Abstract
During the elongation cycle of protein biosynthesis, tRNAs traverse through the ribosome by consecutive binding to the 3 ribosomal binding sites (A-, P-, and E- sites). While the ribosomal A- and P-sites have been functionally well characterized in the past, the contribution of the E-site to protein biosynthesis is still poorly understood in molecular terms. Previous studies suggested an important functional interaction of the terminal residue A76 of E-tRNA with the nucleobase of the universally conserved 23S rRNA residue C2394. Using an atomic mutagenesis approach to introduce non-natural nucleoside analogs into the 23S rRNA, we could show that removal of the nucleobase or the ribose 2'-OH at C2394 had no effect on protein synthesis. On the other hand, our data disclose the importance of the highly conserved E-site base pair G2421-C2395 for effective translation. Ribosomes with a disrupted G2421-C2395 base pair are defective in tRNA binding to the E-site. This results in an impaired translation of genuine mRNAs, while homo-polymeric templates are not affected. Cumulatively our data emphasize the importance of E-site tRNA occupancy and in particular the intactness of the 23S rRNA base pair G2421-C2395 for productive protein biosynthesis.
Introduction
The ribosome, one of the oldest enzymes, is a highly conserved cellular complex and constitutes the center of protein biosynthesis.Citation1 During synthesis of a protein, the ribosome progresses through the elongation cycle which encompasses the initiation, elongation, termination, and recycling phases.Citation2 During bacterial elongation, the aminoacyl-tRNA is brought to the ribosomal A-site in a ternary complex together with elongation factor Tu (EF-Tu) and GTP. Occupation of the A-site occurs according to the mRNA codon displayed in the 30S subunit. After accommodation of the A-site tRNA into the 50S subunit and dissociation of EF-Tu*GDP from the ribosome, a new peptide bond is formed in the peptidyl transferase center (PTC). The PTC consists of universally conserved 23S rRNA residues, suggesting the catalytic activity to originate from the rRNA.Citation3,4 Upon transpeptidation the growing nascent polypeptide of the P-site bound peptidyl-tRNA is transferred to the aminoacyl-tRNA in the A-site, thus leaving a deacylated tRNA in the P-site. After peptide bond formation, the ribosome is in the so-called pre-translocation (PRE) state, which is characterized by hybrid tRNA state formation of both the deacylated (P/E-state) and peptidyl-tRNAs (A/P state).Citation5 In order to add another amino acid to the growing peptide chain, the tRNA-mRNA complex has to move one codon respective to the small ribosomal subunit to the so-called post-translocation (POST) state. In the POST state the deacylated tRNA and the peptidyl-tRNA are positioned in the canonical ribosomal E/E- and P/P-site, respectively. This translocation is mediated by elongation factor G (EF-G).Citation6 Upon binding of the next incoming ternary complex at the A-site, the deacylated tRNA leaves the ribosome from the E-site. Most of the steps of the elongation cycle are structurally and biochemically well characterized.Citation2,7 Although the existence of a third tRNA binding site, the E-site, was proposed and experimentally described already 3 decades ago,Citation8 its functional significance in translation is still not fully revealed and continues to be controversially discussed.Citation9,10 The first crystallographic visualization of a tRNA bound to the E-site of the ribosome came from the crystal structure of the 70S from T. thermophilus, with all 3 sites occupied with tRNAs.Citation11 The 5.5 Å resolution structure of the T. thermophilus ribosome unequivocally established the presence of the E-site on both the 30S and 50S subunit.Citation11 The first structure of a cognate E-tRNA, contacting a matching E-codon on the mRNA, has been recently solved.Citation12 This structure confirmed that indeed the E-site tRNA is connected to the mRNA via some sort of codon-anticodon interaction, involving the first 2 nucleotides of the E-codon. The third nucleotide of the codon does not seem to make direct contact with the anticodon, but interacts via a water molecule.Citation12 In variance to the A- and P-sites, the E-site is exclusively occupied by deacylated tRNA.Citation13
What is known about the functional features of the E-site tRNA during protein synthesis? Several reports suggest a role of the E-site in reading frame maintenance. Marquez et al.Citation14 have shown that a tRNA bound to the E-site is required to maintain the reading frame during synthesis of RF2. This idea is supported by an in vivo analysis showing the essential role of codon-anticodon interactions at the E-site for prevention of frame shifting.Citation15 The allosteric 3-site-model postulates a reciprocal relationship between the A- and E-sites,Citation16,17 stating a higher activation energy for A-site occupation when the E-site carries a tRNA. This coupling was suggested to result in a higher selectivity for the cognate A-site tRNA and thereby contributing to the observed low misincorporation rate during translation.Citation18 Besides codon-anticodon interaction, which appear to contribute to E-site binding,Citation19 several other interactions of E-tRNA and 23S rRNA have been shown to be crucial for E-site occupancy. Modifications of the 3′-terminal tRNA residue or its removal dramatically affect E-site binding, indicating the importance of the 3′-adenosine.Citation20 Despite the contacts the E-tRNA makes with the small subunit, most of the free energy of E-tRNA binding comes from its interaction with the large subunit, and therefore deacylated tRNA can also bind to isolated 50S, although fairly weak.Citation21,22 Chemical probing revealed the 23S rRNA residues C2394, G2112, and G2116 to contact the E-tRNA,Citation5,23 and later on it has been shown that the interaction with the nucleobase of C2394 is essential for tRNA binding to the E-site.Citation24 Mutation of C2394 destabilizes E-tRNA binding, leads to translocation defects in vitro and promotes frame shifting and misreading of stop codons in vivo.Citation25 This critical E-tRNA/23S rRNA interaction is also represented in more detail in the crystal structure, suggesting the terminal A76 of E-tRNA making H-bond interactions from its N3 and 2′-OH group to the N4 and N3 of C2394 of the 23S rRNA.Citation26 The precise H-bonding pattern however is not clear, as in a different crystal structure, the distance of A76 N3 to C2394 is too far to establish an H-bond.Citation27 Despite these differences, it is commonly accepted that the nucleobase of C2394 is involved in H-bond interactions with E-tRNA A76 (), as chemical modification of the base interferes with E-tRNA binding.Citation24 Besides, other possible interactions between 23S rRNA and E-tRNA, like the H-bond between G71 of the tRNA with U1851 of rRNA (), have been observed in 70S crystal structures.Citation27 The putative importance of A76 and G71 for E-site binding has previously been shown by Joseph and colleagues,Citation28 demonstrating that replacing the ribose 2′-OH at A76 or G71 of the P-tRNA by 2′-F or 2'-H, significantly inhibits the EF-G-driven translocation into the E-site.
Figure 1. Secondary structure of the 23S rRNA from T.aq. and circular permutated (cp) constructs used for atomic mutagenesis of the ribosome to modify 23S rRNA residues suggested to be required for E-site function. A. Schematic representation of the secondary structure of the 23S rRNA from T.aq. Regions of interest for investigation of E-site function are highlighted in green, blue, orange and yellow. To chemically engineer the ribosome the natural 5′- and 3′- ends of 23S rRNA are covalently linked to create a circularly permuted molecule (indicated by the red circle). B. Interaction of residue 71 of the E-site tRNA (green) with U1851 of helix 68 in 23S rRNA (red) (left panel) and A76 of the acceptor end of the E-site tRNA intercalates between 23S rRNA residues G2421 and A2422 and interacts with C2394 and the highly conserved base pair G2421-C2395. Potential hydrogen bond interactions are indicated by dashed lines. Based on the structure from Jenner et al.Citation27 Figures were generated using the pdb files 3I8I and 3I8H. C. Detailed view of 23S rRNA candidates that might be crucial for E-site function. Positions of new 5′- and 3′- ends for the cp-23S rRNA are shown, and the constructs are named according to their new ends. The compensating synthetic RNA oligo, which could carry any modification desired, is shown in pink. The RNA fragment is added in trans during in vitro reconstitution and held in its position by Watson-Crick base pairing. 23S rRNA prime candidates U1851, C2394, G2421, and A2422 are highlighted in colors.
