Abstract
tRNA modifications are crucial for efficient and accurate protein translation, with defects often linked to disease. There are 7 cytoplasmic tRNA modifications in the yeast Saccharomyces cerevisiae that are formed by an enzyme consisting of a catalytic subunit and an auxiliary protein, 5 of which require only a single subunit in bacteria, and 2 of which are not found in bacteria. These enzymes include the deaminase Tad2-Tad3, and the methyltransferases Trm6-Trm61, Trm8-Trm82, Trm7-Trm732, and Trm7-Trm734, Trm9-Trm112, and Trm11-Trm112. We describe the occurrence and biological role of each modification, evidence for a required partner protein in S. cerevisiae and other eukaryotes, evidence for a single subunit in bacteria, and evidence for the role of the non-catalytic binding partner. Although it is unclear why these eukaryotic enzymes require partner proteins, studies of some 2-subunit modification enzymes suggest that the partner proteins help expand substrate range or allow integration of cellular activities.
Introduction
The complexity of the post-transcriptional tRNA modification machinery is remarkable, with 63 genes known to be required for synthesis of the 25 chemically distinct modifications found in the cytosolic tRNA of the yeast Saccharomyces cerevisiae.Citation1 Over the past decade it has become apparent that formation of a significant number of these modifications requires a complex comprised of 2 different subunits in eukaryotes, but where known, only one protein subunit in bacteria.Citation1,2 Remarkably, these complexes include 2 tRNA methyltransferases that share the same scaffold (along with 2 other methyltransferases), each catalyzing formation of different modifications,Citation3-8 as well as one methyltransferase that uses 2 different partner proteins for the same modification on different residues.Citation9-11Here we discuss these modifications and the corresponding complexes, to shed light on the origin and conservation of the complexes and the functions of the subunits.
Post-transcriptional tRNA modifications occur in all domains of life, including the simplest organisms.Citation12,13 Modifications in and around the anticodon loop are often critical for translational fidelity and efficiency,Citation14 whereas modifications in the body of the tRNA often contribute to tRNA folding and stability.Citation15-17 The importance of tRNA modifications is underscored by their high conservation in different organisms,Citation18 by the frequent occurrence of a defined growth defect due to deletion of genes required for modifications,Citation2,19 and by the increasingly frequent association of human diseases with defects in modification.Citation20
Several modifications on specific tRNA residues are known to be formed by a conserved enzyme family within all 3 domains of life.Citation13,18 Some examples include formation of Ψ38 and Ψ39 (pseudouridine, standard tRNA numbering system) by the Escherichia coli TrmA/S. cerevisiae Pus3 family of pseudouridylases,Citation22-24 formation of Ψ13 by the Ec TruD/Sc Pus7 family of pseudouridylases,Citation25-27 and formation of t6A37 (N6-threonylcarbamoyladenosine) by the core Sc Sua5/Ec YrdC and Sc KaeI/Ec YgjD families of proteins, along with other components depending on the organism.Citation28-32 By contrast, the highly conserved m1G37 (1-methylguanosine) modification is catalyzed by the Sc Trm5 family of Rossman fold methyltransferases in eukaryotes and archaea, and by the unrelated Ec TrmD family of SPOUT methyltransferases in prokaryotes.Citation33,34
Of particular interest are the modifications that are catalyzed by enzymes comprised of a catalytic subunit and a partner subunit in eukaryotes, but not, apparently, in prokaryotes. In the yeast S. cerevisiae, there are 5 tRNA modification enzymes that are composed of 2 distinct subunits (, ), whereas available evidence suggests only a single gene product is required in bacteria. Moreover, there are 2 additional modifications found in S. cerevisiae and other eukaryotes but not in bacteria that are catalyzed by 2-subunit enzymes.Citation1,2 Here we discuss the 7 S. cerevisiae tRNA modification enzymes that modify cytoplasmic tRNAs and are comprised of 2 distinct protein subunits. These two-subunit modification enzymes fall into 2 classes, one of which can be further subdivided into 3 subclasses based on common themes, as described below. For each enzyme, we will discuss the occurrence and biological role of the corresponding modification, evidence for the requirement of a partner protein for catalytic activity of the S. cerevisiae protein, evidence for the conservation of this requirement in other eukaryotes, evidence for a single catalytic component in bacteria, and what is known about the function of the non-catalytic binding partner.
Table 1. Eukaryotic tRNA modifications that require 2-subunit enzymes.
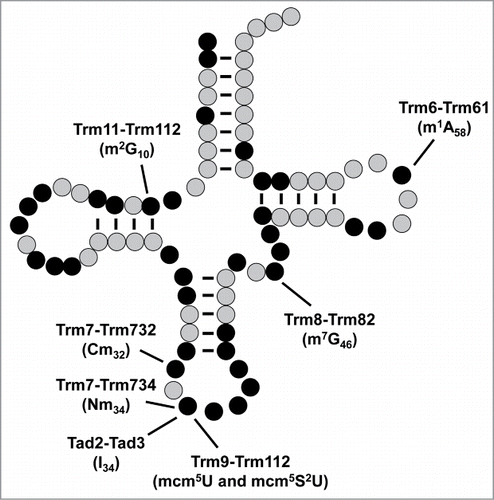
Figure 1. S. cerevisiae tRNA modifications formed by 2-subunit enzymes are located throughout the tRNA. Cloverleaf schematic depicting tRNA residues known to be modified (black) in S. cerevisiae, and those that are not modified (gray). Modifications (in brackets) formed by 2-subunit enzymes are labeled.
Heterodimers by Duplication and Divergence
As detailed below, there are 2 examples of eukaryotic 2-subunit enzymes that seem to have arisen by gene duplication events. For both of these complexes, it appears that the eukaryotic non-catalytic subunit has taken the place of one of the catalytic subunits of the bacterial homodimer,Citation35–37 perhaps to increase the substrate repertoire of the enzyme.Citation38,39
Tad2-Tad3, the A34 deaminase
Conversion of the wobble residue A34 to I34 (inosine) is thought to occur on the majority of tRNA species that encode an A34 residue in bacteria and eukaryotes, but is not known to occur in archaea, which lack tRNA genes encoding A34.Citation13,40 In prokaryotes, I34 is only known to occur on tRNAArg(ACG), and all 3 bacterial tRNAArg(ACG) species that have been examined contain I34. All 6 (of 7) S. cerevisiae tRNA species with an encoded A34 residue that have been examined have I34, and all eukaryotic cytoplasmic tRNAs with an encoded A34 that have been examined have the I34 modificationCitation13, with the exception of wheat tRNAArg(ACG).Citation41 I34 increases the reading capacity of tRNAs, allowing decoding of codons ending in U and C, and often A,Citation42,43 and lack of the modification gene is lethal in S. cerevisiae, S. pombe, and E. coli.Citation44-46
In S. cerevisiae, I34 is formed by the Sc Tad2-Tad3 protein pair.Citation44 Sc TAD2 was identified as being homologous to adenosine deaminases acting on RNA, the gene was determined to be essential, a temperature sensitive mutant was generated, extracts from the temperature sensitive mutant were shown to lack A34 deaminase activity, and the activity was complemented by additional recombinant Sc Tad2 purified from E. coli.Citation44 However, purified recombinant Sc Tad2 itself lacked deaminase activity, and purification of Sc Tad2 from yeast cells resulted in co-purification of an additional polypeptide, called Sc Tad3. A34 deaminase was concluded to be comprised of an Sc Tad2-Tad3 heterodimer, since Sc Tad2 and Sc Tad3 co-purified in stoichiometric amounts, and biochemical fractions containing both Sc Tad2 and Sc Tad3, but neither alone, could convert A34 to I on a synthetic tRNAAla construct.Citation44
It is likely that TAD2 and TAD3 arose by a gene duplication event, followed by subsequent sequence divergence, since the 120 amino acid C-terminus of Sc Tad3 is 26% identical and 45% similar to Sc Tad2, and since both Sc Tad2 and Sc Tad3 have a conserved Zn2+ coordination motif, as well as a conserved proline that is generally required for ammonium group binding.Citation44 Although both proteins are required for binding to tRNA, mutational analysis showed that Sc Tad2 is almost certainly the catalytic subunit of the deaminase. An Sc Tad2 variant with an alanine substitution in the predicted catalytic residue E56 was not active, whereas Sc Tad3 has a valine (V218) in this position and is presumably inactive; moreover, a complex of the Sc Tad3-V218E variant and Sc Tad2 had wild type activity, but the Sc Tad3-V218E variant did not restore activity to the Sc Tad2-E56A variant.Citation44
Tad2 and Tad3 homologs are widely found in eukaryotes,Citation47 and evidence from eukaryotes other than S. cerevisiae further suggests that the Tad2-Tad3 complex is required for A34 deaminase activity. Thus, a screen for temperature sensitive S. pombe mutants identified a mutant encoding an Sp Tad3 variant with greatly reduced binding of Sp Tad2 and associated with reduced levels of inosine in tRNA.Citation48 Moreover, the temperature sensitive phenotype of the Sp tad3 mutant could be suppressed by additional copies of Sp tad2+, suggesting that overexpression drove formation of catalytically active complexes.Citation48 Similarly, analysis of Trypanosoma brucei A34 deaminase activity suggests the requirement of a Tad2-Tad3 complex, since a Tb Tad2-Tad3 complex has activity, but a Tb Tad2 homodimer lacks catalytic activity.Citation39
