Abstract
Tandem alternative splice sites (TASS) form a defined class of alternative splicing and give rise to mRNA insertion/deletion variants with only small size differences. Previous work has confirmed evolutionary conservation of TASS elements while many cases show only low tissue specificity of isoform ratios. We pinpoint stochasticity and noise as important methodological issues for the dissection of TASS isoform patterns. Resolving such uncertainties, a recent report showed regulation in a cell culture system, with shifts of alternative splicing isoform ratios dependent on cell density. This novel type of regulation affects not only multiple TASS isoforms, but also other alternative splicing classes, in a concerted manner. Here, we discuss how specific regulatory network architectures may be realized through the novel regulation type and highlight the role of differential isoform functions as a key step in order to better understand the functional role of TASS.
Abbreviations
| AS | = | alternative splicing |
| TASS | = | tandem alternative splice sites |
| PCAS | = | physiologically triggered, concerted shift in alternative splicing |
It is widely appreciated that most mammalian genes give rise to multiple distinct isoforms due to alternative splicing (AS).Citation40 AS-derived protein isoforms can display structural and functional differences,Citation4,33 contribute to cell type-specific phenotypes or respond to environmental stimuli.Citation4,5,33 Through coupling to surveillance mechanisms like nonsense-mediated decay, AS additionally represents an important regulatory step in gene expression.Citation24 One explanation for AS being so frequent is that splice site signals, the RNA sequence elements which mark the boundaries of introns (5′ and 3′ splice sites, respectively; ), do often occur in redundant and ambiguous arrangements. Such arrangements were classified according to transcript structure and outcome. A so-called cassette exon, flanked by weak splice site signals, may result in alternative mRNA isoforms with included or skipped exon. An intron flanked by weak splice site signals may be spliced out from or retained in the mature mRNA. In addition, competing splice site signals at the upstream or downstream termini of the intron result in a shorter or longer exon contributing to the mRNA. Such a competition is the more frequent the closer the splice sites are, and this justifies grouping cases with 2 to 12 nucleotides distance into a core class termed tandem alternative splice sites (TASS). TASS make up the second-largest AS class in mammals.Citation1,13,17,31 TASS result in a longer or a shorter isoform depending on which splice site is used, and, in case they affect the coding sequence and maintain the reading frame, these mRNAs mostly translate into protein isoforms with subtle differences of just few amino acids.Citation16,17,36 TASS can occur at the 5′ or the 3′ splice site, but the vast majority of cases are 3′ TASS with splice sites only 3 nucleotides apart. Giving reference to their common sequence motif, these latter were termed NAGNAG 3′ splice sites, previously.Citation1,13,16,17 About 15–25% of mammalian genes harbor TASS, and TASS occur ubiquitously throughout the eukaryote kingdom.Citation17,30,31 Even though TASS-derived isoform differences are subtle, evolutionary patterns suggest that purifying selection acts to maintain their alternative splicing.Citation2,18
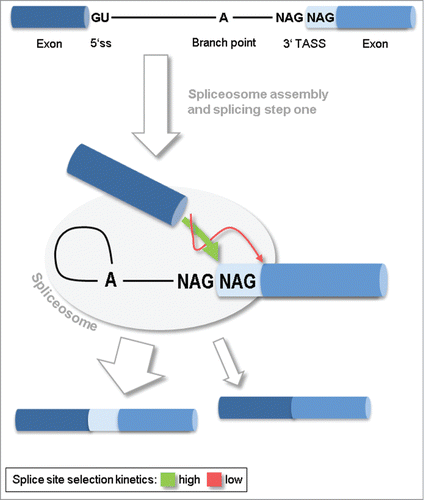
Figure 1. Model of tandem alternative splice sites (TASS) selection and splicing outcome. Splicing at TASS essentially takes place after the assembly of the enzymatic spliceosome complex and splicing step I, and precedes splicing step II. The first NAG sequence is preferably taken as the 3' splice site. An alternative NAG located further downstream may compete, but is kinetically disfavored.
Tissue-specific splicing has often been explored as a proxy for functional relevance of TASS.Citation16,17,30,35,36 Though it may seem that AS classes are structurally distinct and exist side by side, they do share different relations with respect to AS mechanism and, probably, AS regulation. Here, we review splice regulation of TASS in contrast to other AS classes, both on the ground of models of the splicing mechanism as well as in the view of recent experimental findings.