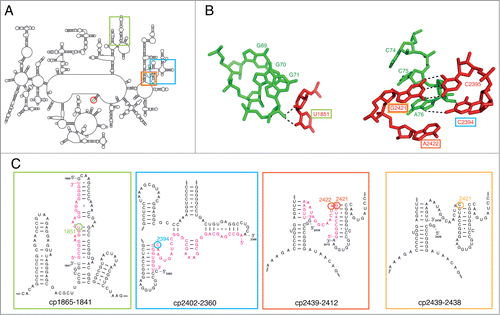
In order to more precisely characterize the 23S rRNA functional groups involved in E-tRNA binding, we used the atomic mutagenesis approach.Citation29 This approach allows introducing non-natural nucleoside analogs at specific positions of the 23S rRNA from Thermus aquaticus (T.aq.) () in the context of the entire 70S ribosome and subsequent analysis of the consequences on ribosome functions. Our data demonstrate that none of the modification at U1851 does result in translation defects, concluding that this residue is not crucial for E-site functioning. Unexpectedly, also the removal of the universally conserved nucleobase at the E-site position C2394 was tolerated by the ribosome since no significant negative effect on in vitro translation was evident. The most drastic effects on protein synthesis were observed upon the removal of the nucleobase at G2421 or the introduction of the G2421C mutation. In both cases the highly conserved Watson-Crick base pair G2421-C2395 was destroyed. The reason for this translation defect was a dramatically decreased E-site tRNA binding. Our data are compatible with the view that the G2421-C2395 23S rRNA base pair, rather than the prime suspect C2394, is pivotal for proper E-site functioning.
Results
Involvement of U1851 and the universally conserved C2394 in ribosomal E-site function
To investigate the potential role of U1851 and the universally conserved C2394 in ribosomal E-site function, an atomic mutagenesis approach was employed, which allows the site-specific incorporation of non-natural nucleosides into any desired position within the 23S rRNA ().Citation29 Ribosomes carrying the modified chemical group in the 23S rRNA were reconstituted in vitro and subsequently tested in in vitro translation or in different sub-steps or aspects of protein synthesis.
U1851 of helix 68 (H68) of the 23S rRNA is involved in a ribose-zipper interaction, with its 2′-OH group and its nucleobase, with the 2′-OH group of G71 of the E-tRNA ().Citation27,30 Furthermore both of the 2′-OH groups of G71 and A76 of the E-tRNA have been shown to be crucial for tRNA translocation.Citation28 Based on these data, we used reconstituted ribosomes carrying deoxy-U1851 () in the 23S rRNA and performed in vitro translation. We reasoned that if any of the introduced changes hamper the 50S E-site, these ribosomes should be inactive or at least less competent during in vitro translation. Although these ribosomes showed a slightly decreased activity in poly(Phe) synthesis (), they were fully active in in vitro translation of a ribosomal protein (), therefore indicating that the interaction of the 2′-OH group of U1851 with G71 is not crucial for protein synthesis. To address the function of the interaction of E-tRNA position G71 with the nucleobase of U1851 we introduced an abasic nucleoside analog at this position (). This modification affected neither poly(Phe) synthesis () nor in vitro translation of a genuine mRNA (). Even the simultaneous removal of the nucleobase and most parts of the ribose at position 1851, by introducing the C3-linker analog (), had no or only mild effects on poly(U) and r-protein mRNA translation, respectively (). All together, these data suggest U1851 not to be critical for E-site function during in vitro translation.
Figure 2. In vitro translation of poly(U) mRNA or S8 mRNA with ribosomes carrying modifications at U1851 or C2394. A. Modifications inserted at U1851 or C2394. wt: reconstituted wild type ribosomes, dN: 2'-deoxy U or C, rab: ribose-abasic analog, C3-linker: lacks the entire base as well as C1, C2 and O4 of the ribose. B. Ribosomes modified at U1851 were tested in poly(U)-directed poly(Phe) synthesis. Means and standard deviation of 2 independent experiments are shown. C. Ribosomes modified at U1851 were used for in vitro translation of genuine mRNA for ribosomal protein S8. Products of S8 mRNA translation were separated via SDS-PAGE and visualized using a phosphorimager. 30S: translation reaction in the absence of reconstituted 50S subunits, w/o mRNA: translation reaction in the absence of the S8 mRNA. One in vitro translation gel is shown exemplarily. D. Poly(Phe) synthesis using ribosomes modified at C2394. Means and standard deviation of 3 independent experiments are shown. E. In vitro translation of S8 mRNA using ribosomes modified at C2394 (see C). One representative in vitro translation gel is shown. In (B) and (D) the relative activities using wt ribosomes were set to 1.0.
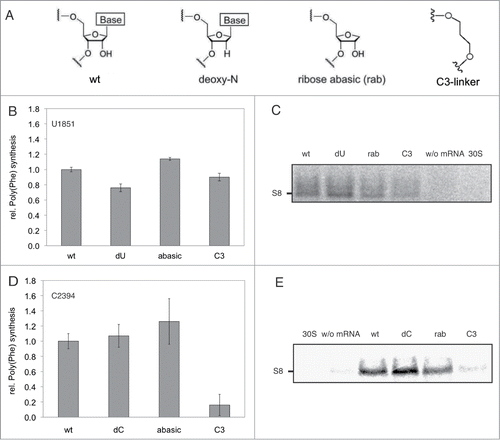
The base of the universally conserved C2394 has been shown to be in H-bonding distance to the 2′-OH group of A76 ().Citation26,27,31 Additionally, C2394 is protected from chemical modification when an E-tRNA is bound to the ribosome.Citation5 Unexpectedly, replacing the ribose with a deoxyribose or removal of the nucleobase at position 2394 did not affect the activity of these ribosomes in poly(Phe) synthesis or in vitro translation of a hetero-polymeric mRNA, respectively (). Only the introduction of a C3-linker at this position turned ribosomes inactive in in vitro translation. Summarizing, these data suggest that nucleotide C2394 contributes to a productive E-site conformation, without direct involvement of the ribose 2'-OH group or its nucleobase.
The base of G2421 is crucial for in vitro translation
Besides the interaction with C2394, the CCA-end of the E-tRNA also interacts with other 23S rRNA residues, namely G2421 and A2422.Citation26,27,31 The adenine of A76 of the E-tRNA intercalates between the nucleobases of G2421 and A2422 (). To investigate the significance of this possible base stacking interaction, ribosomes lacking the nucleobase at position 2421, or 2422, or at both positions simultaneously were generated.
In order to verify the correct overall assembly of these modified ribosomes, a puromycin reaction, showing the ability of the ribosome to catalyze peptide bond formation, was performed. Ribosomes carrying a single deletion of the base either at position 2421, or 2422, respectively, as well as base deletions at both sites simultaneously were as active as wild type (wt) ribosomes in the puromycin reaction (). Since peptide bond formation proceeds as a single turnover reaction under these conditions,Citation32 one can conclude from these data that comparable amounts of correctly assembled 50S particles were generated in all cases.
