Abstract
Background: Berberine (BBR) is a natural alkaloid derived from a traditional Chinese herbal medicine. However, the exact mechanisms underlying the different effects of berberine on MM cells have not been fully elucidated. Methods: A systematic analysis assay integrated common signaling pathways modulated by the 3 miRNA clusters and mRNAs in MM cells after BBR treatment. The role of the mir-99a∼125b cluster, an important oncomir in MM, was identified by comparing the effects of t-anti-mirs with complete complementary antisense locked nucleic acids (LNAs) against mature mir-125b (anti-mir-125b). Results: Three miRNAs clusters (miR-99a∼125b, miR-17∼92 and miR-106∼25) were significantly down-regulated in BBR-treated MM cells and are involved in multiple cancer-related signaling pathways. Furthermore, the top 5 differentially regulated genes, RAC1, NFκB1, MYC, JUN and CCND1 might play key roles in the progression of MM. Systematic integration revealed that 3 common signaling pathways (TP53, Erb and MAPK) link the 3 miRNA clusters and the 5 key mRNAs. Meanwhile, both BBR and seed-targeting t-anti-mir-99a∼125b cluster LNAs significantly induced apoptosis, G2-phase cell cycle arrest and colony inhibition. Conclusions: our results suggest that BBR suppresses multiple myeloma cells, partly by down-regulating the 3 miRNA clusters and many mRNAs, possibly through TP53, Erb and MAPK signaling pathways. The mir-99a∼125b cluster might be a novel target for MM treatment. These findings provide new mechanistic insight into the anticancer effects of certain traditional Chinese herbal medicine compounds.
Introduction
Berberine (BBR), a clinically important natural isoquinoline alkaloid derived from the Berberis species, has been reported to exhibit multiple pharmacological activities including anti-cancer effects.Citation1-3 Recent studies indicated that BBR down-regulates both the mRNA and protein levels of IL-6, which is a key factor in the proliferation of multiple myeloma (MM) cells.Citation4,5 Thus, it is possible that BBR may be effective in suppressing MM cells. However, the exact mechanisms underlying the different effects of berberine on MM cells have not been fully elucidated.
MM is a malignant clonal B cell proliferation of bone marrow plasma cells, characterized by profound genomic instability involving both numerical and structural chromosomal aberrations of potential prognostic relevance.Citation6 Nearly half of MM tumors are hyperdiploid with multiple trisomies of nonrandomly odd-numbered chromosomes and a low prevalence chromosome 13 deletion and chromosomal translocations involving the immunoglobulin heavy chain (IgH) locus at 14q32.Citation7 It has been suggested that chromosomal abnormalities and other types of genetic or epigenetic alterations might contribute to miRNA deregulation in cancer.Citation8-10
Accumulating evidence suggests that miRNAs that are significantly overexpressed in tumors may represent a novel class of oncogene. Termed “oncomirs,” these oncogenic miRNAs usually promote tumor development by negatively regulating tumor suppressor genes that control various biological processes. Therefore, altering oncomir expression is a promising cancer treatment strategy.Citation11,12
Recently, differential expression of miRNA clusters has been found in MM. These miRNA clusters are usually oncomirs with high levels of expression, including miR-17∼92, mir-99a∼125b and miR-106∼25.Citation10
To characterize miRNAs in the context of the major MM molecular types, Lionetti et al.Citation10 generated miRNA expression profiles of highly purified malignant plasma cells from 40 primary tumors. Multiclass analysis identified a set of 26 miRNAs showing highly significant differential expression across the 5 TC (translocation/cyclin D) groups. In particular, 10 (38%) miRNAs (including miR-99a, miR-125b, and let-7c) were expressed at higher levels in TC5 than in the other classes and belong to a paralogous cluster on 21q21.1.
We show that BBR significantly down-regulates the expression level of some members of the miRNAs clusters, including miR-17∼92, mir-99a∼125b and miR-106∼25 (). Bioinformatic analysis shows that these 3 clusters are involved in multiple cancer-related signaling pathways in BBR-treated MM cells, and mir-125b participates in all signaling pathways. Therefore, the mir-99a∼125b cluster can be considered as one of the most important oncomirs in MM cells treated with BBR. Furthermore, 5 differentially expressed genes after BBR treatment (RAC1, NFκB1 MYC, JUN and CCND1) might play a key role in pathogenesis and progression of MM.
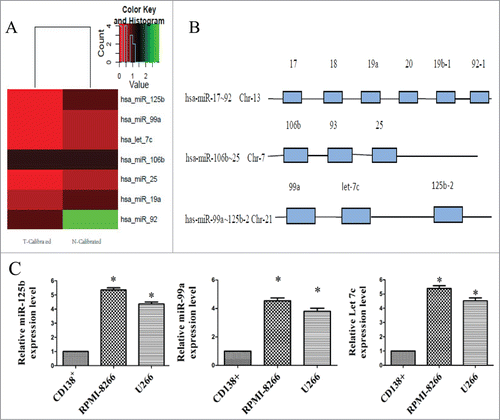
Figure 1. Unsupervised clustering analysis of miRNA cluster expression in BBR-treated MM cells and miRNA cluster chromosomal loci. (A) Heat maps illustrating unsupervised clustering of the 3 miRNA clusters that were weakly expressed after BBR treatment. The red and green colors indicate relatively low and high fold expression changes, respectively. (B) The 3 miRNA clusters (miR-17∼92, 105∼25 and 99a∼125b) are located on different chromosomes. (C) Real-time RT-PCR analysis of miR-99a∼125b cluster was performed in multiple myeloma cell lines (RPMI-8266 and U266) and normal bone marrow-derived CD138+ plasma cells from healthy donors. The data are shown as mean ± SD. *P < 0.05 vs control group.