AS Regulation Probably Does Not Function Through a Universal Mechanism
The spliceosome, a multimeric enzyme complex which performs intron splicing on pre-mRNA is composed of 5 small nuclear ribonucleoproteins (snRNPs), U1, U2, U4, U5 and U6 snRNP.Citation41 First, the U1 snRNP binds to the 5′ splice site of an intron, and the U2 snRNP, in reinforcing interaction with the U2-snRNP accessory factor (U2AF), binds to a ca. 50‑nucleotide region comprising the branchpoint, a polypyrimidine tract and the 3′ splice site downstream of the intron (). Stable interaction between the pre-mRNA-bound U1 and U2 snRNPs, after intron folding, then recruit the remaining snRNPs and, by a series of phosphorylation steps, commit the complex for the enzymatic reaction. Models of AS regulation, principally referring to this complex formation between pre-mRNA and spliceosomal snRNPs, bring a group of proteins into the play, accessory splicing factors. These belong to the SR and SR-related protein families as well as other families RNA-binding proteins.Citation4,25,40 The factors bind to pre-mRNA in a sequence-specific manner and contact spliceosomal components by protein-protein interactions. Depending on the mode of interaction, positive or negative, they promote or hinder the formation of the multimeric spliceosomal complex, respectively. Hundreds of different splicing factors mediate their regulatory potential through cell type- and development-specific expression patterns or signal-specific phosphorylation status, whereby each affects splicing of a subset of introns, under defined circumstances.Citation4,5 This model was very useful to explain AS among the cassette exons class,Citation4,40 and transfers easily to the intron retention class. However, the model of splicing factor-mediated AS regulation might well be restricted to these structural AS classes because cassette exons were by far the most extensively studied group in this respect.Citation4,40
The actual splicing reaction itself takes place in 2 steps. In step I, the RNA polymer chain is disrupted at the 5′ splice site, and a new diester bond is formed between the 5′ splice site and the branchpoint (Fig. 1). In step II, the free end of the upstream exon attacks the downstream exon and displaces the intron. Prior to splicing step II, 3′ splice site choice in 3′ TASS appears to work according to a “scanning mechanism”Citation12: In search for a splice site sequence, preferably YAG (Y: = C,U), the spliceosome scans along the polypyrimidine tract situated between the branchpoint and the 3′ splice site. The first such sequence is likely to be chosen in the splicing step II but, another splice site sequence located further downstream may compete with the first one. The outcome of this splice site competition is ruled by distance and quality of the splice site motifs, as well as their distance to the branchpoint.Citation12,37 The scanning mechanism leaves little room for regulation of the splice site choice, as was pointed out earlier.Citation9 The spliceosome had a phase of multi-factorial assembly and interaction phase before commitment and entry into splicing step I, in splicing step II it enrolls its enzymatic program for removal of the intron. This way, the spliceosome just behaves analogous to multi-step biosynthesis pathways in implementing regulatory connections in the first step. This is most economic because, once, a substrate molecule is committed to the pathway it is desirable to pass it through as fast as possible (without regulation), given that intermediates hav no use and may occupy intermediary enzymes. According to the scanning model, accessory splicing factors certainly do not function the same way as they do on cassette exons, if at all.Citation9 Since the spliceosome has a stable and active conformation, reinforcing intereactions between pre-mRNA, splicing factors and snRNPs are unlikely to have the effect observed in assembly phase. The possibility that splicing factors physically block the scanning process is low, because splicing factor binding affinities to RNA are typically weak. Finally, splicing factors may interfere with the scanning process by stabilizing hairpin loops of the branchpoint-3′ splice site region of the pre-mRNA and thereby masking a region from splice site search. Even though introns from fungi and flies have been reported where secondary structure significantly contributes to 3′ splice site choice, none of these examples have been found to be protein-regulated.Citation8
Consistent with the scanning mechanism, the AS propensity of 3′ TASS cases can be predicted based on the sequence of the splice site region alone.Citation2,10,32 Such predictions were successful across different animal species and on plants, and models may even be transferred between distant organisms.Citation30,32 Moreover, 3′ TASS predictions could be trained and validated using mRNA from a variety of organs and cell types.Citation2,10,30,32 These findings disfavor a model of splicing factor-mediated TASS regulation. On the other hand, those computational studies which implemented quantitative prediction of 3′ TASS ratios have obtained 3-fold differences against experimental data, which leaves a resolution gap for putative regulatory effects (ref. Citation30 and unpublished results). In fact, one group reported an overabundance of putative cis-regulatory elements in proximity of alternatively spliced NAGNAGs,Citation1,2 which points to a regulatory involvement of splicing factors.