Figure 3. Puromycin reaction and in vitro protein synthesis using ribosomes carrying ribose abasic (rab) modifications at G2421 and/or A2422. A. Peptide bond formation activities of modified ribosomes determined by the puromycin reaction. Means and standard deviation from 3 independent experiments are shown. B. Time course over 90 minutes of poly(U)-directed poly(Phe) synthesis; means and standard deviation from at least 2 independent experiments are shown. C. In vitro translation of mRNA encoding the ribosomal protein L12, one representative in vitro translation gel is shown. 30S: native 30S subunit alone; w/o oligo: reconstitution reaction of 50S without the compensating RNA oligo, rab: ribose abasic site at 2421 and/ or 2422. w/o oligo, and 30S show the background level of in vitro translation and serve as negative controls. In vitro translation products were separated via SDS-PAGE and visualized by phosphor-imaging. D. Poly(A)-directed poly(Lys) synthesis using modified ribosomes, means and standard deviation of 3 independent experiments are shown. In (A), (B), and (D), relative activities using wt reconstituted ribosomes were set to 1.0.
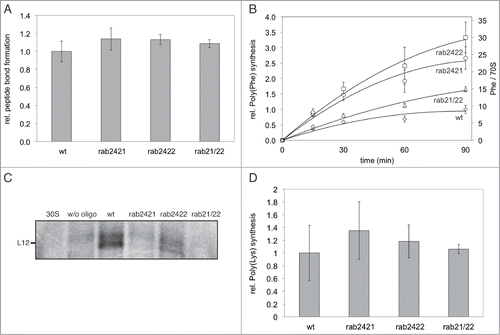
The next step was to investigate the impact of G2421 and A2422 on the ability of ribosomes to perform in vitro protein synthesis, therefore these modified ribosomes were first tested in poly(Phe) synthesis. All tested mutants were fully active in the poly(Phe) assay, their activity even exceeded the one of wt ribosomes (). However, when a genuine hetero-polymeric mRNA coding for an r-protein was used as template, this pattern changed drastically (). Ribosomes with a 2421 abasic site were unable to synthesize a full-length protein in vitro, both in the single abasic version as well as in the context of the 2422 nucleobase deletion. Removal of the nucleobase at position 2422 however, still allowed full-length protein synthesis in vitro. These data suggest that the interaction of A76 of E-tRNA with G2421 is essential for proper function of the ribosome, whereas the interaction with A2422 is less critical.
To investigate the apparent discrepancy between the activity of ribosomes lacking the nucleobase at position 2421 in poly(Phe) synthesis and their inability to synthesize a full-length r-protein in vitro, a poly(Lys) assay was performed. The rationale behind this was to investigate whether the physicochemical peculiarity of the synthesized poly(Phe) peptide or the employed homo-polymeric mRNA analog are responsible for the observed discrepancies. Indeed poly(Phe) peptides have been demonstrated before to behave differently on the ribosome than natural polypeptides and thus may represent a special case.Citation33 As shown in ribosomes lacking the nucleobase at residue 2421, or 2422, or both simultaneously were still able to synthesize poly(Lys) peptides in vitro. Like for poly(Phe) synthesis they exhibit even a slightly higher product formation than wt ribosomes. Thus, we have shown here, that the base at position 2421 is dispensable for translation of a homo-polymeric mRNA, whereas these ribosomes fail to synthesize polypeptides encoded in a genuine mRNA.
G2421 is part of a critical base pair
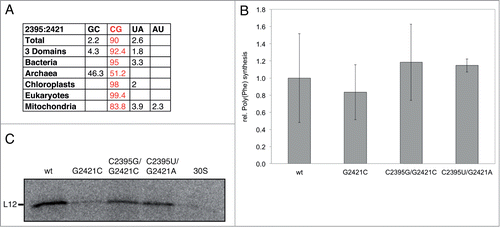
We wondered if the lacking base stacking interaction of G2421 with A76 of E-tRNA or the disrupted G2421-C2395 base pair are the cause for the disability of abasic 2421 ribosomes to translate a genuine mRNA. Phylogenetic analyses clearly demonstrate that this base pair is highly conserved and present among all domains of life ().Citation34 We destroyed this base pair by mutating G2421 to C (resulting in a C2421:C2395 mismatch) in order to test its importance. As expected, poly(Phe) synthesis was unaffected by this mutation (). In contrast, these ribosomes were unable to efficiently synthesize an r-protein in vitro (). However, introducing the complementary mutation at position 2395, resulting in a C2421-G2395 base pair, successfully restored in vitro translation almost up to wt levels. Similarly, introducing a canonical A-U base pair also yielded fully active ribosomes (). These data suggest that the presence of a Watson-Crick base pair at 2421–2395 in the ribosomal E-site, regardless of its sequence composition, is essential for in vitro translation.
Figure 4. Effects of disrupting the G2421-C2395 base pair in the 23S rRNA on poly(Phe) synthesis and on L12 in vitro translation. A. Table showing the conservation of the base pair G2421-C2395 in all 3 domains of life (in%). Data adapted from ref. 34. B. Relative poly(Phe) synthesis, using in vitro reconstituted ribosomes carrying mutations at positions 2421 and 2395 of 23S rRNA; the amount of poly(Phe) product using wt reconstituted ribosomes (cp2439-2438) was set to 1.0. Means and standard deviation of 4 independent experiments are shown. C. In vitro translation products (one gel is shown exemplarily) were separated via SDS-PAGE and visualized using a phosphorimager. 30S: native 30S subunit alone.
Ribosomes lacking the guanine at 2421 are not prone to errors
After having shown that destruction of the base pair G2421-C2395 renders ribosomes inactive in in vitro synthesis of a full-length protein but failed to affect poly(U) translation, we wanted to reveal the origin of this unexpected observation. We first addressed the question, whether these E-site modified ribosomes possess an elevated error rate during A-site tRNA decoding. Therefore, ribosomes carrying an abasic site at position 2421 were subjected to a misincorporation assay. Nierhaus and colleagues reported that E-site tRNA occupancy positively correlates with decoding accuracy at the A-site.Citation35,36 In our assay we measured the misincorporation of leucine during poly(U)-directed poly(Phe) synthesis.Citation37 Misincorporation events were monitored and normalized to total poly(Phe) synthesis (). To assure the proper functionality of this assay neomycin, an aminoglycoside antibiotic known to increase the error rate during the decoding process,Citation38 was added to one reaction. Neither the single nucleobase deletion at 2421 nor the simultaneous base removal at 2421 and 2422 resulted in a higher rate of leucine misincorporation (), concluding that ribosomes lacking the crucial G2421-C2395 base pair in the E-site do not possess a markedly altered decoding accuracy.
Figure 5. Functional assays using modified ribosomes carrying an abasic site either at position 2421, 2422, or both. A. Leucine misincorporation assay based on the poly(Phe) system. The relative amount of misincorporated leucine into the poly(Phe) peptide is shown (means and standard deviation of 6 independent experiments). Wt + Neomycin (Neo): an antibiotic known to increase the misincorporation rateCitation59 served as misincorporation control. B. Reading-frame maintenance assayed by the use of poly(UUC) instead of poly(U) mRNA for poly(Phe) synthesis; relative poly(Phe) synthesis (wt was set 1), means and standard deviation of at least 2 independent experiments, is shown. The ribosomal mutant G2252C known to lose the reading frameCitation39 served as control for defects in reading-frame maintenance. C. EF-G dependent tRNA translocation efficiencies of modified ribosomes were addressed by toeprinting analysis. Modified ribosomes were programmed with an mRNA analog and used to construct tRNA/70S/mRNA complexes. Pre-translocation complexes (Pre) were established by binding of deacylated tRNAfMet into the P-site, followed by binding of Ac-Phe-tRNAPhe into the A-site. Post-translocation complexes (Post) were created by addition of EF-G*GTP. Lanes designated Pi represent control complexes carrying only deacylated tRNAfMet in the P-site. Pi complexes result in a stop at position +16 in the primer extension (first nucleotide of the AUG codon is assigned as +1). The additional stop at +17 in Pre complexes has been observed also in other toeprinting assays using native 70S particles.Citation60 Percentage of ribosomal complexes in the Pre and Post state of at least 3 independent toeprinting experiments are given underneath the respective lanes. Relative translocation efficiency of the G2421C mutant was calculated from percentage of ribosomal complexes, translocated to the Post state through addition of EF-G*GTP.
