase structures that are now available in complex with the critical cofactor SAM or SAH and by comparison to related enzymes.Citation6
Methylation at carbon (m5C and m5U)
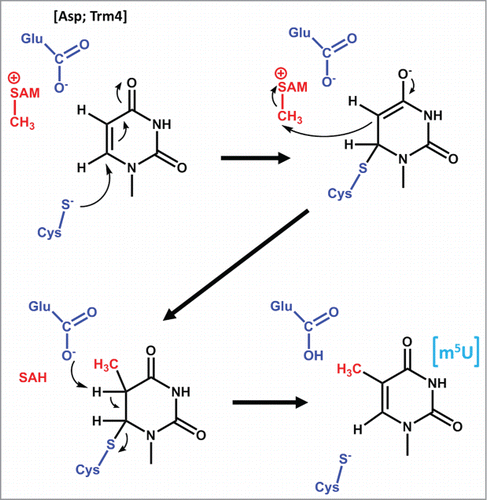
Methylation at C5 of pyrimidines is a common modification found throughout life (). m5U has been identified in tRNA and rRNA in all 3 domains, while m5C has been identified in mRNA, tRNA and rRNA in Archaea and EukaryaCitation2. tRNA methyltransferases responsible for m5U or m5C formation are predominantly Class I methyltransferases utilizing SAM as the methyl donor, although an alternative methyltransferase (TrmFO) that utilizes methylene-tetrahydrofolate as the carbon donor has been observed in some bacteria. In this case, the change in oxidation state required for the methylene group also necessitates participation of a flavin cofactor.Citation33-35 The crystal structure of E. coli TrmA, which catalyzes m5U54 formation, bound to a T-stem tRNA analog has been solved and this enzyme has been extensively studied, revealing the participation of 2 key residues in the methylation reaction ().Citation31 One of these, a conserved cysteine, acts as a nucleophile to covalently bind C6 via Michael addition, which promotes the attack of C5 on the SAM methyl group, and in turn allows for abstraction of the C5 proton by a glutamate general base.Citation31,36-38 Interestingly, 2 other m5U/m5C tRNA methyltransferases, TrmFO (catalyzing m5U54) and Trm4 (catalyzing m5C at multiple positions), also utilize a conserved cysteine nucleophile, although other aspects of their mechanisms differ, as described above for TrmFO and in terms of the identity of the putative general base for Trm4, which is likely an aspartate ().Citation33,39,40
Figure 2. Proposed mechanism for m5U methylation catalyzed by TrmA. Two active site residues (shown in blue) are proposed to act as a nucleophile to attack C5 and create a covalent enzyme-nucleotide intermediate and a general base to remove the proton from C6 following methylation. The methyl donor (SAM) and resulting SAH are shown in red. Although other C5 methyltransferases maintain the cysteine nucleophile, the identity of the general base is not the same for other enzymes; i.e. for Trm4, an Asp is proposed to catalyze this function.
Methylation at endocyclic nitrogen (m1A, m1G, m3U, m3C, m1Ψ and m7G)
As with m5C/m5U, methylation occurring on endocyclic nitrogen atoms is abundant in all domains of life, with m3C and m1Ψ absent so far only from Archaea and Bacteria, respectively ()Citation2. General features of mechanisms employed by these enzymes have been revealed from comprehensive biochemical and structural studies carried out with 2 representative enzymes that catalyze the universal N1 methylation of G37, which are a Class I methyltransferase (Trm5) in Eukarya and Archaea and a SPOUT methyltransferase (TrmD) in Bacteria. Archaeal Trm5 is the only example of a tRNA methyltransferase that has been co-crystallized in the presence of a complete tRNA and this, combined with extensive kinetic characterization and mutagenesis has resulted in a relatively good understanding of the overall catalytic mechanism.Citation32,41,42 As expected, a glutamate general base (proposed to remove the N1 proton prior to attack of N1 on the SAM methyl group) was recently identified that appears to function in both archaeal and human Trm5 ().Citation41 However, acid-base catalysis in the Trm5 reaction is not rate-limiting for the overall reaction, and instead, the removal of the N1 proton appears to occur during a rate-limiting induced fit step, which is consistent with isotope effect experiments.Citation41 Class I methyltransferases also catalyze the ubiquitous N1 methylation of A58, including the enzymes Trm6/Trm61 in Eukarya (Trm61 is the catalytic subunit), which is known as TrmI in Bacteria.Citation6 Structures are available for representatives of both families, albeit in the absence of tRNA but with a bound SAM or SAH cofactor, and models suggest that an aspartate residue is positioned to function as a general base ().Citation43-46 For m1A formation, however, the proton appears to be removed from N6, thus promoting attack of the adjacent N1 on the SAM methyl group.
Figure 3. Proposed mechanisms for m1G and m1A formation. (A) Mechanism of m1G37 formation catalyzed by Trm5, with the active site glutamate that serves as the general base indicated in blue. For other m1G methyltransferases, the general base is proposed to be an aspartate, as indicated. (B) Mechanism of m1A58 formation catalyzed by TrmI. The suggested mechanism (based on modeled structures with the target adenosine) involves the action of the aspartate general base (blue) as shown, which may occur during the same elemental step as the attack of N1 on the SAM methyl donor (proposed based on the lower pKa for deprotonation of m1A than for deprotonation of adenosine).
The precise nature of the enzymatic mechanism employed for other endocyclic nitrogen methyltransferases is less clear. Although there are crystal structures reported for the SPOUT methyltransferases TrmD (m1G37), Trm10 (m1G9) and TrmY (m1Ψ54), only some details of catalysis have been identified.Citation14-16,28,47,48 Crystal structures of Trm10 (m1G9) and TrmD (m1G37) suggest that for each enzyme, a conserved aspartate residue may act as the general base to abstract a proton from N1 and promote methylation,Citation14,28 but general base residue(s) that function in TrmY have not been identified. Likewise, for m7G formation, crystal structures of eukaryal Trm8/Trm82 (Trm8 is catalytic) and bacterial TrmB (both Class I enzymes) in complex with SAM have been determined.Citation49,50 From these structures, highly conserved glutamate and aspartate residues are suggested to participate in neutralizing the positive charge of the solfonium ion and other important residues involved in catalysis by TrmB have been described.Citation51 Nonetheless, the catalytic mechanism for these representative m7G methyltransferases has not been conclusively shown. A recent addition to the endocyclic nitrogen methyltransferase family was the identification of the first family members that catalyze methylation of pyrimidines to form m3C and m3U. The m3C methyltransferase Trm140 is a distantly-related member of the SPOUT enzyme family, but the basis for catalysis awaits further structural and mechanistic characterization.Citation52,53 Interestingly, an orthologous enzyme from T. brucei has also been characterized and may exhibit both m3C and m3U methylation activities, raising the possibility of further complexity in the reactions characterized by this branch of the methyltransferase family (I. Fleming and J. Alfonzo, personal communication).
Methylation at exocyclic nitrogen (m6A, m6,6A, m2G and m2,2G)
The enzymes that methylate the exocyclic amines found at N6 of adenosine and N2 of guanosine have so far all been classified as members of the Class I family, and between these 2 types of modification, N2 methylation is by far more common in tRNA ().Citation1,2 Although other sites of modification are known, N2 methylation of guanosines is typically found at position 10 (catalyzed by Trm11/Trm112 in eukaryotes where Trm11 is catalytic, and by TrmG10 in Archaea) and/or positions 26 and 27 (catalyzed by Trm1) and can consist of either monomethylation (m2G) or dimethylation (m2,2G), depending on the tRNA and organism that is modified.Citation54-59 Archaeal Trm1 from P. horikoshii and bacterial Trm1 from A. aeolicus have been characterized structurally and share similar N-terminal domains that contain the methyltransferase active site, including a characteristic DPFG/DPPY sequence motif that contains the aspartate that is proposed to act as a general base to remove a proton from the target N2 amino group, according to structural and mutational studies ().Citation60,61 Recently, the first tRNA methyltransferase that catalyzes exocyclic N6 methylation of adenosine was identified and, as predicted bioinformatically, was shown to correspond to E.coli YfiC, which catalyzes m6A37 modification of tRNAVal.Citation62,63 This modification is more commonly found in mRNA, rRNA and DNA, and the details of the catalytic mechanism await further characterization.
Methylation at ribose oxygen (Gm, Am, Um, Cm)
One of the founding members of the SPOUT methyltransferase family is the highly conserved TrmH (originally SpoU) enzyme that catalyzes 2′-O methylation of G18 ().Citation25 This enzyme (known as Trm3 in eukaryotes) has been extensively characterized structurally and biochemically, and these data were the basis for the proposed mechanism in which a conserved arginine residue (attracted by the phosphate group of the tRNA substrate) acts as a general base to abstract the proton from the 2′-hydroxyl, thus activating the oxygen for attack on the SAM methyl group ().Citation64,65 There is strong mutational evidence in support of this proposed mechanism, although again, additional structures obtained in the presence of bound tRNA will be important for complete evaluation of the reaction mechanism.
Lessons from tRNA methyltransferase homologs: alternative substrate recognition and catalytic features
Enzymes that catalyze known tRNA methylations have nearly all been identified, and sequence and structural similarity between family members have been extensively relied upon to suggest common mechanistic features and functions for related enzymes. In general, tRNA modification is thought to be a precise process, with a specific type of modification occurring predictably at certain sites on a given set of tRNAs. However, as diverse members of some tRNA methyltransferase enzyme families have been identified and characterized, several instances of unexpected catalytic properties have been described, yielding differences in either RNA substrate recognition and/or the chemistry of the methylation reaction itself. Several recent examples of alternative recognition and chemistry are highlighted below.
