Abstract
In higher eukaryotes, eIF4A, eIF4E and eIF4G homologues interact to enable mRNA recruitment to the ribosome. eIF4G acts as a scaffold for these interactions and also interacts with other proteins of the translational machinery. Trypanosomatid protozoa have multiple homologues of eIF4E and eIF4G and the precise function of each remains unclear. Here, 2 previously described eIF4G homologues, EIF4G3 and EIF4G4, were further investigated. In vitro, both homologues bound EIF4AI, but with different interaction properties. Binding to distinct eIF4Es was also confirmed; EIF4G3 bound EIF4E4 while EIF4G4 bound EIF4E3, both these interactions required similar binding motifs. EIF4G3, but not EIF4G4, interacted with PABP1, a poly-A binding protein homolog. Work in vivo with Trypanosoma brucei showed that both EIF4G3 and EIF4G4 are cytoplasmic and essential for viability. Depletion of EIF4G3 caused a rapid reduction in total translation while EIF4G4 depletion led to changes in morphology but no substantial inhibition of translation. Site-directed mutagenesis was used to disrupt interactions of the eIF4Gs with either eIF4E or eIF4A, causing different levels of growth inhibition. Overall the results show that only EIF4G3, with its cap binding partner EIF4E4, plays a major role in translational initiation.
Abbreviations
| eIF | = | eukaryotic Initiation Factor |
| PABP | = | Poly(A) Binding Protein |
| MIF4G | = | Middle domain of eukaryotic Initiation Factor 4G |
| HEAT domain | = | Huntingtin, elongation factor 3 (EF3) |
| = | protein phosphatase 2A (PP2A) and the yeast kinase TOR1 | |
| GST | = | Glutathione-S-Transferase |
Introduction
The trypanosomatids are pathogenic protozoa responsible for human diseases of worldwide impact, caused by species belonging to the genera Leishmania and Trypanosoma.Citation1 One feature of these organisms is the dominance of post-transcriptional mechanisms for the control of gene expression. Protein coding genes are arranged in tandem arrays that are co-transcribed from a distant promoter and co-trancriptionally processed to individual monocistronic mRNAs by linked trans-splicing and polyadenylation reactions. Transcription start sites for protein coding genes appear to be marked epigenetically and there is no evidence for selective transcription by RNA polymerase II; Citation2-5 indeed the evidence favors constitutive transcription with non-selective regulation of the rate of initiation, for example in response to stress.Citation6 Selective regulation of gene expression acts on mRNA levels and efficiency of translation.Citation7-9 There are several examples of regulation of translation in trypanosomatids Citation10-14 and many others are likely to emerge, but the molecular mechanisms associated with this regulation are not well understood. In higher eukaryotes most translation regulation events occurs at the level of translation initiation when selection of the mRNAs by the protein synthesis apparatus takes place.
In plants, yeast and animals, translation initiation is mediated by a number of translation initiation factors (eIFs) which facilitate ribosome binding to the mRNA and identification of the AUG start codon.Citation15-18 The process is initiated by the eIF4F complex, a heterotrimer of eIF4E, eIF4A and eIF4G. To initiate translation, eIF4E binds to the 7-methylguanosine cap at the 5′ end of the mRNA and mediates assembly of eIF4F on the mRNA.Citation19 eIF4A is a RNA helicase and eIF4G acts as a scaffold binding eIF4E and eIF4A as well as the eIF3 complex, which mediates recruitment of the ribosome small subunit.Citation20 Initiation of translation is enhanced by a direct interaction between eIF4G and poly(A) binding protein (PABP) bound to the 3′ poly(A) tail of the mRNA. The ability of eIF4G to bind to proteins associated with both the 5′ (eIF4E) and 3′ (PABP) ends of the mRNAs provides a means by which the integrity of mRNAs can be evaluated prior to translation.Citation21,22
Mammalian eIF4G can be divided into 3 parts: an N-terminal region, which includes the conserved eIF4E binding site;Citation23 a central segment containing the characteristic MIF4G domain, also known as HEAT-1 domain, which binds eIF4A;Citation24 and the C-terminus, containing both the MA3 domain (HEAT-2) also involved in binding to eIF4A and the W2 domain (HEAT-3).Citation25 eIF4G binds to PABP through motifs close to the N-terminus of the mammalian protein and its W2 domain mediates binding to the Mnk kinase that can phosphorylate eIF4E.Citation26 The interaction between eIF4G and eIF3 is critical for their combined role in recruiting the small ribosomal subunit to the mRNA and it requires one or more motifs localized within a linker region between the MIF4G and MA3 domains which is also targeted by phosphorylation events.Citation27,28 eIF4G also binds directly to RNA, an interaction mediated by a segment between the eIF4E binding motif and the MIF4G domain and which enhances binding of eIF4E to the cap structure.Citation29 During translation initiation in mammals, the interaction between eIF4G and eIF4A plays a critical role in the melting of secondary structures along the mRNA 5′ UTR which can interfere with the scanning performed by the small ribosomal subunit in order to identify the translation initiation codon.Citation17,18 This is mediated mainly by the helicase activity of eIF4A but also requires interaction with eIF4G.Citation30,31
The eIF4G tripartite structure is conserved in most metazoans and is also present in proteins unrelated in function but which may share a common evolutionary origin, such as the CBP80 subunit of the nuclear cap binding complex.Citation32 In contrast, budding yeast eIF4G not only lacks a C-terminal region, missing both MA3 and W2 domains, but also has distinct motifs implicated in the interactions with PABP, eIF3 and RNA.Citation18,26 Nevertheless for mammals, yeast and plants 2 different eIF4G homologues are usually present which seem to be differentially involved with the translation of specific mRNA groups.Citation33
In trypanosomatids, 5 putative eIF4G homologues were originally identified in the L. major genome (EIF4G1 to EIF4G5), all sharing a central conserved MIF4G domain. All five are conserved in other trypanosomatid species.Citation34 EIF4G3 and EIF4G4 have conserved sequence features outside the MIF4G domain which are not conserved in EIF4G1, EIF4G2 and EIF4G5. In addition, there are no significant similarities between these three eIF4G homologues outside the MIF4G domain.Citation34 To date, EIF4G3 and EIF4G4 have been shown to form eIF4F-like complexes, through interactions with one eIF4A (EIF4AI) and 2 eIF4E homologues (EIF4E3 and EIF4E4), in both T. brucei and Leishmania.Citation34-37 Different experimental approaches have implicated both EIF4E3 and EIF4E4 in translation initiation,Citation38,39 and it is assumed that this is mediated by their eIF4G partners acting in a manner reminiscent of other eukaryotic eIF4Gs. However, any functional differences between the resulting complexes remain unresolved. More recently EIF4G1, EIF4G2 and EIF4G5 have been shown to form complexes with 2 novel eIF4E homologues, EIF4E5 and EIF4E6, but they have not been linked to the translation initiation process.Citation40,41
Here, the functional properties and individual roles of EIF4G3 and EIF4G4 have been further investigated in both L. major and T. brucei. First, binding to eIF4Es, eIF4A and PABPs were investigated in vitro. Second, the subcellular localizations were determined. Third, the growth and protein synthesis phenotypes after RNAi knockdown were analyzed. Fourth, perturbations of the interactions of the eIF4Gs with the eIF4Es and eIF4A were investigated by expression of transgenes encoding variants unable to bind to eIF4E or eIF4A. The results show that EIF4G3 is necessary for protein synthesis and thus viability, a role that depends on its ability to interact with EIF4E4 and EIF4AI. In contrast, EIF4G4 is also required for viability and growth but its depletion does not affect overall protein synthesis.
Results
Sequence comparison of EIF4G3 and EIF4G4 homologues
A sequence comparison of EIF4G3 and EIF4G4 from different species was first carried out to pinpoint conserved features possibly necessary for function. An alignment of EIF4G3 and EIF4G4 orthologues from L. major, T. brucei and T. cruzi () highlighted several conserved segments in the N- and C-terminal regions as well as within the MIF4G domain. Many of these features were present in both EIF4G3 and EIF4G4, compatible with both proteins’ ability to form eIF4F-like complexes. The single most relevant difference between the EIF4G3 and EIF4G4 sequences are the Q or N/Q rich stretches localized immediately after the MIF4G domain in the EIF4G4 homologues and which are more noticeable in the 2 Trypanosoma sequences. Noteworthy, however, are several conserved elements positioned within the C-terminal halves of all sequences and which are predicted to be mainly α-helical in nature. When compared through pair-wise BLAST analysis, however, no clear cut sequence homology was observed with the MA3 and W2 domains found in mammalian eIF4G. Nevertheless, structure prediction analysis using only the C-terminal region of the different EIF4G3 and EIF4G4 homologues predicted with high confidence (greater than 75% confidence) a MA3 domain similar to one of the domains from the mammalian translation regulator Pdcd4, whose structure has been solved.Citation42 Furthermore, 2 conserved tryptophan residues, plus neighboring amino acids, in the C-terminus of the trypanosomatid proteins (highlighted in ) resemble the 2 conserved aromatic and acidic boxes previously observed within the W2 domain of mammalian eIF4G and related proteins.Citation25,43 It is likely then that both MA3 and W2 domains, divergent in sequence due to the greater evolutionary distance between trypanosomatids and higher eukaryotes, are present in the EIF4G3 and EIF4G4 homologues.
