Abstract
Myocyte enhancer factor 2c (MEF2C) is the MADS-box type transcription factor involved in the differentiation of cardiac and skeletal muscle and synaptic formation. Alternatively spliced transcripts of the MEF2C gene were proven to encode isoforms which exert distinct functions in transcriptional regulation. During the differentiation of brown adipocytes, upregulated RBM4 enhanced skipping of the MEF2Cγ region which functions as a transcriptional repressor. The presence of an overexpressed MEF2Cγ- isoform in turn induced transcriptional activity of the RBM4 promoter, constituting a positive feedback circuit in differentiating brown adipocytes. The RBM4-MEF2Cγ- network induced the expression of "myogenic" miR-1 to a greater extent than did PRDM17, BMP7 C/EBPβ, or UCP1 transcripts in C3H10T1/2 cells. Overexpression of miR-1 independently exerted the same activity as RBM4 and the MEF2Cγ- isoform of upregulating brown adipocyte-specific factors in C3H10T1/2 cells, which suggests a potential effect of miR-1 on brown adipocytes. These results indicated that the RBM4-MEF2C-miR-1 network constitutes a novel mechanism which programs the gene expression profile toward the development of brown adipocytes.
Introduction
Imbalanced energy homeostasis has led to a substantial rise in the worldwide incidence of obesity, a common origin of many metabolic diseases. Adipose tissue is an endocrine organ that plays an important role in energy homeostasis. Two major types of adipocytes, white and brown adipocytes (BAs), were identified in mammals based on their macroscopic appearance.Citation1 Obesity depends on an increase in the mass of white adipose tissues (WATs) that store energy in the form of triglycerides (TGs). In contrast, brown adipose tissues (BATs) dissipate fatty acids in the form of heat to maintain the body temperature,Citation2 which implies their therapeutic potential for combating obesity. The developmental process of BATs is still being debated. Results of transcriptomic analyses and lineage tracing suggest the existence of the same precursor cells for both myocytes and BAs.Citation3 Classical BAs were demonstrated to originate from Myf5-positive progenitors in the prenatal stage.Citation3,4 However, BA-like beige/brite cells are occasionally found as abundant clusters in WATs of adult animals under cold exposure conditions, which was proposed as a potential mechanism to fulfill the demands of emergency thermogenesis.Citation5 A protein network associated with BAs was uncovered,Citation6,7 but the comprehensive mechanism is largely unknown. Interest is building in pursuing the distinct mechanism involved in the development of classical BAs and brite cells in terms of fat metabolism and energy homeostasis.
The myocyte enhancer factor 2C (MEF2C) protein is a MADS-box transcription factor that contains a conserved MADS-box and Mef2 domain in its N-terminusCitation8,9 and was demonstrated to participate in muscle differentiation and synapse formation.Citation10,11 N-terminal domains are responsible for DNA binding and protein-protein interactions,Citation12 and the C-terminal region of MEF2C contains transregulation domains.Citation9 The human MEF2C gene consists of 10 constitutive and 3 alternatively spliced exons. Mutual splicing takes place in exons α1 and α2 which encode the region adjacent to the Mef2 domain. The selection of the β region influences the length of the transcriptional domain.Citation13 Use of the alternative 3′ splice site leads to the existence of a γ region which is located in the C-terminal domain of the MEF2C protein.Citation14 The function of mutually spliced exons α1/α2 is largely unknown. It was shown that the β region-encoded fragment substantially enhances the activity of the transactivation domain.Citation13 In contrast, the presence of the γ region-encoded fragment represses the activity of the transactivation domain in a phosphorylation-dependent manner.Citation14 Muscle regulatory factors (MRFs), including Myf5, Mrf4, MyoD, and myogenin, were shown to activate expression of the MEF2C gene,Citation15 which subsequently upregulates levels of late-stage differentiation proteins during myogenesis.Citation16 Moreover, the upregulated MEF2C protein enhances the expression of the so-called myomiR-1 which largely supports late-stage differentiation of myocytes.Citation17 The role that MEF2C plays in the development of skeletal and cardiac muscle has been widely investigated,Citation18 but its influence on differentiating BAs, which share a developmental origin with skeletal muscle, is largely unknown.Citation19
RNA-binding motif protein 4 (RBM4) is a multifunctional protein that regulates alternative splicing and messenger (m)RNA translation.Citation20,21 The reduced mass of interscapular (i) BATs which exhibit a low respiratory rate was observed in RBM4−/− mice.Citation22 RBM4 was shown to reprogram the splicing profile of precursor cells of muscle and BAs, which facilitates the differentiating process of these 2 cell types.Citation21,22 During BA differentiation, RBM4 synergizes its effect on regulating the splicing network by autoregulating its splicing pattern. An increase in RBM4 induces the exon 11-included transcript of the insulin receptor (IR) gene, IR-B, the predominant expression of which enhances the development of insulin-sensitive tissues, including adipocytes.Citation23 These results indicate that RBM4 modulates splicing events which in turn influence the development of BATs.
In this study, a relatively high level of the MEF2Cγ- transcript was observed in adult iBATs compared to embryonic tissues, whereas an increase in the MEF2Cγ- transcript was not noted in RBM4−/− iBATs. These results provide a hint that RBM4 may have the potential to modulate inclusion of the MEF2Cγ region during BA differentiation. The presence of an upregulated MEF2Cγ- transcript subsequently prompted the investigation of its influence on BA-related transcriptional regulation. We present the results which suggest a role of the RBM4-MEF2C network in BA differentiation.
Results
Depletion of RBM4 reduces the level of the MEF2Cγ- transcript in BAs
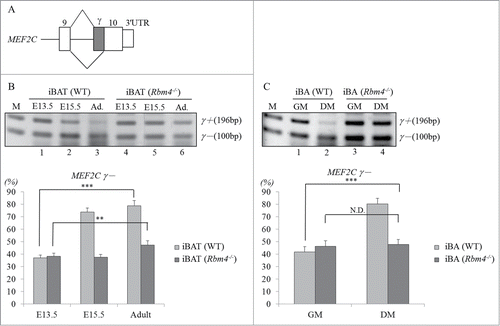
Four vertebrate MEF2 genes produce alternatively spliced isoforms which have highly similar gene structures.Citation9 The presence of an alternative 3′ splice site within MEF2C exon 10 results in the unique exclusion of a γ region which functions as a transcriptional repressorCitation14 (). The majority of MEF2C transcripts are γ region-excluded isoforms in most adult tissues and differentiating myocytes.Citation14 An RT-PCR analysis showed the gradual increase of MEF2Cγ- transcripts during the development of iBATs in WT mice (, lanes 1–3). In contrast, relative levels of MEF2Cγ- transcripts were sustained in RBM4−/− embryonic and adult iBATs (lanes 4–6), of which the mass was substantially reduced compared to that of WT littermates.Citation23 To further examine the splicing pattern of the MEF2C gene, iBAs established from WT and RBM4−/− adipose tissues were subjected to an in vitro differentiation assay. shows that the majority of MEF2C transcripts were γ-included in iBAs cultured in growth medium (GM; lane1), whereas the presence of differentiation medium (DM) enhanced the relative level of the MEF2Cγ- transcript in WT iBAs (, lane 2). In contrast, a variant change in the MEF2C gene was rarely observed in differentiating RBM4−/− iBAs which exhibited differentiating and functional deficiencies, such as a low respiratory rate (lane 4).Citation23 The results suggested that RBM4 may participate in regulating MEF2C gene splicing during BAT development.