Organism-specific features of RNA recognition
Each tRNA species carries its own unique complement of tRNA modifications, including methylation, and therefore tRNA methyltransferases exhibit distinct tRNA substrate specificities.Citation1,2,66 Thus, the molecular basis by which individual tRNA methyltransferases recognize and act on specific tRNA species from among the total tRNA pool has long been an area of interest.Citation67 In many cases, the determinants that allow recognition of certain tRNAs are relatively straightforward to deduce, since the corresponding enzyme will modify essentially any species that encodes the correct target nucleotide at the position to be modified.Citation68,69 For example, m1G37 methylation occurs on all of the tRNA species that encode a G at position 37. Although the 2 unrelated m1G37 methyltransferases (Trm5 and TrmD) each interact with tRNA substrates and recognize G37 in slightly different ways, in both cases the enzymes appear to directly recognize the G37 nucleotide in a specific sequence context for efficient catalysis.Citation11,12,70
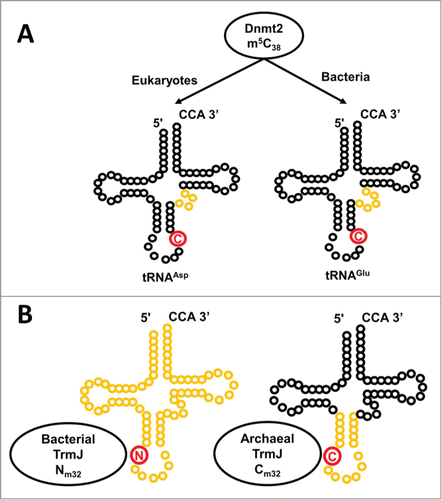
In contrast, there are cases of methylations that are highly tRNA species-specific, such as the m3C32 methylation that is a characteristic feature of tRNAThr, or m5C38 that was first associated with tRNAAsp in eukaryotes.Citation52,53,71,72 However, as additional homologs of the enzyme that catalyzes m5C38 modification (Dnmt2) have been characterized, considerable expansion or alteration of tRNA substrate specificity for some members of this enzyme family has been suggested. Initial reports centered on the identification of additional weakly recognized tRNA substrates for Dnmt2 and its (mainly eukaryotic) homologs, including tRNAGlu, tRNAVal and tRNAGly, and suggested that the additional modification of minor tRNA substrates might be protective against various types of stress.Citation73-75 However, a more complex picture has emerged from the recent investigation of a bacterial homolog of Dnmt2 (Geobacter sulfurreducens Dnmt2) in which the inherent preference for Geobacter tRNAAsp has been usurped by a preference for methylation of tRNAGlu, and indeed tRNAGlu appears to be the predominant m5C38-containing tRNA in Geobacter ().Citation76 Precise elements that are responsible for this completely swapped tRNA substrate specificity are not known, but the nucleotides and size of the variable loop appear to play some role.Citation76 Moreover, even in eukaryotes where the preference for tRNAAsp has not yet been observed to be altered, the identification of non-substrate tRNAs bound to Dictyostelium discoideum Dnmt2 in vivo suggested that there may be additional roles for these enzymes that do not depend on methylation activity.Citation75 Therefore, even in the case of a well-characterized tRNA methyltransferase with demonstrated tRNA substrate specificity, diverse homologs can stray from these well-defined roles.
Figure 6. Alternative substrate specificities exhibited by homologous methyltransferases. (A) Dnmt2 catalyzes m5C38 methylation (the C38 target nucleotide is indicated in red), with tRNAAsp thought to be the major substrate for eukaryotic Dnmt2, while tRNAGlu appears to be the predominant substrate for the G. sulfurreducens enzyme. Differences in the size and identity of nucleotides in the variable loop (highlighted in yellow) are thought to play a role in dictating alternative substrate specificity. (B) 2′-O methylation at position 32 is catalyzed by TrmJ. In Bacteria, TrmJ is capable of methylating any nucleotide at position 32 (indicated in red on each tRNA), whereas the archaeal enzyme is restricted to 2′-O methylation of cytosine. Moreover, the different structures required for tRNA recognition by the 2 enzymes (archaeal TrmJ requires only the anticodon stem-loop structure, while bacterial TrmJ requires a full-length tRNA) are indicated by yellow highlighting.
A third class of tRNA substrate specificity exists that falls somewhere in between the previous 2 classes of enzymes that are relatively non-tRNA specific (modifying any tRNA that contains a target nucleotide) or highly tRNA specific (modifying only one or 2 target tRNAs). These enzymes recognize only a subset of the possible tRNA targets that are expressed in a cell, and therefore must utilize other criteria for selection of substrates. Several examples of tRNA methyltransferases of this type have been described, including some of the enzymes that catalyze Gm18 (Trm3 in S. cerevisiae and TrmH in E. coli and Aquifex aeolicus) and the enzyme that catalyzes m1G9 in S. cerevisiae (Trm10). The tRNA recognition problem faced by these enzymes is somewhat more complex, since it is often difficult to find specific sequence elements that are universally shared by all substrate or non-substrate tRNAs that might explain the observed patterns of substrate specificity.
For Trm3/TrmH, the sequence context of the G18 target nucleotide, as well as the length and composition of the D-stem and loop were all found to play a role in tRNA recognition and the contributions of various important base pairs in specific context could be used to explain most of the observed specificity of A. aeolicus TrmH.Citation69,77,78 However, analysis of the pattern of tRNA substrates that contain Gm18 in E. coli indicated that these rules are not necessarily universal, underscoring the need to investigate the properties of distinct family members to understand their substrate specificities. Kinetic and structural characterization of TrmH suggested that tRNA recognition occurs through an induced fit process that allows an initial non-specific interaction with tRNA followed by conformational changes that are only allowed with substrate tRNAs.Citation77,79 The same type of alternative substrate recognition among related SPOUT methyltransferases was also recently noted for bacterial and archaeal members of the TrmJ family (which each catalyze 2′-O methylation at position 32 of tRNA). In this case, each enzyme utilizes different elements of the tRNA for proper recognition, and even different specificities for the nucleotide target of modification, with archaeal TrmJ only catalyzing efficient Cm formation, while E. coli TrmJ methylates all 4 possible nucleosides ().Citation80 Interestingly, structural comparison of archaeal vs. bacterial TrmJ revealed that the difference in nucleotide specificity may be related to different conformations of the methyl-cofactor in the active sites. However, many questions remain regarding the precise mechanisms by which substrate recognition occurs for either of these examples and further structural characterization of homologs with different substrate specificities will be required to completely address this question.
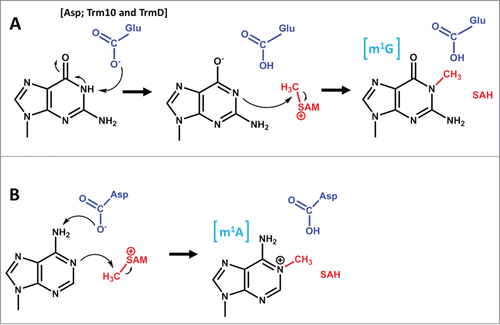
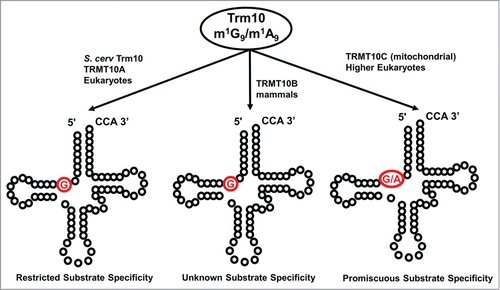
The tRNA m1G9 methyltransferase Trm10 also modifies only a subset of its potential substrates, since only 13 out of the 24 G9-containing tRNAs in S. cerevisiae whose sequences have been determined are modified by Trm10 under normal growth conditions ().Citation81,82 The substrate specificity of Trm10 has been investigated in vitro and shown to depend somewhat on the presence of other modifications on the tRNA in order to enhance methylation of target substrates, but the proficient in vitro methylation activity of S. cerevisiae Trm10 with tRNAs that are not detectably modified to any level in vivo, combined with the ability of some non-substrate tRNAs to acquire methylation upon overexpression of Trm10, suggests that the recognition of Trm10 can also be more flexible than would be expected from the restricted subset of tRNAs that carry the m1G9 modification.Citation81 As with TrmH, the properties that determine substrate recognition in one organism may not necessarily be the same as those that dictate specificity in another; for example, tRNAGlyGCC is a robust substrate for methylation by S. cerevisiae Trm10, both in vitro and in vivo, but in humans, tRNAGlyGCC contains an unmethylated G9.Citation1,2 Although distinct sequence determinants between the yeast and human tRNA that explain this difference in substrate specificity may yet be identified, the sequence conservation between eukaryotic tRNAs and the need for each enzyme to act on multiple substrates diminishes the possibilities for recognition based entirely upon sequence. Interestingly, also like the TrmH/Trm3 family where the T. thermophilus enzyme is non-tRNA specific,Citation69 some members of the Trm10 family appear to exhibit completely promiscuous tRNA methylation activity. Most well-studied is the tRNA methylation activity of one of the 3 human paralogs of Trm10 (TRMT10C). This enzyme, which is part of the unique entirely proteinaceous ribonuclease P complex in mammalian mitochondria, exhibits a broad tRNA specificity that is much less restrictive than its cytosolic counterparts.Citation83,84 The recent crystal structure of fungal Trm10 suggests that the tRNA is recognized by a positively charged surface on the monomeric enzyme, but no co-crystal structure with tRNA is available.Citation28
Figure 7. The Trm10 family of methyltransferases exhibit differences in chemistry and substrate recognition. Trm10 enzymes were originally identified based on their ability to catalyze m1G9 methylation (the G9 target nucleotide is shown in red), which only occurs on a subset of tRNAs in S. cerevisiae (left panel). Subsequent investigation revealed that higher eukaryotes encode up to 3 Trm10 paralogs (TRMT10A, B and C, as indicated) and that the methylation activity and substrate specificities of these enzymes are distinct. TRMT10A (left panel) is most similar to S. cerevisiae Trm10 and exhibits similar biochemical properties. TRMT10B (middle panel), like TRMT10A, is a G9 methyltransferase but substrate specificity has not been well characterized, while TRMT10C (right panel) is a strictly mitochondrial methyltransferase with much broader tRNA substrate specificity and the unusual ability to catalyze both G9 and A9 methylation.