TASS alternative splicing is regulated, though with low dynamic range
In search for signatures of functional relevance of TASS, almost all studies concluded that isoform ratios varied across different tissues.Citation6,16,30,34-36 However, what also seems important for the potential relevance of changes in TASS splicing is the dynamic range imposed by regulation, in comparison to other AS classes. Early case reports of human genes with TASS showed almost constant isoform ratios, for example in NR3C1, ATN1, IGF1R, SCN5A, ING4, whereas some showed differential isoform ratios, for example in EDA and CACNA1A (reviewed in ref 17). Subsequent studies, aware of TASS as a wide-spread phenomenon, have addressed this question still on a selection of TASS cases.Citation16,35 These studies identified a couple of cases with tissue-specific TASS isoform but were less conclusive on a global scale. High resolution measurements of isoform ratios in larger-scale analyses, including a monitoring of observational error, were possible using capillary electrophoresis with laser-induced fluorescence detection.Citation29 A first systematic study by Tsai and Lin, characterizing one 5′ TASS and 9 NAGNAG cases in an array of human tissues, showed an overall low tissue-specific variation of TASS isoforms.Citation38 These variation patterns motivated the authors to conclude that the levels were “almost constant among tissues.” Actually, results of Tsai and Lin showed small tissue-specific variation for most TASS isoforms but, unfortunately, statistical measures were not given. Another study from our group analyzed 11 cases of 5′ and 3′ TASS in multiple human and mouse tissues and found a median cross-tissue variation of TASS isoform ratio with ±2.5%.Citation34 The tissue-associated variation was statistically significant in 90% of the cases, but the variation level was low in comparison to cassette and mutally exclusive exons which showed a 7-fold higher cross-tissue variation of isoform ratio at ±17.0%. Finally, a study that used deep RNA-seq data to analyze NAGNAG isoforms genome-wide, came to the conclusion that 73% of human and 28% mouse NAGNAGs are significantly tissue-specific. Moreover, 42% of human and 8% of mouse NAGNAGs showed tissue-specific differences of ≥25%.Citation6
Together, three studies provided high-resolution evidence that 3′ TASS isoform ratios differ across different mammalian tissues.Citation6,34,38 Even though Tsai and Lin concluded that NAGNAG isoform ratios are “almost constant among tissues,” their presented results do nicely fit other reports which diagnose cross-tissue differences.Citation34,38 Also the results from RNA-seq data support the tissue-dependent differences, with somewhat less resolution on individual TASS cases but clearly higher genome-wide coverage.Citation6 Next, concerning the extent of cross-tissue variation in isoform ratios, two studies are very close in their conclusion that the isoform ratios are “almost constant” or “very small” compared to tissue-specific splicing of cassette exons.Citation34,38 The study of Bradley et al. stands out in reporting higher rates of strong tissue-specific isoforms, at least in human where 42% of NAGNAG cases showed ≥25% differences in isoform fractions.Citation6 The minimum-maximum difference measure used in that study has the drawback that it chronically increases with an increasing number of analyzed tissues. In order to facilitate cross-study comparisons, we suggest use of normalized measures, for example cross-tissue variance.Citation34 In addition, the intriguing discrepancy between human and mouse concerning the frequency of strongly tissue-specific isoforms may suggest that also noise of RNA-seq data contributes to the very high human estimate.6
Remarkably, concerning strongly tissue-specific isoform ratios, conflicting results were reported for several NAGNAG cases. For example, a NAGNAG in human NOXO1 was found differentially spliced based on RT-PCR products separated on polyacrylamide,Citation36 and upon re-examination using capillary electrophoresis, NOXO1 isoforms were found very stable across 16 tissues.Citation38 Similar discrepancies were found when other putatively differential TASS were re-examined. Five of 6 cases were found indifferent across relevant tissues, and the one remaining was inconclusive due to weak RT-PCR signals.Citation34 Possible reasons for such discrepancies are further discussed in the next section.
According to the experimental results, AS of 3′ TASS is governed by rules which apply irrespective of the tissue, highly consistent with the scanning mechanism. Cross-tissue variation is predominantly weak, suggesting that tissue-specific splicing factors only have a minor influence on TASS splicing ratios. The stability of TASS isoform ratios may even suggest a model in which the balance of alternative splice site selection converges to a “defined” set point, a concept termed splico-stat.Citation11,23 This model does particularly support the finding of highly reproducible isoform ratios in inter-individual comparisons, with an underlying variation of the genetic background which could be expected to cause heterogenity in trans-regulatory effects. For example, the 5′ TASS ratio of the Wilms' tumor suppressor gene (WT1) and the 4-isoform pattern of the gene G subunit α S (GNAS; cassette exon plus 3′ TASS) is surprisingly stable in a large set of different individual mouse and human samples.Citation23 This is even more surprising given that gene expression, which is supposed to be under tight regulatory control, showed variation over several orders of magnitude between the samples.
Alternative splicing is a stochastic process and noisy at the molecular level
As illustrated in the previous section, conflicting data on isoform quantities have obscured the picture of tissue-specific TASS isoform patterns in the past, and possibly still do. However, it seems possible to resolve such conflicts. Studies using a classical concept of replicate measurements identified technical aspects as possible error sources in isoform quantification.Citation29,34 A stochastic model of isoform measures is sufficient to explain seemingly conflicting data of TASS isoform quantities reported from different laboratories over the last decade.