Figure 1 (See previous page). Sequence comparison of the EIF4G3 and EIF4G4 homologues from different trypanosomatids. (A) Clustal W alignment of the EIF4G3 and EIF4G4 homologues from L. major (Lm), T. brucei (Tb) and T. cruzi (Tc), highlighting regions of homology outside the HEAT-1/MIF4G domain. Amino acids identical in more than 60% of the sequences are highlighted in dark gray, while amino acids defined as similar, based on the BLOSUM 62 Matrix, on more than 60% of the sequences, are shown in pale gray. When necessary, spaces were inserted within the various sequences (dots) to allow better alignment. The two candidate eIF4E binding segments mentioned in the text are boxed and numbered 1 and 2. The central MIF4G/HEAT-1 domain, the region which defines the various eIF4G homologues, as well as the putative α-helical rich regions which are likely variants of the MA3/HEAT-2 and W2/HEAT-3 domains are underlined. ▾ indicates the conserved amino acid segments which have been mutated in either L. major or T. brucei EIF4G3 and EIF4G4 in order to investigate their interactions with eIF4E and eIF4A homologues. ⇓ indicates the unique isoleucine/leucine residue which has been identified to be relevant for the interactions between both eIF4G homologues with their eIF4E partners. The * highlights the 2 conserved tryptophan/aromatic residues at the proteins’ C-terminus which resemble equivalent residues within the metazoan HEAT-3/W2 domain. (B) Schematic representation comparing the domain organization of the human eIF4GI with those of L. major EIF4G3 and EIF4G4. For the human protein the sequences implicated in the interactions with PABP, eIF4E, RNA and eIF3 are indicated as well as the 3 defined HEAT domains. For the L. major homologues the positions of the 2 candidate eIF4E binding elements as well as the MIF4G and putative MA3 and W2 domains are also shown.

An unusual feature of trypanosomatid EIF4G3 and EIF4G4 sequences is the very short N-terminal regions, 50 or 51 residues in EIF4G3s and 74 to 77 residues in EIF4G4s from different trypanosomatid species, compared to nearly 600 residues in yeast and more than 700 residues in human eIF4G (). This is relevant as this region has been shown to encompass both eIF4E and PABP binding motifs Citation44-46 and ~100 residues of the eIF4G N-terminus, surrounding the consensus eIF4E binding motif, is required for the yeast eIF4G/eIF4E interaction.Citation23 In , 2 conserved motifs shown in the alignment (boxed in the figure) resemble the consensus eIF4E binding peptide,Citation44 defined as YXXXXLΦ (where X can be any amino acid and Φ is usually a hydrophobic amino acid such as L, M or F). These are localized to residues 3 to 9 (Box 1) and 23 to 29 (Box 2) in the Leishmania EIF4G3 sequence, but no single motif fits completely in the described consensus although Box 1 from the 2 Trypanosoma EIF4G3 homologues and Box 2 from all EIF4G4 sequences and Leishmania EIF4G3 contain only conserved substitutions (F for Y and V or I for L).
Comparative analysis of the binding properties of EIF4G3 and EIF4G4 to other EIF4F components and PABP
In vitro co-precipitation assays have been used previously to investigate the interaction of L. major EIF4G3 with EIF4AI and PABP1,Citation34,47 as well as between T. brucei EIF4G3 and EIF4G4 and eIF4E partners.Citation36 Here, co-precipitation assays were performed to better characterize these interactions and investigate how L. major EIF4G3 and EIF4G4 differ in binding to putative or known binding partners. First, interaction with the L. major eIF4A homolog (EIF4AI Citation48) was evaluated. For these assays, a GST fusion of EIF4AI was incubated with 35S-labeled EIF4G3 and EIF4G4, using GST or samples containing only glutathione sepharose beads as controls. As shown in , both full-length EIF4G3 and EIF4G4 were able to bind to EIF4AI and, when the MIF4G domain of either protein (EIF4G3 residues 26–310 and EIF4G4 residues 37–362) were tested in similar assays, the EIF4G3 MIF4G domain still bound efficiently to EIF4AI, as previously reported,Citation34 with no apparent reduction in affinity when compared with the wild type protein. In comparison, little or no binding by the EIF4G4 MIF4G truncation was observed. To confirm that the MIF4G domain in both proteins is still required for efficient binding, the conserved LNK amino acid triplet localized near the N-terminal end of this domain was replaced for 3 alanines in full-length EIF4G3 and EIF4G4. Both variant proteins were unable to bind EIF4AI, confirming a requirement of the MIF4G domain for the eIF4G/eIF4A interaction which, for EIF4G3, but not EIF4G4, is sufficient.
Figure 2. Analysis of the interaction between the L. major EIF4G3 and EIF4G4 and selected partners. (A) Binding interactions between EIF4G3 and EIF4G4 and the EIF4AI homolog. Co-precipitation assays were carried out between a GST-EIF4AI fusion and different [35S]-labeled EIF4G3 and EIF4G4 produced after in vitro transcription and translation reactions. The upper panel shows a representative Coomassie-blue stained gel highlighting the recombinant GST (negative control) or GST fusion protein used in the assays. The panels below show the results from the various assays carried out with the wild-type full-length EIF4G3 and EIF4G4, truncation variants containing only the HEAT-1/MIF4G domain or full-length proteins with the LNK/AAA mutation. “Input” is equivalent to the same amount of labeled proteins (total translation reactions) used in the assays. Numbers on the left indicate the migration of molecular weight markers in kDa. (B) Binding interactions with the 3 Leishmania PABP homologues. Assays were carried out as described in A using recombinant GST-fusioned PABP1, 2 and 3 tested for their ability to bind [35S]-labeled EIF4G3 and EIF4G4. (C) Reverse co-precipitation assays with GST-EIF4G3 and 4 immobilized on the glutathione sepharose beads and incubated with the different [35S]-labeled PABP and eIF4E homologues from L. major.
![Figure 2. Analysis of the interaction between the L. major EIF4G3 and EIF4G4 and selected partners. (A) Binding interactions between EIF4G3 and EIF4G4 and the EIF4AI homolog. Co-precipitation assays were carried out between a GST-EIF4AI fusion and different [35S]-labeled EIF4G3 and EIF4G4 produced after in vitro transcription and translation reactions. The upper panel shows a representative Coomassie-blue stained gel highlighting the recombinant GST (negative control) or GST fusion protein used in the assays. The panels below show the results from the various assays carried out with the wild-type full-length EIF4G3 and EIF4G4, truncation variants containing only the HEAT-1/MIF4G domain or full-length proteins with the LNK/AAA mutation. “Input” is equivalent to the same amount of labeled proteins (total translation reactions) used in the assays. Numbers on the left indicate the migration of molecular weight markers in kDa. (B) Binding interactions with the 3 Leishmania PABP homologues. Assays were carried out as described in A using recombinant GST-fusioned PABP1, 2 and 3 tested for their ability to bind [35S]-labeled EIF4G3 and EIF4G4. (C) Reverse co-precipitation assays with GST-EIF4G3 and 4 immobilized on the glutathione sepharose beads and incubated with the different [35S]-labeled PABP and eIF4E homologues from L. major.](/cms/asset/636bfc5f-c031-41ca-a7a0-df5327ff0d28/krnb_a_1017233_f0002_b.gif)
Next, the interactions between EIF4G3 and EIF4G4 with PABP homologues were evaluated. The interaction between GST-tagged EIF4G3 and the 3 distinct PABPs from L. major (PABPs 1 to 3) was previously evaluated and only PABP1 bound EIF4G3,Citation47 however an independent 2 hybrid assay failed to confirm this interaction.Citation49 Here, the same interactions were evaluated comparing the 2 eIF4G homologues and using both GST-tagged PABPs and 35S-labeled EIF4G3 and EIF4G4 and the reverse assay with GST-tagged eIF4Gs with 35S-labeled PABPs. As shown in , both assays confirmed a reproducible interaction of EIF4G3 with PABP1, but not with PABP2 or PABP3, and no clear interaction between EIF4G4 and any of the PABP homologues. The four GST-tagged eIF4E homologues from L. major were then assayed with the 35S-labeled EIF4G3 and EIF4G4 to confirm previously described interactions seen in Leishmania and Trypanosoma proteins,Citation35–37 in a comparative way. As shown in , EIF4G3 was found to interact only with EIF4E4, while EIF4G4 bound specifically to EIF4E3. Overall, although both EIF4G3 and EIF4G4 interact specifically with their respective eIF4E partners, only EIF4G3 interacts with the PABP homolog and it also seems to bind more efficiently EIF4AI, as would be needed to adequately support translation and form an eIF4F-like complex.