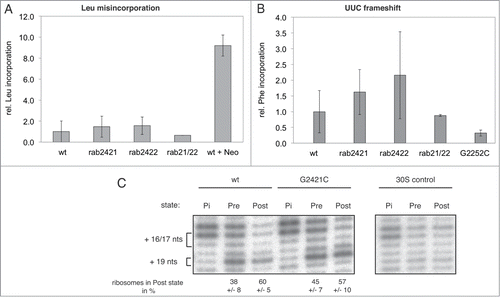
Another possible explanation for the divergent in vitro translation activities of the modified ribosomes in the poly(U) and genuine mRNA systems can be an impaired reading-frame maintenance. Such a defect would not affect translation of the former, but would have detrimental effects on protein synthesis of a hetero-polymeric mRNA. Thus we investigated the ability of these ribosomes to keep the reading-frame, by using a poly(UUC) message instead of poly(U) in the poly(Phe) assay. Both mRNA analogs code for poly-phenylalanine but if the E-site modified ribosomes are defective in reading-frame maintenance, such an impairment would only be detected in the presence of poly(UUC). The G2252C mutant ribosome, which is prone to lose the reading-frame,Citation39 showed a clearly reduced activity utilizing poly(UUC), and served as frame-shift control construct. For both the single nucleobase deletion at 2421 as well as the double base deletion 2421/2422, poly(Phe) synthesis was not inhibited compared to wt reconstituted ribosomes (). Consequently, the reason for the inability of translating a hetero-polymeric mRNA by ribosomes lacking the G2421-C2395 base pair is not due to decreased reading-frame maintenance.
The next step was to analyze the ability of ribosomes carrying a destroyed G2421-C2395 base pair to perform EF-G mediated translocation. In order to translate a genuine mRNA into a full-length protein, the ribosome has to translocate properly in a consecutive manner along the open reading frame to produce a full-length polypeptide chain. Poly(Phe) synthesis on the other hand has been shown to yield detectable translation products even in the absence of EF-G.Citation40,41 Therefore, to assess the EF-G-driven translocation ability of ribosomes harboring a disrupted 2421–2395 base pair, toeprint analysis was performed. This procedure allows determining how efficient ribosomes move from the PRE to the POST state upon addition of EF-G and GTP. When wt ribosomes were incubated with EF-G and GTP, we found 60% of reconstituted ribosomes in the POST state, compared to 38% prior to EF-G driven translocation (). This means that 22% of the wt ribosomes translocated in an EF-G dependent manner. EF-G driven translocation was not as efficient for G-C base pair mutant ribosomes, where we found only 57% of ribosomes in the POST state, compared to 45% prior addition of EF-G and GTP (). Relative high occupation of the POST state (38% wt and 45% G2421C, respectively), even before addition of EF-G*GTP, is probably a result of an inherent instability of the reconstituted ribosomes in the PRE state.Citation42 Our data show only a moderately reduced translocation efficiency with ribosomes carrying a disrupted E-site base pair, which may sum up and account for the severe defect of this mutant in translation of a genuine mRNA ( and ), as this requires iterative rounds of efficient translocation steps. If this slight defect in translocation of ribosomes lacking the G2421-C2395 base pair accumulates over several rounds of translocation, one would expect that the poly(Phe) peptides synthesized by mutant ribosomes to be shorter than the one produced by the corresponding wt ribosomes. In order to test this hypothesis we separated the poly(Phe) chains on a tricine-gel. However, no size differences of the synthesized poly(Phe) peptides were evident (Fig. S1). From these data we conclude that the slight translocation defect does not account for the different activities of ribosomes lacking the G2421-C2395 in translation of a homo-polymeric and a genuine mRNA.
G2421-C2395 base pair is crucial for E-site tRNA binding
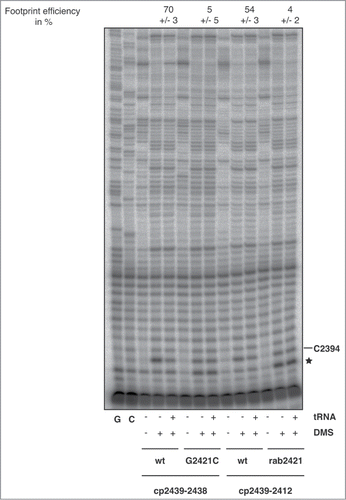
E-site mutations have previously been linked to translocation defects.Citation43 In order to understand the cause of the observed translation deficiencies of ribosomes carrying a disrupted E-site base pair G2421-C2395, we analyzed the E-site tRNA binding properties of these mutant ribosomes by tRNA footprinting (DMS modification and protection from modification by tRNA binding was analyzed by primer extension (Fig. 6; Fig. S2). G2421C as well as 2421 abasic ribosomes were probed with DMS, after tRNA binding of deacylated tRNAPhe under conditions known to establish the P/E hybrid state.Citation23,24 The diagnostic 50S E-tRNA footprint at C2394 was used to monitor E-site occupancy. For both, the G2421C as well as the 2421 abasic ribosome, E-site tRNA footprinting was almost completely abolished, as shown by the footprinting efficiency of only 5 or 4%, respectively (). E-site tRNA binding for wt ribosomes was efficient, with footprint efficiencies of 70 or 54%, respectively. To confirm these data, we additionally analyzed E-site tRNA binding in a system where we first filled the P-site with the peptidyl-tRNA analog Ac-Phe-tRNAPhe, a tRNA substrate known to establish the classical P/P state. To subsequently fill the E-site, deacylated tRNAfMet was added in excess over the ribosome, that displayed a cognate AUG mRNA codon in the 30S subunit. In this scenario the E-site tRNA is expected to adopt the classical E/E state.Citation23 In agreement with the data shown in , ribosomes with a broken 2421–2395 base pair were defective in E-site tRNA binding (Fig. S3).
Figure 6. E-site tRNA binding to G2421C ribosomes and to 2421 abasic ribosomes addressed by footprinting analysis. Deacylated tRNAPhe was bound to sucrose cushion-purified reconstituted ribosomes prior to chemical probing with DMS. DMS modification status of C2394 was analyzed by primer extension. C2394 is known to interact with E-site tRNA and to be protected from DMS modification by a bound tRNA. Under the applied conditions deacylated tRNAPhe adopts the P/E-hybrid state and thereby protects C2394 from DMS modification.Citation5 To unambiguously identify DMS reactive residues, ribosomes without DMS treatment were used as control. Primer extension stop at C2394 for the reaction without bound tRNA was set as 100%. Footprint efficiency was calculated as the percentage of decrease in the band intensity from the primer extension stop at C2394. Means and standard deviation of footprinting efficiencies (in%) from 3 independent experiments are given above the respective lanes. G,C indicate sequencing reactions. The increased DMS reactivity of C2395 upon deletion of the base pairing with 2421 (in both mutants G2421C and 2421 abasic ribosomes) is marked with asterisk.