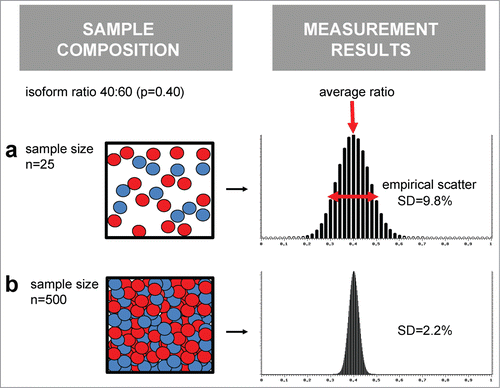
Basically, the biochemical decision event on an alternatively spliced region of a pre-mRNA molecule is categorial. That means, the irreversible splicing reaction can result in only one of possible mRNA isoforms. Only by application of the law of large numbers, we can reliably observe 2 or more AS isoforms, and isoform fraction values that are stable enough to be meaningful. Implicitly, we should keep that in mind, we have turned the perspective to a series of splicing events; often we then refer to AS in a tissue, no longer on the cellular level. In mathematical terms, an isoform mixture produced by alternative splicing is the cumulative outcome of a stochastic process, which is nicely described by the binomial (for 2 isoforms) or multinominal function (for multiple isoforms). These functions form a theoretical ground for a description of noise in measures of isoform ratios. In particular, they predict how the concentration of a particular mRNA species in biological samples relates to errors in quantification (). It is estimated that a eukaryotic cell contains between 20,000 and 300,000 mRNA molecules. Due to an approximate exponential slope of gene expression, only about 9,000 out of 20,000 human genes are expressed by at least one mRNA copy per cell.Citation3,19 While it is true that a typical RT-PCR reaction contains a multitude of cell equivalents as template, these numbers nevertheless illustrate that only a limited number of genes are expressed to an amount sufficient for quantitative isoform analysis. Sufficient here means that the molecule numbers fulfil the law of large numbers as necessary to transform finite molecule counts into an isoform fraction value with desired resolution ().
Figure 2. Sample size impacts on the reliability of isoform ratio measurements. Molecules of 2 isoform species (blue and red balls) exist in a given ratio, here 40:60%. Imagine that repeated isoform measurements are performed multiple times, using either a small sample size (row a, n = 30 molecules) or larger ample size (row b, n = 500 molecules). On average, the measured isoform ratio will reflect the real situation. But, the scatter of measurement results is significantly affected by the sample size.
Transfering this concept to real data, it important to note that many ealier studies that reported tissue-dependent variations in TASS isoform ratios did not monitor measurement variance.Citation16,35,36,38 Subsequent studies noted that the measurement uncertainty rises critically with low concentration of target genes in representative biological samples.Citation29,34 In fact, sub-critical cDNA concentrations and extremely high measurement variance coincide in TASS cases for which re-evaluation failed to reproduce strongly tissue-specific isoform ratios.Citation34 For 5 of 6 re-evaluated TASS cases, there was found no confirming support for tissue-specific isoform ratios (human ITGAM, BTNL2, SMARCA4, BRUNOL4, and murine Ccl20), and one case was still inconclusive. It indicates that reports of strongly tissue-specific isoform ratios are questionable for these genes, and similar reports for other genes should be taken with care. It also suggests to perform replicate measurements in TASS isoform quantification in order to determine the measurement uncertainty, routinely. In order to avoid backlash late in experimentation, we recommend to check, at the very beginning of a quantitative study, the expression levels of targeted genes in the given biological samples.Citation34,35 Public databases may help to verify that gene expression rank is in the top 10,000 to 12,000 genes in mammalian samples, for example using EST counts via UniGene (http://www.ncbi.nlm.nih.gov/UniGene/, in gene overview choose “EST Profile”). If gene expression is critically low, validate experimentally that it does not affect the quantification of isoforms. And if, for any reason, noise affects isoform measurements, it may be compensated by using larger amounts of RNA template in RT-PCR.
Stochasticity is also an issue of next-generation sequencing data. RNA-seq library preparation may lead through bottlenecks of very low cDNA amounts which is often obscured by subsequent polymerase chain amplification steps. The variance of count data obtained from data analysis will correspond to the molecule mixture in the bottleneck stage. Thus, high sequencing depth does not necessarily reflect high resolution of the biological signals, an effect called overdispersion.Citation27 It is recommended to test RNA-seq data accordingly and chose appropriate statistical models.
A novel physiologically induced splicing-regulatory mechanism
Interestingly, in mammalian cell culture, a significant and reproducible regulation of TASS isoforms was observed.Citation34 With increasing cell density most studied TASS cases showed an increased fraction of short isoform some responded with the opposite effect. Not only was this response homogeneously found across 5′ and 3′ TASS cases, it was also observed in cassette exons and a case of mutually exclusive exons. This indicated a novel sequence-independent regulatory mechanism which we term “physiologically triggered, concerted shift in alternative splicing” (PCAS). Even though PCAS seems to act on all AS classes, it is overlaid on the well-known splicing factor-mediated regulation pathways found for cassette exons and intron retention. TASS appears special in that it displays PCAS in an almost pure form.Citation34 Therefore, it is reasonable to focus on TASS while attempting to characterize PCAS in more detail, and for simplicity, we do not mention its analogous function on other AS classes, further on. Based on the finding that the AS shift is readily reversible and conditioned culture medium accellerates PCAS, it is likely that the effect is mediated by a diffusable molecule.Citation34 Reasonable candidates are sparse nutrients like glucose or some metals, oxygen, or accumulating metabolic products like carbonate or lactate. Since the experimental cell system allows for a systematic identification of the factor in the near future, we do not further speculate about its identity.