The question of RNA substrate specificity has recently been extended beyond tRNA itself by the discovery of methyltransferases with activities on other substrates apart from those originally associated with the enzyme family. For example, one of the earliest identified tRNA methyltransferases, TrmA, is a well-studied enzyme that catalyzes the highly conserved m5U54 modification in the T-loop; however, the E. coli enzyme was also recently implicated in an m5U modification that occurs in the tRNA-like domain of tmRNA.Citation85 Indeed, a large family of m5U methyltransferases has been identified in which an ancestral bacterial enzyme is thought to have duplicated into multiple groups of related enzymes with distinct substrate specificities, with the TrmA enzymes acting on tRNA (and tRNA-like molecules) and other homologs (RlmC and RlmD family members) acting on rRNA substrates.Citation86,87 But, even among these classes the lines can become blurred, since a recently described homolog of RlmD from Pyrococcus abyssi catalyzes a TrmA-like tRNA methylation, instead of acting on rRNA.Citation88,89 Structural characterization of several m5U54 enzymes from all 3 families suggests that there may be conserved conformational changes that cause refolding of the tRNA target, thus explaining the tRNA specificity of some of these enzymes.Citation31,88,90 Another case of a dual-specificity methyltransferase was recently reported with the identification of the m2A37 methyltransferase from E. coli. In this case, the RlmN enzyme previously associated with catalyzing the m2A modification in rRNA similarly catalyzes the same modification in tRNA, both in vitro and in vivo.Citation91 Finally, there are cases in which the practical substrate pool for a given methyltransferase is defined by its localization or expression patterns in the cell. For example, in Trypanosoma brucei, a paralog of the well-characterized Trm5 enzyme that catalyzes m1G37 formation localizes to the mitochondrion, which was surprising since T. brucei does not encode any tRNAs in its mitochondrial genome and would be presumed to import m1G37-containing tRNA generated by nuclear Trm5-catalyzed modification.Citation92 In this case, the mitochondrial Trm5 appears to function to protect translational fidelity in the mitochondria by methylating any unmethylated tRNA that is imported into the organelle by the apparently non-discriminating import machinery. In sum, the inherent RNA substrate flexibility associated with many tRNA methyltransferases suggests that additional substrates remain to be identified for these enzymes, and elucidation of these will be important for a complete understanding of the biological functions of these enzymes.
Alternative chemistry associated with tRNA methyltransferases
In the examples described above, although the tRNA target for a given methylation might differ, the chemical nature of the methylation reaction is not changed between closely related family members. However, again recent observations have challenged the universality of even this feature of tRNA methyltransferase activity. The Trm10 family is widely distributed among Eukarya and Archaea, reflecting the highly conserved nature of the m1G9 modification in tRNA.Citation82 However, in addition to the questions about RNA substrate specificity described above, recent characterization of several Trm10 homologs demonstrated that some family members are capable of catalyzing m1A9 formation, either in addition to or instead of the m1G9 modification activity.Citation84,93 Although the methylation in this case occurs on the same position (N1) of the purine ring, the difference in the expected protonation state of the target nitrogen for these 2 bases (protonated G9 and deprotonated A9) raises questions about the mechanism, and specifically about the role of acid-base catalysis, in an enzyme that is capable of catalyzing both of these reactions presumably using the same active site (). It is worth noting that residues characterized so far to act as a general base for other m1G vs. m1A methyltransferases () are proposed to remove the proton from different atoms (N1 in the case of guanosine and N6 in the case of adenosine), and the possibility of alternative targets for the putative general base in Trm10 has not been evaluated. Alternatively, a mechanism involving the pre-dissociation of the N1 proton prior to the rate-determining step for chemistry, as has been observed with Trm5 could also explain this dual specificity.
In another example of unusual chemistry, also involving an m1G methyltransferase, an ortholog of the m1G37 enzyme Trm5 from P. abyssi is suggested to catalyze 2 distinct methylation reactions during the production of the hypermodified wyosine derivative mimG.Citation94,95 One of these is the familiar methylation at N1 to produce m1G37, but the second reaction involves a subsequent methylation at N7 of the 4-demethylwyosine intermediate (imG-14) to yield imG2 (). Although the imG-14 methylation reaction involves an entirely different target atom (N7) on the nucleotide base from the well-characterized m1G chemistry, some similarities in terms of the chemical environment of these 2 nitrogens have been noted and future structural and functional characterization will be required to determine the precise mechanism of this dual functional enzyme.Citation94 Finally, the lack of evolutionary relatedness between Trm5 and TrmD m1G37 methyltransferases is underscored by the recent observation that E. coli TrmD, but not Trm5 (or any other known SAM-dependent methyltransferase for that matter) depends on the participation of a divalent metal ion, presumably Mg2+, for catalytic activity. The precise role of this Mg2+ ion remains to be fully determined, but appears to both stabilize the developing negative charge on O6 during methyltransfer and to assist in deprotonation of N1 by the aspartate general base ().Citation96
Figure 8. Unusual chemistry associated with tRNA methyltransferase homologs. (A) An archaeal homolog of Trm5 (aTrm5a, also known as Taw22) catalyzes 2 distinct methylation reactions during the formation of the wyosine derivative, mimG. The first reaction, formation of m1G (added methyl group is highlighted in red) is the same reaction catalyzed by other studied Trm5 enzymes, but m1G is subsequently converted to imG-14 (by the action of Taw1) and this modified nucleotide (not shown) is the substrate for the second methylation reaction catalyzed by aTrm5 to form imG2 (the second methyl group added by aTrm5a is shown in red). (B) An unusual SAM-derivative. The metabolite Cx-SAM, with the carboxymethyl group highlighted in red, is attacked by the hydroxyl oxygen at the 5 position of ho5U to generate cmo5U.
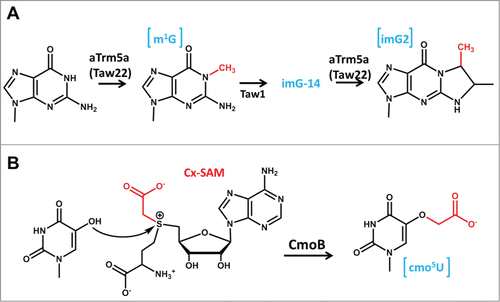
Differences in the chemical nature of the reaction catalyzed by tRNA methyltransferases have been extended to the cofactor molecule itself. The SAM cofactor has seemingly been selected as the preferred methyl donor for most tRNA methyltransferases, likely due to the high reactivity of the methyl group bound to the positively-charged destabilized sulfonium ion, and only a small number of enzymes (bacterial members of the TrmFO family) utilize the alternative methylene-tetrahydrofolate (THF) cofactor for as a methyl donor.Citation33,35,97 However, the recent discovery of a new metabolite, carboxy-S-adenosyl methionine (Cx-SAM) has increased the diversity of cofactors that are associated with even SAM-dependent methyltransferases ().Citation98 In E. coli, 2 members of the SAM methyltransferase superfamily, CmoA and CmoB, act together to synthesize the cmo5U34 modification, with CmoA catalyzing the biosynthesis of Cx-SAM and CmoB acting as the carboxymethyl transferase to generate the modified nucleotide (). This expansion of chemistry raises the possibility of additional versatility associated with the SAM cofactor that remains to be explored.