Identification of a common eIF4E binding motif in both EIF4G3 and EIF4G4 homologues
The interaction between eIF4E and eIF4G is necessary for mRNA recruitment in higher eukaryotes and it is mediated by conserved motifs in the 2 binding partners. In trypanosomatid EIF4G3 and EIF4G4, the regions N-terminal of the MIF4G domain are much shorter than other characterized eukaryotic eIF4Gs. In mammalian eIF4G, the eIF4E binding motif lies toward the end of its very long N-terminal region which also includes the PABP binding motif (see ). In contrast, in trypanosomatids, long N-terminal extensions are seen for the EIF4G3 and EIF4G4 binding partners, EIF4E3 and EIF4E4, which are absent in metazoan eIF4E. The function of these N-terminal extensions is unknown but deletion in EIF4E4 does not impair its binding to EIF4G3.Citation49 Both the EIF4G3/EIF4E4 and EIF4G4/EIF4E3 interactions were previously found to require distinct binding motifs in the respective eIF4Gs, located at different positions within these proteins’ N-terminal regions.Citation35,37 Distinct eIF4E binding motifs, however, are in clear disagreement with the similarities in sequence, structure and functional properties of EIF4E3 and EIF4E4 and which suggest a common mode of interaction with their eIF4G partners.
Here, a re-evaluation of the eIF4E binding motifs in both EIF4G3 and EIF4G4 was performed through parallel experiments with both proteins and using the pull-down strategy previously validated. GST-tagged variants of EIF4G3 and EIF4G4 were expressed in E. coli and equivalent quantities evaluated for binding to the corresponding 35S-labeled eIF4E partners (). First, truncations on both L. major eIF4G homologues were generated to test the involvement of both N-termini in the binding to EIF4E3 and EIF4E4. Site-directed mutagenesis was then carried out, using the full-length proteins and targeting selected residues identified as conserved in both EIF4G3 and EIF4G4 (). The conserved triplets FSL, KLV and LNK, were mutated in turn, each to 3 alanines. The corresponding binding assays showed that mutating the FSL motif in EIF4G3, but not the KLV or LNK triplets (not shown), prevented binding to EIF4E4, but the same mutation in EIF4G4 did not affect the binding to EIF4E3 (). These results are in agreement with previous reports that mutating the F23 and L25 residues within the EIF4G3 FSL motif prevents binding to EIF4E4,Citation35 while removal of a 15 residue long segment encompassing this motif from EIF4G4 does not abolish the interaction with EIF4E3.Citation37 Individual residues were also targeted and likewise replaced by alanines. For EIF4G3, single mutations in 2 neighboring residues placed in conserved positions in the different EIF4G3 and EIF4G4 sequences, I8 and R9, impaired its interaction with EIF4E4. In contrast, for EIF4G4, single mutations in the equivalent positions in the L. major protein (I25 and L26) did not interfere with its binding to EIF4E3, but when both residues were simultaneously replaced by alanines (IL25-26AA) this interaction was nearly abolished. The figure also shows results from mutations in 2 other conserved residues in L. major EIF4G3 and EIF4G4 sequences (R12/R29 and Y15/W32, respectively) which did not affect binding to their eIF4E partners. Other mutations which did not interfere in the binding between these proteins and were not included in the figure are V29 for EIF4G3 and F3, M20 and L46 for EIF4G4.
Figure 3. Fine mapping of the interactions between L. major EIF4G3 and EIF4G4 with their eIF4E partners. (A) Fine mapping of the interactions between L. major EIF4G3 and EIF4E4 and between EIF4G4 and EIF4E3. Co-precipitation assays were carried out as described in using either of the eIF4G homologues fusioned to GST and assayed for their ability to bind [35S]-labeled EIF4E3 and 4. The full-length eIF4G sequences as well as truncations lacking either the N or C-terminal regions of the proteins were used in the assay, as well as site-directed variants where selected residues were replaced by alanines as indicated. The results shown are representative of multiple co-precipitation assays carried out with a minimum of 2 independent sets of GST-tagged proteins. (B) Sequence alignment comparing the eIF4G binding residues (highlighted with a *), previously identified in 3 distinct blocs of the human and yeast eIF4E sequences,Citation23,50 with the equivalent motifs found in different trypanosomatid EIF4E3 and 4 homologues. The alignment was carried out as described in for the eIF4G homologues, but only the segments relevant for the eIF4G interaction are shown. (C) Schematic representation of either known or proposed interactions between different eIF4E/eIF4G homologues. The left scheme summarizes the interactions previously observed, based on the crystal structure,Citation50 between human eIF4E and oligopeptides containing the consensus eIF4E binding motif from human eIF4G and eIF4E-binding proteins (YXXXXLΦ), plus the 3 subsequent residues. The middle and right schemes highlight the likely interactions presumed to occur between the trypanosomatid EIF4E4/EIF4G3 and EIF4E3/EIF4G4 pairs, assuming a conserved mode of binding between the different protein complexes.
![Figure 3. Fine mapping of the interactions between L. major EIF4G3 and EIF4G4 with their eIF4E partners. (A) Fine mapping of the interactions between L. major EIF4G3 and EIF4E4 and between EIF4G4 and EIF4E3. Co-precipitation assays were carried out as described in Fig. 2 using either of the eIF4G homologues fusioned to GST and assayed for their ability to bind [35S]-labeled EIF4E3 and 4. The full-length eIF4G sequences as well as truncations lacking either the N or C-terminal regions of the proteins were used in the assay, as well as site-directed variants where selected residues were replaced by alanines as indicated. The results shown are representative of multiple co-precipitation assays carried out with a minimum of 2 independent sets of GST-tagged proteins. (B) Sequence alignment comparing the eIF4G binding residues (highlighted with a *), previously identified in 3 distinct blocs of the human and yeast eIF4E sequences,Citation23,50 with the equivalent motifs found in different trypanosomatid EIF4E3 and 4 homologues. The alignment was carried out as described in Fig. 1 for the eIF4G homologues, but only the segments relevant for the eIF4G interaction are shown. (C) Schematic representation of either known or proposed interactions between different eIF4E/eIF4G homologues. The left scheme summarizes the interactions previously observed, based on the crystal structure,Citation50 between human eIF4E and oligopeptides containing the consensus eIF4E binding motif from human eIF4G and eIF4E-binding proteins (YXXXXLΦ), plus the 3 subsequent residues. The middle and right schemes highlight the likely interactions presumed to occur between the trypanosomatid EIF4E4/EIF4G3 and EIF4E3/EIF4G4 pairs, assuming a conserved mode of binding between the different protein complexes.](/cms/asset/67c6f27d-1ce6-426d-a390-1025325b34f8/krnb_a_1017233_f0003_b.gif)
Single isoleucine/leucine to alanine or arginine to alanine substitutions should not impact significantly on the structure of the EIF4G3 N-terminus, and even the double substitution EIF4G4 IL25-26AA is unlikely to have a significant effect. In the alignment shown in , the identified doublets for both EIF4G3 and EIF4G4 are localized within the first box (Box 1) found to resemble the proposed eIF4E binding consensus of higher eukaryotes (YXXXXLΦ). Their positions are equivalent to the conserved LΦ residues, both hydrophobic amino acids, as seen in most trypanosomatid EIF4G3 and EIF4G4 sequences. In L. major EIF4G3 the second hydrophobic residue is replaced by an arginine (R9), but the substitution of the Φ residue for other amino acids with long aliphatic side chains, charged or not such as arginine, was predicted to be possible when the structure for the eIF4E/eIF4G interaction was originally solved.Citation50 Nevertheless, the lack of a conserved tyrosine or phenylalanine at the first position in Box 1 of the EIF4G4 homologues questions the assumption that this is indeed a binding motif similar to the one described for higher eukaryotes. To better understand these interactions in the 2 trypanosomatid protein pairs, the putative eIF4G binding sites in EIF4E3 and EIF4E4 were investigated through sequence analysis. highlights the 3 sets of motifs which, in higher eukaryotes, have been shown to be implicated in mediating eIF4E's interaction with eIF4G. Of the residues directly implicated in the interaction with eIF4G, only the tryptophan residue (W73 in the mammalian sequence) is universally conserved in the different trypanosomatid eIF4E sequence although the V69 is found to be replaced only by the similarly hydrophobic isoleucine. Both residues interact directly with the leucine in the eIF4E binding consensus of mammalian eIF4G consensus (as does L135). In contrast, other residues which have been found to interact directly with the aromatic residue in eIF4G, H37 and P38, for example, are not conserved within the different eIF4E homologues, although a proline is present in the same position of Leishmania EIF4E3. summarizes the known interactions involved in mammalian eIF4E/eIF4G binding as well as the ones which would be expected between EIF4G3/EIF4E4 and EIF4G4/EIF4E3, assuming a conserved set of interactions for the different protein pairs. Overall, the evidence presented indicates then that a common eIF4E binding motif exists in both eIF4G homologues studied and that these should include the currently identified residues (I8 and R9 in EIF4G3/I25 and L26 in EIF4G4).