PCAS is remarkable in its aspects of regulatory network structure. The well-studied AS of cassette exons is complex and case-specific. Typically, multiple splicing-regulatory factors bind in a sequence-specific manner to the pre-mRNA, and the splicing outcome is quantitatively dependent of the involved splicing-regulatory factors, sometimes in a non-linear way. Thereby, the spliceosome integrates multiple signals in performing the splicing decision. In the novel splicing regulatory mechanism, PCAS, this is fundamentally different. The data on multiple AS cases indicates that it functions independent of the structure of the pre-mRNAs and is almost inert to the activity of splicing factors,Citation34 suggesting that the spliceosomal core, comprising 5 snRNPs and a number of ubiquitous co-factors, is a direct target of PCAS signaling. According to this model, the role of the spliceosome changes from a decoder/integrator of regulatory information to a mediator of a regulatory signal. As a consequence, all AS events in the cell will become subject of modulation (Fig. 3).
Predominantly stable isoform ratios, but response upon environmental triggers, oscillatory response patterns and reversibility of induced responses, these characteristics of TASS regulation in PCAS do favor a model of a negative feedback regulation or, homeostasis.Citation34 This model is in line with a regulation model, which was previously defined in a larger context of AS, called splico-stat.Citation11,23 One example illustrates that TASS may play a role in a homeostatic regulatory network and, related to the stability of the TASS isoform ratio, also has an impact on a clinical phenotype. Usage of 5′ TASS in Wilm's tumor gene (WT1) results in the insertion/deletion of 9 nucleotides, corresponding to 3 amino acids (KTS).Citation14 The ±KTS isoform ratio is almost constant across many tissues. This appears to be important, since an isoform imbalance is associated with pathological phenotypes, pseudohermaphroditism and progressive glomerulopathy, circumscribed as the Frasier syndrome.Citation15,21 Alterations of the WT1 TASS ratio can be attributed to genetic variations of the splice site region, in particular to intronic mutations 4 and 5 nucleotides downstream of the proximal splice site.Citation21 Nevertheless, the TASS ratio of the human ectodysplasin A gene (EDA) varied tissue-specifically.Citation42 Insertion/deletion of 2 amino acids creates the isoforms EDA-A1 and EDA-A2, which bind exclusively to different receptors. Distinctive temporal and spatial differences in the expression of both isoforms suggest that they have distinct roles, e.g. in the development of the hair follicle. Thus, in the view of current data, differential functions of EDA TASS isoforms are still in agreement with the splico-stat concept, but the observed variability in TASS ratios disfavor its application to EDA.
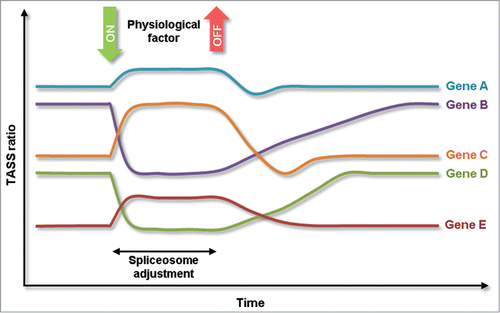
Another interesting aspect of PCAS, and also other modes of signal-induced AS regulation, is that AS-derived mRNA and protein isoforms represent a memory of the physiological conditions at the time of splicing (). Beginning with the time of environmental changes, the isoform ratio starts to shift with the addition of newly spliced mRNA. After restoration of the environmental conditions, the shifted isoform ratio still persists and returns to original levels as a result of continuous turnover of mRNA. Depending on the kinetics of mRNA expression and degradation, this results in gene-characteristic memory traces of the environmental history. For AS cases which are associated with high turnover of the mRNA, this memory is relatively short, for AS cases with slow mRNA turnover, this memory is relatively long-lasting. It is tempting to combine this with homestatic models of splicing regulation because it was shown that negative feedback networks are increasingly effective in regulation if they are fed with scattered history information. Alternative, sparse network structures with response delay may result in oscillatory patterns.Citation26 It is tempting to speculate how this contributes to the regulatory control of certain TASS cases. For example, hnRNP R, whose pre-mRNA carries a conserved 3′ TASS, has been implicated as a regulator of circadian oscillations in mRNA turnover in the pineal gland.Citation20,35
Figure 3. Model of physiologically triggered, concerted shift in alternative splicing (PCAS). PCAS functions through a basic decision rule on TASS splicing at multiple genes simultaneously. Nevertheless, changes in TASS ratios retain individual characteristics with respect to response magnitude and response times.