Figure 1. Relatively high expression of the MEF2Cγ- transcript was observed in mature brown adipocytes (BAs). (A) Schematic of the alternative spliced γ region (diagonal bar) in MEF2C exon 10. Total RNA isolated from the (B) wild-type and RBM4−/− interscapular brown adipose tissues (iBATs), (C) wild-type and RBM4−/− iBAs cultured in growth medium (GM) and differentiating medium (DM) was subjected to an RT-PCR using a specific primer set against MEF2C (Table S1). The bar graph shows the relative ratio of MEF2Cγ- over total MEF2C transcripts using TotalLab Quant Software (*p < 0.05; **p < 0.01; ***p < 0.005).
RBM4 expression correlates with the level of the MEF2Cγ- transcript
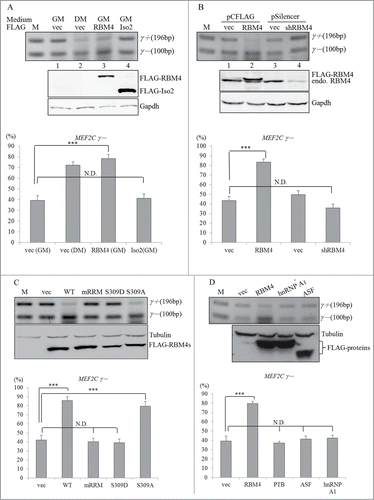
To investigate the effect of RBM4 on utilization of the MEF2C γ region, an in vivo splicing assay was conducted with C3H10T1/2 cells, which can differentiate into BA-like cells.Citation24 An RT-PCR assay was performed with total RNAs extracted from differentiating and transfected C3H10T1/2 cells. Results consistently showed substantial skipping of the MEF2Cγ region in differentiating C3H10T1/2 cells (, lane 2) as previously observed in primary BAs. The presence of overexpressed RBM4 (lane 3), but not its truncated isoform (Iso2, lane 4), increased the relative level of MEF2Cγ- transcripts under a proliferating condition. Depletion of endogenous RBM4 by introducing the RBM4-targeting shRNA-expressing vector contrastingly repressed expression of the MEF2Cγ- transcripts (, lane 4). also shows that the nonphosphorylatable S309A mutant, but not the phosphomimetic S309D or RNA recognition motif mutant (mRRM), exerted a similar effect as did WT RBM4 on upregulating MEF2Cγ- transcripts as demonstrated in other splicing events.Citation26 In addition, the influence of other splicing factors, including hnRNP A1 and ASF/SF1, on the splicing of MEF2C pre-mRNA was evaluated. As shown in , overexpression of FLAG-tagged splicing factors but not RBM4 had no obvious effect on inducing the level of MEF2Cγ- transcripts. These results suggested a specific effect of RBM4 on the splicing pattern of the MEF2C gene.
Figure 2. Relative levels of MEF2Cγ- transcripts correlated with the abundance of RBM4. (A) Mock vector-transfected C3H10T1/2 cells were cultured in growth (GM) or differentiating medium (DM) for 7 days. Cells overexpressing FLAG-RBM4 or its truncated isoform, Iso2, were cultured in GM for 24 h. An RT-PCR analysis was performed as described in . Extracted total lysates from the same batch of cells were subjected to an immunoblotting assay by probing with the indicated antibodies. (B) C3H10T1/2 cells were transiently transfected with a mock vector, or a FLAG-RBM4-expressing vector, or a RBM4-targeting shRNA expressing vector and cultured in GM for 24 h. RT-PCR and immunoblotting assays were performed as described in panel A. Total RNA was extracted from cells overexpressing (C) wild-type or mutant FLAG-RBM4 proteins (D) or distinct FLAG-tagged RNA-binding proteins. The RT-PCR and immunoblotting assay were performed as described in the previous panel. The bar graph showed the relative ratio of MEF2Cγ- over total MEF2C transcripts using TotalLab Quant Software (*p < 0.05; **p < 0.01; ***p < 0.005).
RBM4 facilitates utilization of the alternative 3′ splice site in MEF2C exon 10 via an exonic CU-element
RBM4 was demonstrated to enhance the selection of α-tropomyosin exon 9a by binding to the CU-rich element in the downstream intron, which promotes utilization of the 5′ splice site within it.Citation25 However, the simultaneous binding of RBM4 to CU-elements within the regulated exon and upstream or downstream intron results in its exclusion.Citation21,26 A CU-rich element is present in MEF2C exon 10 adjacent to the alternative 3′ splice site (, underlined characters), which may participate in RBM4-mediated skipping of the γ region. To demonstrate this hypothesis, the MEF2C minigene and a derived mutant containing the cytosine-to-guanine nucleotide substitution in the CU element were constructed (). Quantitative results of the RT-PCR assay showed that the relative ratio of MEF2Cγ- over all MEF2C transcripts changed from 40% to ∼65% in the presence of DM (, DM). Overexpression of WT RBM4, but not the truncated isoform, consistently exerted a repressive effect on inclusion of the γ region in the MEF2C minigene (). shows that cytosine-to-guanine nucleotide substitutions in the CU element (CUgg) abolished the repressive effect of RBM4 on inclusion of the MEF2C γ region, resulting in an unchanged level of MEF2Cγ- transcripts (, lane 4). Even though the isoform shift of the MEF2Cγ+ to the MEF2Cγ- transcript originating from the minigene was less than that of the endogenous MEF2C gene, the statistical analysis of the quantitative results demonstrated the credibility of the in vivo splicing assay (). These results indicate that the exonic CU-rich element acted as a responsive cis-element of RBM4 against utilization of the MEF2C γ region.
Figure 3. RBM4 mediates the exclusion of the MEF2Cγ region by recruiting the U2AF65 subunit in a CU element-dependent manner. (A) The diagram presents the wild-type and mutant MEF2C minigenes in an overexpressing plasmid. (B) C3H10T1/2 cells were transfected with the MEF2C minigene vector and cultured in growth (GM) or differentiating medium (DM). (C) C3H10T1/2 cells were cotransfected with the MEF2C minigene vector and a mock vector, or the FLAG-RBM4-expressing vector, or the FLAG-RBM4 Iso2-expressing vector. Transfected cells were cultured in GM for 24 h. (D) The wild-type (CU) or mutant (CUgg) MEF2C minigene vector was cotransfected with the mock vector, or the FLAG-RBM4-expressing vector into C3H10T1/2 cells. RT-PCR and immunoblotting assays were performed as described in a previous panel. The number below the gel indicates the relative ratio of MEF2Cγ- over the total MEF2C transcripts using TotalLab Quant Software. (E) The schematic shows the probe sequence flanking (italics) and within the MEF2Cγ region, with the putative polypyrimidine tract and splice site underlined. (F) The mock eluate (E) from the Ni2+ agarose resin or recombinant His-tagged RBM4 protein (1, 2, or 4 μg) was incubated with 10 nM DIG-labeled wild-type (CU) or mutant (CUgg) probes. The mixtures were fractionated in an 8% native acrylamide gel and transferred to a Nylon membrane. The membrane was probed using an HRP-conjugated anti-DIG Fab fragment. (G) Nuclear extracts prepared from C3H10T1/2 cells were incubated with the DIG-labeled CU or CUgg probes and recombinant RBM4 protein, followed by UV crosslinking. Reactions were analyzed using an immunoblotting assay with the indicated antibody. The bar graph shows the relative ratio of MEF2Cγ- over total MEF2C transcripts and the relative level of UV-crosslinked proteins using TotalLab Quant Software (ND, no difference; * p < 0.05; ** p < 0.01; *** p < 0.005).
