Abstract
The U2AF heterodimer is generally accepted to play a vital role in defining functional 3′ splice sites in pre-mRNA splicing. Given prevalent mutations in U2AF, particularly in the U2AF1 gene (which encodes for the U2AF35 subunit) in blood disorders and other human cancers, there are renewed interests in these classic splicing factors to further understand their regulatory functions in RNA metabolism in both physiological and disease settings. We recently reported that U2AF has a maximal capacity to directly bind ˜88% of functional 3′ splice sites in the human genome and that numerous U2AF binding events also occur in various exonic and intronic locations, thus providing additional mechanisms for the regulation of alternative splicing besides their traditional role in titrating weak splice sites in the cell. These findings, coupled with the existence of multiple related proteins to both U2AF65 and U2AF35, beg a series of questions on the universal role of U2AF in functional 3′ splice site definition, their binding specificities in vivo, potential mechanisms to bypass their requirement for certain intron removal events, contribution of splicing-independent functions of U2AF to important cellular functions, and the mechanism for U2AF mutations to invoke specific diseases in humans.
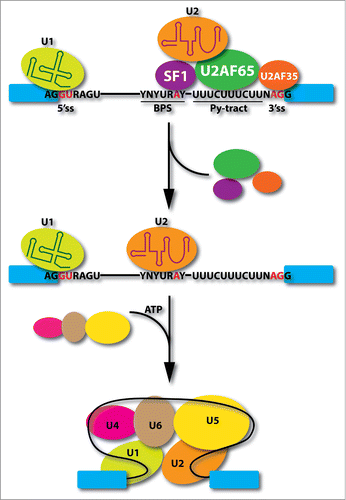
Pre-mRNA splicing takes place in a large RNA processing machine known as the spliceosome, which consists of U1, U2, and U4/U6.U5 small nuclear ribonucleoprotein particles (snRNPs).Citation1 In addition to the removal of the vast majority of introns by this machinery in mammals, there also exists of minor class of introns, which are excised by the minor spliceosome consisting of U11, U12, U4atac/U6atac, and the shared U5 with the major spliceosome.Citation2 According to the textbook version for the spliceosome assembly pathway in early steps (), U1 defines the functional 5′ splice site (5′ss) whereas U2 recognizes the functional 3′ splice site (3′ss), both via base pairing with specific splicing signals at the ends of the intron. Because the branchpoint sequence (BPS) as part of the functional 3′ss is quite degenerate in higher eukaryotic cells, the addition of U2 snRNP requires multiple auxiliary factors, the most important one being the U2AF heterodimer consisting of a 65kD and a 35kD subunit, which binds the polypyrimidine tract (Py-tract) immediate downstream of the BPS and contacts the AG dinucleotide, respectively. Following a series of ATP-dependent steps, the U4/U6.U5 tri-snRNP complex joins the initial pre-spliceosome to convert it into the mature spliceosome. The U2AF heterodimer is one of the best-characterized splicing factors in higher eukaryotic cells. Numerous studies have been done about its function in defining functional 3' splice sites (3′ss) as if no room has left for further investigation. However, this is clearly not the case. This Point-of-View commentary focuses on some outstanding questions on the function of U2AF in both physiological and disease states in the context of our recently published genomic analysis of U2AF.Citation3
Figure 1. Critical events in early spliceosome assembly steps. U1 snRNP interacts with the functional 5' splice site (5'ss), SF1 recognizes branchpoint sequence (BPS), and U2 snRNP auxiliary factors bind the polypyrimidine tract (Py-tract) between the BPS and AG dinucleotide. After U2 snRNP binding at the 3' splice site (3'ss), SF1 and U2AF heterodimer detach from the spliceosome followed by the joining of the U4/U6.U5 tri-snRNPs in an ATP-dependent manner to convert pre-spliceosome to mature spliceosome.