Both EIF4G3 and EIF4G4 are moderately expressed in T. brucei procyclic cells and localize strictly to the cytoplasm
To continue in vivo the functional characterization of EIF4G3 and EIF4G4, a second trypanosomatid system was studied, T. brucei, so as to fully exploit the tools available for its genetic manipulation. Antibodies against T. brucei EIF4G3 and EIF4G4 were first used to evaluate expression in procyclic cells (). EIF4G3 was detected as a single band of the expected size while EIF4G4 was seen to be represented by 2 bands of very similar sizes, both compatible with the predicted molecular weight for this protein. Western blots were used to estimate the proteins’ copy number using a procedure previously described for T. brucei eIF4E homologues.Citation36 Although the procedure only produces an estimate, the results for EIF4G3 (1 to 2 × 104 molecules/cell) shows that it is 3 to 10-fold more abundant than EIF4G4 (2 to 3 × 103 molecules/cell). When compared with the previous copy number estimates for the corresponding eIF4E partners, EIF4G3 is similar to EIF4E4, while EIF4G4 is present at levels at least 10-fold lower than EIF4E3.
Figure 4. Expression analysis and subcellular localization of the T. brucei EIF4G3 and EIF4G4. (A) Quantitation and expression analysis of T. brucei EIF4G3 and EIF4G4 in wild type, exponentially grown, procyclic cells. Recombinant His-tagged EIF4G3 and EIF4G4 were quantitated, diluted to defined concentrations (in fentomoles) and ran on SDS-PAGE gels with whole parasite extract obtained from known number of cells. The gels were then transferred to PVDF membranes and blotted with antibodies directed against T. brucei EIF4G3 or EIF4G4. Densitometric analyses of the results allowed for a rough estimation of the intracellular levels of both proteins, as described.Citation36 (B) Subcellular localization of EIF4G3 and EIF4G4 through indirect immunofluorescence. Procyclic cells were incubated with the same antibodies used in A followed by Alexa Fluor 488-conjugated secondary antibody. DNA was stained with DAPI to identify the nucleus and kinetoplast. (C) The localization of EIF4G3 and EIF4G4 was also confirmed through the expression of eYFP fusion proteins in transfected T. brucei procyclic cells examined under the confocal microscope.
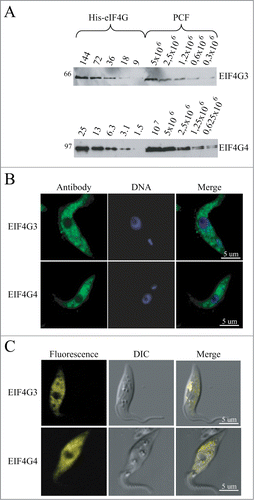
The antibodies were then used for immunofluorescence experiments to investigate subcellular localization (). Both proteins localized predominantly to the cytoplasm with very little or no localization within the nucleus and with no apparent sublocalization within the cytoplasm. This subcellular localization was also investigated through the expression of enhanced yellow fluorescent fusion proteins (eYFP) in transgenic cells, this confirmed the strict cytoplasmic localization of the 2 proteins (). The cytoplasmic localization of both proteins argues against any role in mRNA transcription or processing in the nucleus and is consistent with major or minor roles in mRNA metabolism and translation in the cytoplasm.
Both EIF4G3 and EIF4G4 are essential for viability of T. brucei procyclic cells and depletion of EIF4G3, but not EIF4G4, directly impacts on protein synthesis
To investigate any differences in the functions of EIF4G3 and EIF4G4, we analyzed phenotype after RNAi knockdown. First, the growth phenotypes are summarized in ; both proteins were essential for viability, on EIF4G3 knockdown cell proliferation stopped very soon after RNAi induction and cells began to die within 24h (). Despite this immediate onset of growth arrest it took 24 hours for the protein to decrease substantially and this observation suggests that synthesis of EIF4G3 may be necessary for proliferation. In contrast, knockdown of EIF4G4 showed slowed growth after 24 to 48h but with cell death occurring only after 120h of RNAi, despite no detection of the protein after 48h of the induction ().
Figure 5. RNA interference of EIF4G3 and EIF4G4. Procyclic T. brucei cells were transfected with the p2T7-177 derived plasmid containing either of the eIF4G genes (EIF4G3 – (A), EIF4G4 – (B). Transfected cells were selected after growth in the presence of phleomycin and RNA interference induced after tetracycline addition. At regular intervals, cellular growth was monitored by counting the number of viable cells of cultures with and without tetracycline and the resulting values used to plot the curves shown (plus tetracycline – gray; minus tetracycline – black). Below each graph are Western-blot analyses of the proteins being targeted, using the affinity-purified antisera specific for each protein. The same blots were then reprobed with affinity purified antibodies against the endogenous control EIF4AI as loading control. All RNAi results shown are representative of multiple experiments performed with distinct transfection events. (C) Effect on cell morphology of the EIF4G4 RNAi preceding cell death. At selected time points after induction of the EIF4G4 RNAi, the cells were visualized at the confocal microscope to monitor for abnormalities in cell morphology. All RNAi results shown in the figure are representative of multiple experiments carried out with a minimum of 2 independently transfected cell lines.
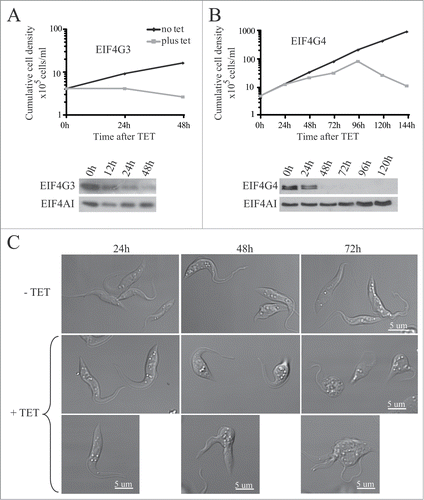
The effect of knockdown on cell morphology differed; on EIF4G3 knockdown no major changes in cell morphology was observed during the growth curves shown. In contrast, on EIF4G4 knockdown the cells developed multiple abnormalities in their morphology, some assuming a more rounded shape while other arresting during cytokinesis and other still displaying multiple flagella. These changes increased with time just up to the point where cells lost viability and the total cell count started to decline ().
To evaluate the effect of knockdown on protein synthesis, we performed metabolic labeling of the cells with [35S]-methionine at selected time points after RNAi. Samples were investigated for changes in total protein synthesis as well as in overall synthesis profile. No significant differences were observed in protein synthesis for the RNAi of EIF4G4 even up to 72 h after tetracycline addition. However, the knock down of EIF4G3 inhibited protein synthesis as soon as 6 h after induction with ~80% inhibition after 24 h. No selective changes in protein expression were observed (). So far, these results are consistent with a major role for EIF4G3 in protein synthesis whereas EIF4G4, despite being essential for cell viability, may not be directly required for protein synthesis or may perform minor roles in this process.
Figure 6. Metabolic labeling of procyclic cells from the RNAi curves of EIF4G3 and EIF4G4. (A) Qualitative analysis of the proteins being synthesized after RNAi. At selected time points after RNAi aliquots of the cells were incubated with [35S]-methionine followed by harvesting and analysis of total protein through SDS–PAGE and autoradiography. (B) Total protein synthesis was also estimated after RNAi for each eIF4G homolog by TCA precipitation and quantitation of the incorporated radioactivity. The results shown are normalized by comparing the incorporation in the RNAi treated cells with the minus tetracycline controls.
![Figure 6. Metabolic labeling of procyclic cells from the RNAi curves of EIF4G3 and EIF4G4. (A) Qualitative analysis of the proteins being synthesized after RNAi. At selected time points after RNAi aliquots of the cells were incubated with [35S]-methionine followed by harvesting and analysis of total protein through SDS–PAGE and autoradiography. (B) Total protein synthesis was also estimated after RNAi for each eIF4G homolog by TCA precipitation and quantitation of the incorporated radioactivity. The results shown are normalized by comparing the incorporation in the RNAi treated cells with the minus tetracycline controls.](/cms/asset/325d1251-d68b-4857-b0f8-7d02851877ab/krnb_a_1017233_f0006_b.gif)
Phenotype after overexpression of EIF4G3 and EIF4G4 wild type and variants
Cell lines were generated containing tetracycline-inducible transgenes encoding either wild-type EIF4G3 and EIF4G4 or variants in specific motifs selected based on the results shown in . All transgenes encoded proteins with an N-terminal TY-tag and expression and co-precipitation experiments used either anti-TY monoclonal antibody or the polyclonal sera available against T. brucei EIF4G3 and EIF4G4, EIF4E3 and EIF4E4 and EIF4AI.