MicroRNAs (miRNAs) have critical roles in the regulation of gene expression. miRNA activity requires base pairing with only 7–8 nucleotides of a mRNA; therefore, the 8-mer seed sequence of miRNAs is a key feature in mRNA target recognition Citation13 and plays a critical role in miRNA activities. Therefore, the seed sequence is a novel target of anti-miRNAs (anti-mirs).Citation14 A seed-directed 8-mer antisense locked nucleotide acid (LNA), termed t-anti-mir, has been successfully applied to the functional identification of miRNAs.Citation14
In this work, we present a systematic integration of common signaling pathways modulated by the 3 miRNA clusters and mRNAs identified in MM cells after BBR treatment. We also identify the role of the mir-99a∼125b cluster, an important oncomir in MM, by comparing the effects of t-anti-mirs with complete complementary antisense LNAs against mature mir-125b (anti-mir-125b). Our findings provide new insight into anti-cancer mechanisms of traditional Chinese herbal medicines.
Results
BBR modulates miRNA cluster expression in MM
Among the 1152 probes, 87 miRNAs were differentially expressed between control and BBR-treated cells. Of these, 49 were downregulated and 38 were up-regulated compared with the control. Further analysis revealed that the miR-17∼92, 105∼25 and 99a∼125b clusters [located on different chromosomes ()] were down-regulated by BBR treatment (). Among these clusters, mirFocus software identified 7 miRNAs that are involved in p53 signaling, the cell cycle and other cancer pathways. Of these, miR-125b has the most identified target genes and participates in all of the above signaling pathways (Table S1). Thus the mir-99a∼125b cluster can be considered as one of the most important oncomirs. showed that miR- 99a∼125b cluster expression level were moderately higher in MM cells (RPMI-8266 and U266) than in normal CD138+ plasma cells. These results imply that miR-99a∼125b cluster might also function as an oncomir in MM. These results suggest that BBR suppression of MM cells might involve miRNA cluster-mediated gene expression.
BBR modulates the mRNA expression profile in MM cells
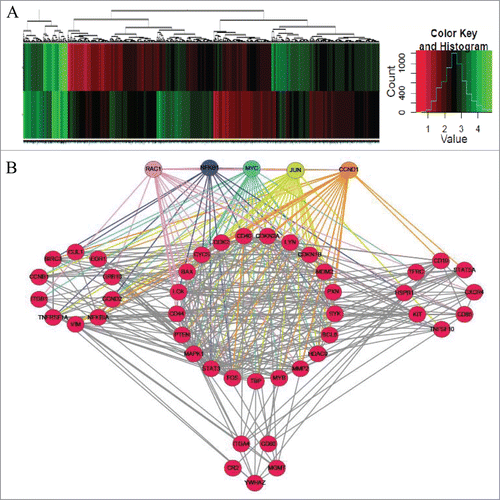
Of the 25,100 genes represented on the microarray, 1893 were differentially expressed between BB-treated and control cells (); 820 were downregulated, and 1073 were upregulated compared with the control. Integrative analysis of the 1893 mRNA expression profiles was used to reconstruct a gene regulatory network using R software with the gplots package (). Among these genes, Rac1, NFκB1, MYC, JUN and CCND1 had the most nodes and are, therefore, indicated as key genes in BBR-induced pathways. Integrative analysis showed that the 5 genes are involved in p53 signaling, the cell cycle and other cancer pathways (Table S2).
Figure 2. Unsupervised clustering analysis of mRNA expression in BBR-treated MM cells. (A) Heat maps illustrating unsupervised clustering of mRNAs that were differentially expressed after BBR treatment. The red and green colors indicate relatively low and high fold expression changes, respectively. (B) Integrative analysis of mRNA expression profiles to reconstruct a gene regulatory network. Among all genes, these 5, each having the most nodes, are considered key genes in BBR-induced pathways: (Rac1, NFκB1, MYC, JUN and CCND1).
Integration of signaling pathways common to the 3 differentially expressed miRNA clusters and key mRNAs
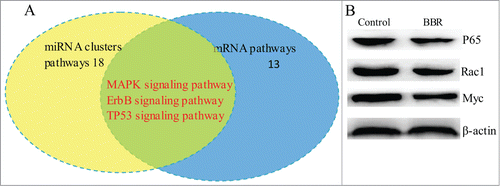
To explore the connections among miRNAs and mRNAs, we integrated signaling pathways in which the 3 miRNA clusters and the differentially expressed mRNAs are involved. The results showed 3 common pathways (TP53, ErbB and MAPK signaling pathways) for these 2 groups (). Western blot confirmed that BBR downregulated P65, Rac1 and Myc protein level().
Figure 3. Overlap of signaling pathways among those affected by differential expression of mRNAs and miRNA clusters in BBR-treated MM cells. (A) The signaling pathways in which the 3 differentially expressed miRNA clusters and mRNA profiles are involved were integrated. The results showed that TP53, ErbB and MAPK signaling pathways are common in BRB-treated MM cells. (B) Western blot confirmed that BBR suppressed protein expression of TP53, ErbB and MAPK.