U2AF: A conserved auxiliary factor vital for 3′ss recognition
U2AF was originally identified as a splicing factor required for facilitating U2 snRNP's interaction with BPS in Michael Green's laboratory.Citation4 In follow-up studies, they found U2AF consists of 65kD and 35kD subunits, and intriguingly, the initial evidence based on in vitro depletion and reconstitution suggested that U2AF65 was both necessary and sufficient for spliceosome assembly and the splicing reaction.Citation5 U2AF65's homologs were identified in other organisms and shown to be required for both in vivo splicing and survival in fission yeast and Drosophila.Citation6,7 The involvement of U2AF65 in alternative splicing was first characterized by Valcarcel et al on the Sxl gene model, observing its ability to bind and modulate the polypyrimidine tract,Citation8 which was quickly substantiated in a series of studies.Citation9-12
The functional importance of U2AF35 was subsequently appreciated by studies in Drosophila and C. elegans where it was found to be essential for survival.Citation13,14 In fission yeast Schizosaccharomyces pombe, the U2AF35 homolog U2AFsm could be functionally substituted by human U2AF35 (ref.15), underscoring both structural and functional conservation of U2AF35 from fission yeast to humans. The apparent dispensable role of U2AF35 in in vitro splicing was re-visited by Zuo et al., finding that both subunits were required for efficient splicing, and interestingly, one subunit could enhance the splicing activity of the other in U2AF-depleted nuclear extracts.Citation16 Using site-specific crosslinking, Wu et al. demonstrated the ability of U2AF35 in direct contact with the 3′ss AG dinucleotide.Citation17 This finding emphasizes the cooperation between the large and small U2AF subunits in 3'ss recognition, especially on introns associated with weak polypyrimidine tract,Citation17-19 which is in line with their requirement for viability in Drosophila.Citation20
A universal role of U2AF in 3′ss definition?
One of the remaining questions on the function of U2AF is whether it is responsible for defining all functional 3' splice sites in mammalian genomes. In fact, this question has been partially addressed by the observation that U2AF is not required for splicing of the U12-type of introns,Citation21 and thus, the question is narrowed down to whether U2AF is required for splicing of all introns in the major U2 class, which may further be divided into 2 related questions. The first is whether U2AF has the capacity to directly bind 3'ss of all U2-type introns and the second is whether U2AF is functionally required for the removal of these introns.
We have some clues to the first question, as the data from both fission yeast and mammalian cells indicate that not all introns contain a classic Py-tract as part of the functional 3′ss in U2-type introns. Interestingly, in fission yeast, splicing of those introns with atypical Py-tracts appears to depend on sequences near the 5′ss in the intron, but excision of those introns seems to still require the function of U2AF.Citation22 In HeLa cells, our recent maximal likelihood analysis indicates that ˜12% introns may not be directly bound by U2AF.Citation3 Therefore, U2AF may not be able to directly bind on a fraction of U2-type introns in higher eukaryotic cells, but importantly, binding may be decoupled with functional requirement at least in certain cases.
Bypassing the requirement for U2AF binding versus function
In principle, U2AF may not be able to directly bind some functional 3′ss, but still act on those sites through the recruitment by some other factors. A recent study showed that the apparently weak 3′ss of alternative CD44 ex5 contains CAUC motifs between the BPS and 3'AG, which have been characterized as the binding site for YB-1 to help recruit U2AF.Citation23 Therefore, the lack of binding is not equivalent to the lack of functional requirement, and it is quite challenging at this point to address the question of the functional requirement for U2AF in the genome.
In unicellular organisms, such as budding yeast, there is a U2AF65 homolog called Mud2, but Mud2 is a non-essential gene, which may be due to the fact that the splicing signals in budding yeast are largely invariant.Citation24 However, no one has been able to design a perfect pre-mRNA to splice without U2AF in mammalian cells or their nuclear extracts. The only exception is the splicing reaction in U2AF-depleted nuclear extracts supplemented with an excessive amount of SR proteins.Citation25 However, it has been unclear whether SR proteins merely dramatically reduce the required threshold for U2AF to an undetectable level or are able to truly bypass it. One way to test the idea is to determine whether U2AF is still present in purified pre-spliceosome from biochemically depleted extracts supplemented with SR proteins.