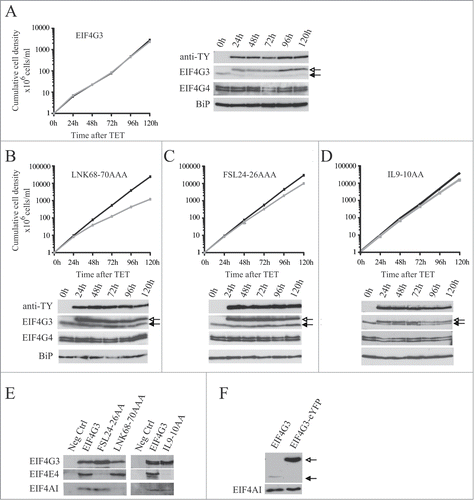
First, the growth of the cell lines was investigated after EIF4G3 transgene induction (). First, cells expressing the wild-type gene were evaluated and no changes in growth were observed after induction (). Expression was confirmed by Western blot analysis using either the anti-TY or anti-EIF4G3 antibodies (). Next, the effect of expressing EIF4G3 LNK68-70AAA, with a mutation in the EIF4A binding site, was investigated (). Induction led to a decrease in growth rate compatible with a dominant negative effect. Two further EIF4G3 variants were tested, each with a mutation affecting EIF4E4 binding, IL9-10AA and FSL24-26AAA. Upon induction, both variants induced a minor but reproducible reduction in proliferation rate (). Co-immunoprecipitation was then used to confirm the effect of the different mutations upon interactions. As expected, EIF4AI and EIF4E4 co-precipitated with the wild type protein, while the LNK68-70AAA mutation specifically impaired the interaction with EIF4AI and both the IL9-10AA and FSL24-26AAA mutations prevented the interaction with EIF4E4 ().
Figure 7. Expression of TY-tagged EIF4G3 and variants in procyclic cells. Growth curves and Western blot analysis of the expression of the TY-EIF4G3 wild-type (A) or the TY-EIF4G3 LNK68-70AAA (B), TY-EIF4G3 FSL24-26AAA (C) and TY-EIF4G3 IL9-10AA (D) variants in transfected cells in the presence or absence of tetracycline (plus tetracycline – gray; minus tetracycline – black). In each case the expression was detected using monoclonal anti-TY and antibodies specific to each of the eIF4G homologues. The blot was simultaneously probed with anti-BiPCitation73 as a loading control. All curves shown are representative of multiple experiments performed with distinct transfection events. The arrows highlight the expression of either the tagged-EIF4G3 proteins (white-filled arrow) or the endogenous EIF4G3 (black-filled arrow). (E) Interaction profile of wild-type TY-EIF4G3, or selected variants, with its EIF4AI and EIF4E4 binding partners. (F) Expression of EIF4G3-eYFP in transfected cells after 48h of tetracycline induction in comparison with the endogenous protein in control cells.
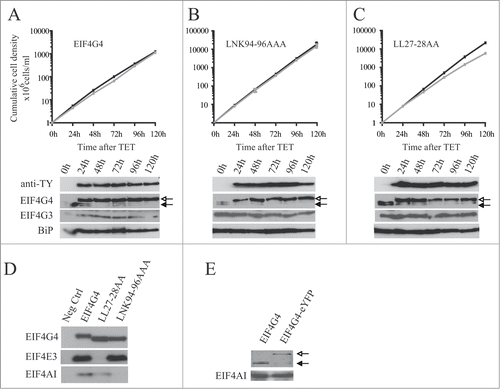
Similar experiments were carried out using cell lines containing inducible eIF4G4 transgenes (). Induction of the wild-type protein had no effect on proliferation and, in contrast to EIF4G3, the expression of the LNK94-96AAA variant did not impact on growth (). Expression of EIF4G4 with mutations in the 2 leucines at the end of Box 1, LL27-28AA (), caused a slight reduction in proliferation rate, a similar effect to that seen with the EIF4G3 IL9-10AA variant (). Co-immunoprecipitations followed by Western blots from cell lysates were then carried out to confirm the effect of the mutations and, as expected, mutations in the LNK motif specifically impaired the EIF4G4 interaction with EIF4AI while mutation in the LL dipeptide specifically prevented its interaction with EIF4E3 ().
Figure 8. Expression of TY-tagged EIF4G4 and variants in procyclic cells. Growth curves and Western blot analysis of the expression of the TY-EIF4G4 wild-type (A) or the TY-EIF4G4 LNK94-96AAA (B) and TY-EIF4G4 LL27-28AA (C) variants in transfected cells as shown for . (D) Interaction profile of wild-type TY-EIF4G4, or selected variants, with its EIF4AI and EIF4E3 binding partners. (E) Expression of EIF4G4-eYFP in transfected cells after 48 h of tetracycline induction in comparison with the endogenous protein in control cells.
The endogenous levels of EIF4G3 and EIF4G4 are tightly regulated and for EIF4G3 this regulation requires its interaction with EIF4AI and EIF4E4
Expression of the ectopic EIF4G3 or EIF4G4 suppressed the levels of the respective endogenous proteins (). The effect was specific, expression of eIF4G3 did not suppress endogenous eIF4G4 and vice versa. The suppression was also observed after induction of the EIF4G3/4-eYFP for the subcellular localization experiments (). For EIF4G3, the suppression did not occur when any of the variants described above were expressed and indeed the expression of these variants was higher than that seen for the wild type protein. In contrast, for EIF4G4, the suppression was also observed when the equivalent variants were overexpressed. These results imply the existence of a regulatory mechanism which tightly controls the endogenous levels of both EIF4G3 and EIF4G4. For EIF4G3 this requires binding to both EIF4AI as well as EIF4E4, forming a functional eIF4F-like complex. In contrast, for EIF4G4, this mechanism is independent of its ability to bind EIF4AI or EIF4E3.
Discussion
The data presented here builds upon previous published works Citation38,39 which have highlighted complex and unique events associated with translation initiation in trypanosomatids and which may have implications into how they regulate their gene expression. The new results highlight the overall conservation in tructure and sequence between EIF4G3 and EIF4G4 and in both EIF4E and EIF4A binding motifs. This conservation is in striking contrast with the divergent N and C-terminal regions of the other 3 trypanosomatid eIF4G homologues (EIF4G1, EIF4G2 and EIF4G5) which have recently been seen to participate in novel eIF4F-like complexes that do not seem to have primary roles in translation.Citation40,41 The presence of putative MA3/HEAT-2 and W2/HEAT-3 domains in at least 2 trypanosomatid eIF4G homologues confirms the likely ancient origin of the eIF4G tripartite structure, despite the fact that both MA3 and W2 are missing from yeast eIF4G and the W2 domain is not found in plant eIF4G homologues.Citation32
The evidence presented clearly distinguishes EIF4G3 and EIF4G4 functionally. Knockdown of T. brucei EIF4G3 shows a strong effect on cell growth and protein synthesis, a result which is consistent with the requirement for this protein to support mRNA recruitment through its interaction with eIF4E and eIF4A partners. Indeed the clear dominant negative phenotype observed for the EIF4G3 variant impaired in its binding to its eIF4A partner is similar to the one observed upon the induction of an EIF4AI variant (DEAD/DQAD) known to negatively impact on translation, as previously shown.Citation48 The critical role seen for EIF4G3 in overall protein synthesis in T. brucei is also in agreement with data from Leishmania which shows it co-migrating in sucrose gradients with EIF4E4 and EIF4AI in polysome-associated fractions.Citation35 In other organisms regulation of eIF4G activity/function is strictly controlled and can be mediated through different means, such as direct phosphorylation, expression of different isoforms and through regulation of its association with binding partners, such as eIF4E and PABP.Citation51–53 In T. brucei regulation of the endogenous levels of EIF4G3 seem to be strictly controlled and minor depletions induced by the knockdown procedure seemed to be sufficient to substantially interfere with translation and impact on cellular growth. The fact that the levels of both EIF4G3 and its partner EIF4E4 are roughly equivalent also might have implications for the control of EIF4G3 function, although as previously shown Citation36 not all of it is necessarily found in a state bound to EIF4E4.
In contrast to EIF4G3, T. brucei EIF4G4 does not seem to play a major role in translation initiation, or function as a true translational initiation factor, since its knockdown does not impact significantly on protein synthesis and its interaction with EIF4AI may not be required for its function. Again this is in agreement with data from Leishmania which does not show an association of EIF4G4 with polysomes in sucrose gradients.Citation37 Nevertheless, the loss of cellular viability upon knockdown and the similarities observed with EIF4G3 both in sequence/structure as well as in binding to related eIF4E partners are indicative of an essential function for cell survival. In yeast, the 2 known eIF4G homologues were shown to functionally overlapCitation54 and both in mammals and plants the 2 typical eIF4G homologues found in each organism were shown to be directly associated with mRNA translation.Citation55,56 The observation that knockdown of EIF4G4 induces a change in morphology of the cells prior to their death may indicate a selective role in the translation of specific mRNAs, whose products, however, are not visible through SDS-PAGE after metabolically labeling. Indeed, in different organisms divergent eIF4G homologues or isoforms, lacking typical eIF4E binding domains for example, have been shown to be associated with the translation of specific mRNAsCitation52,57,58 reinforcing the possibility that this may also be the case for EIF4G4. It remains to be seen what are the functional implications for the great differences in levels between EIF4G4 and its partner EIF4E3, with the latter being present well in excess of EIF4G4 and apparently being able to participate in other roles which do not require binding to this eIF4G homolog. Also it is important to highlight that 2 bands are observed for EIF4G4 on Western blots which probably reflect post-translational modification events which target this protein and which are less clear for EIF4G3, as previously reported,Citation59,60 or variations in sequence derived from the presence of 2 distinct alleles.