BBR and t-anti-miR-99a∼125b cluster LNAs induce apoptosis and G2-phase arrest
To investigate whether BBR treatment or miR-99a∼125b cluster inhibition could induce apoptosis in MM cells, RPMI-8266 cells were treated with BBR or transfected with t-anti-miR-99a∼125b cluster LNAs. The cells were then stained with annexin V and propidium iodide (PI), and assessed with flow cytometry. As shown in , treatment with 75 μM BBR significantly induced cell apoptosis. Similar results were also observed when miR-125b was knocked down by t-anti-miR-125b or anti-miR-125b, suggesting that seed-targeting with t-anti-miR-125b has a similar effect to anti-miR-125b, whereas t-anti-mir-99a and t-anti-mir-let7c did not have such an effect.
Figure 4. BBR and t-anti-miR-99a∼125b cluster LNA-induced cell apoptosis and G2 phase arrest. (A) RPMI-8266 cells were treated with 75 μM BBR or 0.5 μM t-anti-miR-99a∼125b cluster LNAs for 48 h, then stained with FITC-conjugated annexin V and PtdIns, followed by flow cytometry analysis. (B) RPMI-8266 cells were collected 48 h after treatment and stained with PI solution. Cell cycle was analyzed using flow cytometry. (C) The results are shown as the average of 3 replicates for apoptosis and cell cycle. One asterisk (*) denotes a P-value < 0.01 vs. blank or SCR groups.
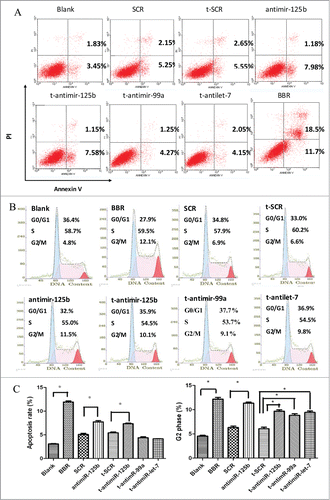
To elucidate the influence of BBR treatment or inhibition of the miR-99a∼125b cluster on the cell cycle, RPMI-8266 cells treated with BBR or transfected with t-anti-mir LNAs against the miR-99a∼125b cluster were subjected to flow cytometry cell cycle analysis. As shown in , BBR treatment resulted in an accumulation of cells in the G2/M phase. Interestingly, miR-99a∼125b cluster inhibition by t-anti-mir LNAs produced almost identical effects on the cell cycle.
Effect of miR-99a∼125b cluster inhibition on cell colony formation
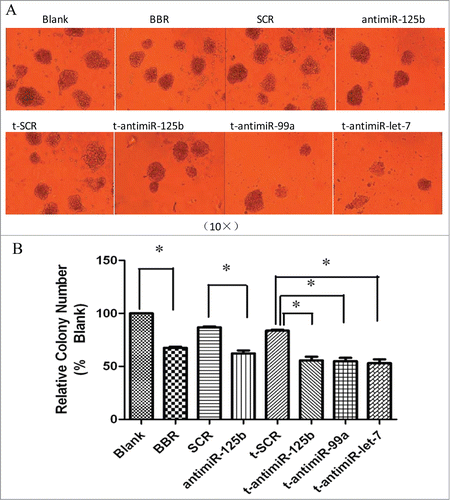
Cell colony growth is closely related to neoplastic capacity. The effect on neoplastic capacity of the miR-99a∼125b cluster in RPMI-8266 cells was assessed by the methylcellulose colony formation assay. Inhibition of the miR-99a∼125b cluster significantly reduced colony number 1 week post-transfection in comparison to the control t-SCR groups (). An identical result also was seen for the BBR and anti-miR-125b groups.
Figure 5. BBR and anti-miR-99a∼125b cluster LNAs suppress colony formation. (A) Schematics of representative colony formation. Colony growth capacity was assessed by the methylcellulose colony formation assay. RPMI-8266 cells were treated with 75 μM BBR or 0.5 μM anti-miR-99a∼125b LNAs and the number of colonies were observed and counted under light microscopic observation after incubation for one week (B) The results are shown as the average of 3 replicates. One asterisk (*) denotes the P-value < 0.01 vs. blank or SCR groups.
Inhibition of the miR-99a∼125b cluster in RPMI-8266 cells promotes B-lineage cell differentiation
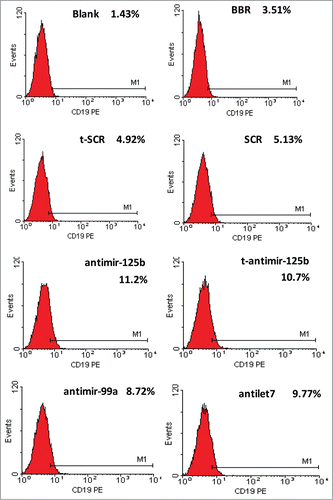
To investigate the effect of anti-miR-125b and t-anti-miR-99a∼125b cluster LNAs toward B-lineage differentiation capacity, CD19 expression on the cell surface was measured in RPMI-8266 cells. There was a significant increase in CD19-positive cells transfected with antimiR-125b and t-antimiR-99a∼125b cluster LNAs, compared with cells in the control t-SCR or SCR groups. BBR had no such effect ().