The strongest evidence for some U2AF-independent introns to date is from genetic analysis in fission yeast by characterizing a conditional (ts) U2AF mutant on global splicing, which revealed ˜8% U2AF-insensitive introns.Citation26 Again, until the ts mutant is proven to be an absolute functional null mutation at the restrictive temperature, one cannot rule out the possibility that some of those apparent U2AF-insenstive introns may merely have much reduced dependence on U2AF. Therefore, the question has remained, which may be pursued by using biochemical approaches employed to demonstrate U2AF-independent splicing of the U12 type of introns.Citation21 Analysis of those introns may reveal alternative mechanisms for 3′ss definition in higher eukaryotic cells, which has been showcased by the demonstrated requirement for Urp, an U2AF35-related protein,Citation27 in splicing U12 introns.Citation28
Roles of U2AF related proteins
Eukaryotic cells often evolve additional pathways by using gene family members with related, yet distinct functions. This also applies to U2AF because of the existence of multiple proteins with related structures to both U2AF65 and U2AF35 (). There are 3 factors with structural similarities to U2AF65 and U2AF35, respectively.Citation29 The U2AF65 related protein PUF60 appears to have similar binding specificity for the Py-tract; however, the existing evidence suggests that it still functions in conjunction with, rather than independent of, the U2AF heterodimer in in vitro splicing.Citation30 Whether PUF60 has a distinct RNA binding profile from U2AF65, especially among those that do not enlist U2AF65 to splice, awaits future studies. RBM39 (a.k.a. HCC1 or CAPERα) and RBM23 (a.k.a. CAPERβ) are highly related to U2AF65, and interestingly, besides their roles in splicing, these RNA binding proteins are also involved in transcriptional control,Citation31,32 which is important to keep in mind in understanding the in vivo function of U2AF and its family members (see below). Systematic investigation of the RNA maps and their potential overlapping and distinct functions in splicing will be an important subject for future studies.
Figure 2. Domain structures of human U2AF-related proteins. (A), Human U2AF65 (NCBI Accession No. NP_009210) and its paralogues RBM39/CAPERα (NP_004893), RBM23/CAPERβ (NP_060577) and PUF60 (NP_001258027). (B), Human U2AF35 (NP_006749) and its paralogues U2AF26 (NP_659424), U2AF35R1 (Q15695) and Urp/U2AF35R2 (NP_005080). RRM, RNA recognition motif (blue); UHM, U2AF homology motif (green); RS, arginine-serine-rich (deep pink); Zn, zinc knuckle (gold).
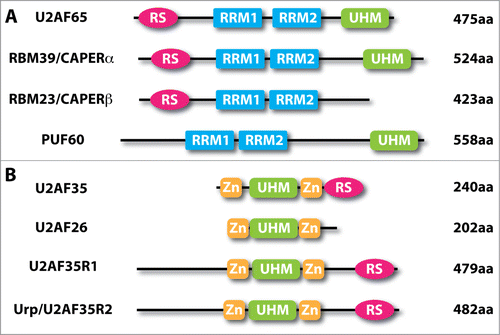
The U2AF35-related proteins are also interesting. U2AF26 appears to have related functions to U2AF35 in splicing.Citation33 Urp, also known as ZRSR2 encoded by the U2AF1L2 gene,Citation34 on the other hand, has been demonstrated to be critical for splicing of U12-type of introns, even though it has the capacity to interact with U2AF65 and appears to be required for the U2-type of introns in the second step of the splicing reaction.Citation27,28 The U2AF1L1 gene encodes for the third family member of U2AF35-related proteins, but little is known about its function in splicing or other gene expression pathways. It will be interesting to determine their prospective partnership of these U2AF35-related molecules with U2AF65, U2AF65-related proteins, and perhaps other RNA binding proteins to understand their potential contributions to constitutive and regulated splicing.
