Abstract
Exposure to oxygen and light generates photooxidative stress by the bacteriochlorophyll a mediated formation of singlet oxygen (1O2) in the facultative photosynthetic bacterium Rhodobacter sphaeroides. We have identified SorY as an sRNA, which is induced under several stress conditions and confers increased resistance against 1O2. SorY by direct interaction affects the takP mRNA, encoding a TRAP-T transporter. We present a model in which SorY reduces the metabolite flux into the tricarboxylic acid cycle (TCA cycle) by reducing malate import through TakP. It was previously shown that oxidative stress in bacteria leads to switch from glycolysis to the pentose phosphate pathway and to reduced activity of the TCA cycle. As a consequence the production of the prooxidant NADH is reduced and production of the protective NADPH is enhanced. In R. sphaeroides enzymes for glycolysis, pentose phosphate pathway, Entner–Doudoroff pathway and gluconeogenesis are induced in response to 1O2 by the alternative sigma factor RpoHII. The same is true for the sRNA SorY. By limiting malate import SorY thus contributes to the balance of the metabolic fluxes under photooxidative stress conditions. This assigns a so far unknown function to an sRNA in oxidative stress response.
Introduction
Microorganisms have to cope with oxidative stress in almost all environments and have therefore evolved sophisticated defense systems. A special type of oxidative stress is the so-called photooxidative stress, which occurs in the presence of light, oxygen and a photosensitizer. In photosynthetic organisms (bacterio-) chlorophylls act as photosensitizers, which transfer energy to ground state triplet oxygen generating highly reactive 1O2. The response to photooxidative stress has been intensively investigated in the facultative photosynthetic alpa-proteobacterium Rhodobacter sphaeroides (reviewed inCitation1,2). Rhodobacter uses oxygen for aerobic respiration but induces the formation of photosynthetic complexes when oxygen concentration decreases. The presence of blue light counteracts the formation of photosynthetic complexes at intermediate oxygen concentrations most likely in order to prevent the formation of 1O2.Citation3-5 1O2 triggers a complex response at transcriptional and post-transcriptional level.Citation6-9 A main regulator of this response is the alternative sigma factor RpoE, which directly activates a small subset of genes including the rpoHII gene. The RpoHII sigma factor subsequently induces a large subset of genes, many of them with known functions in the oxidative stress response.Citation6,10 Under non-stress conditions RpoE is inactivated by the anti-sigma factor ChrR,Citation11 which is proteolytically degraded upon 1O2 stress.Citation12,13 Many genes which are targeted by RpoHII are also targeted by RpoHI, which is independent of RpoE and has a main role in the heat shock response.Citation14,15 1O2 does not only induce transcription of protein coding RNAs in R. sphaeroides but also of small non-coding RNAs (sRNAs).Citation9,16-18 sRNAs have been identified as components of many bacterial regulatory networks and have important roles in the fine-tuning of adaptation processes. Trans-acting sRNAs target mRNAs which are transcribed from other regions of their genome and affect their translation and/or stability.Citation19 The RNA chaperon Hfq is often required for the interaction of trans-acting sRNAs and their target mRNAs.Citation20-22
The sRNA RSs1543 of R. sphaeroides is induced in response to photooxidative stress.Citation9 Here we show that RSs1543 responds to a variety of stresses and influences photooxidative stress resistance of R. sphaeroides. Therefore RSs1543 was renamed SorY for 1O2 resistance RNA Y. We identified the takP mRNA, that encodes the extracytoplasmic receptor subunit of a TRAP-T transporter, as direct target of SorY and provide a model for the role of this sRNA in the photooxidative stress response. According to the model SorY regulates TakP synthesis negatively during photooxidative stress to decrease harmful malate influx at high rates into the cell to favor the pathway shunt from the tricarboxylic acid cycle (TCA cycle) to the pentose phosphate pathway and Entner–Doudoroff pathway.
Results
SorY is induced by different stress factors
SorY was originally identified as an sRNA induced by 1O2 in Rhodobacter sphaeroides.Citation9 The sorY gene is preceded by an RpoHI/RpoHII promoter consensus motif and a transcriptional terminator structure is localized at the 3´end.Citation15 Both sigma factors can activate sorY transcription but at least one of them is required for 1O2-dependent induction. Rhodobacter genes under control of RpoHI/RpoHII promoters often respond to multiple stresses.Citation15,23 We therefore tested the expression of SorY under several conditions in the wild type strain. Indeed SorY is also induced by the organic hydroperoxide tert-butyl hydroperoxide (tBOOH), hydrogen peroxide, the super oxide generating paraquat, by heat, by diamide, by NaCl, by CdCl2 and by SDS/EDTA (). There was no effect of ethanol or polymyxin-B, which alters membrane structures. Thus, SorY is indeed induced by various stresses, but strongest induction was observed in response to singlet oxygen.
Figure 1. Induction of the small RNA SorY under various stress conditions shown by northern blot analysis of total RNA isolated from R. sphaeroides. SorY fold changes (FC) are normalized to the 5S rRNA. The quantification was done with Quantity One Software (Bio-Rad). (A) The following reagents (and their final concentrations) were added to aerobically grown cultures at OD600 of 0.4 and samples were taken immediately before (time 0) or 10 min after addition: 0.2 µM methylene blue in the presence of 800 Wm−2 white light, 300 µM tBOOH, 1 mM H2O2 and 250 µM paraquat were added to aerobically grown cultures at OD 0.4 and samples were taken before and 10 min after the addition. (B) Semiaerobic cultures were shifted to 42°C at time 0 or the following reagents were added: 500 µM diamide, 500 mM NaCl, 10 µM CdCl2, 2.5 % ethanol, 1 µg/mL polymyxin-B or 0.005 % SDS along with 1 mM EDTA. (C) Altered SorY transcript levels in the overexpression strains and the knockout mutant (2.4.1ΔSorY) before and after 1O2 stress in comparison to the wild-type strain harboring the empty vector (2.4.1pBBR). SorY transcription in plasmid born overexpression strains is either constitutive due to the use of a 16S rRNA promoter (2.4.1pBBRSorY) or inducible due to the RpoHI/II dependent promoter of SorY (2.4.1pBBRSorYi).