By contrast, A34 deamination of tRNAArg(ACG) is catalyzed only by TadA (the homolog of Tad2 and Tad3) in prokaryotes,Citation13,40 based on the occurrence of only this homolog in bacteria,Citation47 and the activity of the purified proteins from E. coli and Agrobacterium tumefaciens.Citation38,45
TadA was shown to be a homodimer based on its crystal structure from Staphylococcus aureus, A. aeolicus, and A. tumefaciens.Citation36,38,49 The co-crystal structure of the S. aureus TadA homodimer bound to the 15-mer tRNAArg(AGC) anticodon stem-loop suggests that substrate binding occurs via an induced fit of the anticodon to the rigid interface between the homodimer via specific contacts predominantly with the 5 nucleotides of the anticodon loop and the C32-A38 pair at the top of the loop.Citation36
Many residues shown to be important for tRNA binding in bacterial TadA are not conserved in Tb Tad2, suggesting a role for Tb Tad3 in substrate binding.Citation50 Indeed, some findings suggest that the eukaryotic heterodimer is important for recognition of a larger region on substrate tRNAs to catalyze modification of the multiple different tRNA targets, as opposed to local structural elements of the single tRNA recognized by homodimeric TadA. Thus, although bacterial TadA has activity toward only a stem-loop RNA construct, Sc Tad2-Tad3 requires a full tRNA construct and a proper 3-dimensional structure for activity.Citation38,44,51 Consistent with these findings, TadA can deaminate eukaryotic tRNAArg(ACG), but cannot deaminate other eukaryotic Tad2-Tad3 tRNA substrates.Citation45 Remarkably, the Tb Tad2-Tad3 enzyme has C to U ssDNA deaminase activity both in vivo and in vitro, further demonstrating the increased substrate repertoire of eukaryotic Tad2-Tad3 as compared to bacterial TadA.Citation52 Alfonzo and colleagues have proposed a model wherein binding of the Zn2+ ion occurs intermolecularly, possibly granting increased ability to diversify substrates (as needed for the 7 substrate tRNAs in S. cerevisae) while still maintaining specificity for A34.Citation39
Trm6-Trm61, the m1A58 methyltransferase
The m1A58 (1-methyladenosine) modification is commonly found in eukaryotic tRNAs, including 21 cytoplasmic S. cerevisiae tRNAs. This modification is also found less frequently in bacterial and archaeal tRNAs, with a greater frequency of occurrence in tRNA from thermophilic organismsCitation13. m1A58 likely contributes to tRNA stability,Citation53 and S. cerevisiae mutants lacking this modification are inviable due to degradation of hypomodified tRNAiMet by the TRAMP complex (Trf4/Air2/Mtr4p polyadenylation complex) and the nuclear exosome.Citation16,54-57 This specific degradation of only tRNAiMet is likely due to loss of stability in an important substructure unique to eukaryotic initiator tRNAs, wherein m1A58 is involved in hydrogen bonding interactions with residues A20, A54, and A60.Citation53,58 The requirement of m1A58 for viability does not extend to all eukaryotes, since S. pombe mutants lacking this modification are viable,Citation46 albeit with a slow growth defect.Citation59
In S. cerevisiae, m1A58 is formed by Trm6/Trm61.Citation58 Sc TRM6 (also named GCD10) and Sc TRM61 (GCD14) were first identified in screens selecting for mutations that increased GCN4 expression in an Sc gcn2–101 gcn3–101 mutant.Citation60-62 GCN4 expression is higher when there is less functional eIF2-GTP-tRNAiMet (eIF2: eukaryotic initiation factor 2) initiation ternary complex, and it was found that high copy tRNAiMet suppressed the temperature sensitive phenotype of Sc gcd10–504 (trm6) mutants and the lethality of Sc trm6Δ and Sc trm61Δ mutants by restoring levels of the initiator tRNA.Citation58,63 It was also shown that tRNA from Sc trm6Δ and Sc trm61 mutants lacked m1A58 modification,Citation58,64 that Sc Trm6 and Sc Trm61 form a complex with m1A58 catalytic activity, and that activity was dependent on the S-adenosyl methionine (AdoMet) binding domain of Sc Trm61, which is the catalytic subunit.Citation58,64 The Sc Trm6-Trm61complex appears to be a dimer of heterodimers based on size exclusion chromatography.Citation65
The m1A58 modification enzyme also appears to consist of a Trm6-Trm61 complex in other eukaryotes. Trm6 and Trm61 family proteins are found in yeast, plants, and animals,Citation66,67 and the human methyltransferase appears to require both Trm6 and Trm61. Thus, co-expression of human TRM6 and human TRM61 suppressed the temperature sensitive growth of Sc trm6–504 and Sc trm61–2 mutants, restored levels of m1A58 on tRNA in these mutants, and led to m1A58 formation on human tRNALys introduced on a plasmid, as measured by an altered electrophoretic mobility of the tRNA.Citation67 Furthermore, expression of only human TRM6 or human TRM61 did not lead to a substantial increase in m1A58 modification in mutants, and a complex of human Trm6-Trm61 purified from yeast was able to specifically methylate yeast tRNAiMet.Citation67
It is likely that TRM6 and TRM61 arose from a gene duplication event followed by sequence divergence, based on the sequence similarity between predicted bacterial Trm61 homologs and eukaryotic Trm6, as well as the conservation of predicted secondary structural elements in eukaryotic Trm6, eukaryotic Trm61, and predicted bacterial Trm61 homologs.Citation66 This argument is further strengthened by the finding that many of the residues involved in Mycobacterium tuberculosis TrmI (a bacterial Trm61 homolog) homotetramer formation are also involved in the interaction between Sc Trm6 and Sc Trm61.Citation35,65
By contrast, m1A58 modification of bacterial and archaeal tRNA is formed by only TrmI (the homolog of Trm61), based on the occurrence of only one Trm61 homolog and no obvious Trm6 homolog in bacterial and archaeal species,Citation66,68,69 and the activity of the purified Thermus thermophilus, M. tuberculosis, and Pyrococcus abyssi proteins.Citation68–70 Based on electrospray ionization mass spectrometry of the native complex, Tt TrmI is a homotetramer,Citation37 and it is also likely that the M. tuberculosis protein is homotetrameric, based on gel filtration analysis.Citation70
One of the major functions of Trm6 in the Trm6-Trm61 complex appears to be tRNA binding. Thus, Trm6 contains an RNA recognition motif, and wild type Sc Trm6-Trm61 binds tRNA, as does a complex containing Sc Trm6 and an Sc Trm61 variant with mutations in the AdoMet binding domain, whereas Sc Trm61 by itself does not.Citation64 Moreover, mutations of conserved residues predicted to be involved in the interface between Sc Trm6 and Sc Trm61 abrogate tRNA binding and m1A58 activity (based on the TrmI crystal structureCitation35), but do not appear to disrupt the heterotetrameric complex or AdoMet binding.Citation65 These results have led to the speculation that these conserved residues are required to form a Trm6/Trm61 interface that is required for tRNA binding, rather than for complex formation of the proteins themselves.Citation65
Acquisition of an Unrelated Partner Protein
As detailed below, there are 5 eukaryotic methyltransferases that have acquired partner proteins unrelated to the catalytic subunit. We further divide these methyltransferases into 3 distinct subclasses.
Acquisition of an unrelated subunit by eukaryotes for the same bacterial modification
This class of modification enzymes catalyzes conversion of G46 to m7G46 (7-methylguanosine). The m7G46 modification occurs in bacterial and eukaryotic tRNAs,Citation13 but to date has not been found in tRNA from archaea, although m7G49 is found on tRNALeu(UAG) from Thermoplasma acidophilum.Citation71 m7G46 plays a role in stabilizing the tertiary fold of the tRNA, and is part of a commonly occurring base triple with N13 and N22.Citation72,73 S. cerevisiae strains lacking m7G46 have a mild growth defect when grown at 38°C on synthetic media containing glycerol,Citation74 and mutants lacking m7G46 and m5C (5-methylcytidine) are temperature-sensitive due to degradation of tRNAVal(AAC) by the rapid tRNA decay pathway, which also affects strains lacking m7G46 in combination with lack of any of several other modifications in the body of the tRNA.Citation17,75,76 Additionally, m7G46 was shown to be required for growth of T. thermophilus at high temperature, and to be required for subsequent Gm18 (2′-O-methylguanosine) and m1G37 modification on tRNAPhe as part of a tRNA modification network.Citation77
In S. cerevisiae, m7G46 is formed by the Sc Trm8-Trm82 protein pair.Citation78 Sc Trm8-Trm82 was discovered using a biochemical genomics approachCitation79 when it was found that protein purified from S. cerevisiae strains expressing tagged open reading frames for either Sc TRM8 or Sc TRM82 yielded m7G formation activity on pre-tRNAPhe.Citation78 Evidence indicating that the enzyme is composed of the Sc Trm8-Trm82 complex includes the observation that deletion of either gene results in lack of m7G modification on tRNA, that the 2 proteins form a stoichiometric complex, that recombinant Sc Trm8 purified from E. coli has low in vitro m7G formation activity that is increased 250-fold when co-expressed with Sc Trm82, and that co-translation is required for an active complex.Citation78 Sc Trm8 contains a methyltransferase domain and is the catalytic subunit of the enzyme, whereas Sc Trm82 is a WD-repeat protein.