Genomic landscape of U2AF binding and regulation
The U2AF heterodimer binds RNA mainly through the U2AF65 subunit, whose preference for pyrimidine-rich sequences has been extensively demonstrated by a series of biochemical studies.Citation35,36 Its in vivo binding profile determined by UV-induced crosslinking coupled with deep sequencing (CLIP-seq) has confirmed such binding specificity on functional 3′ss.Citation3,37 The question is why it has such exceptional capacity to avoid many related sequences abundantly present in both exonic and intronic regions of many pre-mRNAs. Early studies suggest that DEKCitation38 and hnRNP A1Citation39 may help orient U2AF on authentic 3'ss that contains a flanking AG dinucleotide; however, it has remained to be determined how extensively these mechanisms are utilized to prevent U2AF binding elsewhere in mammalian genomes. In fact, 2 published genome-wide analysis of U2AF65 showed numerous binding events beyond functional 3′ss.Citation3,37 Still, there are just as many pyrimidine-rich sequences without evidence for U2AF binding.
In principle, the RNA binding specificity of a given RNA binding protein may be influenced by both cooperative and antagonist proteins besides their intrinsic RNA binding preference. The former may enhance the binding specificity whereas the latter may prevent binding on certain locations. This principle has been demonstrated with U2AF35, which appears to guide U2AF65 to the Py-tract flanked by the AG dinucleotide.Citation17,40 As spliceosome assembly involves extensive protein-protein interaction networks across the exon and then across the intron, we may further envision that such interaction networks may help confine U2AF to its intended target sites in mammalian genomes. These positive enhancement mechanisms may be further enforced by other RNA binding proteins that also show strong preference for pyrimidine-rich sequences, such as hnRNP C, TIA-1/TIAR, and PTB,Citation37,41-43 which many effectively compete U2AF off on many non-functional 3′ss. A more recent study revealed a critical role of hnRNP C, which appear to act to prevent accidental activation of potential 3′ss associated with a set of expressed Alu repeats in the human genome.Citation37 Therefore, both positive and negative enforcements may together provide critical regulatory mechanisms for U2AF to interact with a unique set of functional RNA elements in mammalian genomes.
Distinct mechanisms for U2AF to participate in regulated splicing
Alternative splicing is widespread in higher eukaryotic cells, which in many cases take advantage of evolutionarily conserved weak splice sites in which U2AF has been a key target for modulation by other RNA binding splicing regulators.Citation19,44 However, recent bioinformatics analyses and genomic mapping studies suggest additional mechanisms for more active involvement of U2AF in regulated splicing. For example, various potential U2AF binding motifs were found in some exonic regions. By engineering those motifs in model genes, it has been demonstrated that U2AF binding in exons cause exon skipping.Citation45,46
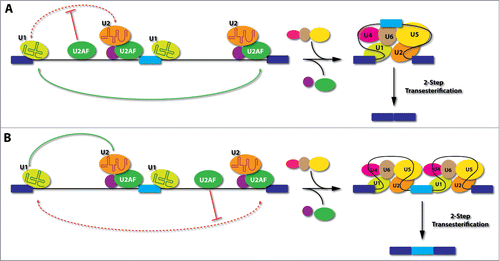
In addition to such interference on exons, recent genomic mapping analysis also revealed extensive U2AF binding in many intronic locations.Citation3,37 Our mechanistic studies further showed that such intronic U2AF binding events have position-dependent effects on the splicing outcomes (), a general trend for nearly all splicing regulators characterized to date.Citation47 When U2AF binds to an intronic location upstream of the alternative exon, it tends to cause exon skipping, which likely results from interference with the communication between the splicing complexes assembled on the upstream 5'ss and those on the 3'ss of the alternative exon. On the other hand, if U2AF binds to an intronic location downstream of the alternative exon, it appears to interfere with the recognition or pairing of the downstream 3'ss, thus offering certain advantage for the upstream competing 3'ss associated with the alternative exon to engage in productive recognition or pairing interactions, thereby causing increased inclusion of the alternative exon.