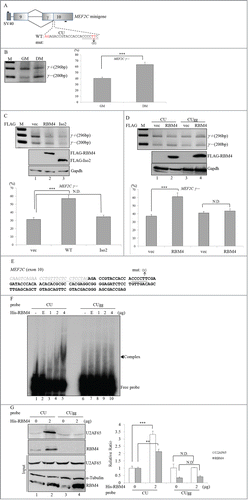
The binding of U2AF65 to the polypyrimidine tract is a required step in the recruitment of U2 snRNP onto the branch point, which initiates intronic recognition and splicing. Thus, we investigated whether the presence of RBM4 could affect binding of U2AF65 to the putative polypyrimidine tract. To demonstrate this hypothesis, the interaction between RBM4 and the exonic CU-element was first evaluated. DIG-labeled probes that originated from the CU-element in MEF2C exon 10 (, 140 nt) were incubated with the mock eluate or the recombinant His-tagged RBM4 protein under a splicing condition, and the mixtures were subjected to an REMSA. The DIG-labeled probes formed ribonucleoprotein (RNP) complexes with the recombinant RBM4 protein in a dose-dependent manner (, lanes 3–5), whereas the cytosine-to-guanine nucleotide substitutions within the CU-rich element (, mut) substantially reduced RNP complex formation (, CUgg). To investigate the impact of RBM4 on recruitment of the U2AF65 subunit to the putative polypyrimidine tract (, italicized and underlined characters), a UV cross-linking assay was conducted by incubating DIG-labeled probes with the C3H10T1/2 nuclear extract in the absence and presence of the recombinant His-tagged RBM4 protein. The blot probed with antibodies against U2AF65 and RBM4 showed an elevated RNA-protein complex of around 65 and 40 kDa, which indicated that the presence of recombinant RBM4 protein enhanced the association of RBM4 and the U2AF65 protein with the MEF2C exon 10 fragments (, lane 2). In contrast, the association between U2AF65 and the mutant probe, which contained the cytosine-to-guanine nucleotide substitution, being unchanged was noted with a concomitant decline in RBM4 binding (lanes 3 and 4). The crosslinking efficiency of RBM4 and the U2AF65 protein with the MEF2C exon 10 fragments was normalized, and a statistical analysis of the quantitative results proved the credibility of the crosslinking assay (, right panel). These results showed that the interaction between RBM4 and the exonic CU-element facilitated the recruitment of U2AF65 to the alternative polypyrimidine tract in MEF2C exon 10.
Overexpression of the MEF2Cγ- isoform induces BA differentiation
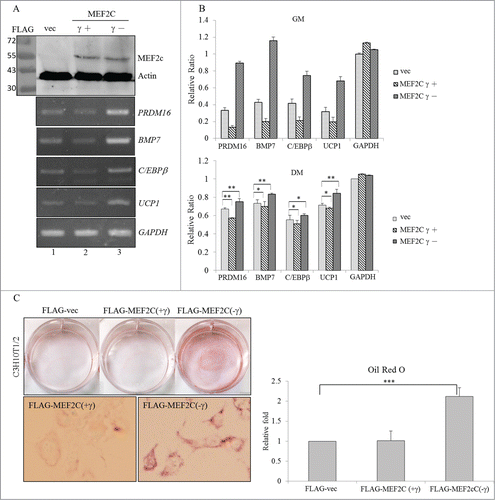
We next investigated the potential effect of the MEF2Cγ- isoform on the development of BAs due to its relatively high level (). The γ region of the MEF2C protein was demonstrated to act as a transcriptional repressor in a phosphorylation-dependent manner.Citation14 Results of the RT-PCR analysis showed that the presence of the overexpressed MEF2Cγ- isoform resulted in increases in the PRDM16, BMP7, and C/EBPβ transcripts which participate in the BA-specific transcriptional network (, lane 3). In addition, upregulation of UCP1 mRNA suggested that the presence of the MEF2Cγ- isoform directed differentiation of C3H10T1/2 cells to BA-like cells. In contrast, overexpression of the MEF2Cγ+ isoform reduced the expression of these BA-specific factors compared to those in empty vector-transfected cells (, lane 2). Distinct effects of MEF2C variants on the expression profiles of BA-specific factors were clearly dependent on their biological activities since expression levels were equal (, top row). The expression profile of BA-related genes was further quantified using a qRT-PCR in the presence of distinct MEF2C isoforms (). A statistical analysis of the qRT-PCR results showed that the MEF2C isoforms possessed similar trends to the expression profile of BA-related factors even though the differentiation-mediated signaling pathways lessened their effect (, lower panel). Oil-red-O staining was conducted to evaluate impacts of the MEF2C isoforms on physiological alterations of C3H10T1/2 cells. The accumulation of lipid droplets was clearly noted in cells which overexpressed the MEF2Cγ- isoform, whereas the presence of the MEF2Cγ+ isoform exhibited a faint influence on this phenomenon (). Effects of MEF2C isoforms on lipid accumulation were quantitatively evaluated in terms of the extracted Oil-red-O by a spectrophotometric analysis (, right row). The results indicated opposite effects of MEF2C isoforms on BA development.
Figure 4. Overexpressed MEF2C variants exerted opposite effects on the differentiation of BAs. (A) C3H10T1/2 cells were mock-transfected or transfected with FLAG-MEF2C variant-expressing plasmids. Total proteins were extracted and subjected to an immunoblotting assay using the indicated antibodies. Total RNAs were subjected to an RT-PCR using specific primer sets against BA-specific factors. (B) Total RNAs extracted from the proliferating (in growth medium; GM) or differentiating (in differentiation medium; DM) cells were subjected to a qRT-PCR using specific primer sets (Table S2). (C) The mock vector or FLAG-MEF2C variant vector-transfected C3H10T1/2 cells were cultured in GM for 48 h and then subjected to oil-red-O staining. The bar graph shows the spectrophotometric analysis of oil-red-O optical densities (ODs) at 550 nm after being extracted from stained cells in the previous panel (*p < 0.05; **p < 0.01; ***p < 0.005).