In other eukaryotes, the m7G46 modification enzyme also appears to consist of the Trm8-Trm82 protein pair. Trm8 and Trm82 homologs are found in yeast, plants, and animals,Citation78 and co-expression of human METTL1 (human TRM8) and WDR4 (human TRM82) complemented the m7G46 modification defect in trm8Δ or trm82Δ mutant cells, whereas expression of only human METTL1 or human WDR4 did not.Citation78
In humans, it was also recently reported that HeLa cells with reduced levels of human METTL1 and human NSUN2 (required for m5C) are sensitive to 5-fluorouracil, resulting in a decrease in tRNAVal(AAC) levels.Citation80 Interestingly, human METTL1 was found to be inactivated by phosphorylation at Ser27 by protein kinase B, suggesting a possible mechanism to regulate m7G modification levels in the cell.Citation81
By contrast, in bacteria m7G46 is formed by the Trm8 homolog TrmB alone, based on the apparent absence of Trm82 homologs,Citation82 the activity of purified E. coli TrmB,Citation83 and the ability of E. coli TrmB to complement the lack of Sc Trm8-Trm82 in S. cerevisiae trm8Δ trm82Δ double mutants.Citation74 Ec TrmB is monomeric,Citation83 whereas Bacillus subtilis TrmB is homodimeric in solution and in its crystal structure, which has been proposed as a first evolutionary step in the requirement for a dimeric enzyme.Citation84
It is clear that part of the role of Sc Trm82 is to maintain levels of Sc Trm8 in yeast, and that Sc Trm82 is also required for other reasons, since Sc Trm8 levels are greatly reduced in Sc trm82Δ mutants, but restoration of levels in S. cerevisiae through Sc Trm8 overexpression only marginally restores m7G46 activity.Citation74 In the crystal structures of members of the Ec TrmB/Sc Trm8 family, the B4-αD loop region of unbound Sc Trm8 is in a much different conformation than that of the Sc Trm8-Trm82 complex and that of the B. subtilis TrmB, which have a similar conformation to one another.Citation84,85 In Sc Trm8, the distinct conformation of the unbound form is stabilized by a salt bridge between R195 and E204, which is unable to form in the Sc Trm8-Trm82 dimer due to steric constraints imposed by Sc Trm82.Citation85 The equivalent residue to Sc Trm8 R195 (R129) in B. subtilis TrmB points in the opposite direction compared to unbound R195 in Sc Trm8,Citation84 and alanine substitution of the equivalent arginine residue in E. coli TrmB results in loss of more than 90% of its methyltransferase activity,Citation85,86 suggesting that this residue is important for Trm8/TrmB activity.
Sc Trm82 does not appear to be involved in tRNA binding since tRNAPhe cross-links only to Sc Trm8, and not to Sc Trm82,Citation74 and since the best fit small-angle X-ray scattering (SAXS) model of the Sc Trm8-Trm82 complex bound to tRNA suggests that only Sc Trm8 is involved in tRNA binding.Citation85 This SAXS model also suggests that Sc Trm8 binds the tRNA through the local structure around the variable region, especially the D-stem and T-stem,Citation85 which is consistent with the finding that deletion of these stems leads to complete loss of methylation activity by Sc Trm8-Trm82, whereas deletion of the acceptor or anticodon stems does not.Citation87
Acquisition of 2 different unrelated subunits by eukaryotes for the same modification at different locations
The Trm7 methyltransferase is an example of this subclass, wherein a catalytic subunit engages 2 distinct partner proteins to direct the same 2′-O-methylation activity to different residues: to C32 and N34 to form Cm32 and Nm34. Nm32 and Nm34 modifications are found in tRNAs from eukaryotes, bacteria, and archaea.Citation13 Cm32 and Nm34 occur in tandem on 3 tRNA species from S. cerevisiae, and Cm32 and Gm34 modification of tRNAPhe appears to be highly conserved in eukaryotes, occurring in 16 of 17 eukaryotic tRNAPhe species that have been examined.Citation13 Although Cm32 and Gm34 are found in tandem on tRNAPhe, they are in chemically distinct environments from each other.Citation72 Other Nm32 and Nm34 modifications are found in mammalian, insect, plant, bacterial, and archaeal tRNAs, although not always in tandem,Citation13 and Nm32 and Nm34 modifications appear to be important in eukaryotes, although their roles are not well understood. Thus, S. cerevisiae and S. pombe mutants lacking Cm32 and Gm34, and S. pombe mutants lacking Gm34, have a severe growth defect due to reduced function of tRNAPhe, whereas S. cerevisiae or S. pombe mutants lacking only Cm32 are healthy.Citation9-11 Furthermore, Cm32 and Gm34 on tRNAPhe are also important for the formation of wybutosine (yW37) from m1G37 in S. cerevisiae and S. pombeCitation10,11 (). In bacteria and archaea, there are no reported deleterious phenotypes associated with lack of Nm32,Citation88,89 but lack of Nm34 in E. coli causes a defect in amber (UAG) suppression by tRNALeu(CUA), which was suggested to implicate Nm34 in wobble codon:anticodon pairing.Citation90
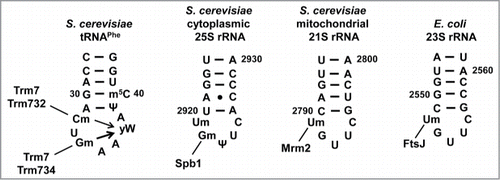
Figure 2. Schematic of FtsJ methyltransferase stem-loop substrates. S. cerevisiae Trm7 requires Trm732 for Cm32 modification and Trm734 for Gm34 modification of the anticodon loop of tRNAPhe which, as for all tRNAs, has 7 bases. These modifications then drive formation of yW37 from its m1G precursor. The thicker arrow from Gm34 indicates that yW formation is more dependent on this modification. Other FtsJ family members modify the 5-base A-loop in the rRNA large subunit in different organisms and organelles, as indicated.
In S. cerevisiae, Cm32 is formed by the Sc Trm7-Trm732 protein pair, and Nm34 is formed by the Sc Trm7-Trm734 pair.Citation9,10 Sc TRM7 was identified by searching for S. cerevisiae homologs of the E. coli 2′-O-methyltransferase FtsJ (RrmJ),Citation9 which 2′-O-methylates U2552 residue of the 23S rRNA subunit.Citation91,92 It was found that tRNAPhe, tRNALeu(UAA), and tRNATrp from S. cerevisiae trm7Δ mutants lacked Cm32 and Nm34, and that tagged Sc Trm7 purified from yeast cells was able to form Cm32, and to a lesser extent, Gm34 on tRNAPhe.Citation9 It was later shown that Sc Trm732 is required for Cm32 formation and that Sc Trm734 is required for Nm34 formation by showing that all 3 Sc Trm7 tRNA substrates from Sc trm732Δ or Sc trm734Δ mutants completely lacked their respective Cm32 or Nm34 modifications, that extracts from Sc trm732Δ or Sc trm734Δ mutants were unable to form their respective Cm32 or Nm34 modifications on synthetic substrates, and that Sc Trm7 forms a distinct complex with Sc Trm732, and a distinct separate complex with Sc Trm734.Citation10 The requirement of Sc Trm732 for Cm32 modification on tRNAPhe and of Sc Trm734 for Gm34 modification was further demonstrated by the failure of overexpressed Sc Trm7 to suppress the slow growth phenotype of Sc trm732Δ trm734Δ mutants.Citation10 Sc Trm732 is an armadillo repeat protein that contains a domain of unknown function (DUF2428), and Sc Trm734 is a WD-repeat protein.
The requirement for the Trm7-Trm732 protein pair for Nm32 formation, and of the Trm7-Trm734 protein pair for Nm34 formation is likely conserved throughout eukaryotes. Thus, analysis of 25 eukaryotic genomes comprising all 5 eukaryotic supergroups readily identified Trm7 homologs in all 25 organisms, Trm732 homologs in 22 organisms, and Trm734 homologs in 14 organisms.Citation11,93 In S. pombe, Sp trm7Δ mutants lack Cm32, Gm34, and yW37 on their tRNAPhe, Sp trm732Δ mutants lack Cm32, and Sp trm734Δ mutants lack Gm34.Citation11 Furthermore, expression of human FTSJ1 (the predicted TRM7 human homolog) suppressed the growth defect of Sc trm7Δ mutants by forming Cm32 on tRNAPhe, and suppression and modification by human FTSJ1 required either Sc TRM732 or human THADA (the predicted TRM732 human homolog).Citation11 These findings implicate defective Nm32 and Nm34 modifications in non-syndromic X-linked intellectual disability, since mutations in FTSJ1 are strongly linked to this disease.Citation94–97
The formation of Nm32 in E. coli and in the archaeon Sulfolobus acidocaldarius requires members of the homodimeric TrmJ SPOUT methyltransferase family,Citation88,89 which are not obviously related to Trm7, Trm732, or Trm734. Ec TrmJ appears to require elements in the D-stem and loop for modification activity, whereas Sa TrmJ appears to require elements solely in the anticodon loop.Citation89
The formation of Nm34 requires distinct genes in bacteria and archaea, neither of which are related to components of the TRM7 modification machinery. Thus, in E. coli, Cm34 and Um34 on certain tRNALeu species are formed by the SPOUT methyltransferase TrmL, which recognizes its substrates by interactions with specific residues, including the N6-(isopentenyl)-2-methylthioadenosine modification formed from A37 in the anticodon loop of substrate tRNAs.Citation90,98 By contrast, in the archaeon Haloferax volcanii, Cm34 formation on tRNATrp and on elongator tRNAMet require box C/D snoRNPs (small nucleolar ribonucleoprotein) specific to each corresponding pre-tRNA.Citation99,100
Comparison of Trm7 to other FtsJ family proteins suggests a possible reason for the requirement of additional proteins for Trm7 activity, since the stem-loop tRNA substrates modified by Trm7 are slightly different than the rRNA stem-loop substrates modified by other proteins in this family (). Thus, E. coli FtsJ (which is ∼34% identical to Sc Trm7) methylates the first residue (Um2552) of the 5-base A-loop in the 23S rRNA,Citation91,92 S. cerevisiae Mrm2 (∼29% identical to Sc Trm7) methylates the first residue (Um2791) of the 5-base A-loop in the the 21S mitochondrial rRNA,Citation101 and S. cerevisiae Spb1 (∼34% identical to Sc Trm7) methylates the second residue (Gm2992) of the same 5-base A-loop of cytoplasmic 27S pre-rRNA,Citation102 each apparently acting alone. We therefore speculate that Sc Trm732 may help Sc Trm7 to recognize and modify the first residue of the 7-base loop of tRNA (as opposed to the 5-base loop in rRNA), and that Sc Trm734 may help Sc Trm7 recognize the N34 residue, which is the third residue in the anticodon loop, and chemically distinct from the substrate residues of the other known FtsJ family members.