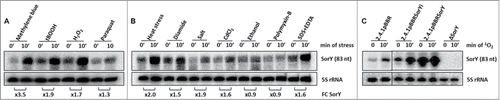
To learn more about the function of SorY we constructed a strain lacking this sRNA (ΔSorY) and strains overexpressing the sRNA from its own promoter (2.4.1pBBRSorYi) or from the strong, constitutive 16S rRNA promoter (2.4.1pBBRSorY). The northern blot shown in verifies the lack of SorY in the deletion mutant and its strong overexpression in the other strains, which depends on 1O2, when the native promoter is present (2.4.1pBBRSorYi) but is very high even in absence of any stress in 2.4.1pBBRSorY.
Prediction and validation of targets for SorY
Many sRNAs act on gene expression by base pairing to target mRNAs and influencing either the stability of these targets or the rate of translation or both. To get a hint on possible targets of SorY we compared the transcriptomes of a control strain harbouring an empty vector (2.4.1pBBR) and of the overexpressing strain (2.4.1pBBRSorYi) after 10 min of 1O2 stress by microarray analysis (Fig. S1). From 3976 genes with a reliable A-value 17 protein coding genes showed a log2 fold change of > 0.6 or < −0.6 between the 2 strains (). Putative interaction of the mRNAs transcribed from these genes and SorY were predicted by the IntaRNA tool.Citation24 shows the predicted parameters of genes with expression changes for which an energy value of less than −10 kcal/mol was calculated for a putative interaction. The effect of SorY on mRNA levels was confirmed by real time RT-PCR for RSP_2400, RSP_0097, sufC, and adhI (). The microarray data for flgF could not be confirmed and RSP_0432 was not tested, since no function for this gene is predicted. Real time RT-PCR also demonstrated that deletion of SorY had an opposite effect on the transcript levels of these mRNAs than overexpression of SorY ().
Figure 2. (A) Validation of microarray results by real-time RT-PCR and expression changes in a SorY deletion strain for putative SorY target genes. Relative expression of strain 2.4.1pBBRSorY is shown in relation to expression in the control strain 2.4.1pBBR, whereas the relative expression of the SorY deletion strain is shown in relation to the parental wild-type strain. Real-time RT-PCR data represents the mean of 3 independent experiments. Error bars indicate the standard deviation. (B) Effect of SorY overexpression on takP mRNA stability. The half-life of the takP mRNA in the control strain pBBR and strain pBBRSorY was calculated based on real-time RT-PCR with total RNA isolated from cultures at several time points after the addition of rifampicin. Values are normalized to the 16S rRNA. The data represents the mean of 3 independent experiments. (C) Binding of takP and SorY in vitro. Band shift experiment with 150 fmolCitation32 P-labeled takP (15 nM) fragment of 92 nucleotides length incubated with increasing concentrations of unlabeled SorY at 32°C for 20 min. Unlabeled sRNA SorY was added in a equimolar concentration (15 nM) to the labeled takP fragment or in 10 fold (150 nM) or 100 fold (1.5 µM) molar excess. As control 150 fmol of Citation32P-labeled takP fragment were incubated with water, the sRNA PcrZ (1.5 µM) or with an mRNA fragment of RSP_2591 (1.5 µM) in 100 fold excess.
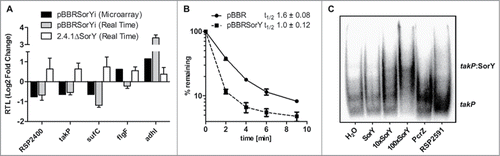
Table 1. Expression level of genes passing the selection criteria of microarray analysis, which is a reliable A-value ≥ 12 and a log2 fold change of > 0.6 or < −0.6 between the 2 strains 2.4.1pBBRSorYi and 2.4.1pBBR. We used the IntaRNA tool to predict a putative interaction of the mRNAs transcribed from these genes and SorY. Predicted energy values and locations of the interaction sites are listed for genes deceeding a predicted energy value of −10 kcal/mol
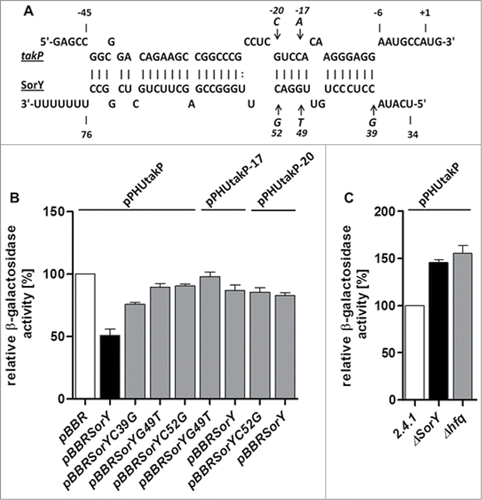
To test whether the mRNAs are direct targets of SorY we applied an in vivo reporter system suitable for α-proteobacteria.Citation4 Sequences predicted to bind SorY were cloned behind a constitutive 16S promoter and translationally fused to the lacZ gene. This reporter construct was conjugated into cells either containing a vector control, or having SorY expressed from the 16S rRNA promoter. For reporter fusions harbouring sequences of RSP_2400, sufC, adhI and flgF the differences in β-galactosidase activity to the control strain were not significant, indicating no direct interaction of SorY with these mRNAs (Fig. S2). For RSP_0097, however, a 2-fold decrease of β-galactosidase activity was observed when SorY was overexpressed, indicating a direct interaction of the 2 RNAs (). RSP_0097 encodes TakP (previously named SmoM), an extracytoplasmic solute receptor of a TRAP transporter suggested to transport α-keto acids.Citation25
Figure 3. (A) SorY-takP seed region as predicted by IntaRNA. Single nucleotide exchanges of SorY and takP are indicated by arrows. Position +1 of the mRNA refers to the translational start site. (B) Relative β-galactosidase activity of the lacZ-based in vivo reporter system. Activity in the control strains containing the takP::lacZ, takP-17::lacZ or takP-20::lacZ reporter fusions and plasmid pBBR is set to 100% (open bar). Activities for all other combinations are calculated in relation to their respective control. Activity in the strain containing the reporter fusion and plasmid pBBRSorY is indicated by a solid bar. Gray bars represent strains overexpressing SorY with a single mutation at 3 different locations and the compensatory mutations of takP in the reporter plasmid. (C) Relative β-galactosidase activity of the in vivo reporter takP::lacZ in wild type R. sphaeroides compared to ΔSorY and Δhfq knockout strains. Activity in the control strain containing the reporter fusion is set to 100 % (open bar). For each strain, 3 independent biological experiments with technical duplicates were performed. Error bars indicate standard deviations.