Methylation reactions influenced by partner proteins
An interesting feature of some tRNA methyltransferases is their requirement for a partner protein for efficient catalysis. To date, partner proteins have been shown to interact with a catalytic subunit of several (so far, strictly eukaryotic) tRNA methyltransferases to organize the methyltransferase active site and/or participate in substrate recognition (). The S. cerevisiae m7G46 methyltransferase complex Trm8/Trm82 is one such example in which the catalytic subunit (Trm8) requires Trm82 for function.Citation50,99 Structural characterization, mutational analyses and domain deletions demonstrated that recognition of the substrate tRNA by Trm8/82 and its bacterial homolog, TrmB are similar in that both enzymes recognize their substrate base from the T-stem side.Citation49-51,100 Yet, while the bacterial and eukaryotic enzymes are similar in terms of tRNA recognition, they differ in that the eukaryotic enzyme requires Trm82 for catalysis, both in vitro and in vivo.Citation101 Trm82 binds to Trm8 and induces a conformational change resulting in formation of the optimal active site.Citation50
Table 1. Eukaryotic tRNA methyltransferases requiring a partner for efficient catalysis
The Trm6/Trm61 complex responsible for m1A58 formation in S. cerevisiae (also known as Gcd10/Gcd14) is similar to Trm8/Trm82 in that only one of the subunits (Trm61) contains a functional methyltransferase active site.Citation102,103 However, the role of the partner subunit in this complex is slightly different. Initial characterization revealed that Trm6 is primarily responsible for the functional interaction with tRNA, although more recent work demonstrated that residues from both subunits participate in tRNA binding.Citation103,104 This contrasts with Trm82, which does not have inherent tRNA affinity on its own. Interestingly, the participation of a separate partner protein is not necessarily conserved throughout evolution, since the TrmI enzyme (a homolog of Trm6/Trm61 that catalyzes m1A58 modification in Archaea and Bacteria) exists as a homotetramer, although TrmI similarly utilizes 2 subunits to interact with tRNA.Citation43-45 Furthermore, using the TrmI structure, attempts to abolish Trm6/Trm61 formation and enzyme activity through mutational analysis were unsuccessful, further underscoring differences between the Trm6/Trm61 and its bacterial/archaeal counterparts.Citation104
Another protein partnership discovered to be involved in tRNA methylation was Trm11/Trm112, which catalyzes m2G10 formation in eukaryotes.Citation54 It was determined, as with Trm8 and Trm61, that Trm11 is the catalytic subunit and requires the Trm112 protein subunit to efficiently catalyze m2G10 formation in vivo, although for this enzyme, additional partner proteins may participate in the reaction, since efficient in vitro activity of the Trm11/Trm112 complex has not yet been detected.Citation54 Moreover, Trm112 is a promiscuous methyltransferase partner that is also required for the activities of Trm9 and Mtq2p, which catalyze the last step of mcm5U34 and methylation of eRF1, respectively.Citation54,105-107 The discovery of the non-tRNA related Mtq2p/Trm112 partnership is particularly striking and underscores the involvement of Trm112 in other biological activities such as cell division in Arabidopsis, methylation of 18S rRNA, ribosome biogenesis and translation.Citation108-110
In contrast to Trm11/Trm112 where the partner protein interacts with multiple different methyltransferases, the Trm7 methyltransferase represents an alternative example where the catalytic subunit interacts with multiple different partner proteins. Trm7, a homolog of human FtsJ, is responsible for 2′O methylation of C32 and C34 in the anticodon loop of tRNAPhe.Citation111,112 Loss of Trm7 results in a slow growth phenotype due to a nonfunctional tRNAPhe.Citation111,113 What is striking is that this methyltransferase is directed to each of these positions by either of 2 protein partners, Trm732 or Trm734, which are each responsible for interacting with Trm7 to target the indicated modification.Citation113
The discovery of these diverse protein partnerships gives rise to the possibility that other unknown protein partners for tRNA methyltransferases may remain to be identified. For example, in the case of Trm10, organism-specific patterns of tRNA substrate specificity, as described above, have not been satisfactorily explained by the tRNA recognition properties of the purified enzyme in vitro or in vivo, and it remains possible that additional subunits are involved.Citation81 In the future it will be interesting to understand eukaryotic tRNA methyltranferase partners and how they differ from their homooligomeric bacterial counterparts, possibly leading to a new group of drug targets.
tRNA methylation reactions in human health and disease
The function of a single methyl group added to RNA has in many cases been difficult to discern. This is largely due to the frequent lack of obvious consequences for the cell upon loss of the modification. However, at least 4 possible molecular functions have been associated with tRNA methylation to date. First, methyl groups can play a structural role that prevents formation of alternative secondary or tertiary structures (usually by interfering with Watson-Crick base pairing). For example, the presence of m1A9 methylation in human mitochondrial tRNALys, blocks base pairing between A9 and U64, and in turn destabilizes a non-functional base pairing between residues 8:65 and 10:63, thus promoting the canonical secondary structure of the tRNA.Citation23,114 Second, the presence of methyl groups (particularly 2′-O methylation) can affect the thermodynamic stability of tRNA. This is most well-substantiated by the case of tRNA species from thermophiles, which contain significantly more 2′-O methylation than their mesophilic or psychrophilic counterparts.Citation115 Third, methylation of tRNA (among other modifications) contributes to the overall stability of tRNA in vivo by protecting it from degradation by various pathways, including the nuclear TRAMP complex and/or rapid tRNA decay (RTD).Citation20,116 RTD was initially associated with the simultaneous loss of methylation at multiple sites on the tRNA, such as at positions 46 and 49 in trm8Δ/trm4Δ strains that leads to degradation of tRNAValAAC, but loss of even a single methylation can cause susceptibility of the tRNA to RTD.Citation17-19 Moreover, the new discovery that RTD is a widespread tRNA surveillance mechanism that acts on a large number of tRNA species in S. cerevisiae opens the door to many other connections between RTD and tRNA methylation.Citation117 Finally, methylation (among other modifications) in the anticodon loop is shown to exert important effects on translation. An example of this is the previously described m1G37 modification, which prevents frameshifting, particularly at codons that start with a C, and the slow growth phenotype associated with deletion of TRM5 in S. cerevisiae is likely due to decreased translational fidelity.Citation9,118 Despite remaining unanswered questions regarding the molecular function(s) of tRNA methylation, defects in tRNA methyltransferases have been increasingly associated with significant biological consequences, including a number of human diseases.
Dynamics of methylation in stress and disease
The modification status of tRNA is often considered to be a static feature of tRNA biology, with each tRNA species in the cell carrying a uniform complement of modified nucleotides. However, the modification state of tRNA can change in response to cell growth conditions and the mol/mol quantity of a particular methylation on a single tRNA isotype can vary. Interestingly, some methyltransferases may function in a network where changes in one enzyme allow for altered activity of another. For example, deletion of TRM82 () in S. cerevisiae results in a detectable increase in other tRNA methylations, suggesting that Trm82 could negatively affect other methyltransferases or modification enzymes.Citation119
Rearrangements of tRNA methylation in response to stress have been associated with translational control. In S. cerevisiae, tRNALeuCAA contains m5C at positions 34 and 48, which are both introduced by the methyltransferase Trm4, and the trm4Δ strain is hypersensitive to growth in the presence of H2O2.Citation119 Analysis of the methylation status of tRNALeuCAA derived from wild-type cells grown in the presence of H2O2 revealed a substantial increase in levels of m5C34 (by 70%) and a smaller (20%) decrease in the amount of m5C48.Citation120 Since tRNALeuCAA is the only tRNA in S. cerevisiae that contains the m5C34 modification, it was proposed that this anticodon methylation could stimulate translation of UUG-enriched mRNA. Interestingly, a protein of the 60S ribosome, Rpl22A, contains an overrepresentation of UUG codons in its mRNA transcript and growth of an rpl22aΔ strain is also hypersensitive to growth in the presence of H2O2.Citation120 Together, these results suggest that Rpl22A expression is advantageous for survival under oxidative stress conditions, and that Trm4 plays a regulatory role in the expression of this protein under these stress conditions, due to the increase in its production of m5C34 in tRNALeuCAA.
Trm4 and its corresponding m5C methylation are also involved in another example of altered levels of tRNA modification in response to stress. In a separate investigation of the G-1 nucleotidyl transferase Thg1, it was observed that tRNAHis, the substrate of Thg1 in S. cerevisiae, accumulates additional m5C methylations at positions 48 and 50 when Thg1 function (and hence the G-1 residue that is required for efficient aminoacylation) is lost due to repressed expression of the gene.Citation121 However, other challenging growth conditions that are not directly related to histidine or tRNAHis, such as starvation for other amino acids or glucose, also lead to accumulation of additional m5C on the same tRNA.Citation122 The fact that the additional m5C modification only occurs on tRNAHis and not on other tRNAs that also contain unmodified C nucleotides at the same positions is not understood and may have to do with localization or other particular chemical features of this tRNA. Although the biological function of m5C accumulation in response to stress in S. cerevisiae has not been deduced, it is posited that the additional m5C could confer a protective element to the tRNA, perhaps by decreasing its susceptibility to degradation or by preventing stress-associated cleavage events.Citation122 More work is required to fully understand the function of altered methylation levels in biological systems, but these data suggest that tRNA methylation is much more complex and dynamic than previously understood.
The previous examples all demonstrate phenotypic responses related to loss of tRNA methylations in unicellular eukaryotes, but tRNA modification also has important effects on human biology. A growing number of human disease syndromes are associated with genetic defects in tRNA methylation enzymes, and although the specific mechanism(s) by which mutations in human tRNA methyltransferases can cause these effects are not well-understood, it appears that loss of tRNA methylation can have devastating consequences for higher eukaryotes. Multiple associations between tRNA defects (including modifications) and human mitochondrial disease are well-established (seeCitation123), and here we focus instead on recent developments specifically associated with mutations in individual tRNA methyltransferases.