The RBM4-MEF2C network enhances the expression of myogenic microRNAs (myomiRs) in differentiating BAs
Expressions of so-called myomiRs including miR-1 and miR-206 were observed in canonical BAs which were traced and found to share a developmental origin with skeletal muscle.Citation27 In addition, PRDM16 was demonstrated to be the target gene of miR-133a, which subsequently represses the differentiation of BAs.Citation28 Interestingly, miR-1 and miR133a often originated together from 2 bicistronic clusters (miR-1-1/miR133a-2 and miR-1-2/miR133a-1) but execute opposite functions during muscle cell differentiations.Citation29 Expression profiles of miR-1-1/2 and miR-133a-1/2 were evaluated by an RT-PCR analysis. Notably, a relatively high expression of miR-1 was observed in BATs isolated from adult mice compared to those from embryos (, lanes 1 and 2). In contrast, a gradual reduction in miR-133 was noted in BATs of adult mice (, lane 2). Conversely, slight alterations in expression profiles of miR-1 and miR-133 were observed in RBM4−/− embryonic and adult BATs (, lanes 3 and 4). Expression profiles of these myomiRs were also evaluated in an in vitro differentiation system. An increase in miR-1 expression was consistently observed in differentiating BAs isolated from WT mice, whereas the level of miR-133 was inversely downregulated (, lane 2) compared to proliferating cells (lane 1). In contrast, expression profiles of miR-1 and miR-133 in RBM4−/− BAs showed a faint response to DM (lane 4). shows that the presence of overexpressed RBM4, but not the truncated isoform, resulted in an increase in miR-1 concomitant with a decrease in the level of miR-133 (lane 2). The presence of overexpressed MEF2Cγ- exerted the same effect as RBM4 of enhancing the expression of miR-1 with a concomitant reduction in miR-133 (, lane 3), whereas overexpression of the MEF2Cγ+ isoform exerted slight effects on the expression profiles of these myomiRs (lane 2). Results of previous assays were quantitated using the qRT-PCR assay followed by a statistical analysis (, right panel). These results suggested that the RBM4-MEF2C network modulates the expression profiles of myomiRs during the differentiation of BAs.
Figure 5. The RBM4-MEF2C network reprograms the myomiR profile during the development of BAs. Total RNAs extracted from (A) wild-type or RBM4−/− iBAT (B) primary wild-type or RBM4−/− (D7) interscapular (i) BAs under a proliferating (D 0) or differentiating condition were polyadenylated using poly(A) polymerase, followed by RT-PCR (left panel) and qRT-PCR analyses (right panel) by specific primer sets against endogenous miR-1/133. C3H10T1/2 cells were transfected with (C) a mock-vector or FLAG-RBM4 variant expression vector or (D) FLAG-MEF2C variant expression vector. The expression level of miR-1/133 in transfected cells was assayed as described in the previous panel (ND, no difference; * p < 0.05; ** p < 0.01; *** p < 0.005).
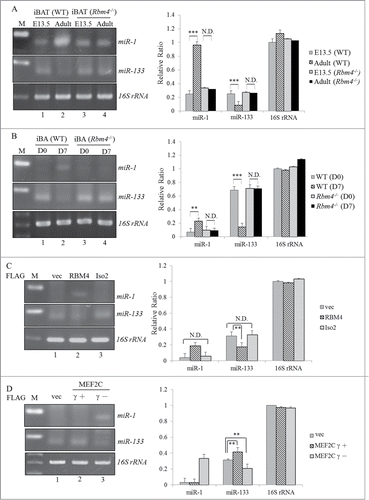
Overexpressing miR-1 enhances BA development
The miR-1 expression vector or its derived mutant was introduced into C3H10T1/2 cells to investigate their potential effects on BA differentiation. RT-PCR results showed that the presence of overexpressed miR-1 (, WT), but not that of the mutant miR-1 (mut), substantially induced transcription of BA-specific factors. shows reproducible results as revealed in the previous panel using the qRT-PCR analysis. Distinct impacts of WT and mutant miR-1 on expression profiles of BA-specific factors were clearly dependent on their biological activities since expression levels were equal (, panel miR-1). Moreover, overexpression of the WT, but not the mutant, miR-1 substantially exhibited the accumulation of lipid droplets in C3H10T1/2 cells (). The effect of WT or mutant miR-1 on lipid accumulation was quantitatively evaluated in terms of the extracted oil-red-O by a spectrophotometric analysis (, right row). These results indicated that the presence of miR-1 exerted a positive effect as did RBM4 and the MEF2Cγ- isoform on BA development.
Figure 6. Overexpression of miR-1 enhances the differentiation of BAs. (A) Total RNAs extracted from transfected C3H10T1/2 cells which overexpressed the wild-type or mutant miR-1 were subjected to an RT-PCR analysis using specific primer sets as described in . (B) Expression levels of BA-specific factors in the extracted RNAs were analyzed using a qRT-PCR with specific primer sets (Table S2). (C) The mock vector or miR-1 expression vector-transfected C3H10T1/2 cells were cultured in growth medium (GM) for 48 h and then subjected to oil-red-O staining. The bar graph shows the spectrophotometric analysis of oil-red-O optical densities (ODs) at 550 nm after being extracted from stained cells in the previous panel (*p < 0.05; **p < 0.01; ***p < 0.005).
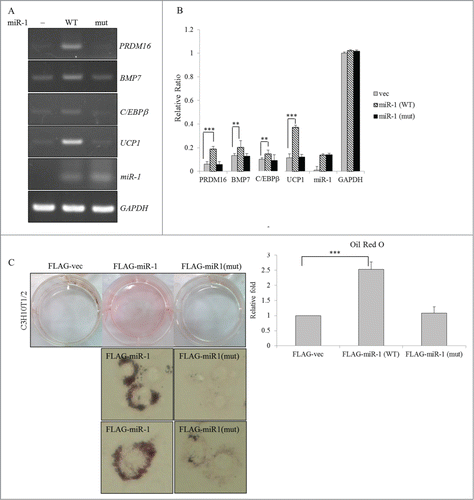
MEF2C isoforms modulate transcriptional activity of the RBM4 promoter
Gene expressions in differentiating cells are meticulously controlled by a complex network composed of transcriptional and posttranscriptional regulation. The autoregulated alternative splicing of RBM4 was demonstrated to be one mechanism that results in the upregulation of differentiating BAs.Citation22 The presence of putative C/EBPα- and -β-responsive elements in the proximal region of the RBM4 promoter () implies the potential influence of a BA-specific pathway on RBM4 expression. Moreover, previous results showed that the MEF2Cγ- isoform substantially induced expression of the C/EBPβ transcript (). Thus, we wondered whether MEF2C isoforms modulate the transcriptional activity of the RBM4 gene during adipogenesis. Using a firefly luciferase reporter driven by the human RBM4 promoter, results showed that the full RBM4 promoter was more active in differentiating cells than in proliferating cells (, pGL3-FL, DM). The proximal region exerted activity similar to that of the full RBM4 promoter in driving the reporter gene (, pGL3-Prox.), whereas the distal region of the RBM4 promoter showed a faint response to DM (, Dist.). Overexpression of the MEF2Cγ- isoform substantially enhanced the activity of the full RBM4 promoter and its proximal region (, checkered bar). On the contrary, overexpression of the MEF2Cγ+ isoform exerted a repressive effect on the activity of the full RBM4 promoter and its proximal region (diagonal bar). Nevertheless, the distal region of the RBM4 promoter showed no response to either MEF2C variant (pGL3-Dist.). Surprisingly, the result of the ChIP-analysis showed that the FLAG-tagged MEF2Cγ- was specifically associated with the proximal region (, lane 4), but not the distal region or open reading frame (lanes 2 and 6) of the RBM4 gene. Although no authentic MEF2C-binding sites were noted within the proximal region of the RBM4 promoter, MEF2Cγ- may be indirectly associated with the RBM4 promoter via interacting with other transcriptional factors, including C/EBPβ. These results showed that the RBM4-MEF2C network may constitute a feed-forward circuit that contributes to the upregulation of RBM4 in differentiating adipocytes.