Figure 3. Model for positional-dependent effects of U2AF in the regulation of alternative splicing. (A), If U2AF binds to an intronic location upstream of the alternative exon, it will interfere with the communication between the splicing complexes assembled on the upstream 5'ss and those formed on the 3'ss of the alternative exon (upstream 3'ss). This would provide competitive advantage in selecting the 3'ss of the downstream intron (downstream 3'ss), thus causing skipping of the alternative exon. (B), If U2AF binds to an intronic location downstream of the alternative exon, it will interfere with the recognition or pairing of the downstream 3'ss, thus offering certain advantage for the upstream 3'ss to engage in productive communication with splicing complexes assembled on the upstream 5'ss. This would favor the inclusion of the alternative exon. Light blue: alternative exon; Dark blue: constitutive exons; Red dashed lines: interference of splice site pairing; Green lines: preferred pairing.
U2AF in disease
For many of us conducting basic research, we love to see the connection of our favorable genes to some human diseases to justify funding from NIH, and contribution of our research to the continuous battle against incurable disease, such as cancer. This is also the case for U2AF, as multiple mutations in both U2AF65 and U2AF35 have been reported to associate with blood disorders, particularly the Myelodysplasia Syndrome (MDS).Citation48 The current battleground is to understand how such mutations may induce specific disease pathways. The obvious possibility is the compromised function of U2AF in splicing regulation. Interestingly, our recent results indicate that the mutations in U2AF35 (which is encoded by the U2AF1 gene) cause widespread changes in alternative splicing, which is linked to decreased cell proliferation, consistent with the disease phenotype in MDS.Citation3 In contrast, we were unable to detect any measurable defect with mutations in U2AF65. Together, these observations raise a possibility that the mutations in U2AF35 may drive the disease whereas those in U2AF65 may be passenger mutations.
While it is quite logical to hypothesize disease mechanisms in the splicing pathway by mutations in specific splicing factors, we may be side tracked by this assumption, because accumulating evidence suggests that many RNA binding splicing regulators have splicing independent functions. This is best exemplified by U1 splicing-independent functions in cryptic transcription terminationCitation49 and by the roles of SR proteins in transcription initiation and elongation.Citation50,51 In fact, U2AF has also been reported to have other functions, such as mRNA export, by binding intronless transcripts in Drosophila Citation52 and certain spliced mRNAs in mammalian cells.Citation48,53 Given the tight coupling between transcription and co-transcriptional RNA processing in the nucleus of mammalian cells, we should not be surprised if U2AF were also found to have a role in transcriptional control. This and other questions are certainly interesting directions to be pursued in future studies.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Our work on U2AF has been supported by NIH grants (GM049369, HG004659 and DK098808).
References
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell 2009; 136:701-18; PMID:19239890; http://dx.doi.org/10.1016/j.cell.2009.02.009
- Will CL, Luhrmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem 2005; 386:713-24; PMID:16201866; http://dx.doi.org/10.1515/BC.2005.084