Two distinct eIF4E binding motifs have been proposed to exist in EIF4G3 and EIF4G4, based on 2-hybrid assays used to investigate the binding interactions with their corresponding eIF4E partners, but neither agrees with the consensus previously defined for the eIF4E binding motif.Citation35,37 Non-canonical eIF4E binding motifs have been identified previously in metazoans in regulatory factors which bind to atypical eIF4E homologues,Citation61,62 but so far these have not been identified in eIF4G or eIF4G-like sequences. In view of the similarities observed between the 2 sets of eIF4E and eIF4G subunits and the possibility of a common evolutionary origin for them (see below), differences in the eIF4E binding motifs to the extent proposed seem unlikely. The results described here confirm the previously described interactions between EIF4G3/EIF4E4 and EIF4G4/EIF4E3 but are consistent with these interactions being mediated by residues positioned in equivalent positions in the 2 sets of eIF4E and eIF4G homologues. Residues adjacent to the binding sites, such as the FSL triplet common to both eIF4Gs but only required for the binding of EIF4G3 to EIF4E4, may play contributory roles in either interaction, perhaps facilitating proper protein folding, but these would need to be investigated further.
Another interaction which is critical for eIF4G function is with PABP, mapped to non-conserved motifs positioned near the N-terminus of both metazoan and yeast eIF4G homologues.Citation45,46,63 The very short N-terminus of the trypanosomatid eIF4Gs are not likely to be sufficient for PABP binding and indeed no interaction was observed between Leishmania EIF4G3 and PABP1 through 2-hybrid assays.Citation49 Nevertheless, wheat PABP binds the plant eIFiso4G homolog through its MIF4G domainCitation64 and the results shown here, with the GST-based co-precipitation assays, confirm a previously reported interactionCitation47 between EIF4G3, but not EIF4G4, and PABP1. It remains to be seen to what extent this interaction contributes to the EIF4G3 function.
The similarities in sequence and structure observed between EIF4G3 and EIF4G4 and also between their eIF4E binding partners, EIF4E3 and EIF4E4, are indicative of gene duplication events preceding the split of the Trypanosoma and Leishmania lineages and which led to a single original complex evolving into 2 distinct complexes, with acquisition of new functions for at least one of these. The primordial complex would be then based on the EIF4G3/EIF4E4 pair, with more general roles in translation, with the second complex, EIF4G4/EIF4E3, evolving later to fulfil more specific roles. These novel roles in mRNA translation, and their impact on overall gene expression regulation, still need to be better defined but the evidence presented highlights once more the unique aspects of the basic molecular biology of trypanosomatids and how its study can have broad implications into the understanding of equivalent processes in other eukaryotes, their conservation and the potential to evolve novel alternatives in different organisms.
Materials and Methods
Sequence analysis
Sequence analysis and alignments were carried out as described,Citation34 using the sequences for the selected trypanosomatid eIF4G and eIF4E homologues available at the TriTrypDB Webpage. Structure prediction analysis was carried out using the Phyre2 (Protein Homology/analogY Recognition Engine V 2.0) Web server available at http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index which uses previously described prediction methods.Citation65
Cloning procedures
The cloning of the L. major EIF4AI and the various EIF4E and PABP genes into the pET21 (Novagen) and pGEX4T3 (GE Healthcare) vectors has been described previously.Citation34,36,47 Constructs expressing the L. major EIF4G3 MIF4G domain and its N-terminus plus MIF4G were also generated previously.Citation34 All cloning procedures carried out here with both sets of L. major and T. brucei EIF4G3 and EIF4G4 genes and their mutagenesis are detailed in the Supplementary Methods section. For the L. major EIF4G3, due to the cloning strategy used, the protein's second residue, a glutamine, was replaced by a glutamate for all variants and wild type proteins evaluated in the co-precipitation assays. For T. brucei, to generate the eYFP fusions, both EIF4G3 and EIF4G4 genes were cloned into the p2216,Citation66 while the vector p2T7-177 Citation67 was used for RNAi. The TY-tagged proteins, were all generated using a modified version of the p3927 plasmid,Citation68 containing a DNA segment encoding for a 44 residue sequence consisting of the λN-22 peptideCitation69 followed by the TY epitope.Citation70 All constructs derived from PCR or mutagenesis were fully sequenced to confirm their identity and, except when stated, seen to be identical to the L. major Friendlin and T. brucei Lister 427 genomic sequences available at GenBank and TriTrypDb.
Co-precipitation assays
Co-precipitation/pull-down assays were essentially performed as described previouslyCitation34 using Glutathione-Sepharose 4B beads (GE Healthcare) and affinity purified GST-tagged recombinant proteins expressed in Escherichia coli using the pGEX4T3 vector. GST or GST-tagged proteins including EIF4AI and both sets of EIF4G3 and EIF4G4 constructs (wild-type, truncations and variants) were immobilized on the beads and assayed for their ability to bind to the [35S]-labeled proteins. The labeled proteins, including all the EIF4G3 and EIF4G4 constructs and the various PABP and EIF4E homologues, were obtained through the linearization of the corresponding pET21d or pET21a derived plasmids with Not I (with the exception of the PABP1 construct, linearized with Hind III), followed by transcription with T7 RNA polymerase in the presence of the cap analog and translation in the rabbit reticulocyte lysate (Promega or Ambion) supplemented with [35S]-methionine (Perkin Elmer).
Parasite growth and transfection
Procyclic T. brucei cells Lister 427 were cultivated at 27°C in medium SDM-79 supplemented with haemin, fetal calf serum and antibiotics. Cultures were grown to mid-log phase for harvesting and production of total protein extract as previously described.Citation36 Transfection procedures were performed using standard conditions. For the p2T7-177 (RNAi) and p2216 (eYFP fusions) derived plasmids, the procyclic cell line Lister 427 29–13 was usedCitation71 and transfected cells were selected after growth in the presence of phleomycin (2.5 μg/ml). For transfections of plasmids derived from p3927,Citation68 a 427 KG cell line expressing a modified version of pSMOxCitation72 as background was used and transfected cells were selected after growth in the presence of blasticidin (10 μg/ml). Expression of TY-tagged proteins was induced after tetracycline addition (1 μg/ml) and cellular growth was monitored by counting the number of viable cells at regular intervals.
Protein expression analysis
The recombinant T. brucei His-tagged EIF4G3 and EIF4G4 proteins were quantified by serial dilution in SDS-PAGE compared with known concentrations of BSA. Then these quantified proteins were used in Western blots with serial dilutions of total protein extract of procyclic form of T. brucei. The Western blots generally used primary immunopurified antibodies and peroxidase-conjugated goat anti-rabbit IgG (Jackson Immunoresearch) as secondary antibody. The levels of endogenous protein were estimated by densitometry analysis using the program Kodak 1D v.3.5.2 (Eastman Kodak Company).
Fluorescence microscopy
For the indirect immunofluorescence assay, wild type procyclic cells grown to mid-log phase were harvested and washed with SDM-79 minus serum and haemin. The cells were fixed at a density of 5 × 106/ml with 3% paraformaldehyde, washed once in PBS and adhered to poly-L-lysine coated slides. Permeabilization was carried out with 0.1% Triton X-100 followed by blocking with 1% BSA and DNA was stained using Hoescht 33258 (Sigma). Antibody detection of EIF4G3 and 4 followed standard procedures using primary antibodies immunopurified from isoform specific antisera prepared as previously describedCitation36 and goat anti-rabbit IgG Alexa Fluor 488 (Invitrogen). For the eYFP fusions, after 24h of induction with tetracycline (1 μg/ml), over-expressing transfected cells were harvested and washed with PBS, then directly observed in 35 mm culture plates using a Leica SPII-AOBS confocal microscope.
RNA interference and metabolic labeling
RNA interference was induced after tetracycline addition (1 μg/ml) to cultures of cells transfected with the p2T7-177 derived plasmids. At regular intervals, cellular growth was monitored by counting the number of viable cells. For morphology analysis, cells were harvested and washed with PBS, then directly observed in 35 mm culture plates using a Leica SPII-AOBS confocal microscope. To measure the rate of protein synthesis, mid-log cultures were washed twice with methionine-free medium and then resuspended at 1 × 107 cells/ml in the methionine-free medium containing 50 μCi [35S]-methionine and incubated for one hour prior to the determination of trichloroacetic acid precipitable incorporation into protein, as previously described.Citation36 For qualitative analysis equivalent labeling experiments were carried out prior to harvesting of the cells, ressuspension in SDS-PAGE sample buffer and analysis through electrophoresis and autoradiography.
Co-immunoprecipitation assays
Immunoprecipitations (IPs) were carried out with T. brucei cytoplasmic extracts produced from exponentially grown procyclic cells lysed in lysis buffer (20 mM HEPES–KOH, pH7.4, 75 mM potassium acetate, 4mM magnesium acetate, 2 mM DTT), through freeze-thawing, at a concentration of 2 × 108 cells/ml. IPs were carried out at 4°C using standard procedures. For the assays with the TY-tagged proteins, the BB2 monoclonal antibodyCitation70 was initially conjugated to GammaBind G Sepharose beads (GE Healthcare) and then incubated with cytoplasmic extracts from transfected cells expressing each recombinant protein. In both instances, proteins were eluted in SDS-PAGE samples buffer and assayed in western blots with the same anti-TY antibody as well as the affinity purified antibodies directed against the different T. brucei eIF4E, eIF4G or eIF4A homologues, as described in the text. For a negative control, the anti-TY antibody conjugated to the beads was incubated with lysis buffer and probed with the same antibodies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
KRNB 1017233 Supplemental Methods.docx
Download MS Word (36.6 KB)Acknowledgments
The authors thank the Program for Technical Development of Health Inputs-PDTIS-FIOCRUZ for the use of its facility, the automatic sequencing facility RPT01C, at the Research Center Aggeu Magalhaes – Fiocruz Pernambuco. The anti-BiP and anti-TY antibodies were kind gifts from Jay Bangs and Keith Gull, respectively.