Figure 6. miR-99a∼125b cluster inhibition promotes B-lineage differentiation in RPMI-8266 cells. The cells were immunostained for 30 min with PE-conjugated CD19 antibody. B-lineage differentiation was evaluated by expression of CD19-PE. Inhibition of miR-99a∼125b cluster promotes B-lineage differentiation in RPMI-8266 cells.
Discussion
In our study, systematic integration analysis showed that 3 common signaling pathways (TP53, Erb and MAPK) are affected by the differential expression of miRNA clusters and their target genes in response to BBR (). MAPK signaling pathway affected by BBR has been comfirmed in breast cancer.Citation15 NFκB, Rac1 and MYC in 3 common signaling pathways have been validated to be significantly downregulated using western blot ().
Among three pathways, TP53 acts as a potent transcription factor and can be activated in response to diverse stresses, leading to cell cycle arrest, apoptosis and senescence in MM.Citation16 Monoallelic deletion of TP53 in MM, which often occurs without mutation to the other allele, is associated with an extremely poor prognosis.Citation17 This supports the idea that a decrease in TP53 levels is associated with tumor progression and numerous reports have shown that therapeutic induction of TP53 might be particularly suitable for the treatment of hematological malignancies,Citation18 including MM.Citation19,20
Recent research shows that alteration of miRNA expression during progression from monoclonal gammopathy of undetermined significance (MGUS) to newly diagnosed MM could be partially responsible for TP53 inactivation in MM patients.Citation21 Some reports have revealed that several miRNAs are important components of the TP53 tumor suppressor network with miR-125b directly targeting TP53.Citation22,23 Dexamethasone is a key front-line chemotherapeutic for B-cell malignant MM that modulates MM cell survival signaling, but which fails to induce marked cytotoxicity as a monotherapy. However, use of antisense miR-125b transcripts enhanced expression of pro-apoptotic p53 and significantly promoted dexamethasone-induced cell death responses in MM. Indeed, epigenetic cross-talk between miR-125b and TP53 has also been demonstrated.Citation24
It is well known that the bone marrow microenvironment plays a prominent role in the biology of MM. Of the 5 identified key genes (NFκB, Rac1, MYC, JUN and CCND1), 3 are down-graded by BBR: NFκB, Rac1 and MYC (, Table S2). These three genes act a critical role in the MM microenvironment. NFκB activation plays a crucial role in anti-apoptotic responses following apoptotic signaling during tumor necrosis factor (TNFα) stimulation in MM. Specific drugs, used alone or in combinations to treat MM have inhibitory effects on the NFκB pathway.Citation25,26
The interaction of MM cells with the bone marrow milieu plays a crucial role in MM pathogenesis. Stromal cell-derived factor-1 (SDF1) regulates homing of MM cells to the bone marrow. We found that Rac1 acts key roles in SDF1-induced adhesion of MM cells to bone marrow stromal cells, and Rac1 inhibitors reduced SDF1-induced polymerization of actin and activation of LIMK, SRC, FAK, and cofilin.Citation27 Therefore, Rac1 GTPases might be novel MM therapeutic targets by inducing the homing of MM cells to bone marrow nichesCitation27 Furthermore, the DOCK2–Rac1 pathway activated by CXCL12 and S1P was required for stimulation of myeloma cell adhesion involving α4β1.Citation28
Recent studies have suggested that c-Myc over-expression may be a factor indicating poor prognosis in MM.Citation29 Myc rearrangements are found in nearly 50% of MM, including smoldering MM, and they are heterogeneous in some cases. Rearrangements reposition Myc near a limited number of genes associated with conventional enhancers, but mostly with super-enhancers. These data suggest that Myc rearrangements, regardless of when they occur during MM pathogenesis, contribute to tumor autonomy.Citation30
miRNAs from the mir-99a∼125b cluster are transcribed together on the same miRNA polycistron, and are over-expressed in multiple hematological malignancies. The mir-99a∼125b cluster is able to function as an oncomir in leukemiaCitation31,32 and prostate cancer.Citation33 Recent studies report that miR-125b is multifaceted and able to function as an oncomir or a tumor suppressor, depending on the cellular context.Citation34 Our work shows that BBR could silence mir-125b, mir-99a and let 7c in MM cells. However, in response to BBR, the exact role of the mir-99a∼125b cluster in MM is elusive. Here, seed sequence-targeting t-anti-mirs were used to antagonize mir-99a∼125b cluster function.
In the present study, we showed that t-anti-mir-125b, like BBR, induced apoptosis, whereas t-anti-mir-99a and t-anti-mir-let7c did not. t-anti- mir-99a∼125b cluster LNAs, similar to BBR, lead to G2 phase arrest (). Colony growth was closely related to neoplastic capacity. We found that RPMI-8266 cells treated with mir-99a∼125b cluster LNAs, or BBR produced fewer colonies compared with the control groups (). Interestingly, inhibiting miR-99a and let-7c does not induce apoptosis, but induces G2 arrest. Perhaps their target genes are different from that of mir-125b.