Figure 7. The MEF2Cγ- isoform constitutes an alternative splicing-coupled transcriptional control of RBM4 expression. (A) The diagram shows putative binding sites of C/EBP proteins within the human RBM4 promoter. (B) The pGL3-basic or firefly luciferase reporter driven by the intact or truncated RBM4 promoter (proximal and distal regions) was cotransfected with a pRL-SV40 reference vector into C3H10T1/2 cells cultured in GM or differentiation medium (DM). (C) The intact pGL3-hRBM4 reporter or its truncated mutant was cotransfected with the expression vector of the MEF2C variants and the pRL-SV40 reference vector into C3H10T1/2 cells. The bar graph shows the relative firefly luciferase activity normalized to Renilla luciferase activity. (D) Genomic DNA prepared from cells overexpressing FLAG-MEF2Cγ- was sheared and subjected to a chromatin immunoprecipitation analysis. The FLAG-MEF2Cγ- bound DNA fragment was examined using a PCR analysis with specific primer sets targeting the promoter and open reading frame of the human RBM4 gene (Table S1) (ND, no difference; * p < 0.05; ** p < 0.01; *** p < 0.005).
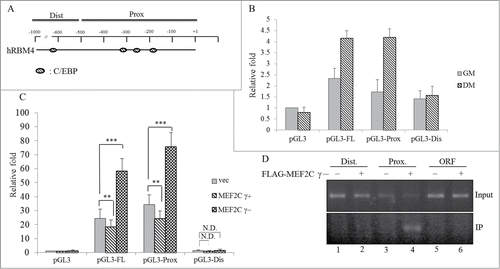
Figure 8. RBM4 and MEF2C constitute a feed-forward circuit which enhances BA-specific signaling. Upregulated RBM4 mediates skipping of the MEF2Cγ region, which substantially enhances its transactivation activity on BA-specific factors and RBM4. Moreover, the MEF2Cγ- isoform reprograms the myomiR-1/133 profile which also independently enhances the expression of BA-specific factors.
Discussion
The MEF2C protein was demonstrated to play a pivotal role in initiating and maintaining late-stage differentiation of striated muscle and neurons.Citation11,30 Despite the splicing pattern of the MEF2C gene being characterized as highly conserved in distinct species,Citation9,13,14 there is little information regarding its expression profiles or biological functions during the development of distinct tissues. Herein, we first present results relevant to the function of the γ domain-excluded MEF2C isoform which is involved in BA development. The alternative spliced variant of MEF2C and RBM4 constitute a feed-forward circuit which facilitates the expression of BA-associated transcription factors and miRNAs in differentiating BAs.
The MEF2C gene contains 10 constitutive and 3 alternatively spliced exons, which generate 5 variants through alternative splicings.Citation31 It was revealed that the α2 exon-included and β and γ region-excluded transcript is the major MEF2C variant in mature muscle and neuron cells of different speciesCitation13,32,33 (Fig. S1). The transactivation activity of the γ region was demonstrated to be highly conserved in MEF2 members,Citation13 even though a previous transcriptional assay showed that the effect of the γ region on the transactivation activity of the MEF2C protein was less than that of γ region exclusion.Citation13 The presence of the γ region probably constitutes a specific mechanism that principally manipulates the transactivation activity of the MEF2C protein, whereas the β region mainly modulates the activities of other Mef2 members. Opposite effects of γ-included and -excluded MEF2C isoforms are likely behind the controversy about the role that MEF2C plays in BA differentiation.Citation19,28 The present study showed reduced expressions of BA-specific factors with overexpression of the γ-included MEF2C isoform (, lane 2). The relatively high level of the γ region-included variant in embryonic iBATs also suggested that the γ region-included MEF2C isoform may exert a repressive effect at the onset stage of BA development as reported in a previous study.Citation28 Nevertheless, RBM4-mediated variant changes in the MEF2C gene may be one mechanism that initiates or facilitates the differentiating course of BAs.
Posttranscriptional control, including miRNA-mediated regulation, plays an important role in reprogramming gene expression profiles during tissue development.Citation34,35 Influences of the myomiRs, miR-1, -206, and -133a, on the development of striated muscles were comprehensively investigated.Citation36 Exclusive expressions of miR-1, -206, and -133 in differentiating BAs were recently noted, which suggested that myomiRs might also participate in the BA differentiation process.Citation27 The relatively high expression of miR-133 may halt the commitment step and exhibit the proliferation of premature adipocytes, whereas elevated miR-1 probably facilitates the BA differentiating process (). Increases in myomiR-1 and other miRNAs, including miR-455 and -26, were highly correlated with BA-specific proteins in differentiating BAs in the present and previous studies.Citation27,37 For instance, expression profiles of miR-1/206 and miR-133 were highly correlated with the MEF2C protein during myogenesis,Citation38,39 and the upregulated MEF2C in turn initiated expression of miR-1 at early stage of myogenesis by binding to the intragenic of miR-1-2 cluster.Citation39
miR-1 and -133 were often generated together from 2 bicistronic clusters but execute opposite functions during the differentiation of muscle cells.Citation29 The muscle-specific long noncoding RNA, linc-MD1, was demonstrated to “sponge” miR-133 and subsequently relieve miR-133-mediated repression of the expression of MEF2C at an early stage of myogenesis.Citation38 However, it was also demonstrated that the transcription of 2 clustered miRNAs can be independently controlled in a spatiotemporal manner.Citation40 Our results clarified a distinct molecular mechanism regarding dynamic expression profiles of myomiRs in differentiating BAs. Nevertheless, the detailed mechanism involved in regulating expression profiles of BA-associated miRNAs and exploration of myomiR-specific targets are worthy of further investigation.
Modulating expression profiles and cellular localization constitutes important mechanisms in determining the influence of splicing factors on alternative splicing events.Citation41,,42 Splicing profiles are modulated in a spatiotemporal manner by a complex network which is dynamically composed of various splicing factors. Expression of the MEF2Cγ- transcript is induced in the presence of the BA-specific complex in which RBM4 is involved in differentiating BAs. In the present study, the MEF2Cγ- isoform enhanced transcriptional activity of the hRbm4 promoter, which may lead to the generation of more RBM4 isoforms. Although shuttling of the RBM4-full isoform is influenced by its hyperphosphorylation, elevation of RBM4 could continually increase the level of the RBM4-full transcript through an autoregulatory mechanism. Conversely, cytoplasmic accumulation of the RBM4-truncated isoform abolishes its possible influence on the autoregulatory mechanism. It is consistent that the nuclear nonphosphorylatable S309A, but not the cytoplasmic phosphomimetic S309D mutant, possesses activity similar to that of WT RBM4 of modulating alternative splicing events in differentiating cells.Citation22,26 However, SRPK1-mediated hyperphosphorylation of RBM4 and other SR proteins may be another molecular mechanism that modulates tissue- or stage-specific splicing events. Nevertheless, the detailed mechanism involved in BA-specific splicing events is worthy of further investigation through distinct approaches.