Funding
Work in OPDMN's lab was more recently funded with grants provided by the Brazilian funding agencies FACEPE (APQ-0239-2.02/12) and CNPq (475471/2008-3 and 480899/2013-4) and CAPES (23038.007656/2011-92). Work in MC's lab was funded by the Wellcome Trust, grant number 085956. DMNM, CRSR, CCX, TDCL and RPL received studentships from CNPq, FACEPE or CAPES.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Stuart K, Brun R, Croft S, Fairlamb A, Gurtler RE, McKerrow J, Reed S, Tarleton R. Kinetoplastids: related protozoan pathogens, different diseases. J Clin Invest 2008; 118: 1301–10; PMID:18382742; http://dx.doi.org/10.1172/JCI33945
- Daniels JP, Gull K, Wickstead B. Cell biology of the trypanosome genome. Microbiol Mol Biol Rev 2010; 74: 552–69; PMID:21119017; http://dx.doi.org/10.1128/MMBR.00024-10
- Gunzl A. The pre-mRNA splicing machinery of trypanosomes: complex or simplified? Eukaryot Cell 2010; 9: 1159–70; PMID:20581293; http://dx.doi.org/10.1128/EC.00113-10
- Martinez-Calvillo S, Vizuet-de-Rueda JC, Florencio-Martinez LE, Manning-Cela RG, Figueroa-Angulo EE. Gene expression in trypanosomatid parasites. J Biomed Biotechnol 2010; 2010: 525241; PMID:20169133; http://dx.doi.org/10.1155/2010/525241
- Alsford S, duBois K, Horn D, Field MC. Epigenetic mechanisms, nuclear architecture and the control of gene expression in trypanosomes. Expert Rev Mol Med 2012; 14: e13; PMID:22640744
- Schwede A, Kramer S, Carrington M. How do trypanosomes change gene expression in response to the environment? Protoplasma 2012; 249: 223–38; PMID: 21594757; http://dx.doi.org/10.1007/s00709-011-0282-5
- Ouellette M, Papadopoulou B. Coordinated gene expression by post-transcriptional regulons in African trypanosomes. J Biol 2009; 8: 100; PMID:20017896; http://dx.doi.org/10.1186/jbiol203
- Kramer S. Developmental regulation of gene expression in the absence of transcriptional control: the case of kinetoplastids. Mol BiochemParasitol 2012; 181: 61–72; PMID:22019385; http://dx.doi.org/10.1016/j.molbiopara.2011.10.002
- Clayton CE. Networks of gene expression regulation in Trypanosoma brucei. Mol Biochem Parasitol 2014; 195:96-106.
- Mayho M, Fenn K, Craddy P, Crosthwaite S, Matthews K. Post-transcriptional control of nuclear-encoded cytochrome oxidase subunits in Trypanosoma brucei: evidence for genome-wide conservation of life-cycle stage-specific regulatory elements. Nucleic Acids Res. 2006; 34: 5312–24; PMID:17012283
- Abanades DR, Ramirez L, Iborra S, Soteriadou K, Gonzalez VM, Bonay P, Alonso C, Soto M. Key role of the 3' untranslated region in the cell cycle regulated expression of the Leishmania infantum histone H2A genes: minor synergistic effect of the 5' untranslated region. BMC Mol Biol 2009; 10: 48; PMID:19460148
- Walrad P, Paterou A, Costa-Serrano A, Matthews KR. Differential trypanosome surface coat regulation by a CCCH protein that co-associates with procyclin mRNA cis-elements. PLoS Pathog 2009; 5: e1000317; PMID:19247446
- David M, Gabdank I, Ben-David M, Zilka A, Orr I, Barash D, Shapira M. Preferential translation of Hsp83 in Leishmania requires a thermosensitive polypyrimidine-rich element in the 3' UTR and involves scanning of the 5' UTR. RNA 2010; 16: 364–74; PMID:20040590
- MacGregor P, Matthews KR. Identification of the regulatory elements controlling the transmission stage-specific gene expression of PAD1 in Trypanosoma brucei. Nucleic Acids Res. 2012; 40: 7705–17; PMID:22684509
- Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11: 113–27; PMID:20094052; http://dx.doi.org/10.1038/nrm2838
- Aitken CE, Lorsch JR. A mechanistic overview of translation initiation in eukaryotes. Nat Struct Mol Biol 2012; 19: 568–76; PMID:22664984; http://dx.doi.org/10.1038/nsmb.2303
- Valasek LS. 'Ribozoomin'–translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr Protein Pept Sci 2012; 13: 305–30; PMID:22708493; http://dx.doi.org/10.2174/138920312801619385
- Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Annu RevBiochem 2014; 83: 779–812; http://dx.doi.org/10.1146/annurev-biochem-060713-035802
- von der Haar T, Gross JD, Wagner G, McCarthy JE. The mRNA cap-binding protein eIF4E in post-transcriptional gene expression. Nat Struct Mol Biol 2004; 11: 503–11; PMID:15164008; http://dx.doi.org/10.1038/nsmb779
- Prevot D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol Cell 2003; 95: 141–56; PMID:12867079
- Gallie DR. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene 1998; 216: 1–11; PMID:9714706; http://dx.doi.org/10.1016/S0378-1119(98)00318-7
- Svitkin YV, Sonenberg N. Translational control by the poly(A) binding protein: a check for mRNA integrity. Mol Biol(Mosk) 2006; 40: 684–93; PMID:16913227
- Gross JD, Moerke NJ, von der HT, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 2003; 115: 739–50; PMID:14675538; http://dx.doi.org/10.1016/S0092-8674(03)00975-9
- Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol Cell 2001; 7: 193–203; PMID:11172724
- Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK. Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure 2006; 14: 913–23; PMID:16698552; http://dx.doi.org/10.1016/j.str.2006.03.012
- Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q Rev Biophys 2004; 37: 197–284; PMID:16194295; http://dx.doi.org/10.1017/S0033583505004026
- Dobrikov MI, Dobrikova EY, Gromeier M. Dynamic regulation of the translation initiation helicase complex by mitogenic signal transduction to eukaryotic translation initiation factor 4G. Mol Cell Biol 2013; 33: 937–46; PMID:23263986
- Villa N, Do A, Hershey JW, Fraser CS. Human Eukaryotic Initiation Factor 4G (eIF4G) Protein Binds to eIF3c, -d, and -e to Promote mRNA Recruitment to the Ribosome. J Biol Chem 2013; 288: 32932–40; PMID:24092755; http://dx.doi.org/10.1074/jbc.M113.517011
- Yanagiya A, Svitkin YV, Shibata S, Mikami S, Imataka H, Sonenberg N. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol Cell Biol 2009; 29: 1661–9; PMID:19114555; http://dx.doi.org/10.1128/MCB.01187-08
- Schutz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A 2008; 105: 9564–9; PMID:18606994;http://dx.doi.org/10.1073/pnas.0800418105
- Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 2009; 136: 447–60; PMID:19203580; http://dx.doi.org/10.1016/j.cell.2009.01.014
- Marintchev A, Wagner G. eIF4G and CBP80 share a common origin and similar domain organization: implications for the structure and function of eIF4G. Biochemistry 2005; 44: 12265–72; PMID:16156639; http://dx.doi.org/10.1021/bi051271v
- Hernandez G, Vazquez-Pianzola P. Functional diversity of the eukaryotic translation initiation factors belonging to eIF4 families. Mech Dev 2005; 122: 865–76; PMID:15922571
- Dhalia R, Reis CR, Freire ER, Rocha PO, Katz R, Muniz JR, Standart N, de Melo Neto OP. Translation initiation in Leishmania major: characterisation of multiple eIF4F subunit homologues. Mol Biochem Parasitol 2005; 140: 23–41; PMID:15694484
- Yoffe Y, Leger M, Zinoviev A, Zuberek J, Darzynkiewicz E, Wagner G, Shapira M. Evolutionary changes in the Leishmania eIF4F complex involve variations in the eIF4E-eIF4G interactions. Nucleic Acids Res 2009; 37: 3243–53; PMID:19321500; http://dx.doi.org/10.1093/nar/gkp190
- Freire ER, Dhalia R, Moura DM, da Costa Lima TD, Lima RP, Reis CR, Hughes K, Figueiredo RC, Standart N, Carrington M, et al. The four trypanosomatid eIF4E homologues fall into two separate groups, with distinct features in primary sequence and biological properties. Mol Biochem Parasitol 2011; 176: 25–36; PMID:21111007; http://dx.doi.org/10.1016/j.molbiopara.2010.11.011
- Zinoviev A, Manor S, Shapira M. Nutritional stress affects an atypical cap-binding protein in Leishmania. RNA Biol 2012; 9:1450-60.