SorY affects the stability of the takP mRNA
Trans-acting sRNAs can have a direct effect on translation of an mRNA or its stability. However in most cases mRNA stability and consequently the mRNA level is also affected if the rate of translation is altered by an sRNA. Therefore the half-life of the takP mRNA was determined in a control strain (2.4.1pBBR) and in strain 2.4.1pBBRSorY, which constitutively overexpressed the sRNA. A clear difference in the decay rates was observed for the 2 strains (). When only taking into account the initial phase of decay, which followed a logarithmic kinetic, the half-life was about 1.6 min for the control strain but only about 1 min for the strain overexpressing SorY. This effect of SorY on takP mRNA half-life hints at a direct interaction of the 2 RNAs, which leads to destabilization of the target mRNA.
Single point mutations in SorY partially release its effect on takP
The reporter gene study and the effect of SorY on takP stability indicated a direct interaction between SorY and takP. This assumption was supported by an in vitro interaction study. Addition of SorY to a radio-labeled takP mRNA segment resulted in retardation in a native gel. Addition of the unrelated sRNA PcrZCitation4 or a segment of RSP_2591 mRNA did not cause a retardation of takP (). To further prove a direct interaction between SorY and takP mRNA in vivo we exchanged single nucleotides in the SorY sequence and tested the effect of the mutations on takP expression in the reporter system. SorY sequences with point mutations at positions +39, +49 or +52 had a significantly smaller effect on takP expression than the wild type sequence (). While overexpression of wild type SorY decreased expression levels of the takP reporter to about 50%, expression in the single mutants was only reduced to 75-85 %. When compensatory mutations complementary to G49T or C52G are inserted into takP at positions −20 or –17, respectively, binding to the corresponding mutated SorY counterparts could not be restored as we did not observe any significant decrease in β-galactosidase activity. The lack of restoration is most likely due to altered secondary structure of the sRNA after single nucleotide exchanges, which in all cases leads to an extended stem-loop in the sRNAs seed region (Fig. S3). Moreover, predicted energy values for the interaction of mutated SorY with corresponding compensatory mutated takP do not reach the value predicted for the original sequence (Table S1). We also tested the effect of the wild type SorY on the takP reporter with point mutations at position −20 or −17. Both point mutations led to a significantly smaller effect of SorY on reporter expression in comparison to the reporter plasmid with the original takP sequence.To further confirm the interaction of SorY with takP, the takP::lacZ reporter plasmid was used to measure β-galactosidase activity in a SorY and hfq deletion strain in relation to their isogenic wild type strain. In both strains expression levels of the takP reporter increased about 50% (). In total these results support the view that SorY reduces takP mRNA levels in an Hfq-dependent manner. SorY was previously shown to interact with Hfq.Citation26
Effect of SorY and takP on resistance to 1O2
We performed zone of inhibition assays, to further characterize the role of SorY in the photooxidative stress response. Constitutive overexpression of SorY conferred increased resistance, while deletion of SorY resulted in higher sensitivity against 1O2 (). However, SorY overexpression had no effect on resistance in strains lacking Hfq. These data indicate an important role of SorY in the stress response which is dependent on Hfq.
Figure 4. Effect of SorY expression levels on resistance against 1O2 stress visualized by inhibition zones. Comparison of strains with different SorY levels, in wild type, ΔtakP- or Δhfq- background. Zones of inhibition for the different strains are shown in relation to the zones determined for their respective control strain set to 100% (open bar). Assays were performed on media with malate (A) or lactate (B) as main carbon source. The asterisk highlights the parental wild type to the ΔtakP mutant. Error bars indicate the standard deviation of biological triplicates.
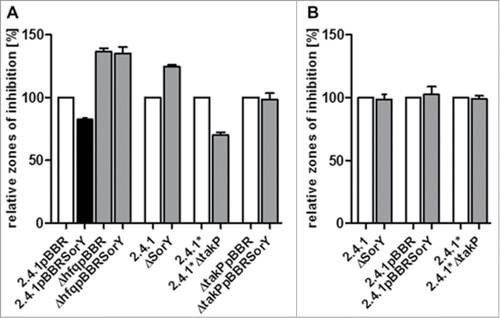
If the increased stress resistance of a strain overexpressing SorY was due to the negative regulation of the sRNA on the expression of takP, a mutant lacking the takP gene should show a similar phenotype. Indeed a takP deletion strain also showed increased resistance to 1O2 in comparison to its isogenic wild type (). Although the mutant was derived from a 2.4.1 strain, we used the original parental strain from which the mutant was generated as a control, since originally identical strains often show some genetic diversification when maintained in different laboratories. We designate the parental strain to the takP mutant 2.4.1*. Interestingly, overexpression of SorY in strain 2.4.1*ΔtakP did not affect resistance at all in comparison to the same strain harbouring the empty vector (). In summary these results suggest that SorY supports the 1O2 defense primarily through takP under these experimental conditions.
The SorY phenotype depends on growth on malate
Two homologues of TakP in R. capsulatus were previously characterized. DctP2 (RCAP_rcc02417; 81% sequence identity) was shown to bind α-keto acids in vitroCitation27 whereas DctP3 (RCAP_rcc03024; 45% sequence identity) was needed for the uptake of the C4-dicarboxylates malate, succinate, and fumarate in vivo.Citation28,29 In this study malate was used as main carbon source. To confirm that resistance to 1O2 conferred by an overexpression of SorY or a deletion of takP (), is due to altered malate import, we performed zone of inhibition assays with lactate as main carbon source (). Smaller zones of inhibition were observed on media containing lactate compared to malate media (data not shown). Therefore the methylene blue concentration was increased for further assays on lactate media to achieve evaluable inhibition zones. On lactate there was no difference in size of inhibition zones, when SorY overexpression, SorY deletion or takP deletion were compared to their respective control strains (), strongly suggesting that TakP is involved in malate uptake.