CD19 is an antigen expressed at an early stage of B differentiation and is always present on the majority of normal plasma cells. Reactive plasma cells also express CD19; however, neoplastic plasma cells show no or only a low level of CD19 expression.Citation35,36 Enforced expression of CD19 in human myeloma cell lines resulted in growth inhibition and reduced tumorigenicity,Citation26 indicating that CD19 has an inhibitory effect on MM. Our study validated that CD19 is weakly expressed on the RPMI-8266 cell surface, in agreement with a previous report.Citation35,36 After transfection, t-anti-miR-99a∼125b cluster LNAs significantly increased numbers of positive CD19 cells (). Because of the loss of CD19 expression in MM, CD19 is considered a most useful biomarker to discriminate neoplastic plasma cells from reactive plasma cells.Citation37 These data support the utility of tiny LNAs in elucidating the functions of miRNA families in vitro and miRNAs as molecular targets for natural product anticancer agents.Citation38-40
In conclusion, BBR modulates the expression profile of miRNAs and mRNAs in MM cells, and the mir-99a∼125b cluster functions as an oncomir in MM cells. BBR suppresses MM cells, in part by down-regulating 3 miRNAs clusters and many mRNAs, possibly through TP53, ErbB and MAPK signaling pathways. These findings may also provide a new mechanistic insight into the anticancer effects of certain traditional Chinese herbal medicine compounds.
Materials and Methods
Cell lines and normal control samples
MM cell line RPMI-8266 and U266, were obtained from the Shanghai Institute of Cell Biology. The cells were cultured in RPMI containing 25 mM HEPES, 10% fetal bovine serum (FBS), 0.05 mM 2-mercaptoethanol, 1 mM sodium pyruvate, 2 mM L-glutamine, 100 U/mL penicillin, and 50 U/mL streptomycin at 37°C in a 5% CO2 humidified atmosphere (Thermo FORMA 3110, USA). Normal control samples were obtained from 3 healthy donors. Plasma cells were purified from BM aspiration using CD138 immunomagenetic microbeads (MidiMACS; Miltenyi Biotec). The purity of the positively selected plasma cells (≥ 90% ) was assessed by flow cytometry.
Antisense LNAs and transfection
The sequences of anti-mir-99a∼125b cluster LNAs were designed according to the principles of sequences complementary to mature miRNAs. The LNA sequences used in this study were as follows: anti-miR-125b, 5′-AGG GAC TCT GGG ATTT GAA CAC T-3′ (22 bp); t-anti-miR-125b, 5′-AGG GAC TC -3′; t-anti-miR-99a, 5′-TTG GGC AT -3′; t-anti-miR-let-7, 5′-ACT CCA TC-3′; Scramble (SCR), 5′ -TCATACTA-3′ (8 bp) (Fig. S1). All LNAs were chemically synthesized and/or modified with fluorescein isothiocyanate (FITC) by the Shanghai Sangon Bio-engineering Company.
BBR was purchased from Sigma-Aldrich. RPMI-8266 cells in the exponential phase of growth were seeded in 96- or 24-well plates (Costar) and transfected with 0.5 μM t-anti-miR-99a∼125b cluster LNAs using Lipofectamine 2000 reagent (Invitrogen) in serum-free RPMI-1640.
Microarray analysis of miRNA and mRNA expression
Based on our preliminary study, 75 μM BBR was used to treat RPMI-8266 cells for 48 h. Total miRNA from 1 × 108 cells was isolated using mirVANA™ miRNA Isolation kits according to the manufacturer's instructions. A total of 4 μg of miRNA was labeled with Cy3/Cy5 using mirVANA miRNA labeling kits and hybridized on an miRNA microarray (CSC-GE-3, Chipscreen Biosciences, Shenzhen, China). Similarly, RNA Samples (4 μg) labeled with Cy3/Cy5 were hybridized on an mRNA microarray (CSC-GE-30, Chipscreen Biosciences) containing 39,557 oligonucleotide probes. Each chip was scanned with a Generation III array scanner (Amersham Pharmacia). Data analyses were performed using Imagequant 5.0 (Array Vision 6.0).
Real time qRT-PCR analysis of miR-99a∼125b cluster expression level
Total RNA was isolated from RPMI-8266, U266 cells and normal control cells using ENgeneTM RNA Miniprep Kit (BioMIGA, USA) according to the manufacturer's instructions. cDNA was prepared from total RNA using a Hight Capacity cDNA Reverse Transcription Kit (Genepharma, shanghai, China). The expression of mature of miR-99a∼125b cluster was quantified via real-time PCR using the Hairpin-itTM miRNAs qPCR Quantitaion Kit (GenePharma, shanghai, China). Quantization of U6 was used as the endogenous control to normalize miRNA expression level. qPCR was performed in the ABI 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Independent experiments were performed in triplicate. The quantity of RNA expression was calculated using the 2−ΔΔCt method of relative quantification.
Bioinformatic analysis
miRFocus software (http://mirfocus.org), developed by LC Science USA, was used for miRNA-target gene pathway analysis and the related miRNA annotations. R software with gplots package was used to study PPI (protein-protein interaction) and network construction. the Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway is integrated by the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 tools as the official gene symbol.