S. cerevisiae Trm734 has also been implicated in endoplasmic recycling, seemingly unrelated to tRNA modification. Sc TRM734 (ERE2) was identified in a genome-wide screen for deletion mutants with increased canavanine resistance due to defects in endoplasmic recycling.Citation103 It was suggested that Sc Trm734 regulates the function of Sc Ere1, which was identified in the same screen, since it interacts with Trm734 in membrane bound fractions on a glycerol gradient, co-immunoprecipitates with Sc Trm734, and since some Trm734 colocalized with Sc Ere1 in endosomal compartments in ESCRT (endosomal sorting complexes required for transport) mutant cells. Thus, it may be that cytosolic Sc Trm734 functions for tRNA modification, and that membrane-bound Sc Trm734 plays a role in endoplasmic recycling.Citation103
Sc TRM734 (RTT10) was also identified in a screen for S. cerevisiae deletion mutants that showed increased Ty1 retrotransposition.Citation104 In retrospect, however, this defect is likely due to decreased expression of an important protein(s) involved in repression of Ty1 transposition caused by lack of Nm34 modification, since Sc TRM7 was identified in the same screen.
Acquisition of a common unrelated partner protein for different methyltransferases
The Trm9 and Trm12 methyltransferases are examples of proteins in this subclass, wherein different catalytic subunits engage the same partner protein Trm112 to direct different chemical modifications on different residues. The first tRNA methyltransferase that requires Trm112 is responsible for formation of the terminal methyl group of mcm5U34 (5-methoxycarbonylmethyluridine) and related modifications. The mcm5U family of modifications is found only in eukaryotes and is implicated in efficient reading of AGA and AAG codons by tRNAArg(UCU) and tRNAGlu(UUC), respectively in S. cerevisiae.Citation105 Lack of the modification is associated with sensitivity to DNA damaging agents in yeast and in humans,Citation105,106 as well as sensitivity to aminoglycosides at high temperature and resistance to zymocin-mediated tRNA cleavage and cell death in yeast.Citation107-109
In S. cerevisiae, the terminal methyl group of mcm5U34 and mcm5S2U34 (5-methoxycarbonylmethyl-2-thiouridine) is formed by the Sc Trm9-Trm112 protein pair.Citation5,6,107,109,110 Sc TRM9 was identified as a putative methyltransferase by bioinformatics, tRNA from S. cerevisiae trm9Δ mutants was shown to lack the mcm5U and mcm5S2U modifications, and tagged Sc Trm9 protein purified from yeast was shown to methylate a saponified tRNA extract, demonstrating that Sc Trm9 is required for formation of the terminal methyl group of mcm5U.Citation107 Subsequently, several suppressors of zymocin toxicity in S. cerevisiae were identified as mutants encoding Sc Trm9 variants lacking the C-terminal domain required for interaction with Sc Trm112.Citation109,110 The requirement of Sc Trm112 for mcm5U and mcm5S2U was later determined explicitly by showing that tRNA from Sc trm112Δ mutants lacked the terminal methyl group of the mcm5U modification, and that purified Sc Trm9- Trm112 from E. coli had this methylation activity, whereas Sc Trm9 alone did not.Citation5,6 Interestingly, lack of either Sc Trm9 or Sc Trm112 gives rise to ncm5U (5-carbamoylmethyluridine) instead of the expected cm5U modification in yeast, raising questions about the biochemistry of mcm5U formation.Citation6
The requirement of Trm9-Trm112 for mcm5U is likely conserved in other eukaryotes. Thus, predicted Trm9 proteins are found in yeast, worms, flies, and humans,Citation107 and predicted Trm112 proteins are found in fungi, animals, and plants.Citation4 Additionally, the Mus musculus Trm9 and Trm112 homologs are required for mcm5U formation in mice,Citation111 and the Trm9 homolog is required for this modification in human cells.Citation106 However, the role that Trm112 plays for the methyltransferase activity of Trm9 is not yet clear.
Trm112-Trm11, the m2G10 methyltransferase
The second tRNA methyltransferase that requires Trm112 is responsible for formation of the m2G10 (N2-methylguanosine) modification, which is found in eukaryotes and archaea, but not in bacteria.Citation13 Although the precise role of this modification is not clear, it is likely involved in tRNA stability since lack of this modification in combination with lack of m2,2G26 (N2,N2,-dimethylguanosine) in S. cerevisiae results in slow growth.Citation3
In S. cerevisiae, m2G10 is formed by the Sc Trm11-Trm112 pair.Citation3 Sc TRM11 was identified by bioinformatics approaches as a putative methyltransferase, and it was found that extracts from S. cerevisiae cells lacking Sc TRM11 were unable to catalyze formation of m2G on synthetic tRNAIle(UAU). However, although purified Sc Trm11 from yeast yielded m2G activity, Sc Trm11 purified from E. coli did not, and it was shown that Sc trm112Δ mutants lacked m2G activity, that tagged Sc Trm112 purified from yeast exhibited m2G activity, and that Sc Trm11 and Sc Trm112 formed a complex.Citation3 Sc Trm112 was later shown explicitly to be required for Sc Trm11 activity using a wheat germ cell free system.Citation112
Since Sc Trm11 and Sc Trm112 homologs are found widely in eukaryotes and in archaea,Citation3 it is likely that they are required for m2G10 activity in other organisms, although to our knowledge this has not been tested experimentally. The precise role of Sc Trm112 for Sc Trm11 activity is not known, but Sc Trm11 stability does not appear to be affected by Sc Trm112 levels.Citation3
Trm112 has distinct roles in other methyltransferase complexes
Trm112 is also required for the function of 2 other 2-subunit methyltransferase enzymes. We will briefly discuss these enzymes because studies on their structure and function suggest several possible roles for Trm112 in Trm9 and Trm11 function, and because the relative amounts of each complex may affect the methylation activity of the other Trm112-containing complexes, since Trm112 appears to be limiting in S. cerevisiae.Citation8,113
Analysis of the Mtq2-Trm112 complex suggests that Trm112 functions to solubilize Mtq2 and to place it in an active conformation.Citation4,114 In S. cerevisiae, Sc Mtq2-Trm112 is the enzyme responsible for N5 methylation of the glutamine residue of the GGQ tripeptide motif of eukaryotic release factor 1 (eRF1).Citation4,115,116 The crystal structure of Mtq2-Trm112 from Encephalitozoon cuniculi consists of a single heterodimer, and suggests that E. cuniculi Trm112 may function to solubilize E. cuniculi Mtq2 by masking a hydrophobic patch involved in the dimer interface.Citation114 Indeed, the presence of Sc Trm112 increases the solubility of Sc Mtq2 when the proteins are expressed in E. coli.Citation4 Analysis of the crystal structure also suggests that E. cuniculi Trm112 may be important for the conformation of the AdoMet binding domain of E. cuniculi Mtq2, since a loop motif is involved in both interaction with E. cuniculi Trm112 and with the AdoMet cofactor.Citation114
Analysis of the overall structure of the E. cuniculi Mtq2-Trm112 complex suggests that Trm112 may also function in substrate binding.Citation114 Although the structure of the methyltransferase domain of E. cuniculi Mtq2 is most similar to that of E. coli PrmC (the protein that N5 methylates the Gln residue of the GGQ motif of bacterial release factors RF1 and RF2Citation117,118), the overall structure of the E. cuniculi Mtq2-Trm112 complex most closely resembles that of E. coli RlmA(I),Citation119 which methylates 23S rRNA residue G745.Citation120,121 The Zn2+ binding domain of E. cuniculi Trm112 superimposes well with the Zn2+ binding domain of E. coli RlmA(1), which has been implicated in rRNA binding,Citation119 suggesting that Trm112 may be important for substrate recognition or binding.Citation114
In addition, analysis of RlmA(I) structure suggested a role for Trm112 in binding and activity of Trm9, since RlmA(I) is the closest E. coli homolog of S. cerevisiae Trm9, and a model of Trm112 binding to Trm9 correctly predicted residues important for both Trm9-Trm112 protein-protein interactions and activity.Citation114
Analysis of the Bud23-Trm112 complex indicates that Trm112 maintains Bud23 protein levels.Citation7 In S. cerevisiae, the Sc Bud23-Trm112 complex is required for the m7G modification of G1575 on the 18S subunit of rRNA in yeast.Citation7,8,122 Sc trm112Δ mutants have severely reduced levels of Sc Bud23,Citation7 and surprisingly, the Sc bud23Δ slow growth phenotype can be rescued by expression of a catalytic null Sc Bud23 variant,Citation122 further indicating that the role of Sc Trm112 in this complex is not for catalysis. Moreover, the slow growth phenotype of the Sc trm112Δ mutant is almost completely due to Bud23 defects, since the Sc bud23Δ trm112Δ double mutant grew as poorly as the Sc bud23Δ or Sc trm112Δ single mutants.Citation5,8
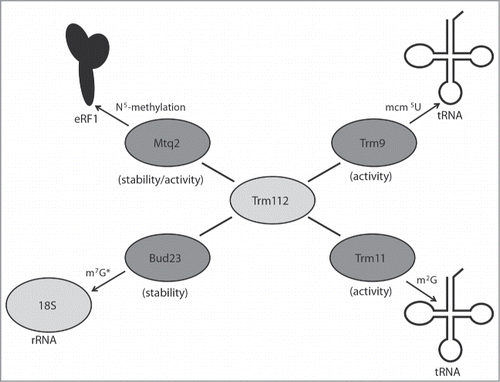
It appears that Sc Trm112 may also act to integrate methylation signals between tRNA, the ribosome, and release factors ().Citation114 Because Sc Trm112 is likely present in non-stoichiometric amounts compared to its methyltransferase partners,Citation8,113 the activity of each methyltransferase partner is sensitive to the relative amounts of the other Sc Trm112 methyltransferase partners. For instance, overexpression of Sc Trm11 or Sc Mtq2 results in resistance to zymocin toxicity and reduced binding between Sc Trm112 and Sc Trm9. Furthermore, overexpression of Sc Mtq2 leads to cell growth defects, presumably by titrating Sc Trm112 away from Sc Bud23, and this defect is partially rescued by overexpression of Sc Bud23.Citation7
Figure 3. Trm112 partners with several methyltransferases involved in diverse translational processes. Schematic of the S. cerevisiae Trm112 physical interactions known to affect the activity or stability (in brackets) of its partner methyltransferases. (*) The Sc Bud23 protein, but not its methyltransferase activity, is required for m7G1575 formation on the cytoplasmic 18S rRNA subunit.