Discussion
RSs1543, now designated SorY, was originally identified as an sRNA, which is induced in R. sphaeroides by 1O2. Transcription from an RpoHI/RpoHII dependent promoterCitation26 suggested the influence of several stress factors on SorY accumulation which was confirmed in this study. Agents leading to membrane stress like ethanol and polymyxin B had no effect on SorY expression. Addition of ROS or reagents leading to ROS formation resulted in SorY induction, which was also observed in response to heat, salt or CdCl2. Non-redox-reactive heavy metals like cadmium can bind thiol groups causing glutathione depletion and oxidation of sulfhydryl groups in proteins and consequently leading to oxidative stress. RpoHI/RpoHII of R. sphaeroides were previously shown to be activated by photooxidative stress caused by strong blue light,Citation30 the presence of methylene blue and light,Citation9,15,26 or by heat shock.Citation10
The focus of this study is on the role of SorY in the photooxidative stress response. To understand how SorY can affect this response, we searched for putative interaction partners and confirmed the takP mRNA as a direct target of SorY. The functional interaction of SorY and takP was confirmed by i) a decreased half-life of the takP mRNA when SorY is overexpressed, ii) the effect of SorY on takP expression in a reporter gene system in vivo, iii) a dose dependent shift caused by direct binding in electrophoretic mobility shift assays in vitro, iv) a similar phenotype of SorY overexpression and a takP deletion strain and v) failure of SorY to influence stress resistance in a takP deletion mutant. Whether a decreased takP half-life in the presence of SorY overexpression is due to a recruitment and cleavage of RNases or due to fewer protective ribosomes present on the mRNA, was not further investigated. The results achieved with reporter assays suggest that SorY decreases takP stability/translation in an Hfq dependent manner, since the reporter activity is strongly decreased by SorY overexpression but strongly increased in SorY or Hfq deletion strains. Single nucleotide exchanges in SorY, predicted to weaken the interaction between the 2 RNAs, indeed diminished the effect of SorY on takP expression in those assays, further supporting the view that both RNAs interact directly. It was however not possible to reverse the effect of the mutations by introducing compensatory mutations, which is most likely due to an extended stem-loop in the sRNA seed regions after each single nucleotide exchange and lower predicted energy values for the interaction of mutated SorY with corresponding compensatory mutated takP. SorY overexpression led to an increased resistance, whereas deletion of SorY resulted in higher sensitivity against 1O2. This effect was clearly dependent on presence of Hfq and TakP. Thus SorY confers resistance to 1O2 stress by a negative regulation of takP expression in an Hfq-dependent manner. Since overexpression of SorY in the takP deletion strain did not affect resistance at all, we conclude that takP is the primary target of SorY promoting resistance under these experimental conditions.
Our results raised the question how SorY influences stress resistance by affecting takP expression. In a wild type strain expression of takP is about 2-fold down-regulated upon exposure to 1O2.Citation17 Studies on R. sphaeroides TakP were initiated due to the higher selenite resistance of a TakP mutant strain. Binding of α-keto acids to this protein was confirmed in vitro and the structure of TakP binding pyruvate was solved.Citation25 The natural ligand of TakP is still unknownCitation25 and lack of pyruvate in our growth medium excluded the possibility that the phenotype of the takP mutant was due to altered pyruvate import. Therefore we considered malate to be the possible ligand for takP, which was the main carbon source in our medium. One of the TakP homologues in Rhodobacter capsulatus, DctP3 (RCAP_rcc03024), was shown in vivo to be needed for the uptake of the C4-dicarboxylates malate, succinate, and fumarate.Citation28,29 To confirm that resistance to 1O2 conferred by an overexpression of SorY or a deletion of takP, is due to altered malate import, we performed zone of inhibition assays with growth media containing an equimolar concentration of lactate as main carbon source. Lactate medium led to slower cell growth and higher resistance towards 1O2 in comparison to malate medium. Using lactate as main carbon source abolished the effects of takP deletion, SorY overexpression or SorY deletion on resistance against 1O2. We conclude that resistance to 1O2 is conferred by down-regulation of TakP levels by SorY and consequently decreased malate import.
Malate is an intermediate of the TCA cycle. A tight regulation of its uptake might be necessary, since very high amounts of malate could be dispensable or even harmful during oxidative stress. Several studies reported during the last years that oxidative stress redirects the metabolic flux from the glycolytic pathway to the oxidative pentose phosphate pathway (PPP) and Entner–Doudoroff pathway (ED), while the fluxes associated with the TCA cycle are decreased. This metabolic shift leads to decreased production of NADH which can increase oxidative stress by transferring electrons to oxygen and increased production of NADPH in the PPP and ED, an important component in the oxidative stress defense.Citation31-35 Also up-regulation of some glycolytic enzymes, i.e. fructose-bisphosphate aldolase and glucose-6-P-isomerase was observed in E. coli during oxidative stress, which then leads to a glyoxylate shunt and subsequent decrease of NADH production.Citation32,36,37
During menadione-induced ROS generation accumulation of pyruvate and malate, intermediates of the TCA cycle, was observed.Citation38 We reanalyzed a previous transcriptome study in R. sphaeroides (Table S2) with respect to changes in the central carbon metabolism in response to 1O2, which has not been reported for this organism so far. Our data revealed 1O2 to reduce expression of genes for some components of the TCA cycle, like genes for succinyl-CoA synthetase (sucC, sucD) and fumarate hydratase (fumC). At the same time genes for the pentose phosphate pathway and Entner–Doudoroff pathway like glucose-6-P-dehydrogenase (zwf), 6-phosphogluconolactonase (pgl), 2-keto-3-deoxy-phosphogluconate aldolase (eda) are induced (Table S2). Additional expression of glucose-6-phosphate isomerase (pgi), fructose-1,6-bisphosphatase (glpX) and fructose-bisphosphate aldolase (fbaB) is induced to produce glucose-6-phosphate in gluconeogenesis, which then can be metabolized in ED and PPP to produce NADPH.Citation8,17,39 Interestingly, it was also shown that all the induced genes of ED, PPP and gluconeogenesis except glpX are part of the RpoHII regulon as is SorY.Citation10,40 Moreover downregulation of sucC and sucD is also reliant on RpoHII.Citation10