It is well possible that future knowledge of the PCAS factor alone will not elucidate the functional role of this mechanism. What is currently lacking is a functional keyword that connects TASS-harboring genes. Previous works have noted that genes that harbor TASS do significantly often code for splicing-regulatory proteins containing the RNA recognition motif (Pfam PF00076).Citation16,34 This is interesting in the light of what has been found out about the regulatory characteristics of TASS. TASS, forming the AS class which is least affected by accesssory splicing factors, stand out as an enriched building block that acts to diversify splicing factors. This suggests that PCAS mediates a primary input to the regulation of splicing factors, which are so far known as ruled by a dense auto- and cross-regulatory network.Citation24,28 This changes a previous suggestion that TASS may integrate into the auto- and cross-regulatory network of RNA-binding proteins.Citation16
In order to better understand the consequences of PCAS and the functional role of TASS, we need to learn more about the functional differences of the AS isoforms. So far, only very few TASS cases were studied in order to resolve the differential functions of the isoforms in detail. To give some examples, the TASS isoforms of human atrophin 1 (DRPLA), a protein associated with myoclonic dystrophy and dementia, were reported to show different subcellular localization.Citation36 Earlier work reported that TASS isoforms of mouse paired-box transcription factors, PAX3 and PAX8, show differential DNA target affinities.Citation22,39 PAX8 functions in cellular differentiation during embryonal development of the thyroid gland and the kidney. PAX3 plays a role in ear, eye and facial development in early embryonal phase, postembryonically it is associated with striated muscle growth and has been linked to rhabdomyosarcoma formation.Citation7 Remarkably, the structurally related transcription factor PAX7, also involved in myogenesis, contains at least 2 functional TASS sites which are conserved between human and mouse (ref. Citation39 and unpublished results). The clear impact of TASS on the molecular phenotypes and the availability of experimental systems for cell differentiation make the paired-box transcription factors promising candidates for the study of functional impact and relevance of their TASS isoforms.
Summary and Conclusions
Consistent over several bioinformatics studies, TASS isoforms are highly predictable based on sequence of the splice site region. Tissue-dependent differences exist, for the majority of cases experimentally analyzed. Albeit, the dynamic range is relatively small with ±2.5% between tissues, about 7-fold lower than is seen for cassette exons. The study of TASS isoform regulation in cell culture has revealed a surprising sequence-independent splicing-regulatory effect, PCAS, which affects AS of cassette exons as well. The suggested central role of the spliceosome as a mediator of regulatory signal separates this novel regulatory pathway from the splicing factor-mediated regulatory mechanism known for cassette exons. The multitude of regulatory output connections associated with PCAS is remarkable and let us ask for the common functional signature of the affected genes. The characterization of differential isoform functions is considered a key step in order to (better) understand the functional role of PCAS.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank many colleagues for their contributions, fruitful collaboration and discussions over many years of research on TASS: Rolf Backofen (University Freiburg), Klemens Hertel (University of California, Irvine) and his group, Michael Hiller (Max Planck Insitute of Molecular Cell Biology and Genetics, Dresden), Klaus Huse and Matthias Platzer (Fritz Lipmann Institute, Jena), Stefanie Schindler, and Rileen Sinha (Memorial Sloan Kettering Cancer Center, New York).
Funding
One author (M.K.) was funded by the Federal Ministry of Education and Research (BMBF, Germany; FKZ 01E01002).