Two independent reports recently identified human familial syndromes involving microcephaly, intellectual disability and defects in glucose metabolism with mutations in TRMT10A, which is one of the 3 paralogs of S. cerevisiae Trm10 that are encoded by mammals, including humans.Citation82 Two separate mutations (either nonsense or missense) in TRMT10A were identified in multiple members of different large families and shown to correlate with the observed patient abnormalities.Citation124,125 Biochemical studies of the missense mutant of TRMT10A (with an G208R substitution in the putative SAM binding site) demonstrated a complete loss of function associated with the mutation, which suggests that loss of tRNA methylation could be the direct cause of the disease symptoms.Citation124 Notably, TRMT10A is ubiquitously expressed, but particularly high concentrations are observed in embryonic and adult brains as well as the pancreas.Citation125 However, knockdown of TRMT10A in β-cells did not impair insulin secretion or levels, but instead induced apoptosis, which is perhaps connected to the observation that tRNAs prevent formation of the apoptosome by inhibiting binding of cytochrome c to Apaf-1.Citation126,127
In the same vein, multiple mutations in NSun2, a human homolog of the m5C tRNA methyltransferase Trm4, are correlated with intellectual disability and a Dubowitz-like syndrome characterized by distinct facial features and other physical abnormalities.Citation128-130 In the case of NSun2, tRNAs lacking m5C are cleaved endonucleolytically by angiogenin causing the resulting 5′ tRNA fragments to accumulate.Citation131 It is noteworthy that angiogenin cleaves tRNA at the variable loop, while NSun2 also methylates at multiple positions in the variable loop.Citation132,133 An exciting hypothesis that connects the loss of tRNA methylation to disease postulates that, when under stress, such as the oxidative stress that has been linked to neurodevelopmental disorders, accumulation of 5′ tRNA fragments would repress translation, and the concomitant lower protein levels could then explain decreased neuronal size.Citation131 In support of this, the inhibition of angiogenin via RNAi inhibits cleavage of non-methylated tRNAs and rescues brain size of NSun2-/- mice.Citation131
Other genetic mutations in tRNA methyltransferases that cause human disease phenotypes may yet be described, since tRNA and various associated modification enzymes have been implicated in a number of other diseases, such as cancer and metabolic syndromes.Citation6,134 Further characterization of the molecular basis for the specific effects of loss of methylation on human biology will be important for a complete understanding of these diseases, and therefore for developing potential therapies. The recent resurgence in interest in tRNA biology and processing events and in the enzymes that catalyze these reactions will likely yield new discoveries that could improve human health.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J. tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 2009; 37:D159-62; PMID:18957446; http://dx.doi.org/10.1093/nar/gkn772
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262-7; PMID:23118484; http://dx.doi.org/10.1093/nar/gks1007
- Jackman JE, Alfonzo JD. Transfer RNA modifications: nature's combinatorial chemistry playground. Wiley Interdiscip Reviews RNA 2013; 4:35-48; PMID:23139145; http://dx.doi.org/10.1002/wrna.1144
- Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 2002; 8:1189-232; PMID:12403461; http://dx.doi.org/10.1017/S1355838202022021
- Grosjean H. DNA and RNA modification enzymes: structure, mechanism, function and evolution. Austin, Tex.: Landes Bioscience, 2009.
- Hori H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front Genet 2014; 5:144; PMID:24904644; http://dx.doi.org/10.3389/fgene.2014.00144
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010; 24:1832-60; PMID:20810645; http://dx.doi.org/10.1101/gad.1956510
- Gustavsson M, Ronne H. Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA 2008; 14:666-74; PMID:18314501; http://dx.doi.org/10.1261/rna.966208
- Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? Embo J 2001; 20:231-9; PMID:11226173; http://dx.doi.org/10.1093/emboj/20.1.231
- Bystrom AS, Bjork GR. The structural gene (trmD) for the tRNA(m1G)methyltransferase is part of a four polypeptide operon in Escherichia coli K-12. Mol Gen Genet 1982; 188:447-54; PMID:6298574; http://dx.doi.org/10.1007/BF00330047
- Takeda H, Toyooka T, Ikeuchi Y, Yokobori S, Okadome K, Takano F, Oshima T, Suzuki T, Endo Y, Hori H. The substrate specificity of tRNA (m1G37) methyltransferase (TrmD) from Aquifex aeolicus. Genes Cells 2006; 11:1353-65; PMID:17121543; http://dx.doi.org/10.1111/j.1365-2443.2006.01022.x
- Christian T, Hou YM. Distinct determinants of tRNA recognition by the TrmD and Trm5 methyl transferases. J Mol Biol 2007; 373:623-32; PMID:17868690; http://dx.doi.org/10.1016/j.jmb.2007.08.010
- Goto-Ito S, Ito T, Ishii R, Muto Y, Bessho Y, Yokoyama S. Crystal structure of archaeal tRNA(m(1)G37)methyltransferase aTrm5. Proteins 2008; 72:1274-89; PMID:18384044; http://dx.doi.org/10.1002/prot.22019
- Ahn HJ, Kim HW, Yoon HJ, Lee BI, Suh SW, Yang JK. Crystal structure of tRNA(m1G37)methyltransferase: insights into tRNA recognition. EMBO J 2003; 22:2593-603; PMID:12773376; http://dx.doi.org/10.1093/emboj/cdg269
- Elkins PA, Watts JM, Zalacain M, van Thiel A, Vitazka PR, Redlak M, Andraos-Selim C, Rastinejad F, Holmes WM. Insights into catalysis by a knotted TrmD tRNA methyltransferase. J Mol Biol 2003; 333:931-49; PMID:14583191; http://dx.doi.org/10.1016/j.jmb.2003.09.011
- Liu J, Wang W, Shin DH, Yokota H, Kim R, Kim SH. Crystal structure of tRNA (m1G37) methyltransferase from Aquifex aeolicus at 2.6 A resolution: a novel methyltransferase fold. Proteins 2003; 53:326-8; PMID:14517984; http://dx.doi.org/10.1002/prot.10479
- Whipple JM, Lane EA, Chernyakov I, D'Silva S, Phizicky EM. The yeast rapid tRNA decay pathway primarily monitors the structural integrity of the acceptor and T-stems of mature tRNA. Genes Dev 2011; 25:1173-84; PMID:21632824; http://dx.doi.org/10.1101/gad.2050711
- Dewe JM, Whipple JM, Chernyakov I, Jaramillo LN, Phizicky EM. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 2012; 18:1886-96; PMID:22895820; http://dx.doi.org/10.1261/rna.033654.112
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'-3' exonucleases Rat1 and Xrn1. Genes Dev 2008; 22:1369-80; PMID:18443146; http://dx.doi.org/10.1101/gad.1654308
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 2006; 21:87-96; PMID:16387656; http://dx.doi.org/10.1016/j.molcel.2005.10.036
- Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. Embo J 2001; 20:4863-73; PMID:11532950; http://dx.doi.org/10.1093/emboj/20.17.4863
- Motorin Y, Helm M. tRNA stabilization by modified nucleotides. Biochemistry 2010; 49:4934-44; PMID:20459084; http://dx.doi.org/10.1021/bi100408z
- Voigts-Hoffmann F, Hengesbach M, Kobitski AY, van Aerschot A, Herdewijn P, Nienhaus GU, et al. A methyl group controls conformational equilibrium in human mitochondrial tRNA(Lys). J Am Chem Soc 2007; 129:13382-3; PMID:17941640; http://dx.doi.org/10.1021/ja075520+
- Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: a chronicle of convergence. Trends Bioch Sci 2003; 28:329-35; PMID:12826405; http://dx.doi.org/10.1016/S0968-0004(03)00090-2
- Anantharaman V, Koonin EV, Aravind L. SPOUT: a class of methyltransferases that includes spoU and trmD RNA methylase superfamilies, and novel superfamilies of predicted prokaryotic RNA methylases. J Mol Microbiol Biotechnol 2002; 4:71-5; PMID:11763972
- Tkaczuk KL, Dunin-Horkawicz S, Purta E, Bujnicki JM. Structural and evolutionary bioinformatics of the SPOUT superfamily of methyltransferases. BMC Bioinformatics 2007; 8:73; PMID:17338813; http://dx.doi.org/10.1186/1471-2105-8-73
- Kimura S, Miyauchi K, Ikeuchi Y, Thiaville PC, Crecy-Lagard V, Suzuki T. Discovery of the beta-barrel-type RNA methyltransferase responsible for N6-methylation of N6-threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res 2014; 42:9350-65; PMID:25063302; http://dx.doi.org/10.1093/nar/gku618
- Shao Z, Yan W, Peng J, Zuo X, Zou Y, Li F, Gong D, Ma R, Wu J, Shi Y, et al. Crystal structure of tRNA m1G9 methyltransferase Trm10: insight into the catalytic mechanism and recognition of tRNA substrate. Nucleic Acids Res 2014; 42:509-25; PMID:24081582; http://dx.doi.org/10.1093/nar/gkt869