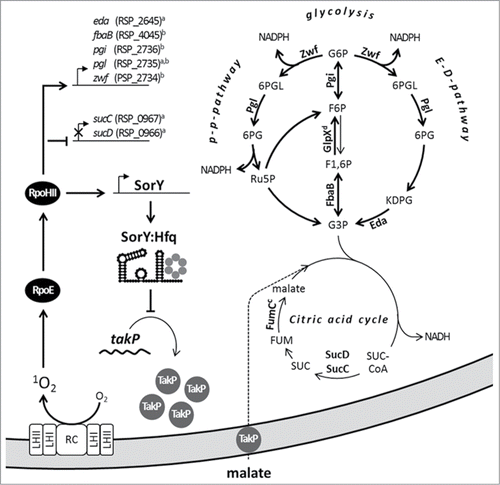
In we present a model for the effect of SorY on the metabolic flux and consequently on stress resistance in R. sphaeroides. To combat oxidative stress the alternative sigma factor RpoE and subsequently RpoHII are activated. RpoHII induces the genes zwf, pgl, eda to switch from glycolysis to PPP and ED to reduce production of NADH and increase NADPH levels. In addition RpoHII triggers pgi and fbaB expression to strengthen gluconeogenesis. At the same time flux through the TCA cycle, which generates excessive amounts of the prooxidant NADH, is decreased by an RpoHII dependent down-regulation of the genes sucC and sucD. In addition RpoHII also induces expression of the sRNA SorY, which regulates TakP synthesis negatively to decrease high but harmful malate influx into the cell and the TCA cycle respectively. This represents a novel mechanism by which an sRNA confers resistance to 1O2. Other sRNA were reported to contribute in the oxidative stress response by mechanisms differing from that of SorY function. In R. sphaeroides PcrZ counteracts the strong increase of photosynthetic gene expression upon a drop in oxygen tensionCitation4 and therefore also bacteriochlorophyl a mediated generation of 1O2. In Enterobacteria the sRNA OxyS is induced upon exposure to H2O2 and negatively influences translation of the transcription factor FhlA and of the stationary phase specific alternative sigma factor RpoS for the benefit of the oxidative stress response.Citation41,42 Furthermore, a tight regulation of iron availability in E. coli by the sRNA RyhBCitation43 and its homologues, e.g. in Pseudomonas aeruginosaCitation44 and Erwinia chrysanthemiCitation45 contributes to the defense against ROS, which are generated in the Fenton reaction.Citation46,47 In Enterobacteria sRNAs like MicF, MicC and CyaR are involved in changing the outer membrane composition in response to oxidative stress.Citation36,48,49 However, SorY is the first example of an sRNA providing resistance during photooxidative stress by decreasing import of a carbon source to limit metabolic flux through the TCA cycle.
Figure 5. To combat 1O2 stress the alternative sigma factor RpoE and subsequently RpoHII are activated in Rhodobacter sphaeroides. RpoHII induces expression of the depicted enzymes for pentose phosphate pathway (PPP), Entner–Doudoroff pathway (ED) and gluconeogenesis to generate the antioxidant NADPH and indirectly represses enzymes for the TCA cycle, which generates the prooxidant NADH. To favor this pathway shunt RpoHII also induces the sRNA SorY. SorY regulates TakP synthesis negatively to limit intermediate availability for the TCA cycle by decreasing malate influx into the cell. This assigns a so far unknown function to a sRNA in oxidative stress response. Abbreviations: G6P – Glucose 6-phosphate, F6P – Fructose 6-phosphate, F1,6P – Fructose1,6-bisphosphate, G3P – Glyceraldehyde 3-phosphate, 6PGL – Phosphoglucono-δ-lactone, 6PG – 6-Phosphogluconate, KDPG – 2-Keto-3-deoxy-6-phosphogluconate, Ru5P – Ribulose 5-P, SUC-CoA – Succinyl CoA, SUC – Succinate, RC – reaction center, LH – light harvesting complex,a RpoHII regulon described by Nuss et al. (2009) orb Dufour et al. (2012),c RpoHII independent down- ord upregulation due to 1O2.Citation8,17
Material and Methods
Bacterial strains and growth conditions
Bacterial strains used in this study are listed in Table S3. Details on their construction are given in Supplementary Material and Methods. For cultivation of R. sphaeroides strains at 32°C, malate minimal-salt medium was usedCitation50 or malate was substituted by an equimolar amount of lactate. To grow the cells aerobically, cultures were either gassed with air in Meplat bottles to attain a concentration of 160 to 180 µM of dissolved oxygen or by continuous shaking of Erlenmeyer flasks containing 20% culture by volume at 140 rpm. For semiaerobic growth conditions, having a dissolved oxygen concentration of about 25 µM, Erlenmeyer flasks containing 80% culture by volume were shaken at 140 rpm. When necessary kanamycin (25 µg ml−1), tetracycline (1.5 µg ml−1) or spectinomycin (10 µg ml−1) was added to liquid and solid growth media (1.6% agar). Photooxidative stress conditions were generated as described earlier,Citation51 except the final concentration of methylene blue (0.2 µM) (Sigma-Aldrich; M9140). Other stress conditions were generated by a final concentration of 500 µM diamide (Sigma-Aldrich; D3648), 500 mM NaCl, 10 µM CdCl2, 2,5% Ethanol (Carl Roth; 5054.3), 1 µg/ml polymyxin-B (Sigma-Aldrich; P4932) or 0.005 % SDS along with 1 mM EDTA. Use of antibiotics was avoided in cultures and agar plates used for inhibition zone assays. To culture Escherichia coli strains, cells were continuously shaken at 180 rpm in Luria–Bertani medium at 37°C or grown on solid growth media containing 1.6% (w/v) agar. When necessary kanamycin (25 µg ml−1) or tetracycline (20 µg ml−1) was added to the media.
Microarray analysis
Microarray analysis was performed as described before (Mank et al., 2012). Total RNA, obtained from aerobically grown cultures of strains 2.4.1pBBRSorYi and 2.4.1pBBR after 10 min of photooxidative stress, was chemically labeled with Cy3 and Cy5 (Kreatech; EA-022/EA-023), respectively. Multiarray analysis was performed with the Bio-conductor package Limma for R. On the basis of calculated MA plots, genes were considered reliable if the average signal intensity [A-value: 1/2 log2 (Cy3×Cy5)] was ≥ 12. To filter out potentially insignificant changes among genes that passed the reliability criterion, a cutoff value was applied; i.e., those genes were retained whose average expression value of 2.4.1pBBRSorYi (a) compared with the average value of the control treatment 2.4.1pBBR (b) was either a log2 Fold Change of ≥ 0.6 or a ≤ −0.6. Microarray data are deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE63328).
