Abstract
In prokaryotes, the CRISPR/Cas system is known to target and degrade invading phages and foreign genetic elements upon subsequent infection. However, the structure and function of many Cas proteins remain largely unknown, due to the high diversity of Cas proteins. Here we report 3 crystal structures of Archaeoglobus fulgidus Csx3 (AfCsx3) in free form, in complex with manganese ions and in complex with a single-stranded RNA (ssRNA) fragment, respectively. AfCsx3 harbors a ferredoxin-like fold and forms dimer both in the crystal and in solution. Our structure-based biochemical analysis demonstrates that the RNA binding sites and cleavage sites are located at 2 separate surfaces within the AfCsx3 dimer, suggesting a model to bind, tether and cleave the incoming RNA substrate. In addition, AfCsx3 displays robust 3′-deadenylase activity in the presence of manganese ions, which strongly suggests that AfCsx3 functions as a deadenylation exonuclease. Taken together, our results indicate that AfCsx3 is a Cas protein involved in RNA deadenylation and provide a framework for understanding the role of AfCsx3 in the Type III-B CRISPR/Cas system.
Introduction
The CRISPR/Cas system provides an RNA-based, adaptive immune defense against bacteriophages and plasmids.Citation1,2 DNA repeats were first discovered in Escherichia coli genome,Citation3 and consequently renamed as Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR).Citation4 Within the CRISPR loci, variable sequences of similar size are separated by repeat sequences. These variable sequences, known as spacers, are juxtaposed between 2 neighboring repeats and have been shown to be highly homologous to phages or foreign genetic elements.Citation5-8 Thus, the CRISPR loci are described as the genetic retention of historical infections. In addition, over 45 CRISPR-associated (cas) gene families have been identified. Citation9 Biochemical and genetics analyses demonstrate that both the CRISPR transcripts and Cas proteins participate in the acquired and heritable immunity system against invading genetic elements.Citation9,10
CRISPR/Cas systems are categorized into 3 types, based on phylogeny, locus organization and protein components.Citation11,12 In the Type I system, the CRISPR-associated complex for antiviral defense (CASCADE) is required for CRISPR RNA (crRNA) processing and the recognition of invading foreign DNA for cleavage.Citation13 In the Type II system, the Cas9-crRNA ribonucleoprotein complex plays a crucial role in crRNA-mediated DNA degradation.Citation14,15 In the Type III system, 2 similar ribonucleoprotein complexes, named CMR and CSM, are responsible for the degradation of RNACitation16 and DNACitation17 targets, respectively. The defense system generally consists of 3 key processes, including spacer acquisition, CRISPR expression and the degradation of foreign genetic elements mediated by crRNA.Citation18-20 In the systems targeting foreign DNA, Cas3 is known to be involved in DNA unwinding and degradation as a single-stranded DNA (ssDNA) nuclease.Citation13,21 However, the Type III-B system exclusively targets RNA and little is known about the degradation mechanism of RNA after being cleaved by the CMR complex.Citation22,23
The hyperthermophilic archaea Archaeoglobus fulgidus DSM 4304 possesses 3 CRISPR loci and at least 20 putative cas genes.Citation24 One gene cluster, including cas1–6, cmr1–6, csa1, csa2, csa3, csa5 and csx3, is located adjacent to a CRISPR locus.Citation25 Remarkably, the AfCsx3-encoding gene AF1864 is located between cmr3 and cmr4 within the gene cluster. Although the gene csx3 is exclusively found in the Type III system and is mostly adjacent to other well-characterized cas genes, Csx3 is not considered as the classical Cas protein due to the lack of identifiable contextual patterns and the fact that its function is largely unknown.Citation9,12
In this study, we report the first crystal structures of AfCsx3 in free form, in complex with manganese ion and in complex with an ssRNA fragment at 2.95Å, 3.0Å and 2.9Å, respectively. AfCsx3 protein harbors a ferredoxin-like fold and forms dimer both in the crystal and in solution. The AfCsx3 homodimer contains a distant ssRNA binding groove and two-metal-ion binding sites, and displays robust 3′-deadenylase activity in the presence of manganese ions. We suggest that CRISPR-associated AfCsx3, in conjugation with other ribonucleases, might facilitate and/or accelerate the degradation of crRNA-targeted RNA in Type III-B CRISPR/Cas systems.