- Kurowski MA, Sasin JM, Feder M, Debski J, Bujnicki JM. Characterization of the cofactor-binding site in the SPOUT-fold methyltransferases by computational docking of S-adenosylmethionine to three crystal structures. BMC Bioinformatics 2003; 4:9; PMID:12689347; http://dx.doi.org/10.1186/1471-2105-4-9
- Hou YM, Perona JJ. Stereochemical mechanisms of tRNA methyltransferases. FEBS Lett 2010; 584:278-86; PMID:19944101; http://dx.doi.org/10.1016/j.febslet.2009.11.075
- Alian A, Lee TT, Griner SL, Stroud RM, Finer-Moore J. Structure of a TrmA-RNA complex: A consensus RNA fold contributes to substrate selectivity and catalysis in m5U methyltransferases. Proc Natl Acad Sci U S A 2008; 105:6876-81; PMID:18451029; http://dx.doi.org/10.1073/pnas.0802247105
- Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S. Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat Struct Mol Biol 2009; 16:1109-15; PMID:19749755; http://dx.doi.org/10.1038/nsmb.1653
- Hamdane D, Argentini M, Cornu D, Myllykallio H, Skouloubris S, Hui-Bon-Hoa G, Golinelli-Pimpaneau B. Insights into folate/FAD-dependent tRNA methyltransferase mechanism: role of two highly conserved cysteines in catalysis. J Biol Chem 2011; 286:36268-80; PMID:21846722; http://dx.doi.org/10.1074/jbc.M111.256966
- Hamdane D, Guerineau V, Un S, Golinelli-Pimpaneau B. A catalytic intermediate and several flavin redox states stabilized by folate-dependent tRNA methyltransferase from Bacillus subtilis. Biochemistry 2011; 50:5208-19; PMID:21561081; http://dx.doi.org/10.1021/bi1019463
- Urbonavicius J, Skouloubris S, Myllykallio H, Grosjean H. Identification of a novel gene encoding a flavin-dependent tRNA:m5U methyltransferase in bacteria–evolutionary implications. Nucleic Acids Res 2005; 33:3955-64; PMID:16027442; http://dx.doi.org/10.1093/nar/gki703
- Santi DV, Hardy LW. Catalytic mechanism and inhibition of tRNA (uracil-5-)methyltransferase: evidence for covalent catalysis. Biochemistry 1987; 26:8599-606; PMID:3327525; http://dx.doi.org/10.1021/bi00400a016
- Kealey JT, Santi DV. Identification of the catalytic nucleophile of tRNA (m5U54)methyltransferase. Biochemistry 1991; 30:9724-8; PMID:1911760; http://dx.doi.org/10.1021/bi00104a022
- Kealey JT, Gu X, Santi DV. Enzymatic mechanism of tRNA (m5U54)methyltransferase. Biochimie 1994; 76:1133-42; PMID:7748948; http://dx.doi.org/10.1016/0300-9084(94)90042-6
- Bujnicki JM, Feder M, Ayres CL, Redman KL. Sequence-structure-function studies of tRNA:m5C methyltransferase Trm4p and its relationship to DNA:m5C and RNA:m5U methyltransferases. Nucleic Acids Res 2004; 32:2453-63; PMID:15121902; http://dx.doi.org/10.1093/nar/gkh564
- King MY, Redman KL. RNA methyltransferases utilize two cysteine residues in the formation of 5-methylcytosine. Biochemistry 2002; 41:11218-25; PMID:12220187; http://dx.doi.org/10.1021/bi026055q
- Christian T, Gamper H, Hou YM. Conservation of structure and mechanism by Trm5 enzymes. RNA 2013; 19:1192-9; PMID:23887145; http://dx.doi.org/10.1261/rna.039503.113
- Christian T, Lahoud G, Liu C, Hoffmann K, Perona JJ, Hou YM. Mechanism of N-methylation by the tRNA m1G37 methyltransferase Trm5. RNA 2010; 16:2484-92; PMID:20980671; http://dx.doi.org/10.1261/rna.2376210
- Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res 2004; 32:465-76; PMID:14739239; http://dx.doi.org/10.1093/nar/gkh191
- Guelorget A, Barraud P, Tisne C, Golinelli-Pimpaneau B. Structural comparison of tRNA m(1)A58 methyltransferases revealed different molecular strategies to maintain their oligomeric architecture under extreme conditions. BMC Struct Biol 2011; 11:48; PMID:22168821; http://dx.doi.org/10.1186/1472-6807-11-48
- Barraud P, Golinelli-Pimpaneau B, Atmanene C, Sanglier S, Van Dorsselaer A, Droogmans L, Dardel F, Tisné C. Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J Mol Biol 2008; 377:535-50; PMID:18262540; http://dx.doi.org/10.1016/j.jmb.2008.01.041
- Kuratani M, Yanagisawa T, Ishii R, Matsuno M, Si SY, Katsura K, Ushikoshi-Nakayama R, Shibata R, Shirouzu M, Bessho Y, et al. Crystal structure of tRNA m(1)A58 methyltransferase TrmI from Aquifex aeolicus in complex with S-adenosyl-L-methionine. J Struct Funct Genomics 2014; 15:173-80; PMID:24894648; http://dx.doi.org/10.1007/s10969-014-9183-0
- Chatterjee K, Blaby IK, Thiaville PC, Majumder M, Grosjean H, Yuan YA, Gupta R, de Crécy-Lagard V. The archaeal COG1901/DUF358 SPOUT-methyltransferase members, together with pseudouridine synthase Pus10, catalyze the formation of 1-methylpseudouridine at position 54 of tRNA. RNA 2012; 18:421-33; PMID:22274953; http://dx.doi.org/10.1261/rna.030841.111
- Wurm JP, Griese M, Bahr U, Held M, Heckel A, Karas M, Soppa J, Wöhnert J. Identification of the enzyme responsible for N1-methylation of pseudouridine 54 in archaeal tRNAs. RNA 2012; 18:412-20; PMID:22274954; http://dx.doi.org/10.1261/rna.028498.111
- Zegers I, Gigot D, van Vliet F, Tricot C, Aymerich S, Bujnicki JM, Kosinski J, Droogmans L. Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase. Nucleic Acids Res 2006; 34:1925-34; PMID:16600901; http://dx.doi.org/10.1093/nar/gkl116
- Leulliot N, Chaillet M, Durand D, Ulryck N, Blondeau K, van Tilbeurgh H. Structure of the yeast tRNA m7G methylation complex. Structure 2008; 16:52-61; PMID:18184583; http://dx.doi.org/10.1016/j.str.2007.10.025
- Purta E, van Vliet F, Tricot C, De Bie LG, Feder M, Skowronek K, Droogmans L, Bujnicki JM. Sequence-structure-function relationships of a tRNA (m7G46) methyltransferase studied by homology modeling and site-directed mutagenesis. Proteins 2005; 59:482-8; PMID:15789416; http://dx.doi.org/10.1002/prot.20454
- D'Silva S, Haider SJ, Phizicky EM. A domain of the actin binding protein Abp140 is the yeast methyltransferase responsible for 3-methylcytidine modification in the tRNA anti-codon loop. RNA 2011; 17:1100-10; PMID:21518804; http://dx.doi.org/10.1261/rna.2652611
- Noma A, Yi S, Katoh T, Takai Y, Suzuki T. Actin-binding protein ABP140 is a methyltransferase for 3-methylcytidine at position 32 of tRNAs in Saccharomyces cerevisiae. RNA 2011; 17:1111-9; PMID:21518805; http://dx.doi.org/10.1261/rna.2653411
- Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol Cell Biol 2005; 25:4359-70; PMID:15899842; http://dx.doi.org/10.1128/MCB.25.11.4359-4370.2005
- Constantinesco F, Motorin Y, Grosjean H. Characterisation and enzymatic properties of tRNA(guanine 26, N (2), N (2))-dimethyltransferase (Trm1p) from Pyrococcus furiosus. J Mol Biol 1999; 291:375-92; PMID:10438627; http://dx.doi.org/10.1006/jmbi.1999.2976
- Edqvist J, Straby KB, Grosjean H. Enzymatic formation of N2,N2-dimethylguanosine in eukaryotic tRNA: importance of the tRNA architecture. Biochimie 1995; 77:54-61; PMID:7599276; http://dx.doi.org/10.1016/0300-9084(96)88104-1
- Ellis SR, Morales MJ, Li JM, Hopper AK, Martin NC. Isolation and characterization of the TRM1 locus, a gene essential for the N2,N2-dimethylguanosine modification of both mitochondrial and cytoplasmic tRNA in Saccharomyces cerevisiae. J Biol Chem 1986; 261:9703-9; PMID:2426253
- Armengaud J, Urbonavicius J, Fernandez B, Chaussinand G, Bujnicki JM, Grosjean H. N2-methylation of guanosine at position 10 in tRNA is catalyzed by a THUMP domain-containing, S-adenosylmethionine-dependent methyltransferase, conserved in Archaea and Eukaryota. J Biol Chem 2004; 279:37142-52; PMID:15210688; http://dx.doi.org/10.1074/jbc.M403845200
- Awai T, Kimura S, Tomikawa C, Ochi A, Ihsanawati Y, Bessho Y, Yokoyama S, Ohno S, Nishikawa K, Yokogawa T, et al. Aquifex aeolicus tRNA (N2,N2-guanine)-dimethyltransferase (Trm1) catalyzes transfer of methyl groups not only to guanine 26 but also to guanine 27 in tRNA. J Biol Chem 2009; 284:20467-78; PMID:19491098; http://dx.doi.org/10.1074/jbc.M109.020024
- Awai T, Ochi A, Ihsanawati T, Sengoku T, Hirata A, Bessho Y, Yokoyama S, Hori H. Substrate tRNA recognition mechanism of a multisite-specific tRNA methyltransferase, Aquifex aeolicus Trm1, based on the X-ray crystal structure. J Biol Chem 2011; 286:35236-46; PMID:21844194; http://dx.doi.org/10.1074/jbc.M111.253641
- Ihsanawati M, Nishimoto M, Higashijima K, Shirouzu M, Grosjean H, Bessho Y, Yokoyama S. Crystal structure of tRNA N2,N2-guanosine dimethyltransferase Trm1 from Pyrococcus horikoshii. J Mol Biol 2008; 383:871-84; PMID:18789948; http://dx.doi.org/10.1016/j.jmb.2008.08.068
- Golovina AY, Sergiev PV, Golovin AV, Serebryakova MV, Demina I, Govorun VM, Dontsova OA. The yfiC gene of E. coli encodes an adenine-N6 methyltransferase that specifically modifies A37 of tRNA1Val(cmo5UAC). RNA 2009; 15:1134-41; PMID:19383770; http://dx.doi.org/10.1261/rna.1494409