Concluding Remarks
Several themes emerge upon analysis of the protein pairs involved in eukaryotic tRNA modification. As described above, tRNA modification enzymes have acquired a second subunit either by duplication of a gene or by acquisition of an unrelated partner protein, but with multiple different properties. One functional theme that has emerged is that the addition of a partner protein may allow for an expansion of the substrate repertoire for a given enzyme, as for the eukaryotic Tad2-Tad3 enzyme, which modifies an additional 6 tRNA species (in S. cerevisiae) in contrast to its homodimeric bacterial TadA counterpart, which appears to only modify tRNAArg(ACG). A second theme is the expanded location of modifications facilitated by the acquisition of different partners for eukaryotic Trm7 activity, allowing modification of 2 different residues in chemically distinct environments in the same tRNA species. A third theme that emerges is the distinct possibility of cross-pathway regulation by modification. This is most clear for the different methyltransferases that each require association with limiting amounts of Trm112 for crucial methylations involved in translation termination, tRNA modifications, and ribosome biogenesis. The modulation of several methyltransferase activities by one protein in theory allows the integration of multiple signals to fine-tune translation. Similarly, it is possible that Trm734 could also act to integrate different cellular functions because of the involvement of Trm734 in endoplasmic recycling and retrotransposition.
While we have pointed out some of the common themes in these 2-subunit eukaryotic tRNA modification enzymes, it remains unclear precisely how the non-catalytic subunits function in the modification complexes. Although we have cited evidence describing roles of these proteins in maintaining stability, altering conformations of proteins, expanding substrate repertoire, and promoting substrate binding, in many cases the evidence is indirect and limited. Furthermore, the mechanism by which unrelated partners were acquired is not clear in most cases. Further research will undoubtedly lead to insight into these and other questions regarding the roles of partner proteins in these important and conserved modification reactions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank E. Grayhack for discussions.
Funding
This work is supported by NIH grant GM052347 to EMP.
References
- Hopper AK. Transfer RNA post-transcriptional processing, turnover, and subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics 2013; 194:43-67; PMID:23633143; http://dx.doi.org/10.1534/genetics.112.147470
- El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Ann Rev Genet 2012; 46:69-95; PMID:22905870; http://dx.doi.org/10.1146/annurev-genet-110711-155641
- Purushothaman SK, Bujnicki JM, Grosjean H, Lapeyre B. Trm11p and Trm112p are both required for the formation of 2-methylguanosine at position 10 in yeast tRNA. Mol Cell Biol 2005; 25:4359-70; PMID:15899842; http://dx.doi.org/10.1128/MCB.25.11.4359-4370.2005
- Heurgue-Hamard V, Graille M, Scrima N, Ulryck N, Champ S, van Tilbeurgh H, Buckingham RH. The zinc finger protein Ynr046w is plurifunctional and a component of the eRF1 methyltransferase in yeast. J Biol Chem 2006; 281:36140-8; PMID:17008308; http://dx.doi.org/10.1074/jbc.M608571200
- Mazauric MH, Dirick L, Purushothaman SK, Bjork GR, Lapeyre B. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J Biol Chem 2010; 285:18505-15; PMID:20400505; http://dx.doi.org/10.1074/jbc.M110.113100
- Chen C, Huang B, Anderson JT, Bystrom AS. Unexpected accumulation of ncm(5)U and ncm(5)S(2) (U) in a trm9 mutant suggests an additional step in the synthesis of mcm(5)U and mcm(5)S(2)U. PloS one 2011; 6:e20783; PMID:21687733; http://dx.doi.org/10.1371/journal.pone.0020783
- Figaro S, Wacheul L, Schillewaert S, Graille M, Huvelle E, Mongeard R, Zorbas C, Lafontaine DL, Heurgue-Hamard V. Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol Cell Biol 2012; 32:2254-67; PMID:22493060; http://dx.doi.org/10.1128/MCB.06623-11
- Sardana R, Johnson AW. The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits. Mol Biol Cell 2012; 23:4313-22; PMID:22956767; http://dx.doi.org/10.1091/mbc.E12-05-0370
- Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B. Trm7p catalyses the formation of two 2'-O-methylriboses in yeast tRNA anticodon loop. EMBO J 2002; 21:1811-20; PMID:11927565; http://dx.doi.org/10.1093/emboj/21.7.1811
- Guy MP, Podyma BM, Preston MA, Shaheen HH, Krivos KL, Limbach PA, Hopper AK, Phizicky EM. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 2012; 18:1921-33; PMID:22912484; http://dx.doi.org/10.1261/rna.035287.112
- Guy MP, Phizicky EM. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 2015; 21:61-74; PMID:25404562; http://dx.doi.org/10.1261/rna.047639.114
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. The minimal gene complement of Mycoplasma genitalium. Science 1995; 270:397-403; PMID:7569993; http://dx.doi.org/10.1126/science.270.5235.397
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262-7; PMID:23118484; http://dx.doi.org/10.1093/nar/gks1007
- Agris PF, Vendeix FA, Graham WD. tRNA's wobble decoding of the genome: 40 years of modification. J Mol Biol 2007; 366:1-13; PMID:17187822; http://dx.doi.org/10.1016/j.jmb.2006.11.046
- Helm M, Giege R, Florentz C. A Watson-Crick base-pair-disrupting methyl group (m1A9) is sufficient for cloverleaf folding of human mitochondrial tRNALys. Biochemistry 1999; 38:13338-46; PMID:10529209; http://dx.doi.org/10.1021/bi991061g
- Kadaba S, Krueger A, Trice T, Krecic AM, Hinnebusch AG, Anderson J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae. Genes Dev 2004; 18:1227-40; PMID:15145828; http://dx.doi.org/10.1101/gad.1183804
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 2006; 21:87-96; PMID:16387656; http://dx.doi.org/10.1016/j.molcel.2005.10.036
- Grosjean H. Nucleic acids are not boring long polymers of only four types of nucleotides: a guided tour. In: Grosjean H, ed. DNA and RNA Modification Enzymes: Structure Mechanism, Function and Evolution. Austin, TX: Landes Bioscience, 2009:1-12.