- Jagus R, Bachvaroff TR, Joshi B, Place AR. Diversity of eukaryotic translational initiation factor eIF4E in protists. Comp Funct Genomics 2012; 2012: 134839; PMID:22778692
- Zinoviev A, Shapira M. Evolutionary conservation and diversification of the translation initiation apparatus in trypanosomatids. Comp Funct Genomics 2012; 2012: 813718; PMID:22829751
- Freire ER, Malvezzi AM, Vashisht AA, Zuberek J, Saada EA, Langousis G, Nascimento JD, Moura D, Darzynkiewicz E, Hill K, et al. Trypanosoma brucei translation initiation factor homolog EIF4E6 forms a tripartite cytosolic complex with EIF4G5 and a capping enzyme homolog. Eukaryot Cell 2014; 13: 896–908; PMID:24839125; http://dx.doi.org/10.1128/EC.00071-14
- Freire ER, Vashisht AA, Malvezzi AM, Zuberek J, Langousis G, Saada EA, Nascimento Jde F, Stepinski J, Darzynkiewicz E, Hill K, et al. eIF4F-like complexes formed by cap-binding homolog TbEIF4E5 with TbEIF4G1 or TbEIF4G2 are implicated in post-transcriptional regulation in Trypanosoma brucei. RNA 2014; 20: 1272–86; PMID:24962368
- Chang JH, Cho YH, Sohn SY, Choi JM, Kim A, Kim YC, Jang SK, Cho Y. Crystal structure of the eIF4A-PDCD4 complex. Proc Natl Acad Sci U S A 2009; 106: 3148–53; PMID:19204291; http://dx.doi.org/10.1073/pnas.0808275106
- Liberman N, Dym O, Unger T, Albeck S, Peleg Y, Jacobovitch Y, Branzburg A, Eisenstein M, Marash L, Kimchi A. The crystal structure of the C-terminal DAP5/p97 domain sheds light on the molecular basis for its processing by caspase cleavage. J Mol Biol 2008; 383: 539–48; PMID:18722383; http://dx.doi.org/10.1016/j.jmb.2008.08.013
- Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol 1995; 15: 4990–7; PMID:7651417
- Tarun SZ, Jr., Wells SE, Deardorff JA, Sachs AB. Translation initiation factor eIF4G mediates in vitro poly(A) tail-dependent translation. Proc NatlAcad Sci U S A 1997; 94: 9046–51; PMID:9256432
- Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998; 17: 7480–9; PMID:9857202
- da Costa Lima TD, Moura DM, Reis CR, Vasconcelos JR, Ellis L, Carrington M, Figueiredo RC, de Melo Neto OP. Functional characterization of three leishmania poly(a) binding protein homologues with distinct binding properties to RNA and protein partners. Eukaryot.Cell 2010; 9: 1484–94; PMID:20675580
- Dhalia R, Marinsek N, Reis CR, Katz R, Muniz JR, Standart N, Carrington M, de Melo Neto OP. The two eIF4A helicases in Trypanosoma brucei are functionally distinct. Nucleic Acids Res 2006; 34: 2495–507; PMID:16687655
- Zinoviev A, Leger M, Wagner G, Shapira M. A novel 4E-interacting protein in Leishmania is involved in stage-specific translation pathways. Nucleic Acids Res 2011; 39: 8404–15; PMID:21764780
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol.Cell 1999; 3: 707–16; PMID:10394359
- Fraser CS, Pain VM, Morley SJ. The association of initiation factor 4F with poly(A)-binding protein is enhanced in serum-stimulated Xenopus kidney cells. J Biol Chem 1999; 274: 196–204; PMID:9867830
- Contreras V, Richardson MA, Hao E, Keiper BD. Depletion of the cap-associated isoform of translation factor eIF4G induces germline apoptosis in C. elegans. Cell Death Differ 2008; 15: 1232–42; PMID:18451872
- Hernandez G, Altmann M, Lasko P. Origins and evolution of the mechanisms regulating translation initiation in eukaryotes. Trends Biochem Sci. 2010; 35: 63–73
- Clarkson BK, Gilbert WV, Doudna JA. Functional overlap between eIF4G isoforms in Saccharomyces cerevisiae. PLoSOne 2010; 5: e9114; PMID:20161741
- Castello A, Alvarez E, Carrasco L. Differential cleavage of eIF4GI and eIF4GII in mammalian cells. Effects on translation. J Biol Chem 2006; 281: 33206–16; PMID:16959778; http://dx.doi.org/10.1074/jbc.M604340200
- Gallie DR, Browning KS. eIF4G functionally differs from eIFiso4G in promoting internal initiation, cap-independent translation, and translation of structured mRNAs. J Biol Chem 2001; 276: 36951–60; PMID:11483601; http://dx.doi.org/10.1074/jbc.M103869200
- Lee SH, McCormick F. p97/DAP5 is a ribosome-associated factor that facilitates protein synthesis and cell proliferation by modulating the synthesis of cell cycle proteins. EMBO J 2006; 25: 4008–19; PMID:16932749; http://dx.doi.org/10.1038/sj.emboj.7601268
- Baker CC, Fuller MT. Translational control of meiotic cell cycle progression and spermatid differentiation in male germ cells by a novel eIF4G homolog. Development 2007; 134: 2863–9; PMID:17611220
- Pereira MM, Malvezzi AM, Nascimento LM, da Costa Lima TD, Alves VS, Palma ML, Freire ER, Moura DM, Reis CR, de Melo Neto OP. The eIF4E subunits of two distinct trypanosomatid eIF4F complexes are subjected to differential post-translational modifications associated to distinct growth phases in culture. Mol Biochem Parasitol 2013; 190: 82–6; PMID:23867205; http://dx.doi.org/10.1016/j.molbiopara.2013.06.008
- Urbaniak MD, Martin DM, Ferguson MA. Global quantitative SILAC phosphoproteomics reveals differential phosphorylation is widespread between the procyclic and bloodstream form lifecycle stages of Trypanosoma brucei. J Proteome Res 2013; 12: 2233–44; PMID:23485197; http://dx.doi.org/10.1021/pr400086y
- Cho PF, Poulin F, Cho-Park YA, Cho-Park IB, Chicoine JD, Lasko P, Sonenberg N. A new paradigm for translational control: inhibition via 5'-3' mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell 2005; 121: 411–23; PMID:15882623; http://dx.doi.org/10.1016/j.cell.2005.02.024
- Villaescusa JC, Buratti C, Penkov D, Mathiasen L, Planaguma J, Ferretti E, Blasi F. Cytoplasmic Prep1 interacts with 4EHP inhibiting Hoxb4 translation. PLoS One 2009; 4: e5213; PMID:19365557
- Morales J, Mulner-Lorillon O, Cosson B, Morin E, Belle R, Bradham CA, Beane WS, Cormier P. Translational control genes in the sea urchin genome. Dev Biol 2006; 300: 293–307; PMID:16959243
- Cheng S, Gallie DR. Competitive and noncompetitive binding of eIF4B, eIF4A, and the poly(A) binding protein to wheat translation initiation factor eIFiso4G. Biochemistry 2010; 49: 8251–65; PMID:20795652; http://dx.doi.org/10.1021/bi1008529
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 2009; 4: 363–71; PMID:19247286; http://dx.doi.org/10.1038/nprot.2009.2
- Kelly S, Reed J, Kramer S, Ellis L, Webb H, Sunter J, Salje J, Marinsek N, Gull K, Wickstead B, et al. Functional genomics in Trypanosoma brucei: a collection of vectors for the expression of tagged proteins from endogenous and ectopic gene loci. Mol Biochem Parasitol 2007; 154: 103–9; PMID:17512617
- Wickstead B, Ersfeld K, Gull K. Targeting of a tetracycline-inducible expression system to the transcriptionally silent minichromosomes of Trypanosoma brucei. Mol Biochem Parasitol 2002; 125: 211–6; PMID:12467990
- Sunter J, Wickstead B, Gull K, Carrington M. A new generation of T7 RNA polymerase-independent inducible expression plasmids for Trypanosoma brucei. PLoS One 2012; 7: e35167; PMID:22511983
- Tan R, Frankel AD. Structural variety of arginine-rich RNA-binding peptides. Proc Natl Acad Sci USA 1995; 92: 5282–6; PMID:7777498
- Bastin P, Bagherzadeh Z, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol 1996; 77: 235–9; PMID:8813669
- Wirtz E, Leal S, Ochatt C, Cross GA. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 1999; 99: 89–101
- Poon SK, Peacock L, Gibson W, Gull K, Kelly S. A modular and optimized single marker system for generating Trypanosoma brucei cell lines expressing T7 RNA polymerase and the tetracycline repressor. Open Biol 2012; 2: 110037
- Bangs JD, Uyetake L, Brickman MJ, Balber AE, Boothroyd JC. Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei. Divergent ER retention signals in a lower eukaryote. J Cell Sci 1993; 105( Pt 4): 1101–13