Western blot
Cellular lysates from RPMI-8266 cells were immunoblotted with monoclonal anti-RalA antibody. Breifly, after treatment with 75 μM BBR for 48 h, RPMI-8266 cells were lysed in RIPA buffer in the presence of proteinase inhibitor (Biocolor BioScience & Technology Company, Shanghai, China). Cell lysates (30 μg) were denatured in Laemmli sample buffer (Bio-Rad) for 5 min at 100ºC, electrophoresed on 10% SDS-PAGE gel, and transferred to a polyvinylidene fluoride membrane. The membrane was blocked with 5% (w/v) fat-free milk in Tris Buffered Saline (TBS) and 0.5% (v/v) Tween-20 for 1 h. The blots were then incubated with antibodies at 4°C for overnight. Anti-NF-kappa B p65 and anti-Rac1 antibody were obtained from CST (Cell Signaling Technologies. INC.). Anti-MYC antibody was from Santa Cruz Biotechnology), After washing, the blots were then incubated with horseradish peroxidase-conjugated secondary antibody. The signals were visualized with enhanced chemiluminescence (ECL) (BeyoECL Plus, Beyotime company), and analyzed using a BI-2000 system.
Flow cytometry
Flow cytometry was performed to analyze the cell cycle profile and apoptosis. For analysis of the cell cycle, cells transfected with t-anti-miR-99a∼125b cluster LNAs (0.5 μM) were collected, rinsed twice with phosphate buffered saline, fixed in 70% ethanol for 1 h at 4°C, stained with PtdIns solution (50 μg/ml) containing RNase A (200 μg/ml). The cell cycle was analyzed using flow cytometry according to DNA content. To analyze apoptosis, cells were stained with fluorescein isothiocyanate (FITC)-conjugated annexin V and PI. For each sample, data from approximately 10,000 cells were recorded in the list mode on logarithmic scales. Apoptotic and necrotic cells were analyzed by performing quadrant statistics on PtdIns-negative/annexin V-positive cells and PI/annexin V double-positive cells, respectively.
Colony assay
The colony assay for dispersed single cells was performed to measure the capacity for cell colony formation. After the harvest of cells treated with BBR (75 μM) or anti-miR-99a∼125b cluster LNAs (0.5 μM), cells (1 × 103) were seeded onto a 24-well plate, and thoroughly mixed with 0.9% methylcellulose solution in RPMI-1640 containing 20% FBS, 2 mM L-glutamine and 5 μM 2-mercaptoethanol. Single cells were randomly and evenly distributed throughout the wells. Colonies were formed during incubation for about 1 week at 37°C in an atmosphere of 5% CO2. The colonies containing more than 50 cells were determined by light microscopic observation.
Flow cytometry immunophenotyping
RPMI-8266 cells were processed within 48 h of collection by washing twice with phosphate buffered saline. The cells were immunostained for 30 min with PE-conjugated CD19 antibody. B-lineage differentiation was evaluated by expression of CD19-PE. Specimens were examined using flow cytometry (Coulter Elite, Beckman Coulter, Brea, CA, USA).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Chipscreening Inc. (http://www.chipscreen.com/en/) (Shenzhen, China) for microarray services.
Author Contributions
JF conceived of and designed the experiments. MF, XC and CG performed the experiments. JF and YM analyzed the data. XZ and JF contributed reagents, materials and analytical tools. JF, YM and XZ wrote the paper.
KRNB_A_1017219_Supplements.pdf
Download PDF (257.1 KB)Funding
This work was supported by grants from the National Natural Science Foundation of China (81170496), the Natural Science Foundation of Guangdong Province (S2013010013462) and the Key Discipline Construction Foundation of Jinan University.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Jantova S, Cipak L, Cernakova M, Kost'alova D. Effect of berberine on proliferation, cell cycle and apoptosis in HeLa and L1210 cells. J Pharm Pharmacol 2003; 55:1143-9; PMID:12956905; http://dx.doi.org/10.1211/002235703322277186
- Hwang JM, Kuo HC, Tseng TH, Liu JY, Chu CY. Berberine induces apoptosis through a mitochondria/caspases pathway in human hepatoma cells. Arch Toxicol 2006; 80:62-73; PMID:16189662; http://dx.doi.org/10.1007/s00204-005-0014-8
- Jabbarzadeh Kaboli P, Rahmat A, Ismail P, Ling KH. Targets and mechanisms of berberine, a natural drug with potential to treat cancer with special focus on breast cancer. Eur J Pharmacol 2014; 740:584-95. Epub 2014 Jun 26; PMID:24973693; http://dx.doi.org/10.1016/j.ejphar.2014.06.025