Discussion
The E-site constitutes one of the 3 ribosomal tRNA binding sites that a tRNA occupies during one round of elongation. Whereas the molecular function of the P- and A-site are well characterized, the role of the E-site in translation remains still elusive.Citation44 It has been shown by several groups that occupation of E-site tRNA is required for reading-frame maintenance,Citation14,45,46 and it might also regulate in an allosteric manner the A-site affinity for aminoacyl-tRNA binding.Citation19 More recent structural studies compellingly demonstrated that the E-site is a robust and highly occupied tRNA binding site. Nevertheless, the biological function of E-tRNA for protein synthesis remained unclear and represents a highly controversial topic.Citation9,10,17,47
Unlike to the A- and the P-site, the E-site exclusively binds deacylated tRNA.Citation8 In addition to rRNA, also r-proteins contribute to 50S E-site tRNA binding, which is not the case for A- and P-site binding.Citation48 The major contribution of E-site tRNA binding energy comes from the 50S subunit.Citation21,22 Molecular details about E-tRNA–ribosome interactions were obtained in crystal structures and chemical modification assays.Citation5,23,24,26,27 Whereas the 23S rRNA residues implicated in E-tRNA binding are all in good agreement, the interactions on the atomic level are not always consistent in different structures.
This work aimed to unravel the critical residues of 23S rRNA for E-site function. We used the atomic mutagenesis approach,Citation29 to modify the residues U1851, C2394, G2421, and A2422 of 23S rRNA in order to analyze their contribution to E-tRNA function and the effect on protein synthesis. This in vitro assembly method allows modification or deletion of the nucleobase, the sugar or both moieties of a defined 23S rRNA residue in the context of the complete 70S ribosome. We can thereby investigate the function of single functional groups or even individual atoms of 23S rRNA, which is beyond the scope of conventional mutagenesis.
The 23S rRNA residue most frequently connected to E-site functions is the nucleobase and ribose 2′-OH of the universally conserved C2394. Its pivotal role for ribosome functioning has been underlined by structural data,Citation27 chemical probing,Citation5 modification interference,Citation24 as well as mutational analyses.Citation25 Cumulatively, these studies provide evidence for a direct interaction of the terminal adenosine of E-tRNA with C2394 of rRNA. Unexpectedly, removal of the cytosine nucleobase or the ribose 2′-OH at position 2394 had essentially no negative effects on protein synthesis in the context of in vitro assembled ribosomes of T.aq. (). In E.coli however, mutation of C2394 impairs E-tRNA binding, the translocation step, and increases frame shifting.Citation25,43 Either the E-tRNA interactions are more robust and thus more resistant to alterations at position 2394 in the thermophilic ribosome of T.aq., or the previously described C2394G mutation affects more indirectly crucial E-tRNA-23S rRNA interactions in immediate proximity. Indeed, A76 of E-site tRNA has been shown to insert and stack in between the 2 23S rRNA residues G2421 and A2422Citation27 and our data demonstrate that removal or mutation of the nucleobase at position G2421, but not A2422, almost completely blocks in vitro translation (). Why is the nucleobase of 2421 so crucial for protein synthesis whereas an abasic site at 2422 had only minor effects? The base of G2421 is involved in a Watson-Crick base pairing interaction with C2395 and this is highly conserved in all domains of life.Citation34 The functional importance of this G-C base pair in the E-site was supported by mutational analyses clearly showing that the base pair itself, rather than the identity of the involved nucleobases matters (). Our study therefore identified a hitherto largely undisclosed functional region in the ribosomal E-site contributing to protein biosynthesis. Support for this conclusion is provided by the fact that the natural anti-ribosomal protein synthesis inhibitors mycalamide A and 13-deoxytedanolide have been shown to utilize the G2421-C2395 base pair as a stacking platform for ribosome binding.Citation49,50 Antibiotic binding sites are typically indicative for a functional hot-spot in the ribosome. Despite the huge size of the ribosomes, only a surprisingly small number of antibiotic binding sites have been “chosen” during the course of evolution as interaction sites of natural antibiotics.Citation51,52
Unexpectedly, disruption of this critical E-site base pair G2421-C2395 had bipartite consequences on translation of homo-polymeric, compared to hetero-polymeric mRNAs. While mutant ribosomes were able to translate the homo-polymeric mRNA analogs poly(U), poly(UUC), and poly(A), they failed to translate a genuine mRNA transcript encoding a r-protein ( and ). What challenges the ribosome in translating a hetero-polymeric compared to a homo-polymeric message? Our data do not reveal any gross defects in reading-frame maintenance, A-site miscoding, and only a slight defect in EF-G-driven tRNA translocation in ribosomes carrying a disrupted G2421-C2395 pair (). However, footprinting analysis showed that ribosomes with a disrupted G2421-C2395 base pair have a vastly reduced E-tRNA binding capability ( and S3). This indicates that for an efficient performance during protein biosynthesis it is beneficial for the ribosome to carry always 2 tRNA molecules before (at the A- and P-sites) and after translocation (at the P- and E-sites).Citation44 Obviously the reduced E-tRNA occupancy of ribosomes with a disrupted G2421-C2395 is more critical when translating a genuine mRNA compared to a homo-polymeric mRNA. It is known that in order to produce detectable amounts of a poly(Phe) peptide, only a few rounds of translocation have to occur, whereas the synthesis of a full-length protein is much more challenging, as it requires numerous rounds of translocation. However, analysis of the length of the poly(Phe) peptides showed that ribosomes lacking the G2421-C2395 base pair, are able to synthesize poly(Phe) peptides of the same lengths as wt ribosomes (Fig. S1). Therefore, the divergent activities using homo- versus hetero-polymeric mRNAs do not result from the observed marginal defects of the EF-G-driven translocation process in ribosomes carrying a disturbed E-site base pair ().
A reasonable explanation would be that the absence of E-tRNA in ribosomes with a disrupted G2421-C2395 base pair, affects the ability of the ribosome to handle potential downstream secondary structure mRNA elements. In support of such a scenario, a recent single molecule FRET study provided evidence that mRNA structure melting takes place after translocation but seemingly before E-tRNA dissociation.Citation53 In other words, a filled E-site enables the ribosome to more effectively deal with downstream located steric hindrances at the mRNA level. Since homo-polymeric mRNAs lack stable secondary structure elements, the loss of the E-tRNA during poly(Phe) or poly(Lys) synthesis can be tolerated while it is detrimental during translation of a genuine mRNA.
In summary, our data disclose a so far unknown active site in the ribosome, namely the highly conserved Watson-Crick base pair at 23S rRNA position 2421–2395 in the E-site. Furthermore we demonstrate that ribosomes with a disrupted base pair are still able to perform well in several sub-steps of the elongation cycle, but appear to have a weakened E-tRNA binding capability. Loss of E-tRNA dramatically affects translation of a hetero-polymeric mRNA, but had no effect on the more artificial poly(Phe) and poly(Lys) translation systems.