- de Crecy-Lagard V, Marck C, Brochier-Armanet C, Grosjean H. Comparative RNomics and modomics in Mollicutes: prediction of gene function and evolutionary implications. IUBMB Life 2007; 59:634-58; PMID:17852564; http://dx.doi.org/10.1080/15216540701604632
- Watanabe K, Nureki O, Fukai S, Endo Y, Hori H. Functional categorization of the conserved basic amino acid residues in TrmH (tRNA (Gm18) methyltransferase) enzymes. J Biol Chem 2006; 281:34630-9; PMID:16963456; http://dx.doi.org/10.1074/jbc.M606141200
- Nureki O, Watanabe K, Fukai S, Ishii R, Endo Y, Hori H, Yokoyama S. Deep knot structure for construction of active site and cofactor binding site of tRNA modification enzyme. Structure 2004; 12:593-602; PMID:15062082; http://dx.doi.org/10.1016/j.str.2004.03.003
- Czerwoniec A, Dunin-Horkawicz S, Purta E, Kaminska KH, Kasprzak JM, Bujnicki JM, Grosjean H, Rother K. MODOMICS: a database of RNA modification pathways. 2008 update. Nucleic Acids Res 2009; 37:D118-21; PMID:18854352; http://dx.doi.org/10.1093/nar/gkn710
- Grosjean H, Edqvist J, Straby KB, Giege R. Enzymatic formation of modified nucleosides in tRNA: dependence on tRNA architecture. J Mol Biol 1996; 255:67-85; PMID:8568876; http://dx.doi.org/10.1006/jmbi.1996.0007
- Jackman JE. tRNA biogenesis. Encyclopedia of Life Sciences. Chichester: John Wiley & Sons, Ltd., 2011; http://dx.doi.org/10.1002/9780470015902.a002089
- Hori H, Yamazaki N, Matsumoto T, Watanabe Y, Ueda T, Nishikawa K, Kumagai I, Watanabe K. Substrate recognition of tRNA (Guanosine-2'-)-methyltransferase from Thermus thermophilus HB27. J Biol Chem 1998; 273:25721-7; PMID:9748240; http://dx.doi.org/10.1074/jbc.273.40.25721
- Sakaguchi R, Giessing A, Dai Q, Lahoud G, Liutkeviciute Z, Klimasauskas S, Piccirilli J, Kirpekar F, Hou YM Recognition of guanosine by dissimilar tRNA methyltransferases. RNA 2012; 18:1687-701; PMID:22847817; http://dx.doi.org/10.1261/rna.032029.111
- Jeltsch A, Nellen W, Lyko F. Two substrates are better than one: dual specificities for Dnmt2 methyltransferases. Trends Biochem Sci 2006; 31:306-8; PMID:16679017; http://dx.doi.org/10.1016/j.tibs.2006.04.005
- Goll MG, Kirpekar F, Maggert KA, Yoder JA, Hsieh CL, Zhang X, Golic KG, Jacobsen SE, Bestor TH. Methylation of tRNAAsp by the DNA methyltransferase homolog Dnmt2. Science 2006; 311:395-8; PMID:16424344; http://dx.doi.org/10.1126/science.1120976
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010; 24:1590-5; PMID:20679393; http://dx.doi.org/10.1101/gad.586710
- Becker M, Muller S, Nellen W, Jurkowski TP, Jeltsch A, Ehrenhofer-Murray AE. Pmt1, a Dnmt2 homolog in Schizosaccharomyces pombe, mediates tRNA methylation in response to nutrient signaling. Nucleic Acids Res 2012; 40:11648-58; PMID:23074192; http://dx.doi.org/10.1093/nar/gks956
- Muller S, Windhof IM, Maximov V, Jurkowski T, Jeltsch A, Forstner KU, Sharma CM, Gräf R, Nellen W. Target recognition, RNA methylation activity and transcriptional regulation of the Dictyostelium discoideum Dnmt2-homologue (DnmA). Nucleic Acids Res 2013; 41:8615-27; PMID:23877245; http://dx.doi.org/10.1093/nar/gkt634
- Shanmugam R, Aklujkar M, Schafer M, Reinhardt R, Nickel O, Reuter G, Lovley DR, Ehrenhofer-Murray A, Nellen W, Ankri S, et al. The Dnmt2 RNA methyltransferase homolog of Geobacter sulfurreducens specifically methylates tRNA-Glu. Nucleic Acids Res 2014; 42:6487-96; PMID:24711368; http://dx.doi.org/10.1093/nar/gku256
- Ochi A, Makabe K, Kuwajima K, Hori H. Flexible recognition of the tRNA G18 methylation target site by TrmH methyltransferase through first binding and induced fit processes. J Biol Chem 2010; 285:9018-29; PMID:20053984; http://dx.doi.org/10.1074/jbc.M109.065698
- Hori H, Kubota S, Watanabe K, Kim JM, Ogasawara T, Sawasaki T, Endo Y. Aquifex aeolicus tRNA (Gm18) methyltransferase has unique substrate specificity. TRNA recognition mechanism of the enzyme. J Biol Chem 2003; 278:25081-90; PMID:12704200; http://dx.doi.org/10.1074/jbc.M212577200
- Ochi A, Makabe K, Yamagami R, Hirata A, Sakaguchi R, Hou YM, Watanabe K, Nureki O, Kuwajima K, Hori H. The catalytic domain of topological knot tRNA methyltransferase (TrmH) discriminates between substrate tRNA and nonsubstrate tRNA via an induced-fit process. J Biol Chem 2013; 288:25562-74; PMID:23867454; http://dx.doi.org/10.1074/jbc.M113.485128
- Somme J, Van Laer B, Roovers M, Steyaert J, Versees W, Droogmans L. Characterization of two homologous 2'-O-methyltransferases showing different specificities for their tRNA substrates. RNA 2014; 20:1257-71; PMID:24951554; http://dx.doi.org/10.1261/rna.044503.114
- Swinehart WE, Henderson JC, Jackman JE. Unexpected expansion of tRNA substrate recognition by the yeast m1G9 methyltransferase Trm10. RNA 2013; PMID:23793893
- Jackman JE, Montange RK, Malik HS, Phizicky EM. Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 2003; 9:574-85; PMID:12702816; http://dx.doi.org/10.1261/rna.5070303
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 2008; 135:462-74; PMID:18984158; http://dx.doi.org/10.1016/j.cell.2008.09.013
- Vilardo E, Nachbagauer C, Buzet A, Taschner A, Holzmann J, Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res 2012; 40:11583-93; PMID:23042678; http://dx.doi.org/10.1093/nar/gks910
- Ranaei-Siadat E, Fabret C, Seijo B, Dardel F, Grosjean H, Nonin-Lecomte S. RNA-methyltransferase TrmA is a dual-specific enzyme responsible for C5-methylation of uridine in both tmRNA and tRNA. RNA Biol 2013; 10:572-8; PMID:23603891; http://dx.doi.org/10.4161/rna.24327
- Auxilien S, Rasmussen A, Rose S, Brochier-Armanet C, Husson C, Fourmy D, Grosjean H, Douthwaite S. Specificity shifts in the rRNA and tRNA nucleotide targets of archaeal and bacterial m5U methyltransferases. RNA 2011; 17:45-53; PMID:21051506; http://dx.doi.org/10.1261/rna.2323411
- Gu X, Ofengand J, Santi DV. In vitro methylation of Escherichia coli 16S rRNA by tRNA (m5U54)-methyltransferase. Biochemistry 1994; 33:2255-61; PMID:8117682; http://dx.doi.org/10.1021/bi00174a036
- Walbott H, Leulliot N, Grosjean H, Golinelli-Pimpaneau B. The crystal structure of Pyrococcus abyssi tRNA (uracil-54, C5)-methyltransferase provides insights into its tRNA specificity. Nucleic Acids Res 2008; 36:4929-40; PMID:18653523; http://dx.doi.org/10.1093/nar/gkn437
- Urbonavicius J, Auxilien S, Walbott H, Trachana K, Golinelli-Pimpaneau B, Brochier-Armanet C, Grosjean H. Acquisition of a bacterial RumA-type tRNA(uracil-54, C5)-methyltransferase by Archaea through an ancient horizontal gene transfer. Mol Microbiol 2008; 67:323-35; PMID:18069966; http://dx.doi.org/10.1111/j.1365-2958.2007.06047.x
- Yao LJ, James TL, Kealey JT, Santi DV, Schmitz U. The dynamic NMR structure of the T psi C-loop: implications for the specificity of tRNA methylation. J Biomol NMR 1997; 9:229-44; PMID:9204554; http://dx.doi.org/10.1023/A:1018618606857
- Benitez-Paez A, Villarroya M, Armengod ME. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA 2012; 18:1783-95; PMID:22891362; http://dx.doi.org/10.1261/rna.033266.112
- Paris Z, Horakova E, Rubio MA, Sample P, Fleming IM, Armocida S, Lukes J, Alfonzo JD. The T. brucei TRM5 methyltransferase plays an essential role in mitochondrial protein synthesis and function. RNA 2013; 19:649-58; PMID:23520175; http://dx.doi.org/10.1261/rna.036665.112
- Kempenaers M, Roovers M, Oudjama Y, Tkaczuk KL, Bujnicki JM, Droogmans L. New archaeal methyltransferases forming 1-methyladenosine or 1-methyladenosine and 1-methylguanosine at position 9 of tRNA. Nucleic Acids Res 2010; 38:6533-43; PMID:20525789; http://dx.doi.org/10.1093/nar/gkq451
- Urbonavicius J, Meskys R, Grosjean H. Biosynthesis of wyosine derivatives in tRNA(Phe) of Archaea: role of a remarkable bifunctional tRNA(Phe):m1G/imG2 methyltransferase. RNA 2014; 20:747-53; PMID:24837075; http://dx.doi.org/10.1261/rna.043315.113
- de Crecy-Lagard V, Brochier-Armanet C, Urbonavicius J, Fernandez B, Phillips G, Lyons B, Noma A, Alvarez S, Droogmans L, Armengaud J, Grosjean H. Biosynthesis of wyosine derivatives in tRNA: an ancient and highly diverse pathway in Archaea. Mol Biol Evol 2010; 27:2062-77; PMID:20382657; http://dx.doi.org/10.1093/molbev/msq096
- Sakaguchi R, Lahoud G, Christian T, Gamper H, Hou YM. A divalent metal ion-dependent N(1)-methyl transfer to G37-tRNA. Chem Biol 2014; 21:1351-60; PMID:25219964; http://dx.doi.org/10.1016/j.chembiol.2014.07.023