Our present study demonstrated that alternative splicing events can widely affect transcriptional regulation and downstream targets. The former is regulated by a complex network of cell type-specific splicing factors. A growing body of studies has comprehensively mapped binding sites of splicing factors to reveal a clearer picture of the regulatory mechanism of splicing regulation. RBM4 was shown to modulate splicing patterns of several genes that comprise a muscle- and adipocyte-specific network.Citation21,22 Those studies also stated that RBM4 has a preference for binding to CU-rich sequencesCitation21 (). Nevertheless, the effect of RBM4 on the regulated exon changed with localization of the RBM4-responsive element. The binding of RBM4 to intronic CU-elements substantially enhanced the selection of the upstream regulated exon.Citation22 Conversely, the simultaneous binding of RBM4 to the regulated exon and its upstream or downstream intron via the CU-element resulted in skipping of the cassette exon in several genes, including PTB.Citation21 In this study, the binding of RBM4 to the exonic CU-element close to the alternative 3′ splice site facilitated recruitment of the U2AF65 subunit, which subsequently strengthened utilization of an alternative 3′ splice site in MEF2C exon 10 (). We thus assumed that the binding of RBM4 to the exonic or intronic CU-element alone may facilitate or stabilize the association of the spliceosome with adjacent splice sites, whereas its simultaneous binding to the exonic and intronic CU-elements interfered with the formation of the spliceosome with the surrounding splice site. These hypotheses remain to be verified in more-specific targets of RBM4.
In conclusion, knockout of the RBM4 gene resulted in the sustained expression of γ region-included MEF2C, miR-1, and miR-133 in adult and embryonic iBATs (), whereas elevated RBM4 induced relative levels of the γ region-excluded MEF2C and miR-1 which subsequently promoted BA differentiation (). The effects of RBM4 on the development and function of BATs suggest its role in manipulating metabolic homeostasis and energy expenditure.
Experimental Procedures
Mice dissection
Male RBM4a−/− mice were generated as previously described.Citation43 Adult mice were fed a regular diet for 8 weeks. After euthanasia, interscapular fat tissues were collected from adult and embryonic mice, weighed, and immediately frozen until the RNA and proteins were extracted.
Cell culture and differentiation
Primary preadipocytes were isolated from interscapular (i)BATs of RBM4a−/− mice and wild-type (WT) littermates and cultured as previously described.Citation22 Mouse C2H10T1/2 cells were cultured in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Invitrogen). To induce adipogenesis, primary adipocytes were cultured in medium supplemented with 5 μM dexamethasone, 0.02 μM insulin, 0.5 mM isobutylmethylxanthine, 1 nM T3, 125 μM indomethacin, and 1 μM rosiglitazone. Forty-eight hours after induction, cells were cultured in DMEM supplemented with 0.02 μM insulin and 1 nM T3 which mediated the differentiation of primary adipocytes. Differentiation of C3H10T1/2 cells was achieved by culturing cells in induction medium supplemented with 20% FBS, 0.5 mM IBMX, 12.7 μM dexamethasone, and 10 μg/ml insulin. Forty-eight hours after induction, the induction medium was replaced with differentiation medium (DM) supplemented with 10% FBS and 10 μg/ml insulin, and it was replenished every 2 days.
Plasmid construction, transfection, and a reverse-transcription polymerase chain reaction (RT-PCR) analysis
Complementary (c)DNAs encoding human MEF2C variants were PCR-amplified using a human fetal brain cDNA library as the template and inserted into Hind III/Xho I sites of the pcDNA3.1 FLAG vector (Invitrogen). The MEF2C minigene reporter was constructed by sequentially inserting 2 human MEF2C genomic fragments into the pCH110 vector (Amersham Pharmacia, Uppsala, Sweden). The MEF2C fragments were as follows: 462-bp exon 9∼intron 9 (nucleotides 88023962 ∼ 88024423) and 713-bp intron 9∼exon 10 (88018421 ∼ 88019133; NM_002397). The genomic fragments were PCR-amplified using genomic DNA prepared from MRC5 fibroblasts as the template and inserted into Hind III/BamH I sites to replace the β-galactosidase gene of pCH110. Mutant pCH-MEF2C vectors containing changed nucleotides were constructed using the QuikChange site-directed mutagenesis system (Stratagene, Amsterdam, Netherlands). The pGL3-hRBM4 reporter was constructed by inserting the human RBM4 promoter into the pGL3-basic vector (Promega, Madison, WI, USA). The human RBM4 promoter was PCR-amplified using genomic DNA prepared from MRC5 fibroblasts as the template and then inserted into Kpn I/Xho I sites of the pGL3-basic vector. The distal region-deleted mutant of the RBM4 promoter (pGL3-Prox) was generated by cleaving the promoter with the Hind III followed by self-ligation. The distal fragment of the Rbm4 promoter was then inserted into the Hind III site of the pGL3-basic vector (pGL3-Dist.). All constructs were auto-sequenced. C3H10T1/2 cells were grown to 60% confluence and transfected using PolyJet (SignaGen Laboratories, Ijamsville, MD, USA) with the indicated plasmid. After 24 h, total RNA was extracted using the Trizol reagent (Invitrogen). For the RT-PCR assay, 2 μg of RNA was reverse-transcribed by SuperScriptase III (Invitrogen) in a 10-μl reaction. The PCR analysis of individual genes was performed using gene-specific primer sets (Table S1). Densities of the PCR products were determined using TotalLab Quant Software. A quantitative (q)RT-PCR was performed with SYBR green fluorescent dye and gene-specific primer sets (Table S2) using an ABI One Step™ PCR machine (Applied Biosystems). The relative messenger (m)RNA level was quantitated by the ΔΔ-Ct method, and the level of GAPDH mRNA served as the internal control.
Immunoblotting
Immunoblotting of cell lysates or tissue extracts was performed using an enhanced chemiluminescence system (Millipore, Billerica, MA, USA). Images were analyzed with the LAS-4000 imaging system (Fujifilm, Japan). Primary antibodies applied in this study included polyclonal antibodies (pAbs) against RBM4 and U2AF65 (H-300; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and monoclonal antibodies (mABs) against the FLAG epitope (M2, Sigma-Aldrich, St. Louis, MO, USA), tubulin (EMD, Millipore), and polyclonal anti-GAPDH (MDBio, Taipei, Taiwan).
Oil-red-O staining
Proliferating and differentiating cells were washed twice with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde for 60 min at room temperature. Cells were washed with PBS twice and rinsed with 60% isopropanol for 5 min at room temperature. Equilibrated cells were stained with a 0.3% filtered oil-red-O solution (Sigma-Aldrich) for 10 min at room temperature. Stained cells were washed with distilled water 3 times. To extract the oil-red-O dye, a culture dish with absolute isopropanol was shaken at room temperature for 2 h. The extract was centrifuged and analyzed at 550 nm using a NanoDrop 2000 spectrophotometer (NanoDrop Tech, Wilmington, DE, USA).
RNA electrophoretic mobility shift assay (REMSA)
Recombinant His-tagged proteins were prepared as described previously.Citation25 The exonic element of MEF2C exon 10 was in vitro-transcribed and used as probes.Citation25 For RNA-protein interaction, the vector-eluate and 1, 2, and 4 μg of recombinant RBM4 protein was incubated with 10 nM of DIG-labeled probe in a 20-μl reaction containing 10 mM HEPES (pH 7.9), 50 μM EDTA, 10% glycerol, 1 mM dithiothreitol, 5 mM MgCl2, 0.5 μg/ml bovine serum albumin, and 12.5 ng/ml transfer (t)RNA for 15 min at room temperature. The reactions were analyzed by electrophoresis on a 8% nondenaturing polyacrylamide gel in TBE buffer (45 mM Tris-HCl, 45 mM boric acid, and 1 mM EDTA, at pH 8.0) and transferred to a Nylon membrane (Hybond N, Amersham, NJ, USA). Binding complexes were irradiated under 254-nm light for 60 s. Immunoblotting was conducted by probing the membranes with horseradish peroxidase (HRP)-conjugated anti-DIG Fab fragments (Roche, Mannheim, Germany).