- Phizicky EM, Hopper AK. tRNA biology charges to the front. Genes Dev 2010; 24:1832-60; PMID:20810645; http://dx.doi.org/10.1101/gad.1956510
- Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends in Mol Med 2014; 20:306-14; http://dx.doi.org/10.1016/j.molmed.2014.01.008
- Sprinzl M, Grueter F, Spelzhaus A, Gauss DH. Compilation of tRNA sequences. Nucleic Acids Res 1980; 8:r1-r22; PMID:6986608; http://dx.doi.org/10.1093/nar/8.1.1
- Kammen HO, Marvel CC, Hardy L, Penhoet EE. Purification, structure, and properties of Escherichia coli tRNA pseudouridine synthase I. J Biol Chem 1988; 263:2255-63; PMID:3276686
- Lecointe F, Simos G, Sauer A, Hurt EC, Motorin Y, Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J Biol Chem 1998; 273:1316-23; PMID:9430663; http://dx.doi.org/10.1074/jbc.273.3.1316
- Blaby IK, Majumder M, Chatterjee K, Jana S, Grosjean H, de Crecy-Lagard V, Gupta R. Pseudouridine formation in archaeal RNAs: The case of Haloferax volcanii. RNA 2011; 17:1367-80; PMID:21628430; http://dx.doi.org/10.1261/rna.2712811
- Kaya Y, Ofengand J. A novel unanticipated type of pseudouridine synthase with homologs in bacteria, archaea, and eukarya. RNA 2003; 9:711-21; PMID:12756329; http://dx.doi.org/10.1261/rna.5230603
- Behm-Ansmant I, Urban A, Ma X, Yu YT, Motorin Y, Branlant C. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA 2003; 9:1371-82; PMID:14561887; http://dx.doi.org/10.1261/rna.5520403
- Muller S, Urban A, Hecker A, Leclerc F, Branlant C, Motorin Y. Deficiency of the tRNATyr:Psi 35-synthase aPus7 in Archaea of the Sulfolobales order might be rescued by the H/ACA sRNA-guided machinery. Nucleic Acids Res 2009; 37:1308-22; PMID:19139072; http://dx.doi.org/10.1093/nar/gkn1037
- El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res 2009; 37:2894-909; PMID:19287007; http://dx.doi.org/10.1093/nar/gkp152
- El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crecy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 2011; 30:882-93; PMID:21285948; http://dx.doi.org/10.1038/emboj.2010.363
- Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J 2011; 30:873-81; PMID:21183954; http://dx.doi.org/10.1038/emboj.2010.343
- Deutsch C, El Yacoubi B, de Crecy-Lagard V, Iwata-Reuyl D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J Biol Chem 2012; 287:13666-73; PMID:22378793; http://dx.doi.org/10.1074/jbc.M112.344028
- Perrochia L, Crozat E, Hecker A, Zhang W, Bareille J, Collinet B, van Tilbeurgh H, Forterre P, Basta T. In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya. Nucleic Acids Res 2013; 41:1953-64; PMID:23258706; http://dx.doi.org/10.1093/nar/gks1287
- Bystrom AS, Hjalmarsson KJ, Wikstrom PM, Bjork GR. The nucleotide sequence of an Escherichia coli operon containing genes for the tRNA(m1G)methyltransferase, the ribosomal proteins S16 and L19 and a 21-K polypeptide. EMBO J 1983; 2:899-905; PMID:6357787
- Bjork GR, Jacobsson K, Nilsson K, Johansson MJ, Bystrom AS, Persson OP. A primordial tRNA modification required for the evolution of life? EMBO J 2001; 20:231-9; PMID:11226173; http://dx.doi.org/10.1093/emboj/20.1.231
- Gupta A, Kumar PH, Dineshkumar TK, Varshney U, Subramanya HS. Crystal structure of Rv2118c: an AdoMet-dependent methyltransferase from Mycobacterium tuberculosis H37Rv. J Mol Biol 2001; 312:381-91; PMID:11554794; http://dx.doi.org/10.1006/jmbi.2001.4935
- Losey HC, Ruthenburg AJ, Verdine GL. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat Struct Mol Biol 2006; 13:153-9; PMID:16415880; http://dx.doi.org/10.1038/nsmb1047
- Barraud P, Golinelli-Pimpaneau B, Atmanene C, Sanglier S, Van Dorsselaer A, Droogmans L, Dardel F, Tisne C. Crystal structure of Thermus thermophilus tRNA m1A58 methyltransferase and biophysical characterization of its interaction with tRNA. J Mol Biol 2008; 377:535-50; PMID:18262540; http://dx.doi.org/10.1016/j.jmb.2008.01.041
- Elias Y, Huang RH. Biochemical and structural studies of A-to-I editing by tRNA:A34 deaminases at the wobble position of transfer RNA. Biochemistry 2005; 44:12057-65; PMID:16142903; http://dx.doi.org/10.1021/bi050499f
- Spears JL, Rubio MA, Gaston KW, Wywial E, Strikoudis A, Bujnicki JM, Papavasiliou FN, Alfonzo JD. A single zinc ion is sufficient for an active Trypanosoma brucei tRNA editing deaminase. J Biol Chem 2011; 286:20366-74; PMID:21507956; http://dx.doi.org/10.1074/jbc.M111.243568
- Chan PP, Lowe TM. GtRNAdb: a database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 2009; 37:D93-7; PMID:18984615; http://dx.doi.org/10.1093/nar/gkn787
- Aldinger CA, Leisinger AK, Gaston KW, Limbach PA, Igloi GL. The absence of A-to-I editing in the anticodon of plant cytoplasmic tRNA (Arg) ACG demands a relaxation of the wobble decoding rules. RNA Biol 2012; 9:1239-46; PMID:22922796; http://dx.doi.org/10.4161/rna.21839
- Crick FH. Codon–anticodon pairing: the wobble hypothesis. J Mol Biol 1966; 19:548-55; PMID:5969078; http://dx.doi.org/10.1016/S0022-2836(66)80022-0
- Murphy FVt, Ramakrishnan V. Structure of a purine-purine wobble base pair in the decoding center of the ribosome. Nat Struct Mol Biol 2004; 11:1251-2; PMID:15558050; http://dx.doi.org/10.1038/nsmb866
- Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 1999; 286:1146-9; PMID:10550050; http://dx.doi.org/10.1126/science.286.5442.1146
- Wolf J, Gerber AP, Keller W. tadA, an essential tRNA-specific adenosine deaminase from Escherichia coli. EMBO J 2002; 21:3841-51; PMID:12110595; http://dx.doi.org/10.1093/emboj/cdf362
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotech 2010; 28:617-23; http://dx.doi.org/10.1038/nbt.1628
- Torres AG, Piñeyro D, Filonava L, Stracker TH, Batlle E, Ribas de Pouplana L. A-to-I editing on tRNAs: Biochemical, biological and evolutionary implications. FEBS Lett 2014; 588:4279-86; PMID:25263703; http://dx.doi.org/10.1016/j.febslet.2014.09.025
- Tsutsumi S, Sugiura R, Ma Y, Tokuoka H, Ohta K, Ohte R, Noma A, Suzuki T, Kuno T. Wobble inosine tRNA modification is essential to cell cycle progression in G(1)/S and G(2)/M transitions in fission yeast. J Biol Chem 2007; 282:33459-65; PMID:17875641; http://dx.doi.org/10.1074/jbc.M706869200
- Kuratani M, Ishii R, Bessho Y, Fukunaga R, Sengoku T, Shirouzu M, Sekine S, Yokoyama S. Crystal structure of tRNA adenosine deaminase (TadA) from Aquifex aeolicus. J Biol Chem 2005; 280:16002-8; PMID:15677468; http://dx.doi.org/10.1074/jbc.M414541200
- Ragone FL, Spears JL, Wohlgamuth-Benedum JM, Kreel N, Papavasiliou FN, Alfonzo JD. The C-terminal end of the Trypanosoma brucei editing deaminase plays a critical role in tRNA binding. RNA 2011; 17:1296-306; PMID:21602302; http://dx.doi.org/10.1261/rna.2748211
- Auxilien S, Crain PF, Trewyn RW, Grosjean H. Mechanism, specificity and general properties of the yeast enzyme catalysing the formation of inosine 34 in the anticodon of transfer RNA. J Mol Biol 1996; 262:437-58; PMID:8893855; http://dx.doi.org/10.1006/jmbi.1996.0527
- Rubio MA, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GA, Papavasiliou FN, Alfonzo JD. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc Natl Acad Sci U S A 2007; 104:7821-6; PMID:17483465; http://dx.doi.org/10.1073/pnas.0702394104
- Basavappa R, Sigler PB. The 3 A crystal structure of yeast initiator tRNA: functional implications in initiator/elongator discrimination. EMBO J 1991; 10:3105-11; PMID:1915284
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005; 121:713-24; PMID:15935758; http://dx.doi.org/10.1016/j.cell.2005.04.029
- Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 2005; 3:e189; PMID:15828860; http://dx.doi.org/10.1371/journal.pbio.0030189
- Kadaba S, Wang X, Anderson JT. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 2006; 12:508-21; PMID:16431988; http://dx.doi.org/10.1261/rna.2305406
- Schneider C, Anderson JT, Tollervey D. The exosome subunit Rrp44 plays a direct role in RNA substrate recognition. Mol Cell 2007; 27:324-31; PMID:17643380; http://dx.doi.org/10.1016/j.molcel.2007.06.006