- Chen FL, Yang ZH, Liu Y, Li LX, Liang WC, Wang XC, Zhou WB, Yang YH, Hu RM. Berberine inhibits the expression of TNFalpha, MCP-1, and IL-6 in AcLDL-stimulated macrophages through PPARgamma pathway. Endocrine 2008; 33(3): 331-7; PMID:19034703; http://dx.doi.org/10.1007/s12020-008-9089-3
- Kim S, Kim Y, Kim JE, Cho KH, Chung JH. Berberine inhibits TPA-induced MMP-9 and IL-6 expression in normal human keratinocytes. Phytomedicine 2008; 15:340-7; PMID:17951041; http://dx.doi.org/10.1016/j.phymed.2007.09.011
- Raab MS, Podar K, Breitkreutz I, Richardson PG, Anderson KC. Multiple myeloma. Lancet 2009; 374 (9686): 324-39; PMID:19541364; http://dx.doi.org/10.1016/S0140-6736(09)60221-X
- Fonseca R, Barlogie B, Bataille R, Bergsagel PL, Chesi M, Davies FE, Drach J, Greipp PR, Kirsch IR, Kuehl WM, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res 2004; 64(4): 1546-58; PMID:14989251; http://dx.doi.org/10.1158/0008-5472.CAN-03-2876
- Calin GA, Croce CM. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene 2006a; 25(46): 6202-10; PMID:17028600; http://dx.doi.org/10.1038/sj.onc.1209910
- Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 2006b; 66(15): 7390-4; PMID:16885332; http://dx.doi.org/10.1158/0008-5472.CAN-06-0800
- Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, Sales G, Deliliers GL, Bicciato S, Lombardi L, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood 2009; 114 (25): e20-26; PMID:19846888; http://dx.doi.org/10.1182/blood-2009-08-237495
- Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer 2007; 6:60; PMID:17894887; http://dx.doi.org/10.1186/1476-4598-6-60
- Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 2006; 6:259-69; PMID:16557279; http://dx.doi.org/10.1038/nrc1840
- Wang X. Composition of seed sequence is a major determinant of microRNA targeting patterns. Bioinformatics 2014; 30(10):1377-83. Epub 2014 Jan 26; PMID:24470575; http://dx.doi.org/10.1093/bioinformatics/btu045
- Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat Genet 2011; 43(4): 371-8; PMID:21423181; http://dx.doi.org/10.1038/ng.786
- Rossi M, Amodio N, Di Martino MT, Tagliaferri P, Tassone P, Cho WC. MicroRNA and multiple myeloma: from laboratory findings to translational therapeutic approaches. Curr Pharm Biotechnol 2014; 15(5): 459-67; PMID:24846067; http://dx.doi.org/10.2174/1389201015666140519104743
- Pichiorri F1, De Luca L, Aqeilan RI. MicroRNAs: new players in multiple myeloma. Front Genet 2011; 24:2:22; PMID:22303318
- Chng WJ, Glebov O, Bergsagel PL, Kuehl WM. Genetic events in the pathogenesis of mul-tiple myeloma. Best Pract Res Clin Haematol 2007; 20:571-96; PMID:18070707; http://dx.doi.org/10.1016/j.beha.2007.08.004
- Saha MN, Micallef J, Qiu L, Chang H. Pharmacological activation of the p53 pathway in haematological malignancies. J Clin Pathol 2010; 63(3): 204-9; PMID:19955555; http://dx.doi.org/10.1136/jcp.2009.070961
- Saha MN, Jiang H, Yang Y, Zhu X, Wang X, Schimmer AD, Qiu L, Chang H. Targeting p53 via JNK pathway: a novel role of RITA for apoptotic signaling in multiple myeloma. PLoS One 2012; 7(1): e30215; PMID:22276160; http://dx.doi.org/10.1371/journal.pone.0030215
- Teoh PJ, Chung TH, Sebastian S, Choo SN, Yan J, Ng SB, Fonseca R, Chng WJ. p53 haploinsufficiency and functional abnormalities in multiple myeloma. Leukemia 2014; 28:2066-74; Mar 14. [Epub ahead of print]; PMID:24625551; http://dx.doi.org/10.1038/leu.2014.102
- Pichiorri F, Suh SS, Rocci A, De Luca L, Taccioli C, Santhanam R, Zhou W, Benson DM Jr, Hofmainster C, Alder H, et al. Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell 2010; 18:367-81; PMID:20951946; http://dx.doi.org/10.1016/j.ccr.2010.09.005
- Kumar M, Lu Z, Takwi AA, Chen W, Callander NS, Ramos KS. Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 2011; 30(7): 843-53; PMID:20935678; http://dx.doi.org/10.1038/onc.2010.457
- Gordon MW, Yan F, Zhong X, Mazumder PB, Xu-Monette ZY, Zou D, Young KH, Ramos KS, Li Y. Regulation of p53-targeting microRNAs by polycyclic aromatic hydrocarbons: implications in the etiology of multiple myeloma. Mol Carcinog 2014; May 6. [Epub ahead of print]; PMID:24798859; http://dx.doi.org/10.1002/mc.22175
- Murray MY, Rushworth SA, Zaitseva L, Bowles KM, Macewan DJ. Attenuation of dexamethasone-induced cell death in multiple myeloma is mediated by miR-125b expression. Cell Cycle 2013; 12(13): 2144-53; PMID:23759586; http://dx.doi.org/10.4161/cc.25251
- Peng H, Wen J, Li H, Chang J, Zhou X. Drug inhibition profile prediction for NFκB pathway in multiple myeloma. PLoS One 2011; 6(3): e14750; PMID:21408099; http://dx.doi.org/10.1371/journal.pone.0014750