References
- Akerman M, Mandel-Gutfreund Y. Alternative splicing regulation at tandem 3′ splice sites. Nucleic Acids Res 2006, 34:23-31; PMID:16394021; http://dx.doi.org/10.1093/nar/gkj408
- Akerman M, Mandel-Gutfreund Y. Does distance matter? Variations in alternative 3′ splicing regulation. Nucleic Acids Res 2007, 35:5487-98; PMID:17704130; http://dx.doi.org/10.1093/nar/gkm603
- Bengtsson M, Stahlberg A, Rorsman P, Kubista M. Gene expression profiling in single cells from the pancreatic islets of langerhans reveals lognormal distribution of mrna levels. Genome Res 2005, 15:1388-92; PMID:16204192; http://dx.doi.org/10.1101/gr.3820805
- Black DL. Mechanisms of alternative pre-messenger Rna splicing. Annu Rev Biochem 2003, 72:291-336; PMID:12626338; http://dx.doi.org/10.1146/annurev.biochem.72.121801.161720
- Blaustein M, Pelisch F, Srebrow A. Signals, pathways and splicing regulation. Int J Biochem Cell Biol 2007, 39:2031-48; PMID:17507279; http://dx.doi.org/10.1016/j.biocel.2007.04.004
- Bradley RK, Merkin J, Lambert NJ, Burge CB. Alternative splicing of Rna triplets is often regulated and accelerates proteome evolution. PLoS Biol 2012, 10:e1001229; PMID:22235189; http://dx.doi.org/10.1371/journal.pbio.1001229
- Buckingham M, Rigby PW. Gene regulatory networks and transcriptional mechanisms that control myogenesis. Dev Cell 2014, 28:225-38; PMID:24525185; http://dx.doi.org/10.1016/j.devcel.2013.12.020
- Buratti E, Baralle FE. Influence of Rna secondary structure on the pre-mrna splicing process. Mol Cell Biol 2004, 24:10505-14; PMID:15572659; http://dx.doi.org/10.1128/MCB.24.24.10505-10514.2004
- Busch A, Hertel KJ. Extensive regulation of nagnag alternative splicing: new tricks for the spliceosome?. Genome Biol 2012, 13:143; PMID:22356731; http://dx.doi.org/10.1186/gb3999
- Chern TM, van Nimwegen E, Kai C, Kawai J, Carninci P, Hayashizaki Y, Zavolan M. A simple physical model predicts small exon length variations. PLoS Genet 2006, 2:e45; PMID:16683028; http://dx.doi.org/10.1371/journal.pgen.0020045
- Chisa JL, Burke DT. Mammalian mrna splice-isoform selection is tightly controlled. Genetics 2007, 175:1079-87; PMID:17179090; http://dx.doi.org/10.1534/genetics.106.066183
- Chua K, Reed R. The Rna splicing factor hslu7 is required for correct 3′ splice-site choice. Nature 1999, 402:207-10; PMID:10647016; http://dx.doi.org/10.1038/46086
- Dou Y, Fox-Walsh KL, Baldi PF, Hertel KJ. Genomic splice-site analysis reveals frequent alternative splicing close to the dominant splice site. RNA 2006, 12:2047-56; PMID:17053087; http://dx.doi.org/10.1261/rna.151106
- Haber DA, Sohn RL, Buckler AJ, Pelletier J, Call KM, Housman DE. Alternative splicing and genomic structure of the wilms tumor gene Wt1. Proc Natl Acad Sci U S A 1991, 88:9618-22; PMID:1658787; http://dx.doi.org/10.1073/pnas.88.21.9618
- Hammes A, Guo JK, Lutsch G, Leheste JR, Landrock D, Ziegler U, Gubler MC, Schedl A. Two splice variants of the wilms' tumor 1 gene have distinct functions during sex determination and nephron formation. Cell 2001, 106:319-29; PMID:11509181; http://dx.doi.org/10.1016/S0092-8674(01)00453-6
- Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. Widespread occurrence of alternative splicing at nagnag acceptors contributes to proteome plasticity. Nat Genet 2004, 36:1255-7; PMID:15516930; http://dx.doi.org/10.1038/ng1469
- Hiller M, Platzer M. Widespread and subtle: alternative splicing at short-distance tandem sites. Trends Genet 2008, 24:246-55; PMID:18394746; http://dx.doi.org/10.1016/j.tig.2008.03.003
- Hiller M, Szafranski K, Sinha R, Huse K, Nikolajewa S, Rosenstiel P, Schreiber S, Backofen R, Platzer M. Assessing the fraction of short-distance tandem splice sites under purifying selection. RNA 2008, 14:616-29; PMID:18268022; http://dx.doi.org/10.1261/rna.883908
- Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S. Characterization of the single-cell transcriptional landscape by highly multiplex Rna-Seq. Genome Res 2011, 21:1160-7; PMID:21543516; http://dx.doi.org/10.1101/gr.110882.110
- Kim TD, Kim JS, Kim JH, Myung J, Chae HD, Woo KC, Jang SK, Koh DS, Kim KT. Rhythmic serotonin n-acetyltransferase mrna degradation is essential for the maintenance of its circadian oscillation. Mol Cell Biol 2005, 25:3232-46; PMID:15798208; http://dx.doi.org/10.1128/MCB.25.8.3232-3246.2005
- Klamt B, Koziell A, Poulat F, Wieacker P, Scambler P, Berta P, Gessler M. Frasier syndrome is caused by defective alternative splicing of Wt1 leading to an altered ratio of Wt1 +/-Kts splice isoforms. Hum Mol Genet 1998, 7:709-14; PMID:9499425; http://dx.doi.org/10.1093/hmg/7.4.709
- Kozmik Z, Czerny T, Busslinger M. Alternatively spliced insertions in the paired domain restrict the dna sequence specificity of Pax6 and Pax8. EMBO J 1997, 16:6793-803.
- Kramer M, Huse K, Menzel U, Backhaus O, Rosenstiel P, Schreiber S, Hampe J, Platzer M. Constant splice-isoform ratios in human lymphoblastoid cells support the concept of a splico-stat. Genetics 2011, 187:761-70.