β-galactosidase activity assay
To confirm the predicted mRNA:sRNA interaction in vivo, β-galactosidase activity was measured in all conjugants obtained after transferring the respective reporter plasmids via di-parental conjugation. Three independent liquid cultures, inoculated with equal numbers of colonies, were grown semiaerobically and diluted to OD660 0.2, before reaching stationary phase. Three samples of 0.5 ml were collected in early exponential growth phase (OD660 0.4). Measurements of β-galactosidase activity were carried out as described previously.Citation12
Sensitivity to ROS generating chemicals
To measure the sensitivity of Rhodobacter sphaeroides to 1O2 zone of inhibition assays were performed as described before.Citation12 50 µl of semiaerobically grown culture (OD660 0.5) were added to 5 ml of top agar (minimal salt medium containing 0.8% agarose) and poured on top of an agar plate (1.6% agarose in minimal salt medium). Filter paper disks soaked with 5 µl of 10 or 50 mM methylene blue (Sigma-Aldrich; M9140) were placed on top of the plates. The diameter of the inhibition zones was measured after incubation for 48 h at 32°C in the dark or with illumination by a fluorescent tube in case of methylene blue.
Isolation of total RNA
Total RNA used for both RNA gel blot and real time RT-PCR was isolated by the hot phenol method.Citation52 To remove remaining traces of DNA, samples were treated with 6 U of DNaseI (Invitrogen; #18047019) per 1 µg of RNA. Absence of DNA contamination was confirmed by PCR with primers targeting gloB (RSP_0799) (Table S4).
Half-life determination of the mRNA
RNA samples were prepared as described earlier.Citation9 Half-lifes were calculated based on Real time RT-PCR with 20 ng of total RNA for the target gene while 0.2 ng of the total RNA was used for the standard gene 16S rRNA.
Real Time RT-PCR
Real Time RT-PCR was performed as described earlierCitation50 Measurements were done in a Bio-Rad CFX96 Real Time System and the corresponding program Bio-Rad CFX Manager was used for data analysis. After normalization relatively to the rpoZ gene,Citation53 which encodes the ω-subunit of the RNA polymerase, relative mRNA levels were calculated according to.Citation54 Primers and corresponding efficiencies are listed in Table S4 and Table S5 respectively.
Northern blot analysis
Northern blots were performed as described earlier.Citation9 Oligodeoxynucleotides used for end-labeling with [γ-32P]-ATP (Hartmann Analytic; SRP-301) by T4 polynucleotide kinase (Fermentas; #EK0031) are listed in Table S4. A low stringency Church buffer was used for hybridization.Citation55 Membranes were washed in 5× SCC buffer + 0.1 % SDS. After exposure on phosphoimaging screens (Bio-Rad), images were analyzed by the 1D-Quantity One software (Bio-Rad).
Electrophoretic mobility shift assay
Templates for in vitro transcription by T7 RNA polymerase (New England Biolabs; #M0251) of SorY (86nt), PcrZ (136nt), the takP mRNA (92nt) and the RSP_2591 (177nt) mRNA (both fragments containing 5′ UTR and the beginning of the coding sequence) were generated from corresponding primer pairs listed in Table S4. [α-32P]-UTP (Hartmann Analytic; FP-210) was used to label takP transcripts internally. In vitro transcripts were treated with DNaseI (Promega; M6101) prior to gel purification. RNA band shift experiments were performed in 1× structure buffer (Ambion) in a total reaction mixture volume of 10 µl as follows. Transcripts were heated (90°C; 10 min), cooled down to 4°C and renatured at 32°C separately. 150 fmol of labeled takP RNA was incubated at 32°C for 20 min with unlabeled SorY (ratios 1:1, 1:10, 1:100) or as controls with water, unspecific sRNA (PcrZ) and mRNA (RSP_2591) in 100 fold molar excess. Binding reactions were mixed with 3 µl of native loading dye (50% glycerol, 0.5× TBE, 0.1% bromophenol blue) and ran on native 6% polyacrylamide gels in 0.25× TBE buffer at 150 V for 2 h.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Material.docx
Download MS Word (6.4 MB)Acknowledgments
We thank David Pignol for providing us with the takP deletion strain, Fabian Billenkamp for the control substrate RSP2591 and student assistant Marcel Volk for support during the revision.