RNA-protein interactions
RNA probes were prepared as described in the previous section. The 10-μl nuclear extract prepared from C3H10T1/2 cells was incubated with 5 ng of a DIG-labeled probe and 2 μg of the recombinant His-RBM4 protein in a 20-μl mixture at 30°C for 30 min. The reaction mixtures were irradiated on ice under 254-nm light for 10 min, and the unbound probes were digested with 1 μg of RNase A (Sigma) at 30°C for 20 min. The RNA-protein complex was immunoprecipitated from the mixture by incubating with α-DIG antibody-conjugated PAS beads (Santa Cruz Biotechnology) for 2 h at 25°C. The enriched samples were subjected to 12% sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then electron-transferred to a nitrocellulose membrane. The membranes were probed with a polyclonal anti-RBM4 antibody and monoclonal anti-U2AF65 antibody. An immunoblot analysis was conducted as previously described.
Poly(A) tailing of small RNA
Small RNAs were poly(A)-tailed by A-Plus Poly(A) polymerase (Cellscript, Madison, WI, USA) as per the user's instruction manual. In brief, 50 μg of total RNA was preheated at 65°C for 10 min. The RNAs were then incubated with 8U A-Plus Poly(A) polymerase in a 100-μl mixture at 37°C for 60 min. The reaction was immediately stopped by storing the mixture in a −70°C freezer. Tailed-RNAs were extracted using PCA/ethanol method. The RNA pellet was dissolved in nuclease-free water and subjected to the RT-PCR assay using specific primer sets (Table S1).
In vitro transcription assay
C3H10T1/2 cells were seeded in 6-well plates (2 × 10Citation5 cells/well) 24 h prior to transfection. The transfection reaction mixture contained 0.5 μg of the pGL3 firefly luciferase reporters, which were driven by a WT or truncated hRBM4 promoter, 1 μg of the effector expression vector, and 0.1 μg of the pRL-SV40 vector (Promega) as the control. After 24 h, the transfectants were lysed using passive lysis buffer, and cell debris was removed after centrifugation. Activities of the firefly and Renilla luciferases were measured using a dual-luciferase assay kit (Promega) and the Synergy HT multi-mode microplate reader (BioTek, Winooski, USA).
Chromatin immunoprecipitation (ChIP) assay
C3H10T1/2 cells transfected with the FLAG-MEF2Cγ- vector were crosslinked with 1% formaldehyde for 15 min at room temperature. Collected cells were washed with chilled PBS. Cell pellets were resuspended in 250 μl lysis buffer (50 mM Tris-HCl [pH 8.0], 85 mM KCl, and 0.5% NP-40) with a protease inhibitor (Roche), incubated on ice for 10 min, and centrifuged for 4 min at 2000 ×g. The pellets were resuspended in 250 μl (SDS) lysis buffer (1% SDS, 10 mM EDTA, and 50 mM Tris-HCl; pH 8.1) with a protease inhibitor and sonicated 4 times for 15 s each time followed by centrifugation at 14,000 ×g for 10 min. Supernatants were 10-fold diluted with dilution buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl [pH 8.1], and 167 mM NaCl) with a protease inhibitor followed by preclearing with sheared salmon sperm DNA (2 μg) and protein A Sepharose (50 μl of a 50% slurry in 10 mM Tris-HCl [pH 8.1]-1 mM EDTA) for 2 h at 4°C. Immunoprecipitation with α-M2 beads (Sigma) was performed at 4°C overnight. Immunoprecipitated complexes were sequentially washed in low-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 150 mM NaCl), high-salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], and 500 mM NaCl), LiCl wash buffer (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl; pH 8.1), and Tris-EDTA buffer. Precipitates were eluted with elution buffer (1% SDS and 0.1 M NaHCO3), and 5 M NaCl was added to reverse the crosslinks at 65°C for 6 h. DNA fragments were isolated with a clean-up kit (Viogen-Biotek, Taiwan) and 0.5 ∼ 3 μl of purified DNA was subjected to a PCR analysis using specific primer sets (Table S1).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_material.zip
Download Zip (170.2 KB)Funding
This work was supported by grants (NSC102-2320-B-038-045) from the National Science Council and (MOST103-2320-B-038-047-MY2) from the Ministry of Science and Technology of Taiwan.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Giordano A, Smorlesi A, Frontini A, Barbatelli G, Cinti S. White, brown and pink adipocytes: the extraordinary plasticity of the adipose organ. Eur J Endocrinol 2014; 170:R159-171; PMID:24468979; http://dx.doi.org/10.1530/EJE-13-0945
- Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev 2004; 84:277-359; PMID:14715917; http://dx.doi.org/10.1152/physrev.00015.2003
- Seale P, Bjork B, Yang W Kajimura S, Chin S, Kuang S, Scimè A, Devarakonda S, Conroe HM, Erdjument-Bromage H, et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008; 454:961-7; PMID:18719582; http://dx.doi.org/10.1038/nature07182
- Seale P, Kajimura S, Spiegelman BM. Transcriptional control of brown adipocyte development and physiological function-of mice and men. Genes Dev 2009; 23:788-97; PMID:19339685
- Cousin B, Cinti S, Morroni M, Raimbault S, Ricquier D, Pénicaud L, Casteilla L. Occurrence of brown adipocytes in rat white adipose tissue: Molecular and morphological characterization. J Cell Sci 1992; 103:931-42; PMID:1362571
- Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006; 7:885-96; PMID:17139329; http://dx.doi.org/10.1038/nrm2066
- Wu J, Cohen P, Spiegelman BM. Adaptive thermogenesis in adipocytes: Is beige the new brown? Genes Dev 2013; 27:234-50; PMID:23388824; http://dx.doi.org/10.1101/gad.211649.112
- Leifer D, Krainc D, Yu YT, McDermott J, Breitbart RE, Heng J, Neve RL, Kosofsky B, Nadal-Ginard B, Lipton SA. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci U S A 1993; 90:1546-50; PMID:7679508; http://dx.doi.org/10.1073/pnas.90.4.1546
- Martin JF, Miano JM, Hustad CM, Copeland NG, Jenkins NA, Olson EN. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol Cell Biol 1994; 14:1647-56; PMID:8114702
- Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 1994; 120:1251-63; PMID:8026334
- Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci 1995; 15:5727-38; PMID:7643214
- Black BL, Olson EN. (1998) Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol 1998; 14:167-96; PMID:9891782
- Zhu B, Ramachandran B, Gulick T. Alternative pre-mRNA splicing governs expression of a conserved acidic transactivation domain in myocyte enhancer factor 2 factors of striated muscle and brain. J Biol Chem 2005; 280:28749-60; PMID:15834131; http://dx.doi.org/10.1074/jbc.M502491200