Experimental Procedures
In vitro reconstitution of 50S subunits
Generation of the circularly permuted (cp) 23S rRNA, subsequent in vitro reconstitution of the 50S particles, and reassociation with native 30S subunits was done as previously described.Citation32,54 The following primers were used to generate the different DNA templates for the cp-23S rRNA in vitro transcription (the first number marks the 5′-end of the cp-23S rRNA and the second number indicates the 3′-end): cp2439–2412 – GGATCCTAATACGACTCACTATAG2439GCCCGGGGATAACAG and T2412TCCACACGGGACCACC. Underlined in the forward primer sequence is the T7 promoter sequence and the cp-23S rRNA positions defining the new 5′- and 3′ ends are highlighted in bold. For cp1865–1841 following primers were used: GGATCCTAATACGACTCACTATAGG1865CAAGCCCCAAACCGAAGCCCCGG and A1841CCGGGCAAGCGTCGCTCCC, for cp2402–2360 GGATCCTAATACGACTCACTATAGGC2402CCGTGTGGAAGGGCCATCGATCA and T2360GCAGGCCTCACAGTCAGGC. The full-length cp23S rRNA cp2439–2438 was designed using the primers GGATCCTAATACGACTCACTATAG2439GCCCGGGGATAACAG and A2438ACTTTTATCCGTTGATCGAT, and cp2413–2412 was generated using the primers GGATCCTAATACGACTCACTATAG2413GGCCATCGATCAACGGATA and T2412TCCACACGGGACCACC. For the 2395–2421 base pair mutant cp-23S rRNAs following primers were used: G2421C -GGATCCTAATACGACTCACTATAG2413GGCCATCCATCAACGGATA and T2412TCCACACGGGACCACC, for C2395G/G2421C -GGATCCTAATACGACTCACTATAG2413GGCCATCCATCAACGGATA and T2412CCACACGGGACCACCCGTTCACTAGGCCCGG, and for C2395U/G2421A – GGATCCTAATACGACTCACTATAG2413GGCCATCAATCAACGGATA and T2412CCACACGGGACCACCAGTTCACTAGGCCCGG, respectively. To compensate the missing RNA sequence in the cp-23S rRNA the following RNA oligos, either containing the unmodified wild-type sequences or carrying single nucleoside analogs, were synthesized as described (ref. 55 and references therein) and added during in vitro reconstitution: 1842–1864 – GCCGGAAGGUCAAGGGGAGGGGU, 2361–2401 – AGCCGAGCAGGGGCGAAAGCCGGGCCUAGUGAACCGGUGGU, 2413–2438 – GGGCCAUCGAUCAACGGAUAAAAGUU.
Puromycin reaction
The peptidyltransferase assay was carried out as previously describedCitation32 using 0.75 pmol N-acetyl-[3H]-Phe-tRNAPhe (6,000 cpm/ pmol) as P-site substrate and 2 mM puromycin as acceptor substrate.
In vitro translation of homo-polymeric mRNA analogs
Poly(U)-directed poly(Phe) synthesis was carried out as previously described.Citation56 To monitor the in vitro translation fidelity of reconstituted ribosomes, the leucine misincorporation during the poly(U)-directed poly(Phe) synthesis carried out according to Erlacher et al.Citation57 To analyze the length of the poly(Phe) peptides synthesized in vitro, we performed poly(Phe) synthesis as described above, however 14C-L-Phenylalanine (100 μCi/ml; 448 mCi/mmol; Hartmann Analytic) was used. After synthesis peptides were precipitated by addition of aceton and incubation for 1 h at −20°C. Samples were loaded on a tricine-gel. Gel was run at 70 V for the stacking gel (3% acrylamid/bisacrylamid 29:1, 0.75 M Tris-HCl pH 8.45, 0.075% SDS) and 150 V for the separation gel (11% acrylamid/bisacrylamid 29:1, 11% glycerol, 1 M Tris-HCl pH 8.45, 0.1% SDS), using 0.1 M Tris, 0.1 M Tricine and 0.1% SDS as cathode buffer and 0.2 M Tris-HCl pH 8.9 as anode buffer. Gel was vacuum-dried and exposed to a phosphorimager screen and scanned with the FLA300 (FujiFilm).
To investigate reading frame maintenance of reconstituted ribosomes, the poly(Phe) synthesis was carried out as describedCitation56 using 300 pmol of the poly(UUC) mRNA analog (5′-GCGGCAAGGAGGUAAAUAUUCUUCUUCUUCUUCUUCUUCUUCUUCUUCUUCUUC-3′) instead of poly(U).
Poly(Lys) synthesis using poly(A) mRNA was performed analogous to the poly(Phe) assay, with the following changes. As template 25 μg of poly(A) mRNA was used. In vitro protein synthesis was carried out in the presence of 40 μM L-Lys and 2 μM L-Lys (4,5–3H) (0.037 MBq/ μl, 1.2 MBq/ nmol). After incubation for 3 h at 42°C the reaction was stopped by adding 200 μg of bovine serum albumin, 2 ml of 5% (w/v) trichloracetic acid (TCA) and 7.6 mM Na2WO4 and incubation for 15 min at 95°C. Filtration via Whatman® filters and liquid scintillation counting as detection method was carried out as described for poly(Phe) synthesis.
In vitro translation of ribosomal proteins S8 or L12
In vitro translation of ribosomal proteins S8 or L12 using in vitro reconstituted ribosomes was performed as previously described.Citation29
Toeprinting assay
Toeprinting assay was performed as described elsewhereCitation42 with some minor changes. For primer extension, 9 pmol of (32P)-labeled primer 5′-CGTTAATCTGTGATG-3′ were annealed to 36 pmol in vitro transcribed and purified mRNA analog coding for MFKSIRYV.Citation37 10.45 μl reconstituted 70S (containing 50S particles assembled from 7.5 pmol 23S rRNA) were incubated at 37°C for 15 min, before adding 1.79 μl of the above mentioned mRNA/primer mix and 7.5 pmol deacylated tRNAfMet. Subsequent to a 15 min incubation step at 37°C (P-site tRNA binding), a 4 μl portion was removed (Pi complex). To the remaining 9.14 μl, 4 pmol of Ac-Phe-tRNAPhe was added and incubated for 10 min at 25°C in order to fill the A-site and form the Pre-translocation (PRE) complex. Subsequently two 4 μl aliquots were removed. To the first aliquot, 1 μl GTP (1.5 mM) was added (PRE complex). To the other aliquot, 1 μl GTP/EF-G (1.5 mM GTP, 5 pmol EF-G) was added and the reaction mix incubated at 37°C for 10 min (POST complex). 4 μl of the Pi, PRE or POST complexes were used for primer extension analysis as described.Citation42 Primer extension was terminated by addition of 20 μl of stop solution (5 M NH4OAc, 100 mM EDTA).
tRNA footprint (DMS chemical probing)
Reconstituted ribosomes from 20 pmol 23S rRNA (cp2439–2438 or cp2439–2412) carrying either the G2421C mutation or an abasic site at 2421, complemented with 2 pmol 30S from E.coli, were purified through a 20% sucrose cushion in tRNA binding buffer (20 mM Hepes/KOH pH 7.6, 6 mM MgOAc2, 150 mM NH4Cl, 4 mM 2-mercaptoethanol, 2 mM spermidine, 50 μM spermine) at 120,000 × g for 2.5 h at 4°C. Ribosomal pellets were resuspended in 23.4 μl tRNA binding buffer and incubated with 20 μg poly(U) mRNA in the absence/ presence of 20 pmol tRNAPhe from E.coli in a total volume of 30 μl for 15 min at 37°C. 5 pmol native 70S from T.aq. were used for tRNA binding and DMS probing. DMS (1:3 in ethanol) was added to a final concentration of 202 mM and probing was performed for 15 min at 37°C. Reaction was terminated by addition of 15 μl stop solution (1 M Tris/HCl pH 7.5, 1 M 2-mercaptoethanol, 50 mM EDTA). Ribosomes were ethanol precipitated overnight at −80°C. Ribosomal pellet (resulting from centrifugation at 18,000 × g for 20 min at 4°C) was resuspended in 150 μl TE/SDS (10 mM Tris/HCl pH 7.4, 100 mM NaCl, 5 mM EDTA, 0.5% (w/v) SDS) and rRNA was extracted with phenol-chloroform-isoamylalcohol and again ethanol precipitated overnight at −80°C. rRNA pellets were resuspended in 10 μl ddH2O for the non-DMS-treated controls and 6 μl ddH2O for the DMS treated samples, respectively. 1 μl of rRNA from in vitro reconstituted ribosomes or 3 μl of rRNA from 70S from T.aq. were used for primer extension analysis.