Funding
This work was supported by Deutsche Forschungsgemeinschaft (Kl563/20-2 and International Research Training Group GRK1384) and Deutscher Akademischer Austauschdienst.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Dufour YS, Landick R, Donohue TJ. Organization and evolution of the biological response to singlet oxygen stress. J Mol Biol 2008; 383:713-30; PMID:18723027; http://dx.doi.org/10.1016/j.jmb.2008.08.017
- Glaeser J, Nuss AM, Berghoff BA, Klug G. Singlet oxygen stress in microorganisms. Adv Microb Physiol 2011; 58:141-73; PMID:21722793; http://dx.doi.org/10.1016/B978-0-12-381043-4.00004-0
- Braatsch S, Gomelsky M, Kuphal S, Klug G. A single flavoprotein, AppA, integrates both redox and light signals in Rhodobacter sphaeroides. Mol Microbiol 2002; 45:827-36; PMID:12139627; http://dx.doi.org/10.1046/j.1365-2958.2002.03058.x
- Mank NN, Berghoff BA, Hermanns YN, Klug G. Regulation of bacterial photosynthesis genes by the small noncoding RNA PcrZ. Proc Natl Acad Sci U S A 2012; 109:16306-11; PMID:22988125; http://dx.doi.org/10.1073/pnas.1207067109
- Han Y, Meyer MH, Keusgen M, Klug G. A haem cofactor is required for redox and light signalling by the AppA protein of Rhodobacter sphaeroides. Mol Microbiol 2007; 64:1090-104; PMID:17501930; http://dx.doi.org/10.1111/j.1365-2958.2007.05724.x
- Anthony JR, Warczak KL, Donohue TJ. A transcriptional response to singlet oxygen, a toxic byproduct of photosynthesis. Proc Natl Acad Sci U S A 2005; 102:6502-7; PMID:15855269; http://dx.doi.org/10.1073/pnas.0502225102
- Glaeser J, Klug G. Photo-oxidative stress in Rhodobacter sphaeroides: protective role of carotenoids and expression of selected genes. Microbiology 2005; 151:1927-38; PMID:15942000; http://dx.doi.org/10.1099/mic.0.27789-0
- Glaeser J, Zobawa M, Lottspeich F, Klug G. Protein synthesis patterns reveal a complex regulatory response to singlet oxygen in Rhodobacter. J Proteome Res 2007; 6:2460-71; PMID:17536848; http://dx.doi.org/10.1021/pr060624p
- Berghoff BA, Glaeser J, Sharma CM, Vogel J, Klug G. Photooxidative stress-induced and abundant small RNAs in Rhodobacter sphaeroides. Mol Microbiol 2009; 74:1497-512; PMID:19906181; http://dx.doi.org/10.1111/j.1365-2958.2009.06949.x
- Nuss AM, Glaeser J, Klug G. RpoH(II) activates oxidative-stress defense systems and is controlled by RpoE in the singlet oxygen-dependent response in Rhodobacter sphaeroides. J Bacteriol 2009; 191:220-30; PMID:18978062; http://dx.doi.org/10.1128/JB.00925-08
- Anthony JR, Newman JD, Donohue TJ. Interactions between the Rhodobacter sphaeroides ECF sigma factor, sigma(E), and its anti-sigma factor, ChrR. J Mol Biol 2004; 341:345-60; PMID:15276828; http://dx.doi.org/10.1016/j.jmb.2004.06.018
- Nuss AM, Adnan F, Weber L, Berghoff BA, Glaeser J, Klug G. DegS and RseP homologous proteases are involved in singlet oxygen dependent activation of RpoE in Rhodobacter sphaeroides. PLoS One 2013; 8:e79520; PMID:24223961; http://dx.doi.org/10.1371/journal.pone.0079520
- Nam TW, Ziegelhoffer EC, Lemke RA, Donohue TJ. Proteins needed to activate a transcriptional response to the reactive oxygen species singlet oxygen. MBio 2013; 4:e00541-12; PMID:23300250; http://dx.doi.org/10.1128/mBio.00541-12
- Karls RK, Brooks J, Rossmeissl P, Luedke J, Donohue TJ. Metabolic roles of a Rhodobacter sphaeroides member of the sigma32 family. J Bacteriol 1998; 180:10-9; PMID:9422586
- Nuss AM, Glaeser J, Berghoff BA, Klug G. Overlapping alternative sigma factor regulons in the response to singlet oxygen in Rhodobacter sphaeroides. J Bacteriol 2010; 192:2613-23; PMID:20304993; http://dx.doi.org/10.1128/JB.01605-09
- Berghoff BA, Glaeser J, Nuss AM, Zobawa M, Lottspeich F, Klug G. Anoxygenic photosynthesis and photooxidative stress: a particular challenge for Roseobacter. Environ Microbiol 2011; 13:775-91; PMID:21108722; http://dx.doi.org/10.1111/j.1462-2920.2010.02381.x
- Berghoff BA, Konzer A, Mank NN, Looso M, Rische T, Forstner KU, Kruger M, Klug G. Integrative “omics”-approach discovers dynamic and regulatory features of bacterial stress responses. PLoS Genet 2013; 9:e1003576; PMID:23818867; http://dx.doi.org/10.1371/journal.pgen.1003576
- Hess WR, Berghoff BA, Wilde A, Steglich C, Klug G. Riboregulators and the role of Hfq in photosynthetic bacteria. RNA Biol 2014; 11:413-26; PMID:24651049; http://dx.doi.org/10.4161/rna.28035
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell 2009; 136:615-28; PMID:19239884; http://dx.doi.org/10.1016/j.cell.2009.01.043
- Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu Rev Microbiol 2004; 58:303-28; PMID:15487940; http://dx.doi.org/10.1146/annurev.micro.58.030603.123841
- Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu Rev Biochem 2005; 74:199-217; PMID:15952886; http://dx.doi.org/10.1146/annurev.biochem.74.082803.133136
- Majdalani N, Vanderpool CK, Gottesman S. Bacterial small RNA regulators. Crit Rev Biochem Mol Biol 2005; 40:93-113; PMID:15814430; http://dx.doi.org/10.1080/10409230590918702
- Green HA, Donohue TJ. Activity of Rhodobacter sphaeroides RpoHII, a second member of the heat shock sigma factor family. J Bacteriol 2006; 188:5712-21; PMID:16885439; http://dx.doi.org/10.1128/JB.00405-06
- Busch A, Richter AS, Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 2008; 24:2849-56; PMID:18940824; http://dx.doi.org/10.1093/bioinformatics/btn544
- Gonin S, Arnoux P, Pierru B, Lavergne J, Alonso B, Sabaty M, Pignol D. Crystal structures of an Extracytoplasmic Solute Receptor from a TRAP transporter in its open and closed forms reveal a helix-swapped dimer requiring a cation for alpha-keto acid binding. BMC Struct Biol 2007; 7:11; PMID:17362499; http://dx.doi.org/10.1186/1472-6807-7-11
- Berghoff BA, Glaeser J, Sharma CM, Zobawa M, Lottspeich F, Vogel J, Klug G. Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol Microbiol 2011; 80:1479-95; PMID:21535243; http://dx.doi.org/10.1111/j.1365-2958.2011.07658.x