- Zhu B, Gulick T. Phosphorylation and alternative pre-mRNA splicing converge to regulate myocyte enhancer factor 2C activity. Mol Cell Biol 2004; 24:8264-75; PMID:15340086; http://dx.doi.org/10.1128/MCB.24.18.8264-8275.2004
- Hinits Y, Osborn DP, Hughes SM. Differential requirements for myogenic regulatory factors distinguish medial and lateral somitic, cranial and fin muscle fibre populations. Development 2009; 136:403-14; PMID:19141670; http://dx.doi.org/10.1242/dev.028019
- Penn BH, Bergstrom DA, Dilworth FJ, Bengal E, Tapscott SJ. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev 2004; 18:2348-53; PMID:15466486; http://dx.doi.org/10.1101/gad.1234304
- Gagan J, Dey BK, Layer R, Yan Z, Dutta A. Notch3 and Mef2c proteins are mutually antagonistic via Mkp1 protein and miR-1/206 microRNAs in differentiating myoblasts. J Biol Chem 2012; 287:40360-70; PMID:23055528; http://dx.doi.org/10.1074/jbc.M112.378414
- Liu N, Nelson BR, Bezprozvannaya S, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc Natl Acad Sci U S A 2014; 111:4109-14; PMID:24591619; http://dx.doi.org/10.1073/pnas.1401732111
- Sharma G, Goalstone ML. Dominant negative FTase (DNFTalpha) inhibits ERK5, MEF2C and CREB activation in adipogenesis. Mol Cell Endocrinol 2005; 245:93-104; PMID:16356629; http://dx.doi.org/10.1016/j.mce.2005.10.027
- Lin JC, Tarn WY. RNA-binding motif protein 4 translocates to cytoplasmic granules and suppresses translation via argonaute2 during muscle cell differentiation. J Biol Chem 2009; 284:34658-65; PMID:19801630; http://dx.doi.org/10.1074/jbc.M109.032946
- Lin JC, Tarn WY. RBM4 down-regulates PTB and antagonizes its activity in muscle cell-specific alternative splicing. J Cell Biol 2011; 193:509-20; PMID:21518792; http://dx.doi.org/10.1083/jcb.201007131
- Lin JC, Tarn WY, Hsieh WK. Emerging role for RNA binding motif protein 4 in the development of brown adipocytes. Biochim Biophys Acta Mol Cell Res 2014; 1843:769-79; PMID:24389249; http://dx.doi.org/10.1016/j.bbamcr.2013.12.018
- Entingh AJ, Taniguchi CM, Kahn CR. Bi-directional regulation of brown fat adipogenesis by the insulin receptor. J Biol Chem 2003; 278:33377-83; PMID:12807888; http://dx.doi.org/10.1074/jbc.M303056200
- Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, et al. New role of bonemorphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature 2008; 454:1000-4; PMID:18719589; http://dx.doi.org/10.1038/nature07221
- Lin JC, Tarn WY (2005) Exon selection in α-tropomyosin mRNA is regulated by the antagonistic action of RBM4 and PTB. Mol Cell Biol 2005; 25:10111-21; PMID:16260624
- Lin JC, Lin CY, Tarn WY, Li FY. Elevated SRPK1 lessens apoptosis in the breast cancer cells through RBM4-regulated splicing events. RNA 2014; 20:1621-31; PMID:25140042; http://dx.doi.org/10.1261/rna.045583.114
- Walden TB, Timmons JA, Keller P, Nedergaard J, Cannon B. Distinct expression of muscle-specific microRNAs (myomirs) in brown adipocytes. J Cell Physiol 2009; 218:444-9; PMID:18937285; http://dx.doi.org/10.1002/jcp.21621
- Trajkovski M, Ahmed K, Esau CC, Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat Cell Biol 2012; 14:1330-5; PMID:23143398; http://dx.doi.org/10.1038/ncb2612
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet 2006; 38:228-33; PMID:16380711; http://dx.doi.org/10.1038/ng1725
- Devakanmalai GS, Zumrut HE, Ozbudak EM. Cited3 activates Mef2c to control muscle cell differentiation and survival. Biol Open 2013; 11:505-14; PMID:23789100; http://dx.doi.org/10.1242/bio.20132550
- Bachinski LL, Sirito M, Böhme M, Baggerly KA, Udd B, Krahe R. Altered MEF2 isoforms in myotonic dystrophy and other neuromuscular disorders. Muscle Nerve 2010; 42:856-63; PMID:21104860; http://dx.doi.org/10.1002/mus.21789
- Ganassi M, Badodi S, Polacchini A, Baruffaldi F, Battini R, Hughes SM, Hinits Y, Molinari S. Distinct functions of alternatively spliced isoforms encoded by zebrafish mef2ca and mef2cb. Biochim Biophys Acta 2014; 1839:559-70; PMID:24844180; http://dx.doi.org/10.1016/j.bbagrm.2014.05.003
- Hakim NH, Kounishi T, Alam AH, Tsukahara T, Suzuki H. Alternative splicing of Mef2c promoted by Fox-1 during neural differentiation in P19 cells. Genes Cells 2010; 15:255-67; PMID:20141540; http://dx.doi.org/10.1111/j.1365-2443.2009.01378.x
- Conte I, Merella S, Garcia-Manteiga JM, Migliore C, Lazarevic D, Carrella S, Marco-Ferreres R, Avellino R, Davidson NP, Emmett W, et al. The combination of transcriptomics and informatics identifies pathways targeted by miR-204 during neurogenesis and axon guidance. Nucleic Acids Res 2014; 42:7793-806; PMID:24895435; http://dx.doi.org/10.1093/nar/gku498
- Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol Cell 2014; 53:506-14; PMID:24440503; http://dx.doi.org/10.1016/j.molcel.2013.12.012
- Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, Münsterberg A. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev Biol 2008; 321:491-9; PMID:18619954; http://dx.doi.org/10.1016/j.ydbio.2008.06.019
- Karbiener M, Pisani DF, Frontini A, Oberreiter LM, Lang E, Vegiopoulos A, Mössenböck K, Bernhardt GA, Mayr T, Hildner F, et al. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells 2014; 32:1578-90; PMID:24375761; http://dx.doi.org/10.1002/stem.1603
- Cesana M, Cacchiarelli D, Legnini I, Santini T, Sthandier O, Chinappi M, Tramontano A, Bozzoni I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous. RNA Cell 2011; 147:358-69; PMID:22000014
- Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci U S A 2007; 104:20844-9; PMID:18093911; http://dx.doi.org/10.1073/pnas.0710558105
- Cacchiarelli D, Martone J, Girardi E, Cesana M, Incitti T, Morlando M, Nicoletti C, Santini T, Sthandier O, Barberi L, et al. MicroRNAs involved in molecular circuitries relevant for the Duchenne muscular dystrophy pathogenesis are controlled by the dystrophin/nNOS pathway. Cell Metab 2010; 12:341-51; PMID:20727829; http://dx.doi.org/10.1016/j.cmet.2010.07.008
- Gonçalves V, Henriques A, Pereira J, Neves Costa A, Moyer MP, Moita LF, Gama-Carvalho M, Matos P, Jordan P. Phosphorylation of SRSF1 by SRPK1 regulates alternative splicing of tumor-related Rac1b in colorectal cells. RNA 2014; 20:474-82; PMID: 24550521://dx.doi.org/10.1261/rna.041376.113
- Sun S, Zhang Z, Sinha R, Karni R, Krainer AR. SF2/ASF autoregulation involves multiple layers of post-transcriptional and translational control. Nat Struct Mol Biol 2010; 17:306-12; PMID:20139984; http://dx.doi.org/10.1038/nsmb.1750
- Lin JC, Yan YT, Hsieh WK, Peng PJ, Su CH, Tarn WY. RBM4 promotes pancreas cell differentiation and insulin expression. Mol Cell Biol 2013; 33:319-27; PMID:23129807; http://dx.doi.org/10.1128/MCB.01266-12