Reverse transcription using the DNA 5'-32P-labeled primer 5′-TTCCACACGGGACC-3′ was performed as described.Citation58 cDNA reaction products were loaded on a 10% sequencing gel (7M urea) and run at 40 W for 1 h 45 min. Gel was exposed on a phosphorimager screen over night at −20°C and scanned with the FLA300 (FujiFilm). Aida Image analysis software was used for bands quantification.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Juliana Komuczki for experimental help during a lab rotation, Tobias Santner and Ronald Micura for the synthesis of chemically modified RNA oligos and Kamilla Bakowska-Zywicka for help and discussions. Dr. Jennifer Gebetsberger is acknowledged for comments on the manuscript.
Funding
Grant support derives from the Swiss National Science Foundation (31003A_143388/1), and the Austrian Science Foundation (Y315) to N.P., and the Medical University Innsbruck (MUI Start ST2010022001) to N.C.
References
- Voorhees RM, Ramakrishnan V. Structural basis of the translation elongation cycle. Annu Rev Biochem 2013; 82:203-36; PMID:23746255; http://dx.doi.org/10.1146/annurev-biochem-113009-092313
- Wilson DN, Blaha G, Connell SR, Ivanov PV, Jenke H, Stelzl U, Teraoka Y, Nierhaus KH. Protein synthesis at atomic resolution: mechanistics of translation in the light of highly resolved structures for the ribosome. Current Prot & Pept Sci 2002; 3:1-53; PMID:12370010; http://dx.doi.org/10.2174/1389203023380846
- Erlacher MD, Polacek N. Ribosomal catalysis: the evolution of mechanistic concepts for peptide bond formation and peptidyl-tRNA hydrolysis. RNA Biol 2008; 5:5-12; PMID:18388484; http://dx.doi.org/10.4161/rna.5.1.5922
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science 2000; 289:920-30; PMID:10937990; http://dx.doi.org/10.1126/science.289.5481.920
- Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature 1989; 342:142-8; PMID:2682263; http://dx.doi.org/10.1038/342142a0
- Yamamoto H, Qin Y, Achenbach J, Li C, Kijek J, Spahn CM, Nierhaus KH. EF-G and EF4: translocation and back-translocation on the bacterial ribosome. Nat Rev Microbiol 2014; 12:89-100; PMID:24362468; http://dx.doi.org/10.1038/nrmicro3176
- Voorhees RM, Ramakrishnan V. Structural basis of the translational elongation cycle. Annual Rev Biochem 2013; 82:203-36; PMID:23746255; http://dx.doi.org/10.1146/annurev-biochem-113009-092313
- Rheinberger HJ, Sternbach H, Nierhaus KH. Three tRNA binding sites on Escherichia coli ribosomes. Proc Natl Acad Sci U S A 1981; 78:5310-4; PMID:7029532; http://dx.doi.org/10.1073/pnas.78.9.5310
- Nierhaus KH, Pech M. Problems with the analyses of the ribosomal allosteric three-site model. J Biol Chem 2012; 287:27049; PMID:22865895; http://dx.doi.org/10.1074/jbc.L112.381848
- Petropoulos AD, Green R. Further in vitro exploration fails to support the allosteric three-site model. J Biol Chem 2012; 287:11642-8; PMID:22378789; http://dx.doi.org/10.1074/jbc.C111.330068
- Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science 1999; 285:2095-104; PMID:10497122; http://dx.doi.org/10.1126/science.285.5436.2095
- Feng S, Chen Y, Gao YG. Crystal structure of 70S ribosome with both cognate tRNAs in the E and P sites representing an authentic elongation complex. PloS One 2013; 8:e58829; PMID:23527033; http://dx.doi.org/10.1371/journal.pone.0058829
- Lill R, Robertson JM, Wintermeyer W. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry 1986; 25:3245-55; PMID:3524675; http://dx.doi.org/10.1021/bi00359a025
- Marquez V, Wilson DN, Tate WP, Triana-Alonso F, Nierhaus KH. Maintaining the ribosomal reading frame: the influence of the E site during translational regulation of release factor 2. Cell 2004; 118:45-55; PMID:15242643; http://dx.doi.org/10.1016/j.cell.2004.06.012
- Trimble MJ, Minnicus A, Williams KP. tRNA slippage at the tmRNA resume codon. RNA 2004; 10:805-12; PMID:15100436; http://dx.doi.org/10.1261/rna.7010904
- Triana-Alonso FJ, Chakraburtty K, Nierhaus KH. The elongation factor 3 unique in higher fungi and essential for protein biosynthesis is an E site factor. J Biol Chem 1995; 270:20473-8; PMID:7657623; http://dx.doi.org/10.1074/jbc.270.35.20473
- Rheinberger HJ, Nierhaus KH. Allosteric interactions between the ribosomal transfer RNA-binding sites A and E. J Biol Chem 1986; 261:9133-9; PMID:2424904
- Bouadloun F, Donner D, Kurland CG. Codon-specific missense errors in vivo. EMBO J 1983; 2:1351-6; PMID:10872330
- Rheinberger HJ, Sternbach H, Nierhaus KH. Codon-anticodon interaction at the ribosomal E site. J Biol Chem 1986; 261:9140-3; PMID:2424905
- Lill R, Lepier A, Schwagele F, Sprinzl M, Vogt H, Wintermeyer W. Specific recognition of the 3'-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J Mol Biol 1988; 203:699-705; PMID:2463367; http://dx.doi.org/10.1016/0022-2836(88)90203-3
- Kirillov SV, Makarov EM, Semenkov Yu P. Quantitative study of interaction of deacylated tRNA with Escherichia coli ribosomes. Role of 50 S subunits in formation of the E site. FEBS Lett 1983; 157:91-4; PMID:6345196; http://dx.doi.org/10.1016/0014-5793(83)81122-3
- Gnirke A, Nierhaus KH. tRNA binding sites on the subunits of Escherichia coli ribosomes. J Biol Chem 1986; 261:14506-14; PMID:3533922
- Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 1989; 57:585-97; PMID:2470511; http://dx.doi.org/10.1016/0092-8674(89)90128-1
- Bocchetta M, Xiong L, Shah S, Mankin AS. Interactions between 23S rRNA and tRNA in the ribosomal E site. RNA 2001; 7:54-63; PMID:11214181; http://dx.doi.org/10.1017/S1355838201001650
- Sergiev PV, Lesnyak DV, Kiparisov SV, Burakovsky DE, Leonov AA, Bogdanov AA, Brimacombe R, Dontsova OA. Function of the ribosomal E-site: a mutagenesis study. Nucleic Acids Res 2005; 33:6048-56; PMID:16243787; http://dx.doi.org/10.1093/nar/gki910
- Schmeing TM, Moore PB, Steitz TA. Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit. RNA 2003; 9:1345-52; PMID:14561884; http://dx.doi.org/10.1261/rna.5120503
- Jenner LB, Demeshkina N, Yusupova G, Yusupov M. Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat Struct Mol Biol 2010; 17:555-60; PMID:20400952; http://dx.doi.org/10.1038/nsmb.1790
- Feinberg JS, Joseph S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc Natl Acad Sci U S A 2001; 98:11120-5; PMID:11562497; http://dx.doi.org/10.1073/pnas.211184098
- Erlacher MD, Chirkova A, Voegele P, Polacek N. Generation of chemically engineered ribosomes for atomic mutagenesis studies on protein biosynthesis. Nat Prot 2011; 6:580-92; PMID:21527916; http://dx.doi.org/10.1038/nprot.2011.306
- Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell 2006; 126:1065-77; PMID:16962654; http://dx.doi.org/10.1016/j.cell.2006.08.032