- Lareau LF, Inada M, Green RE, Wengrod JC, Brenner SE. Unproductive splicing of sr genes associated with highly conserved and ultraconserved DNA elements. Nature 2007, 446:926-9; PMID:17361132; http://dx.doi.org/10.1038/nature05676
- Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors Sr proteins. Genes Dev 2010, 24:1073-4; PMID:20516191; http://dx.doi.org/10.1101/gad.1934910
- Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 2008, 135:216-26; PMID:18957198; http://dx.doi.org/10.1016/j.cell.2008.09.050
- Reeb PD, Steibel JP. Evaluating Statistical Analysis Models for Rna Sequencing Experiments. Front Genet 2013, 4:178; PMID:24062766; http://dx.doi.org/10.3389/fgene.2013.00178
- Saltzman AL, Pan Q, Blencowe BJ. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev 2011, 25:373-84; PMID:21325135; http://dx.doi.org/10.1101/gad.2004811
- Schindler S, Heiner M, Platzer M, Szafranski K. Comparison of methods for quantification of subtle splice variants. Electrophoresis 2009, 30:3674-81; PMID:19862747; http://dx.doi.org/10.1002/elps.200900292
- Schindler S, Szafranski K, Hiller M, Ali GS, Palusa SG, Backofen R, Platzer M, Reddy AS. Alternative splicing at nagnag acceptors in arabidopsis thaliana sr and sr-related protein-coding genes. BMC Genomics 2008, 9:159; PMID:18402682; http://dx.doi.org/10.1186/1471-2164-9-159
- Sinha R, Lenser T, Jahn N, Gausmann U, Friedel S, Szafranski K, Huse K, Rosenstiel P, Hampe J, Schuster S, Hiller M, Backofen R, Platzer M. Tassdb2 - a comprehensive database of subtle alternative splicing events. BMC Bioinformatics 2010, 11:216; PMID:20429909; http://dx.doi.org/10.1186/1471-2105-11-216
- Sinha R, Nikolajewa S, Szafranski K, Hiller M, Jahn N, Huse K, Platzer M, Backofen R. Accurate prediction of nagnag alternative splicing. Nucleic Acids Res 2009, 37:3569-79; PMID:19359358; http://dx.doi.org/10.1093/nar/gkp220
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene 2005, 344:1-20; PMID:15656968; http://dx.doi.org/10.1016/j.gene.2004.10.022
- Szafranski K, Fritsch C, Schumann F, Siebel L, Sinha R, Hampe J, Hiller M, Englert C, Huse K, Platzer M. Physiological state co-regulates thousands of mammalian mrna splicing events at tandem splice sites and alternative exons. Nucleic Acids Res 2014, 42:8895-904; PMID:25030907; http://dx.doi.org/10.1093/nar/gku532
- Szafranski K, Schindler S, Taudien S, Hiller M, Huse K, Jahn N, Schreiber S, Backofen R, Platzer M. Violating the splicing rules: Tg dinucleotides function as alternative 3′ splice sites in U2-dependent introns. Genome Biol 2007, 8:R154; PMID:17672918; http://dx.doi.org/10.1186/gb-2007-8-8-r154
- Tadokoro K, Yamazaki-Inoue M, Tachibana M, Fujishiro M, Nagao K, Toyoda M, Ozaki M, Ono M, Miki N, Miyashita T, Yamada M. Frequent occurrence of protein isoforms with or without a single amino acid residue by subtle alternative splicing: The case of gln in drpla affects subcellular localization of the products. J Hum Genet 2005, 50:382-94; PMID:16091834; http://dx.doi.org/10.1007/s10038-005-0261-9
- Tsai KW, Chan WC, Hsu CN, Lin WC. Sequence features involved in the mechanism of 3′ splice junction wobbling. BMC Mol Biol 2010, 11:34; PMID:20459675; http://dx.doi.org/10.1186/1471-2199-11-34
- Tsai KW, Lin WC. Quantitative analysis of wobble splicing indicates that it is not tissue specific. Genomics 2006, 88:855-64; PMID:16920330; http://dx.doi.org/10.1016/j.ygeno.2006.07.004
- Vogan KJ, Underhill DA, Gros P. An alternative splicing event in the Pax-3 paired domain identifies the linker region as a key determinant of paired domain DNA-binding activity. Mol Cell Biol 1996, 16:6677-86.
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 2008, 456:470-6; PMID:18978772; http://dx.doi.org/10.1038/nature07509
- Will CL, Luhrmann R. Spliceosome structure and function. Cold Spring Harb Perspect Biol 2011, 3: a003707; PMID:21441581; http://dx.doi.org/10.1101/cshperspect.a003707.
- Yan M, Wang LC, Hymowitz SG, Schilbach S, Lee J, Goddard A, de Vos AM, Gao WQ, Dixit VM. Two-amino acid molecular switch in an epithelial morphogen that regulates binding to two distinct receptors. Science 2000, 290:523-7; PMID:11039935; http://dx.doi.org/10.1126/science.290.5491.523