- Shao C, Yang B, Wu T, Huang J, Tang P, Zhou Y, Zhou J, Qiu J, Jiang L, Li H, et al. Mechanisms for U2AF to define 3′ splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol 2014; 21:997-1005; PMID:25326705; http://dx.doi.org/10.1038/nsmb.2906
- Ruskin B, Zamore PD, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell 1988; 52:207-19; PMID:2963698; http://dx.doi.org/10.1016/0092-8674(88)90509-0
- Zamore PD, Green MR. Biochemical characterization of U2 snRNP auxiliary factor: an essential pre-mRNA splicing factor with a novel intranuclear distribution. EMBO J 1991; 10:207-14; PMID:1824937
- Potashkin J, Naik K, Wentz-Hunter K. U2AF homolog required for splicing in vivo. Science 1993; 262:573-5; PMID:8211184; http://dx.doi.org/10.1126/science.8211184
- Kanaar R, Roche SE, Beall EL, Green MR, Rio DC. The conserved pre-mRNA splicing factor U2AF from Drosophila: requirement for viability. Science 1993; 262:569-73; PMID:7692602; http://dx.doi.org/10.1126/science.7692602
- Valcarcel J, Singh R, Zamore PD, Green MR. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 1993; 362:171-5; PMID:7680770; http://dx.doi.org/10.1038/362171a0
- Sharma S, Falick AM, Black DL. Polypyrimidine tract binding protein blocks the 5' splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol Cell 2005; 19:485-96; PMID:16109373; http://dx.doi.org/10.1016/j.molcel.2005.07.014
- Sauliere J, Sureau A, Expert-Bezancon A, Marie J. The polypyrimidine tract binding protein (PTB) represses splicing of exon 6B from the beta-tropomyosin pre-mRNA by directly interfering with the binding of the U2AF65 subunit. Mol Cell Biol 2006; 26:8755-69; PMID:16982681; http://dx.doi.org/10.1128/MCB.00893-06
- Martins de Araujo M, Bonnal S, Hastings ML, Krainer AR, Valcarcel J. Differential 3' splice site recognition of SMN1 and SMN2 transcripts by U2AF and U2 snRNP. RNA 2009; 15:515-23; PMID:19244360; http://dx.doi.org/10.1261/rna.1273209
- Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc Natl Acad Sci U S A 2009; 106:9203-8; PMID:19470458; http://dx.doi.org/10.1073/pnas.0900342106
- Rudner DZ, Kanaar R, Breger KS, Rio DC. Mutations in the small subunit of the Drosophila U2AF splicing factor cause lethality and developmental defects. Proc Natl Acad Sci U S A 1996; 93:10333-7; PMID:8816800; http://dx.doi.org/10.1073/pnas.93.19.10333
- Zorio DA, Blumenthal T. U2AF35 is encoded by an essential gene clustered in an operon with RRM/cyclophilin in Caenorhabditis elegans. RNA 1999; 5:487-94; PMID:10199565; http://dx.doi.org/10.1017/S1355838299982225
- Webb CJ, Wise JA. The splicing factor U2AF small subunit is functionally conserved between fission yeast and humans. Mol Cell Biol 2004; 24:4229-40; PMID:15121844; http://dx.doi.org/10.1128/MCB.24.10.4229-4240.2004
- Zuo P, Maniatis T. The splicing factor U2AF35 mediates critical protein-protein interactions in constitutive and enhancer-dependent splicing. Genes Dev 1996; 10:1356-68; PMID:8647433; http://dx.doi.org/10.1101/gad.10.11.1356
- Wu S, Romfo CM, Nilsen TW, Green MR. Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature 1999; 402:832-5; PMID:10617206; http://dx.doi.org/10.1038/45996
- Guth S, Martinez C, Gaur RK, Valcarcel J. Evidence for substrate-specific requirement of the splicing factor U2AF(35) and for its function after polypyrimidine tract recognition by U2AF(65). Mol Cell Biol 1999; 19:8263-71; PMID:10567551
- Pacheco TR, Coelho MB, Desterro JM, Mollet I, Carmo-Fonseca M. In vivo requirement of the small subunit of U2AF for recognition of a weak 3' splice site. Mol Cell Biol 2006; 26:8183-90; PMID:16940179; http://dx.doi.org/10.1128/MCB.00350-06