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006; 313:1935-42; PMID:16959973; http://dx.doi.org/10.1126/science.1131127
- Erlacher MD, Lang K, Shankaran N, Wotzel B, Huttenhofer A, Micura R, Mankin AS, Polacek N. Chemical engineering of the peptidyl transferase center reveals an important role of the 2'-hydroxyl group of A2451. Nucleic Acids Res 2005; 33:1618-27; PMID:15767286; http://dx.doi.org/10.1093/nar/gki308]
- Odom OW, Picking WD, Tsalkova T, Hardesty B. The synthesis of polyphenylalanine on ribosomes to which erythromycin is bound. European J Biochem 1991; 198:713-22; PMID:1904819; http://dx.doi.org/10.1111/j.1432-1033.1991.tb16071.x
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinf 2002; 3:2; PMID:11869452; http://dx.doi.org/10.1186/1471-2105-3-2
- Geigenmuller U, Nierhaus KH. Significance of the third tRNA binding site, the E site, on E. coli ribosomes for the accuracy of translation: an occupied E site prevents the binding of non-cognate aminoacyl-tRNA to the A site. EMBO J 1990; 9:4527-33; PMID:2265616
- Di Giacco V, Marquez V, Qin Y, Pech M, Triana-Alonso FJ, Wilson DN, Nierhaus KH. Shine-Dalgarno interaction prevents incorporation of noncognate amino acids at the codon following the AUG. Proc Natl Acad Sci U S A 2008; 105:10715-20; PMID:18667704; http://dx.doi.org/10.1073/pnas.0801974105
- Szaflarski W, Vesper O, Teraoka Y, Plitta B, Wilson DN, Nierhaus KH. New features of the ribosome and ribosomal inhibitors: non-enzymatic recycling, misreading and back-translocation. J Mol Biol 2008; 380:193-205; PMID:18508080; http://dx.doi.org/10.1016/j.jmb.2008.04.060
- Grise-Miron L, Noreau J, Melancon P, Brakier-Gingras L. Comparison of the misreading induced by streptomycin and neomycin. Biochim Biophys Acta 1981; 656:103-10; PMID:6796121; http://dx.doi.org/10.1016/0005-2787(81)90032-0
- Gregory ST, Lieberman KR, Dahlberg AE. Mutations in the peptidyl transferase region of E. coli 23S rRNA affecting translational accuracy. Nucleic Acids Res 1994; 22:279-84; PMID:8127663; http://dx.doi.org/10.1093/nar/22.3.279
- Gavrilova LP, Spirin AS. Stimulation of “non-enzymic” translocation in ribosomes by p-chloromercuribenzoate. FEBS Lett 1971; 17:324-6; PMID:11946059; http://dx.doi.org/10.1016/0014-5793(71)80177-1
- Pestka S. Studies on the formation of transfer ribonucleic acid-ribosome complexes. VI. Oligopeptide synthesis and translocation on ribosomes in the presence and absence of soluble transfer factors. J Biol Chem 1969; 244:1533-9; PMID:4886309
- Chirkova A, Erlacher MD, Clementi N, Zywicki M, Aigner M, Polacek N. The role of the universally conserved A2450-C2063 base pair in the ribosomal peptidyl transferase center. Nucleic Acids Res 2010; 38:4844-55; PMID:20375101; http://dx.doi.org/10.1093/nar/gkq213
- Walker SE, Shoji S, Pan D, Cooperman BS, Fredrick K. Role of hybrid tRNA-binding states in ribosomal translocation. Proc Natl Acad Sci U S A 2008; 105:9192-7; PMID:18591673; http://dx.doi.org/10.1073/pnas.0710146105
- Wilson DN, Nierhaus KH. The E-site story: the importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell Mol Life Sci 2006; 63:2725-37; PMID:17013564; http://dx.doi.org/10.1007/s00018-006-6125-4
- Devaraj A, Shoji S, Holbrook ED, Fredrick K. A role for the 30S subunit E site in maintenance of the translational reading frame. RNA 2009; 15:255-65; PMID:19095617; http://dx.doi.org/10.1261/rna.1320109
- Sanders CL, Curran JF. Genetic analysis of the E site during RF2 programmed frameshifting. RNA 2007; 13:1483-91; PMID:17660276; http://dx.doi.org/10.1261/rna.638707
- Semenkov YP, Rodnina MV, Wintermeyer W. The “allosteric three-site model” of elongation cannot be confirmed in a well-defined ribosome system from Escherichia coli. Proc Natl Acad Sci U S A 1996; 93:12183-8; PMID:8901554; http://dx.doi.org/10.1073/pnas.93.22.12183
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science 2001; 292:883-96; PMID:11283358; http://dx.doi.org/10.1126/science.1060089
- Gurel G, Blaha G, Steitz TA, Moore PB. Structures of triacetyloleandomycin and mycalamide A bind to the large ribosomal subunit of Haloarcula marismortui. Antimicrob Agents Chemo 2009; 53:5010-4; PMID:19738021; http://dx.doi.org/10.1128/AAC.00817-09
- Schroeder SJ, Blaha G, Tirado-Rives J, Steitz TA, Moore PB. The structures of antibiotics bound to the E site region of the 50 S ribosomal subunit of Haloarcula marismortui: 13-deoxytedanolide and girodazole. J Mol Biol 2007; 367:1471-9; PMID:17321546; http://dx.doi.org/10.1016/j.jmb.2007.01.081
- Poehlsgaard J, Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat Rev Microbiol 2005; 3:870-81; PMID:16261170; http://dx.doi.org/10.1038/nrmicro1265
- Polacek N, Mankin AS. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit Rev Biochem Mol Biol 2005; 40:285-311; PMID:16257828; http://dx.doi.org/10.1080/10409230500326334
- Chen C, Zhang H, Broitman SL, Reiche M, Farrell I, Cooperman BS, Goldman YE. Dynamics of translation by single ribosomes through mRNA secondary structures. Nat Struct Mol Biol 2013; 20:582-8; PMID:23542154; http://dx.doi.org/10.1038/nsmb.2544
- Erlacher MD, Lang K, Wotzel B, Rieder R, Micura R, Polacek N. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2'-OH at A2451 of 23S rRNA. J Am Chem Soc 2006; 128:4453-9; PMID:16569023; http://dx.doi.org/10.1021/ja0588454
- Lang K, Erlacher M, Wilson DN, Micura R, Polacek N. The role of 23S ribosomal RNA residue A2451 in peptide bond synthesis revealed by atomic mutagenesis. Chem Biol 2008; 15:485-92; PMID:18439847; http://dx.doi.org/10.1016/j.chembiol.2008.03.014
- Clementi N, Chirkova A, Puffer B, Micura R, Polacek N. Atomic mutagenesis reveals A2660 of 23S ribosomal RNA as key to EF-G GTPase activation. Nat Chem Biol 2010; 6:344-51; PMID:20348921; http://dx.doi.org/10.1038/nchembio.341
- Erlacher MD, Polacek N. Probing functions of the ribosomal peptidyl transferase center by nucleotide analog interference. Methods Mol Biol 2012; 848:215-26; PMID:22315072; http://dx.doi.org/10.1007/978-1-61779-545-9_14
- Polacek N, Barta A. Metal ion probing of rRNAs: evidence for evolutionarily conserved divalent cation binding pockets. RNA 1998; 4:1282-94; PMID:9769102; http://dx.doi.org/10.1017/S1355838298980347
- Grise-Miron L, Brakier-Gingras L. Effect of neomycin and protein S1 on the binding of streptomycin to the ribosome. European J Biochem 1982; 123:643-6; PMID:6176448; http://dx.doi.org/10.1111/j.1432-1033.1982.tb06580.x
- Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science 2003; 300:1159-62; PMID:12750524; http://dx.doi.org/10.1126/science.1084571