- Anderson J, Phan L, Cuesta R, Carlson BA, Pak M, Asano K, Bjork GR, Tamame M, Hinnebusch AG. The essential Gcd10p-Gcd14p nuclear complex is required for 1- methyladenosine modification and maturation of initiator methionyl-tRNA. Genes Dev 1998; 12:3650-62; PMID:9851972; http://dx.doi.org/10.1101/gad.12.23.3650
- Zuin A, Gabrielli N, Calvo IA, Garcia-Santamarina S, Hoe KL, Kim DU, Park HO, Hayles J, Ayte J, Hidalgo E. Mitochondrial dysfunction increases oxidative stress and decreases chronological life span in fission yeast. PloS One 2008; 3:e2842; PMID:18665268; http://dx.doi.org/10.1371/journal.pone.0002842
- Harashima S, Hinnebusch AG. Multiple GCD genes required for repression of GCN4, a transcriptional activator of amino acid biosynthetic genes in Saccharomyces cerevisiae. Mol Cell Biol 1986; 6:3990-8; PMID:3540603
- Garcia-Barrio MT, Naranda T, Vazquez de Aldana CR, Cuesta R, Hinnebusch AG, Hershey JW, Tamame M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev 1995; 9:1781-96; PMID:7542616; http://dx.doi.org/10.1101/gad.9.14.1781
- Cuesta R, Hinnebusch AG, Tamame M. Identification of GCD14 and GCD15, novel genes required for translational repression of GCN4 mRNA in Saccharomyces cerevisiae. Genetics 1998; 148:1007-20; PMID:9539420
- Calvo O, Cuesta R, Anderson J, Gutierrez N, Garcia-Barrio MT, Hinnebusch AG, Tamame M. GCD14p, a repressor of GCN4 translation, cooperates with Gcd10p and Lhp1p in the maturation of initiator methionyl-tRNA in Saccharomyces cerevisiae. Mol Cell Biol 1999; 19:4167-81; PMID:10330157
- Anderson J, Phan L, Hinnebusch AG. The Gcd10p/Gcd14p complex is the essential two-subunit tRNA(1- methyladenosine) methyltransferase of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2000; 97:5173-8; PMID:10779558; http://dx.doi.org/10.1073/pnas.090102597
- Ozanick SG, Bujnicki JM, Sem DS, Anderson JT. Conserved amino acids in each subunit of the heteroligomeric tRNA m1A58 Mtase from Saccharomyces cerevisiae contribute to tRNA binding. Nucleic Acids Res 2007; 35:6808-19; PMID:17932071; http://dx.doi.org/10.1093/nar/gkm574
- Bujnicki JM. In silico analysis of the tRNA:m1A58 methyltransferase family: homology-based fold prediction and identification of new members from Eubacteria and Archaea. FEBS Lett 2001; 507:123-7; PMID:11684083; http://dx.doi.org/10.1016/S0014-5793(01)02962-3
- Ozanick S, Krecic A, Andersland J, Anderson JT. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA 2005; 11:1281-90; PMID:16043508; http://dx.doi.org/10.1261/rna.5040605
- Droogmans L, Roovers M, Bujnicki JM, Tricot C, Hartsch T, Stalon V, Grosjean H. Cloning and characterization of tRNA (m1A58) methyltransferase (TrmI) from Thermus thermophilus HB27, a protein required for cell growth at extreme temperatures. Nucleic Acids Res 2003; 31:2148-56; PMID:12682365; http://dx.doi.org/10.1093/nar/gkg314
- Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res 2004; 32:465-76; PMID:14739239; http://dx.doi.org/10.1093/nar/gkh191
- Varshney U, Ramesh V, Madabushi A, Gaur R, Subramanya HS, RajBhandary UL. Mycobacterium tuberculosis Rv2118c codes for a single-component homotetrameric m1A58 tRNA methyltransferase. Nucleic Acids Res 2004; 32:1018-27; PMID:14960715; http://dx.doi.org/10.1093/nar/gkh207
- Tomikawa C, Ohira T, Inoue Y, Kawamura T, Yamagishi A, Suzuki T, Hori H. Distinct tRNA modifications in the thermo-acidophilic archaeon, Thermoplasma acidophilum. FEBS Lett 2013; 587:3575-80; PMID:24076028; http://dx.doi.org/10.1016/j.febslet.2013.09.021
- Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science 1974; 185:435-40; PMID:4601792; http://dx.doi.org/10.1126/science.185.4149.435
- Gautheret D, Damberger SH, Gutell RR. Identification of base-triples in RNA using comparative sequence analysis. J Mol Biol 1995; 248:27-43; PMID:7537339; http://dx.doi.org/10.1006/jmbi.1995.0200
- Alexandrov A, Grayhack EJ, Phizicky EM. tRNA m7G methyltransferase Trm8p/Trm82p: evidence linking activity to a growth phenotype and implicating Trm82p in maintaining levels of active Trm8p. RNA 2005; 11:821-30; PMID:15811913; http://dx.doi.org/10.1261/rna.2030705
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5'-3' exonucleases Rat1 and Xrn1. Genes Dev 2008; 22:1369-80; PMID:18443146; http://dx.doi.org/10.1101/gad.1654308
- Dewe JM, Whipple JM, Chernyakov I, Jaramillo LN, Phizicky EM. The yeast rapid tRNA decay pathway competes with elongation factor 1A for substrate tRNAs and acts on tRNAs lacking one or more of several modifications. RNA 2012; 18:1886-96; PMID:22895820; http://dx.doi.org/10.1261/rna.033654.112
- Tomikawa C, Yokogawa T, Kanai T, Hori H. N7-Methylguanine at position 46 (m7G46) in tRNA from Thermus thermophilus is required for cell viability at high temperatures through a tRNA modification network. Nucleic Acids Res 2010; 38:942-57; PMID:19934251; http://dx.doi.org/10.1093/nar/gkp1059
- Alexandrov A, Martzen MR, Phizicky EM. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 2002; 8:1253-66; PMID:12403464; http://dx.doi.org/10.1017/S1355838202024019
- Martzen MR, McCraith SM, Spinelli SL, Torres FM, Fields S, Grayhack EJ, Phizicky EM. A biochemical genomics approach for identifying genes by the activity of their products. Science 1999; 286:1153-5; PMID:10550052; http://dx.doi.org/10.1126/science.286.5442.1153
- Okamoto M, Fujiwara M, Hori M, Okada K, Yazama F, Konishi H, Xiao Y, Qi G, Shimamoto F, Ota T, et al. tRNA Modifying Enzymes, NSUN2 and METTL1, Determine Sensitivity to 5-Fluorouracil in HeLa Cells. PLoS Genet 2014; 10:e1004639; PMID:25233213; http://dx.doi.org/10.1371/journal.pgen.1004639
- Cartlidge RA, Knebel A, Peggie M, Alexandrov A, Phizicky EM, Cohen P. The tRNA methylase METTL1 is phosphorylated and inactivated by PKB and RSK in vitro and in cells. EMBO J 2005; 24:1696-705; PMID:15861136; http://dx.doi.org/10.1038/sj.emboj.7600648
- Michaud J, Kudoh J, Berry A, Bonne-Tamir B, Lalioti MD, Rossier C, Shibuya K, Kawasaki K, Asakawa S, Minoshima S, et al. Isolation and characterization of a human chromosome 21q22.3 gene (WDR4) and its mouse homologue that code for a WD-repeat protein. Genomics 2000; 68:71-9; PMID:10950928; http://dx.doi.org/10.1006/geno.2000.6258
- De Bie LG, Roovers M, Oudjama Y, Wattiez R, Tricot C, Stalon V, Droogmans L, Bujnicki JM. The yggH gene of Escherichia coli encodes a tRNA (m7G46) methyltransferase. J Bact 2003; 185:3238-43; PMID:12730187; http://dx.doi.org/10.1128/JB.185.10.3238-3243.2003
- Zegers I, Gigot D, van Vliet F, Tricot C, Aymerich S, Bujnicki JM, Kosinski J, Droogmans L. Crystal structure of Bacillus subtilis TrmB, the tRNA (m7G46) methyltransferase. Nucleic Acids Res 2006; 34:1925-34; PMID:16600901; http://dx.doi.org/10.1093/nar/gkl116
- Leulliot N, Chaillet M, Durand D, Ulryck N, Blondeau K, van Tilbeurgh H. Structure of the yeast tRNA m7G methylation complex. Structure 2008; 16:52-61; PMID:18184583; http://dx.doi.org/10.1016/j.str.2007.10.025
- Purta E, van Vliet F, Tricot C, De Bie LG, Feder M, Skowronek K, Droogmans L, Bujnicki JM. Sequence-structure-function relationships of a tRNA (m7G46) methyltransferase studied by homology modeling and site-directed mutagenesis. Proteins 2005; 59:482-8; PMID:15789416; http://dx.doi.org/10.1002/prot.20454
- Matsumoto K, Toyooka T, Tomikawa C, Ochi A, Takano Y, Takayanagi N, Endo Y, Hori H. RNA recognition mechanism of eukaryote tRNA (m7G46) methyltransferase (Trm8-Trm82 complex). FEBS Lett 2007; 581:1599-604; PMID:17382321; http://dx.doi.org/10.1016/j.febslet.2007.03.023
- Purta E, van Vliet F, Tkaczuk KL, Dunin-Horkawicz S, Mori H, Droogmans L, Bujnicki JM. The yfhQ gene of Escherichia coli encodes a tRNA:Cm32/Um32 methyltransferase. BMC Mol Biol 2006; 7:23; PMID:16848900; http://dx.doi.org/10.1186/1471-2199-7-23
- Somme J, Van Laer B, Roovers M, Steyaert J, Versees W, Droogmans L. Characterization of two homologous 2'-O-methyltransferases showing different specificities for their tRNA substrates. RNA 2014; 20:1257-71; PMID:24951554; http://dx.doi.org/10.1261/rna.044503.114