- Peng H, Peng T, Wen J, Engler DA, Matsunami RK, Su J, Zhang L, Chang CC, Zhou X. Characterization of p38 MAPK Isoforms for Drug Resistance Study Using Systems Biology Approach. Bioinformatics 2014; 30(13):1899-907. Epub 2014 Mar 10; PMID:24618474; http://dx.doi.org/10.1093/bioinformatics/btu133
- Azab AK, Azab F, Blotta S, Pitsillides CM, Thompson B, Runnels JM, Roccaro AM, Ngo HT, Melhem MR, Sacco A, Jia X, Anderson KC, Lin CP, Rollins BJ, Ghobrial IM. RhoA and Rac1 GTPases play major and differential roles in stromal cell-derived factor-1-induced cell adhesion and chemotaxis in multiple myeloma. Blood 2009; 114(3): 619-29; PMID:19443661; http://dx.doi.org/10.1182/blood-2009-01-199281
- García-Bernal D, Redondo-Muñoz J, Dios-Esponera A, Chèvre R, Bailón E, Garayoa M, Arellano-Sánchez N, Gutierrez NC, Hidalgo A, García-Pardo A, et al. Sphingosine-1-phosphate activates chemokine-promoted myeloma cell adhesion and migration involving α4β1 integrin function. J Pathol 2013; 229(1): 36-48; PMID:22711564; http://dx.doi.org/10.1002/path.4066
- Sekiguchi N, Ootsubo K, Wagatsuma M, Midorikawa K, Nagata A, Noto S, Yamada K, Takezako N. The impact of C-Myc gene-related aberrations in newly diagnosed myeloma with bortezomib / dexamethasone therapy. Int J Hematol 2014; 99(3): 288-95; PMID:24496825; http://dx.doi.org/10.1007/s12185-014-1514-1
- Affer M, Chesi M, Chen WD, Keats JJ, Demchenko YN, Tamizhmani K, Garbitt VM, Riggs DL, Brents LA, Roschke AV, et al. Promiscuous rearrangements of the MYC locus hijack enhancers and super-enhancers to dysregulate MYC expression in multiple myeloma. Leukemia 2014; 28:1725-35; Feb 12. [Epub ahead of print]; PMID:24518206; http://dx.doi.org/10.1038/leu.2014.70
- Schotte D, De Menezes RX, Akbari Moqadam F, Khankahdani LM, Lange-Turenhout E, Chen C, Pieters R, Den Boer ML. MicroRNA characterize genetic diversity and drug resistance in pediatric acute lymphoblastic leukemia. Haematologica 2011; 96(5): 703-11; PMID:21242186; http://dx.doi.org/10.3324/haematol.2010.026138
- Willimott S, Wagner SD. Stromal cells and CD40 ligand (CD154) alter the miRNome and induce miRNA clusters including, miR-125b/miR-99a/let-7c and miR-17-92 in chronic lymphocytic leukaemia. Leukemia 2012; 26(5): 1113-6; PMID:22024720; http://dx.doi.org/10.1038/leu.2011.299
- Sun D, Layer R, Mueller AC, Cichewicz MA, Negishi M, Paschal BM, Dutta A. Regulation of several androgen-induced genes through the repression of the miR-99a/let-7c/miR-125b-2 miRNA cluster in prostate cancer cells. Oncogene 2014; 33(11): 1448-57; PMID:23503464; http://dx.doi.org/10.1038/onc.2013.77
- Gong J, Zhang JP, Li B, Zeng C, You K, Chen MX, Yuan Y, Zhuang SM. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 2013; 32(25): 3071-9; PMID:22824797; http://dx.doi.org/10.1038/onc.2012.318
- Cannizzo E, Carulli G, Del Vecchio L, Ottaviano V, Bellio E, Zenari E, Azzarà A, Petrini M, Preffer F. The role of CD19 and CD27 in the diagnosis of multiple myeloma by flow cytometry: a new statistical model. Am J Clin Pathol 2012; 137(3): 377-86; PMID:22338049; http://dx.doi.org/10.1309/AJCP63TOCFNAMDMS
- Mahmoud MS, Fujii R, Ishikawa H, Kawano MM. Enforced CD19 expression leads to growth inhibition and reduced tumorigenicity. Blood 1999; 94:3551-8; PMID:10552966
- Jeong TD, Park CJ, Shim H, Jang S, Chi HS, Yoon DH, Kim DY, Lee JH, Lee JH, Suh C, et al. Simplified flow cytometric immunophenotyping panel for multiple myeloma, CD56/CD19/ CD138 (CD38)/CD45, to differentiate neoplastic myeloma cells from reactive plasma cells. Korean J Hematol 2012; 47:260-6; PMID:23320004; http://dx.doi.org/10.5045/kjh.2012.47.4.260
- Lo TF, Tsai WC, Chen ST. MicroRNA-21-3p, a berberine-induced miRNA, directly down-regulates human methionine adenosyltransferases 2A and 2B and inhibits hepatoma cell growth. PLoS One 2013; 8(9): e75628. eCollection 2013; PMID:24098708; http://dx.doi.org/10.1371/journal.pone.0075628
- Qian B, Nag SA, Su Y, Voruganti S, Qin JJ, Zhang R, Cho WC. miRNAs in cancer prevention and treatment and as molecular targets for natural product anticancer agents. Curr Cancer Drug Targets 2013; 13(5): 519-41; PMID:23597193; http://dx.doi.org/10.2174/15680096113139990031
- Hu HY, Li KP, Wang XJ, Liu Y, Lu ZG, Dong RH, Guo HB, Zhang MX. Set9, NF-κB, and microRNA-21 mediate berberine-induced apoptosis of human multiple myeloma cells. Acta Pharmacol Sin 2013; 34(1):157-66; PMID:23247593; http://dx.doi.org/10.1038/aps.2012.161