Results
Overall structure of AfCsx3 in free form
The structure of AfCsx3 in free form was determined in space group P3221 using the single anomalous diffraction (SAD) method against a data set collected on a selenium-substituted AfCsx3 at a wavelength of 0.9792 Å, and refined to a resolution of 2.95 Å (Table 1).
Table 1: Data collection and refinement statistics
Figure 1. (See previous page). Overall Structure of AfCsx3. (A). Sequence alignment of AfCsx3 with various Csx3 proteins from different organisms. The secondary structure elements are shown above the sequence. The α-helices are colored in orange, while the β-strands are colored in green. Conserved residues are shaded in yellow (100% identity), blue (> = 75% identity) and green (> = 50% identity), respectively. Asterisks indicate the critical residues involved in RNA recognition and catalysis. Conserved motif is underlined in red. (B). Stereo view cartoon representation of AfCsx3 showing a ferredoxin-like fold with 6 β-stranded anti-parallel β-sheets located at one side and 2 α-helices resided at the other side. The numbered secondary structures are in the order of appearance in the amino acid sequence. The α-helices are shown in yellow while the β-strands and loop regions are shown in green. (C). AfCsx3 dimer is presented in a cartoon view and the 2 molecules are colored in yellow and magenta, respectively. AfCsx3 dimer aligned by 2-fold non-crystallographic symmetry-related axis. Critical residues which might be involved in dimerization are indicated and colored in cyan. SS-bridge is indicated in green dash line. (D). Superimposition of cartoon and electrostatic potential surface view of AfCsx3 dimer. The dimeric interface harbors a continuously positive-charged groove composed of 4 residues from each molecule. The positive-charged residues are shown as sticks and colored in red.
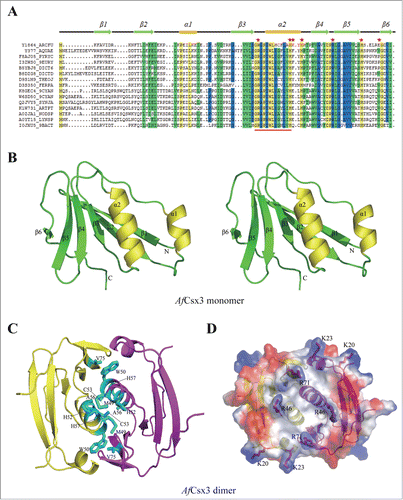
In the crystal, 2 non-crystallographic symmetry-related AfCsx3 molecules show a buried surface area of ∼1,100 Å2/molecule, suggesting that AfCsx3 also forms dimer in its physiological state (). Consistent with structural observations, the apparent molecular mass of AfCsx3 was determined as ˜33.6 kDa in solution, through analytical ultracentrifugation (AUC) experiments. This result corresponds to the approximate molecular mass of AfCsx3 dimer (Fig. S1). Noticeably, the AfCsx3 dimer is formed by a disulfide bond contributed from C53 located at α2 from each molecule (). However, the dimer formation cannot be simply interfered by SS-bridge disruption. The analytical gel filtration experiments showed that alanine-substitution of C53 had little effect on the dimerization (Fig. S2). Although the residue C53 is not a well conserved amino acid among Csx sequences, the residue C53 resides within a highly conserved GRGPIWLY (C/A/G) F/YLV/IH motif (, highlighted in red line underneath). This highly conserved motif forms a well-folded α-helix, which allows the formation of tight dimer via π-stacking and hydrophobic interactions through the conserved aromatic residues, such as W50, H52 and H57 (). Strikingly, the interactions between these 2 monomers were substantially tight that the W50A/H52A/C53A/H57A quadruple mutant still formed dimer in solution according to the analytical gel filtration results (Fig. S2). In addition to all the residues mentioned above, partially conserved hydrophilic and hydrophobic residues surrounding this well-conserved α-helix, such as residues E25, K28, D69, R84, E85, D87 and L27, I49, A56, V67, L72, V75, V77 are also involved in dimer stabilization through the contribution of hydrogen bonds and hydrophobic interactions, respectively. In particular, the residues I49, W50, A56, and V75 form the hydrophobic core along the dimeric interface ().