- Benitez-Paez A, Villarroya M, Douthwaite S, Gabaldon T, Armengod ME. YibK is the 2'-O-methyltransferase TrmL that modifies the wobble nucleotide in Escherichia coli tRNA(Leu) isoacceptors. RNA 2010; 16:2131-43; PMID:20855540; http://dx.doi.org/10.1261/rna.2245910
- Caldas T, Binet E, Bouloc P, Costa A, Desgres J, Richarme G. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J Biol Chem 2000; 275:16414-9; PMID:10748051; http://dx.doi.org/10.1074/jbc.M001854200
- Bugl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U. RNA methylation under heat shock control. Mol Cell 2000; 6:349-60; PMID:10983982; http://dx.doi.org/10.1016/S1097-2765(00)00035-6
- Feder M, Pas J, Wyrwicz LS, Bujnicki JM. Molecular phylogenetics of the RrmJ/fibrillarin superfamily of ribose 2'-O-methyltransferases. Gene 2003; 302:129-38; PMID:12527203; http://dx.doi.org/10.1016/S0378-1119(02)01097-1
- Freude K, Hoffmann K, Jensen LR, Delatycki MB, des Portes V, Moser B, Hamel B, van Bokhoven H, Moraine C, Fryns JP, et al. Mutations in the FTSJ1 gene coding for a novel S-adenosylmethionine-binding protein cause nonsyndromic X-linked mental retardation. Am J Hum Genet 2004; 75:305-9; PMID:15162322; http://dx.doi.org/10.1086/422507
- Ramser J, Winnepenninckx B, Lenski C, Errijgers V, Platzer M, Schwartz CE, Meindl A, Kooy RF. A splice site mutation in the methyltransferase gene FTSJ1 in Xp11.23 is associated with non-syndromic mental retardation in a large Belgian family (MRX9). J Med Genet 2004; 41:679-83; PMID:15342698; http://dx.doi.org/10.1136/jmg.2004.019000
- Froyen G, Bauters M, Boyle J, Van Esch H, Govaerts K, van Bokhoven H, Ropers HH, Moraine C, Chelly J, Fryns JP, et al. Loss of SLC38A5 and FTSJ1 at Xp11.23 in three brothers with non-syndromic mental retardation due to a microdeletion in an unstable genomic region. Human Genet 2007; 121:539-47; http://dx.doi.org/10.1007/s00439-007-0343-1
- Takano K, Nakagawa E, Inoue K, Kamada F, Kure S, Goto Y. A loss-of-function mutation in the FTSJ1 gene causes nonsyndromic X-linked mental retardation in a Japanese family. Am J Med Genet 2008; 147B:479-84; PMID:18081026; http://dx.doi.org/10.1002/ajmg.b.30638
- Liu RJ, Zhou M, Fang ZP, Wang M, Zhou XL, Wang ED. The tRNA recognition mechanism of the minimalist SPOUT methyltransferase, TrmL. Nucleic Acids Res 2013; 41:7828-42; PMID:23804755; http://dx.doi.org/10.1093/nar/gkt568
- Clouet d'Orval B, Bortolin ML, Gaspin C, Bachellerie JP. Box C/D RNA guides for the ribose methylation of archaeal tRNAs. The tRNATrp intron guides the formation of two ribose-methylated nucleosides in the mature tRNATrp. Nucleic Acids Res 2001; 29:4518-29; PMID:11713301; http://dx.doi.org/10.1093/nar/29.22.4518
- Joardar A, Malliahgari SR, Skariah G, Gupta R. 2′-O-methylation of the wobble residue of elongator pre-tRNA(Met) in Haloferax volcanii is guided by a box C/D RNA containing unique features. RNA Biol 2011; 8:782-91; PMID:21654217; http://dx.doi.org/10.4161/rna.8.5.16015
- Pintard L, Bujnicki JM, Lapeyre B, Bonnerot C. MRM2 encodes a novel yeast mitochondrial 21S rRNA methyltransferase. EMBO J 2002; 21:1139-47; PMID:11867542; http://dx.doi.org/10.1093/emboj/21.5.1139
- Lapeyre B, Purushothaman SK. Spb1p-directed formation of Gm2922 in the ribosome catalytic center occurs at a late processing stage. Mol Cell 2004; 16:663-9; PMID:15546625; http://dx.doi.org/10.1016/j.molcel.2004.10.022
- Shi Y, Stefan CJ, Rue SM, Teis D, Emr SD. Two novel WD40 domain-containing proteins, Ere1 and Ere2, function in the retromer-mediated endosomal recycling pathway. Mol Biol Cell 2011; 22:4093-107; PMID:21880895; http://dx.doi.org/10.1091/mbc.E11-05-0440
- Nyswaner KM, Checkley MA, Yi M, Stephens RM, Garfinkel DJ. Chromatin-associated genes protect the yeast genome from Ty1 insertional mutagenesis. Genetics 2008; 178:197-214; PMID:18202368; http://dx.doi.org/10.1534/genetics.107.082602
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 2007; 28:860-70; PMID:18082610; http://dx.doi.org/10.1016/j.molcel.2007.09.021
- Fu D, Brophy JA, Chan CT, Atmore KA, Begley U, Paules RS, Dedon PC, Begley TJ, Samson LD. Human AlkB homolog ABH8 Is a tRNA methyltransferase required for wobble uridine modification and DNA damage survival. Mol Cell Biol 2010; 30:2449-59; PMID:20308323; http://dx.doi.org/10.1128/MCB.01604-09
- Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol 2003; 23:9283-92; PMID:14645538; http://dx.doi.org/10.1128/MCB.23.24.9283-9292.2003
- Lu J, Huang B, Esberg A, Johansson MJ, Bystrom AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA 2005; 11:1648-54; PMID:16244131; http://dx.doi.org/10.1261/rna.2172105
- Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol 2006; 59:677-88; PMID:16390459; http://dx.doi.org/10.1111/j.1365-2958.2005.04972.x
- Studte P, Zink S, Jablonowski D, Bar C, von der Haar T, Tuite MF, Schaffrath R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol Microbiol 2008; 69:1266-77; PMID:18657261; http://dx.doi.org/10.1111/j.1365-2958.2008.06358.x
- Songe-Moller L, van den Born E, Leihne V, Vagbo CB, Kristoffersen T, Krokan HE, Kirpekar F, Falnes PO, Klungland A. Mammalian ALKBH8 possesses tRNA methyltransferase activity required for the biogenesis of multiple wobble uridine modifications implicated in translational decoding. Mol Cell Biol 2010; 30:1814-27; PMID:20123966; http://dx.doi.org/10.1128/MCB.01602-09
- Okada K, Muneyoshi Y, Endo Y, Hori H. Production of yeast (m2G10) methyltransferase (Trm11 and Trm112 complex) in a wheat germ cell-free translation system. Nucleic Acids Symp Ser 2009:303-4; PMID:19749381; http://dx.doi.org/10.1093/nass/nrp152
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature 2003; 425:737-41; PMID:14562106; http://dx.doi.org/10.1038/nature02046
- Liger D, Mora L, Lazar N, Figaro S, Henri J, Scrima N, Buckingham RH, van Tilbeurgh H, Heurgue-Hamard V, Graille M. Mechanism of activation of methyltransferases involved in translation by the Trm112 'hub' protein. Nucleic Acids Res 2011; 39:6249-59; PMID:21478168; http://dx.doi.org/10.1093/nar/gkr176
- Heurgue-Hamard V, Champ S, Mora L, Merkulova-Rainon T, Kisselev LL, Buckingham RH. The glutamine residue of the conserved GGQ motif in Saccharomyces cerevisiae release factor eRF1 is methylated by the product of the YDR140w gene. J Biol Chem 2005; 280:2439-45; PMID:15509572; http://dx.doi.org/10.1074/jbc.M407252200
- Polevoda B, Span L, Sherman F. The yeast translation release factors Mrf1p and Sup45p (eRF1) are methylated, respectively, by the methyltransferases Mtq1p and Mtq2p. J Biol Chem 2006; 281:2562-71; PMID:16321977; http://dx.doi.org/10.1074/jbc.M507651200
- Nakahigashi K, Kubo N, Narita S, Shimaoka T, Goto S, Oshima T, Mori H, Maeda M, Wada C, Inokuchi H. HemK, a class of protein methyl transferase with similarity to DNA methyl transferases, methylates polypeptide chain release factors, and hemK knockout induces defects in translational termination. Proc Natl Acad Sci U S A 2002; 99:1473-8; PMID:11805295; http://dx.doi.org/10.1073/pnas.032488499
- Heurgue-Hamard V, Champ S, Engstrom A, Ehrenberg M, Buckingham RH. The hemK gene in Escherichia coli encodes the N(5)-glutamine methyltransferase that modifies peptide release factors. EMBO J 2002; 21:769-78; PMID:11847124; http://dx.doi.org/10.1093/emboj/21.4.769
- Das K, Acton T, Chiang Y, Shih L, Arnold E, Montelione GT. Crystal structure of RlmAI: implications for understanding the 23S rRNA G745/G748-methylation at the macrolide antibiotic-binding site. Proc Natl Acad Sci U S A 2004; 101:4041-6; PMID:14999102; http://dx.doi.org/10.1073/pnas.0400189101
- Gustafsson C, Persson BC. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J Bact 1998; 180:359-65; PMID:9440525
- Liu M, Douthwaite S. Resistance to the macrolide antibiotic tylosin is conferred by single methylations at 23S rRNA nucleotides G748 and A2058 acting in synergy. Proc Natl Acad Sci U S A 2002; 99:14658-63; PMID:12417742; http://dx.doi.org/10.1073/pnas.232580599
- White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol Cell Biol 2008; 28:3151-61; PMID:18332120; http://dx.doi.org/10.1128/MCB.01674-07