- Urbonavicius J, Brochier-Armanet C, Skouloubris S, Myllykallio H, Grosjean H. In vitro detection of the enzymatic activity of folate-dependent tRNA (Uracil-54,-C5)-methyltransferase: evolutionary implications. Methods Enzymol 2007; 425:103-19; PMID:17673080; http://dx.doi.org/10.1016/S0076-6879(07)25004-9
- Kim J, Xiao H, Bonanno JB, Kalyanaraman C, Brown S, Tang X, Al-Obaidi NF, Patskovsky Y, Babbitt PC, Jacobson MP, et al. Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function. Nature 2013; 498:123-6; PMID:23676670; http://dx.doi.org/10.1038/nature12180
- Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 2002; 8:1253-66; PMID:12403464; http://dx.doi.org/10.1017/S1355838202024019
- Matsumoto K, Toyooka T, Tomikawa C, Ochi A, Takano Y, Takayanagi N, Endo Y, Hori H. RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8-Trm82 complex). FEBS Lett 2007; 581:1599-604; PMID:17382321; http://dx.doi.org/10.1016/j.febslet.2007.03.023
- Alexandrov AV, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. Rna 2002; 8:1253-66; PMID:12403464; http://dx.doi.org/10.1017/S1355838202024019
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Björk GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1- methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 1998; 12:3650-62; PMID:9851972; http://dx.doi.org/10.1101/gad.12.23.3650
- Anderson J, Phan L, Hinnebusch AG. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1- methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2000; 97:5173-8; PMID:10779558; http://dx.doi.org/10.1073/pnas.090102597
- Ozanick SG, Bujnicki JM, Sem DS, Anderson JT. Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 Mtase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res 2007; 35:6808-19; PMID:17932071; http://dx.doi.org/10.1093/nar/gkm574
- Heurgue-Hamard V, Graille M, Scrima N, Ulryck N, Champ S, van Tilbeurgh H, Buckingham RH. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J Biol Chem 2006; 281:36140-8; PMID:17008308; http://dx.doi.org/10.1074/jbc.M608571200
- Liger D, Mora L, Lazar N, Figaro S, Henri J, Scrima N, Buckingham RH, van Tilbeurgh H, Heurgué-Hamard V, Graille M. Mechanism of activation of methyltransferases involved in translation by the Trm112 'hub' protein. Nucleic Acids Res 2011; 39:6249-59; PMID:21478168; http://dx.doi.org/10.1093/nar/gkr176
- Mazauric MH, Dirick L, Purushothaman SK, Bjork GR, Lapeyre B. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J Biol Chem 2010; 285:18505-15; PMID:20400505; http://dx.doi.org/10.1074/jbc.M110.113100
- Sardana R, Johnson AW. The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits. Mol Biol Cell 2012; 23:4313-22; PMID:22956767; http://dx.doi.org/10.1091/mbc.E12-05-0370
- Figaro S, Wacheul L, Schillewaert S, Graille M, Huvelle E, Mongeard R, Zorbas C, Lafontaine DL, Heurgué-Hamard V. Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol Cell Biol 2012; 32:2254-67; PMID:22493060; http://dx.doi.org/10.1128/MCB.06623-11
- Hu Z, Qin Z, Wang M, Xu C, Feng G, Liu J, Meng Z, Hu Y. The Arabidopsis SMO2, a homologue of yeast TRM112, modulates progression of cell division during organ growth. Plant J 2010; 61:600-10; PMID:19929876; http://dx.doi.org/10.1111/j.1365-313X.2009.04085.x
- Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B. Trm7p catalyses the formation of two 2'-O-methylriboses in yeast tRNA anticodon loop. EMBO J 2002; 21:1811-20; PMID:11927565; http://dx.doi.org/10.1093/emboj/21.7.1811
- Tsai EY, Thim S, Baena A, Boussiotis VA, Reynes JM, Sath S, Grosjean P, Yunis EJ, Goldfeld AE. Trm7p catalyses the formation of two 2'-O-methylriboses in yeast tRNA anticodon loop. Proc Natl Acad Sci U S A 2002; 99:7576-81; PMID:12032325; http://dx.doi.org/10.1073/pnas.062056099
- Guy MP, Podyma BM, Preston MA, Shaheen HH, Krivos KL, Limbach PA, Hopper AK, Phizicky EM. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 2012; 18:1921-33; PMID:22912484; http://dx.doi.org/10.1261/rna.035287.112
- Helm M, Attardi G. Nuclear control of cloverleaf structure of human mitochondrial tRNA(Lys). J Mol Biol 2004; 337:545-60; PMID:15019776; http://dx.doi.org/10.1016/j.jmb.2004.01.036
- Edmonds CG, Crain PF, Gupta R, Hashizume T, Hocart CH, Kowalak JA, Pomerantz SC, Stetter KO, McCloskey JA. Posttranscriptional modification of tRNA in thermophilic archaea (Archaebacteria). J Bacteriol 1991; 173:3138-48; PMID:1708763
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 2004; 18:1227-40; PMID:15145828; http://dx.doi.org/10.1101/gad.1183804
- Guy MP, Young DL, Payea MJ, Zhang X, Kon Y, Dean KM, Grayhack EJ, Mathews DH, Fields S, Phizicky EM. Identification of the determinants of tRNA function and susceptibility to rapid tRNA decay by high-throughput in vivo analysis. Genes Dev 2014; 28:1721-32; PMID:25085423; http://dx.doi.org/10.1101/gad.245936.114
- Bjork GR, Wikstrom PM, Bystrom AS. Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 1989; 244:986-9; PMID:2471265; http://dx.doi.org/10.1126/science.2471265
- Chan CT, Dyavaiah M, DeMott MS, Taghizadeh K, Dedon PC, Begley TJ. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet 2010; 6:e1001247; PMID:NOT_FOUND
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 2012; 3:937; PMID:22760636; http://dx.doi.org/10.1038/ncomms1938
- Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol Cell Biol 2005; 25:8191-201; PMID:16135808; http://dx.doi.org/10.1128/MCB.25.18.8191-8201.2005
- Preston MA, D'Silva S, Kon Y, Phizicky EM. tRNAHis 5-methylcytidine levels increase in response to several growth arrest conditions in Saccharomyces cerevisiae. RNA 2013; 19:243-56; PMID:23249748; http://dx.doi.org/10.1261/rna.035808.112
- Suzuki T, Nagao A. Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet 2011; 45:299-329; PMID:21910628; http://dx.doi.org/10.1146/annurev-genet-110410-132531
- Gillis D, Krishnamohan A, Yaacov B, Shaag A, Jackman JE, Elpeleg O. TRMT10A dysfunction is associated with abnormalities in glucose homeostasis, short stature and microcephaly. J Med Genet 2014; 51:581-6; PMID:25053765; http://dx.doi.org/10.1136/jmedgenet-2014-102282
- Igoillo-Esteve M, Genin A, Lambert N, Desir J, Pirson I, Abdulkarim B, Simonis N, Drielsma A, Marselli L, Marchetti P, et al. tRNA methyltransferase homolog gene TRMT10A mutation in young onset diabetes and primary microcephaly in humans. PLoS Genet 2013; 9:e1003888; PMID:24204302; http://dx.doi.org/10.1371/journal.pgen.1003888
- Mei Y, Yong J, Liu H, Shi Y, Meinkoth J, Dreyfuss G, Yang X. tRNA binds to cytochrome c and inhibits caspase activation. Mol Cell 2010; 37:668-78; PMID:20227371; http://dx.doi.org/10.1016/j.molcel.2010.01.023
- Hou YM, Yang X. Regulation of cell death by transfer RNA. Antioxid Redox Signal 2013; 19:583-94; PMID:23350625; http://dx.doi.org/10.1089/ars.2012.5171
- Martinez FJ, Lee JH, Lee JE, Blanco S, Nickerson E, Gabriel S, Frye M, Al-Gazali L, Gleeson JG. Whole exome sequencing identifies a splicing mutation in NSUN2 as a cause of a Dubowitz-like syndrome. J Med Genet 2012; 49:380-5; PMID:22577224; http://dx.doi.org/10.1136/jmedgenet-2011-100686
- Khan MA, Rafiq MA, Noor A, Hussain S, Flores JV, Rupp V, Vincent AK, Malli R, Ali G, Khan FS, et al. Mutation in NSUN2, which encodes an RNA methyltransferase, causes autosomal-recessive intellectual disability. Am J Hum Genet 2012; 90:856-63; PMID:22541562; http://dx.doi.org/10.1016/j.ajhg.2012.03.023
- Abbasi-Moheb L, Mertel S, Gonsior M, Nouri-Vahid L, Kahrizi K, Cirak S, Wieczorek D, Motazacker MM, Esmaeeli-Nieh S, Cremer K, et al. Mutations in NSUN2 cause autosomal-recessive intellectual disability. Am J Human Genet 2012; 90:847-55; PMID:22541559; http://dx.doi.org/10.1016/j.ajhg.2012.03.021
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 2014; 33:2020-39; PMID:25063673; http://dx.doi.org/10.15252/embj.201489282
- Auxilien S, Guerineau V, Szweykowska-Kulinska Z, Golinelli-Pimpaneau B. The human tRNA m (5) C methyltransferase Misu is multisite-specific. RNA Biol 2012; 9:1331-8; PMID:22995836; http://dx.doi.org/10.4161/rna.22180
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 2012; 19:900-5; PMID:22885326; http://dx.doi.org/10.1038/nsmb.2357
- Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med 2014; 20:306-14; PMID:24581449; http://dx.doi.org/10.1016/j.molmed.2014.01.008
- Sierzputowska-Gracz H, Gopal HD, Agris PF. Comparative structural analysis of 1-methyladenosine, 7-methylguanosine, ethenoadenosine and their protonated salts IV: 1H, 13C, and 15N NMR studies at natural isotope abundance. Nucleic Acids Res 1986; 14:7783-801; PMID:3022235; http://dx.doi.org/10.1093/nar/14.19.7783