- Thomas GH, Southworth T, Leon-Kempis MR, Leech A, Kelly DJ. Novel ligands for the extracellular solute receptors of two bacterial TRAP transporters. Microbiology 2006; 152:187-98; PMID:16385129; http://dx.doi.org/10.1099/mic.0.28334-0
- Forward JA, Behrendt MC, Wyborn NR, Cross R, Kelly DJ. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J Bacteriol 1997; 179:5482-93; PMID:9287004
- Shaw JG, Hamblin MJ, Kelly DJ. Purification, characterization and nucleotide sequence of the periplasmic C4-dicarboxylate-binding protein (DctP) from Rhodobacter capsulatus. Mol Microbiol 1991; 5:3055-62; PMID:1809844; http://dx.doi.org/10.1111/j.1365-2958.1991.tb01865.x
- Braatsch S, Moskvin OV, Klug G, Gomelsky M. Responses of the Rhodobacter sphaeroides transcriptome to blue light under semiaerobic conditions. J Bacteriol 2004; 186:7726-35; PMID:15516587; http://dx.doi.org/10.1128/JB.186.22.7726-7735.2004
- Baxter CJ, Redestig H, Schauer N, Repsilber D, Patil KR, Nielsen J, Selbig J, Liu J, Fernie AR, Sweetlove LJ. The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol 2007; 143:312-25; PMID:17122072; http://dx.doi.org/10.1104/pp.106.090431
- Rui B, Shen T, Zhou H, Liu J, Chen J, Pan X, Liu H, Wu J, Zheng H, Shi Y. A systematic investigation of Escherichia coli central carbon metabolism in response to superoxide stress. BMC Syst Biol 2010; 4:122; PMID:20809933; http://dx.doi.org/10.1186/1752-0509-4-122
- Valdivia-Gonzalez M, Perez-Donoso JM, Vasquez CC. Effect of tellurite-mediated oxidative stress on the Escherichia coli glycolytic pathway. Biometals 2012; 25:451-8; PMID:22234496; http://dx.doi.org/10.1007/s10534-012-9518-x
- Bignucolo A, Appanna VP, Thomas SC, Auger C, Han S, Omri A, Appanna VD. Hydrogen peroxide stress provokes a metabolic reprogramming in Pseudomonas fluorescens: enhanced production of pyruvate. J Biotechnol 2013; 167:309-15; PMID:23871654; http://dx.doi.org/10.1016/j.jbiotec.2013.07.002
- Crimmins GT, Schelle MW, Herskovits AA, Ni PP, Kline BC, Meyer-Morse N, Iavarone AT, Portnoy DA. Listeria monocytogenes 6-Phosphogluconolactonase mutants induce increased activation of a host cytosolic surveillance pathway. Infect Immun 2009; 77:3014-22; PMID:19398547; http://dx.doi.org/10.1128/IAI.01511-08
- Blanchard JL, Wholey WY, Conlon EM, Pomposiello PJ. Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS One 2007; 2:e1186; http://dx.doi.org/10.1371/journal.pone.0001186
- Rungrassamee W, Liu X, Pomposiello PJ. Activation of glucose transport under oxidative stress in Escherichia coli. Arch Microbiol 2008; 190:41-9; PMID:18368388; http://dx.doi.org/10.1007/s00203-008-0361-y
- Singh R, Lemire J, Mailloux RJ, Appanna VD. A novel strategy involved in anti-oxidative defense: the conversion of NADH into NADPH by a metabolic network. PLoS One 2008; 3:e2682; PMID:18628998; http://dx.doi.org/10.1371/journal.pone.0002682
- Imam S, Noguera DR, Donohue TJ. Global insights into energetic and metabolic networks in Rhodobacter sphaeroides. BMC Syst Biol 2013; 7:89; PMID:24034347; http://dx.doi.org/10.1186/1752-0509-7-89
- Dufour YS, Imam S, Koo BM, Green HA, Donohue TJ. Convergence of the transcriptional responses to heat shock and singlet oxygen stresses. PLoS Genet 2012; 8:e1002929; PMID:23028346; http://dx.doi.org/10.1371/journal.pgen.1002929
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 1997; 90:43-53; PMID:9230301; http://dx.doi.org/10.1016/S0092-8674(00)80312-8
- Gonzalez-Flecha B, Demple B. Role for the oxyS gene in regulation of intracellular hydrogen peroxide in Escherichia coli. J Bacteriol 1999; 181:3833-6; PMID:10368161
- Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 2002; 99:4620-5; PMID:11917098; http://dx.doi.org/10.1073/pnas.032066599
- Wilderman PJ, Sowa NA, FitzGerald DJ, FitzGerald PC, Gottesman S, Ochsner UA, Vasil ML. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc Natl Acad Sci U S A 2004; 101:9792-7; PMID:15210934; http://dx.doi.org/10.1073/pnas.0403423101
- Boughammoura A, Matzanke BF, Bottger L, Reverchon S, Lesuisse E, Expert D, Franza T. Differential role of ferritins in iron metabolism and virulence of the plant-pathogenic bacterium Erwinia chrysanthemi 3937. J Bacteriol 2008; 190:1518-30; PMID:18165304; http://dx.doi.org/10.1128/JB.01640-07
- Touati D. Iron and oxidative stress in bacteria. Arch Biochem Biophys 2000; 373:1-6; PMID:10620317; http://dx.doi.org/10.1006/abbi.1999.1518
- Varghese S, Tang Y, Imlay JA. Contrasting sensitivities of Escherichia coli aconitases A and B to oxidation and iron depletion. J Bacteriol 2003; 185:221-30; PMID:12486059; http://dx.doi.org/10.1128/JB.185.1.221-230.2003
- Vogel J, Papenfort K. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol 2006; 9:605-11; PMID:17055775; http://dx.doi.org/10.1016/j.mib.2006.10.006
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 2009; 74:139-58; PMID:19732340; http://dx.doi.org/10.1111/j.1365-2958.2009.06857.x
- Remes B, Berghoff BA, Forstner KU, Klug G. Role of oxygen and the OxyR protein in the response to iron limitation in Rhodobacter sphaeroides. BMC Genomics 2014; 15:794; PMID:25220182; http://dx.doi.org/10.1186/1471-2164-15-794
- Glaeser J, Klug G. Photo-oxidative stress in Rhodobacter sphaeroides: Protective role of carotenoids and expression of selected genes. Microbiology 2005; 151:1927-38; PMID:15942000; http://dx.doi.org/10.1099/mic.0.27789-0
- Janzon L, Löfdahl S, Arvidson S. Evidence for a coordinate transcriptional control of alpha-toxin and protein a synthesis in Staphylococcus aureus. FEMS Microbiol Lett 1986; 33:193-8; http://dx.doi.org/10.1111/j.1574-6968.1986.tb01270.x
- Zeller T, Klug G. Detoxification of hydrogen peroxide and expression of catalase genes in Rhodobacter. Microbiology 2004; 150:3451-62; PMID:15470122; http://dx.doi.org/10.1099/mic.0.27308-0
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29:e45; PMID:11328886; http://dx.doi.org/10.1093/nar/29.9.e45
- Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A 1984; 81:1991-5; PMID:6326095; http://dx.doi.org/10.1073/pnas.81.7.1991