- Rudner DZ, Kanaar R, Breger KS, Rio DC. Interaction between subunits of heterodimeric splicing factor U2AF is essential in vivo. Mol Cell Biol 1998; 18:1765-73; PMID:9528748
- Shen H, Green MR. RS domain-splicing signal interactions in splicing of U12-type and U2-type introns. Nat Struct Mol Biol 2007; 14:597-603; PMID:17603499; http://dx.doi.org/10.1038/nsmb1263
- Sridharan V, Singh R. A conditional role of U2AF in splicing of introns with unconventional polypyrimidine tracts. Mol Cell Biol 2007; 27:7334-44; PMID:17709389; http://dx.doi.org/10.1128/MCB.00627-07
- Wei WJ, Mu SR, Heiner M, Fu X, Cao LJ, Gong XF, Bindereif A, Hui J. YB-1 binds to CAUC motifs and stimulates exon inclusion by enhancing the recruitment of U2AF to weak polypyrimidine tracts. Nucleic Acids Res 2012; 40:8622-36; PMID:22730292; http://dx.doi.org/10.1093/nar/gks579
- Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev 1994; 8:843-54; PMID:7926772; http://dx.doi.org/10.1101/gad.8.7.843
- MacMillan AM, McCaw PS, Crispino JD, Sharp PA. SC35-mediated reconstitution of splicing in U2AF-depleted nuclear extract. Proc Natl Acad Sci U S A 1997; 94:133-6; PMID:8990173; http://dx.doi.org/10.1073/pnas.94.1.133
- Sridharan V, Heimiller J, Singh R. Genomic mRNA profiling reveals compensatory mechanisms for the requirement of the essential splicing factor U2AF. Mol Cell Biol 2011; 31:652-61; PMID:21149581; http://dx.doi.org/10.1128/MCB.01000-10
- Tronchere H, Wang J, Fu XD. A protein related to splicing factor U2AF35 that interacts with U2AF65 and SR proteins in splicing of pre-mRNA. Nature 1997; 388:397-400; PMID:9237760; http://dx.doi.org/10.1038/41137
- Shen H, Zheng X, Luecke S, Green MR. The U2AF35-related protein Urp contacts the 3' splice site to promote U12-type intron splicing and the second step of U2-type intron splicing. Genes Dev 2010; 24:2389-94; PMID:21041408; http://dx.doi.org/10.1101/gad.1974810
- Mollet I, Barbosa-Morais NL, Andrade J, Carmo-Fonseca M. Diversity of human U2AF splicing factors. FEBS J 2006; 273:4807-16; PMID:17042780; http://dx.doi.org/10.1111/j.1742-4658.2006.05502.x
- Page-McCaw PS, Amonlirdviman K, Sharp PA. PUF60: a novel U2AF65-related splicing activity. RNA 1999; 5:1548-60; PMID:10606266; http://dx.doi.org/10.1017/S1355838299991938
- Jung DJ, Na SY, Na DS, Lee JW. Molecular cloning and characterization of CAPER, a novel coactivator of activating protein-1 and estrogen receptors. J Biol Chem 2002; 277:1229-34; PMID:11704680; http://dx.doi.org/10.1074/jbc.M110417200
- Dowhan DH, Hong EP, Auboeuf D, Dennis AP, Wilson MM, Berget SM, O'Malley BW. Steroid hormone receptor coactivation and alternative RNA splicing by U2AF65-related proteins CAPERalpha and CAPERbeta. Mol Cell 2005; 17:429-39; PMID:15694343; http://dx.doi.org/10.1016/j.molcel.2004.12.025
- Shepard J, Reick M, Olson S, Graveley BR. Characterization of U2AF(6), a splicing factor related to U2AF(35). Mol Cell Biol 2002; 22:221-30; PMID:11739736; http://dx.doi.org/10.1128/MCB.22.1.221-230.2002
- Kitagawa K, Wang X, Hatada I, Yamaoka T, Nojima H, Inazawa J, Abe T, Mitsuya K, Oshimura M, Murata A. Isolation and mapping of human homologues of an imprinted mouse gene U2af1-rs1. Genomics 1995; 30:257-63; PMID:8586425; http://dx.doi.org/10.1006/geno.1995.9879
- Singh R, Valcarcel J, Green MR. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 1995; 268:1173-6; PMID:7761834; http://dx.doi.org/10.1126/science.7761834
- Valcarcel J, Gaur RK, Singh R, Green MR. Interaction of U2AF65 RS region with pre-mRNA branch point and promotion of base pairing with U2 snRNA [corrected]. Science 1996; 273:1706-9; PMID:8781232
- Zarnack K, Konig J, Tajnik M, Martincorena I, Eustermann S, Stevant I, Reyes A, Anders S, Luscombe NM, Ule J. Direct competition between hnRNP C and U2AF65 protects the transcriptome from the exonization of Alu elements. Cell 2013; 152:453-66; PMID:23374342; http://dx.doi.org/10.1016/j.cell.2012.12.023
- Soares LM, Zanier K, Mackereth C, Sattler M, Valcarcel J. Intron removal requires proofreading of U2AF/3' splice site recognition by DEK. Science 2006; 312:1961-5; PMID:16809543; http://dx.doi.org/10.1126/science.1128659
- Tavanez JP, Madl T, Kooshapur H, Sattler M, Valcarcel J. hnRNP A1 proofreads 3' splice site recognition by U2AF. Mol Cell 2012; 45:314-29; PMID:22325350; http://dx.doi.org/10.1016/j.molcel.2011.11.033
- Merendino L, Guth S, Bilbao D, Martinez C, Valcarcel J. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3' splice site AG. Nature 1999; 402:838-41; PMID:10617208; http://dx.doi.org/10.1038/45602
- Le Guiner C, Lejeune F, Galiana D, Kister L, Breathnach R, Stevenin J, Del Gatto-Konczak F. TIA-1 and TIAR activate splicing of alternative exons with weak 5' splice sites followed by a U-rich stretch on their own pre-mRNAs. J Biol Chem 2001; 276:40638-46; PMID:11514562; http://dx.doi.org/10.1074/jbc.M105642200
- Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Rot G, Zupan B, Curk T, Ule J. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol 2010; 8:e1000530; PMID:21048981; http://dx.doi.org/10.1371/journal.pbio.1000530
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell 2009; 36:996-1006; PMID:20064465; http://dx.doi.org/10.1016/j.molcel.2009.12.003
- Sharma S, Kohlstaedt LA, Damianov A, Rio DC, Black DL. Polypyrimidine tract binding protein controls the transition from exon definition to an intron defined spliceosome. Nat Struct Mol Biol 2008; 15:183-91; PMID:18193060; http://dx.doi.org/10.1038/nsmb.1375
- Lim KH, Ferraris L, Filloux ME, Raphael BJ, Fairbrother WG. Using positional distribution to identify splicing elements and predict pre-mRNA processing defects in human genes. Proc Natl Acad Sci U S A 2011; 108:11093-8; PMID:21685335; http://dx.doi.org/10.1073/pnas.1101135108
- Erkelenz S, Mueller WF, Evans MS, Busch A, Schoneweis K, Hertel KJ, Schaal H. Position-dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA 2013; 19:96-102; PMID:23175589; http://dx.doi.org/10.1261/rna.037044.112
- Fu XD, Ares M, Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 2014; 15:689-701; PMID:25112293; http://dx.doi.org/10.1038/nrg3778
- Cazzola M, Della Porta MG, Malcovati L. The genetic basis of myelodysplasia and its clinical relevance. Blood 2013; 122:4021-34; PMID:24136165; http://dx.doi.org/10.1182/blood-2013-09-381665
- Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, Dreyfuss G. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 2010; 468:664-8; PMID:20881964; http://dx.doi.org/10.1038/nature09479
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 2008; 15:819-26; PMID:18641664; http://dx.doi.org/10.1038/nsmb.1461
- Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 2013; 153:855-68; PMID:23663783; http://dx.doi.org/10.1016/j.cell.2013.04.028
- Blanchette M, Labourier E, Green RE, Brenner SE, Rio DC. Genome-wide analysis reveals an unexpected function for the Drosophila splicing factor U2AF50 in the nuclear export of intronless mRNAs. Mol Cell 2004; 14:775-86; PMID:15200955; http://dx.doi.org/10.1016/j.molcel.2004.06.012
- Gama-Carvalho M, Barbosa-Morais NL, Brodsky AS, Silver PA, Carmo-Fonseca M. Genome-wide identification of functionally distinct subsets of cellular mRNAs associated with two nucleocytoplasmic-shuttling mammalian splicing factors. Genome Biol 2006; 7:R113; PMID:17137510; http://dx.doi.org/10.1186/gb-2006-7-11-r113