To investigate the function of AfCsx3 and reveal its putative mode of action in CRISPR/Cas-mediated host defense, a homolog search was performed using the refined AfCsx3 structure against the Protein Data Bank by Dali server.Citation26 However, no close structural homologues were found and the closest match was an antisigma-factor antagonist (PDBID: 3OIZ) with a Z score of 5.2. On the other hand, the dimeric ferredoxin-like fold revealed by the AfCsx3 dimer structure prompted us to speculate that the Csx3 protein may display RNA binding and cleavage activities, similar to other Cas proteins harboring tandem ferredoxin domains, such as Cas6,Citation27 Cas6e Citation28 and Cas6f Citation29 (Fig. S3). Consistent with the notion that AfCsx3 may bind to nucleic acid, the electrostatic potential map of the AfCsx3 dimer showed a patch of protruding basic residues, including K20, K23, R46 and R71, lining along the dimer interface to form a shallow groove with the dimension of 8 Å × 8 Å × 20 Å (width × length × depth) ().
AfCsx3 is a manganese-dependent ribonuclease
To investigate whether AfCsx3 is a ribonuclease similar to other known Cas6 proteins comprising tandem ferredoxin-like domains, we performed AfCsx3-mediated cleavage assays by using the repeat ssRNA derived from the endogenous A. fulgidus CRISPR sequence as substrate. Taking into consideration that the optimal culture condition of A. fulgidus is 83°C and that many ribonucleases from archaeal strains employ different divalent cations for substrate cleavage, we optimized the reaction conditions of cleavage assays by testing the combination of 3 temperature (25°C, 35°C and 55°C) and 3 divalent cations (Mg2+, Ca2+ and Mn2+). Notably, the repeat ssRNA was cleaved in the presence of 20 mM manganese ions, but not in the presence of magnesium or calcium ions at 25°C, 35°C or 55°C (). By contrast, AfCsx3 failed to cleave repeat ssDNA or unrelated ssRNA under any given condition, indicating that AfCsx3 is a substrate-specific ribonuclease ().
Figure 2. (See previous page). Mn-dependent ribonuclease activity of AfCsx3. (A). Ribonuclease activity displayed by AfCsx3 on repeat ssRNA in the presence of MgCl2, CaCl2 and MnCl2 at 25°C, 35°C and 55°C, respectively. Reaction with no protein added serves as control. The RNA substrates and products are indicated. (B). Ribonuclease activity of AfCsx3 on different nucleic acid substrates. The asterisk indicates the product of cleavable repeat ssRNA. (C).AfCsx3 shows manganese–dependent ribonuclease activity. The manganese-dependent ribonuclease activity is either eliminated by the addition of EDTA or interfered by addition of excessive quantities of Mg2+ or Ca2+ in the reactions. Reaction with no addition of metal ions serves as control. (D). Ribonuclease activity displayed by AfCsx3 on repeat ssRNA in the presence of different divalent cations. (E). Overall cartoon and ribbon view of AfCsx3 in complex with manganese ions. Critical residues involved in the coordination of manganese ions are indicated and colored by element. Manganese ions are indicated as spheres in red mesh contours and colored in wheat. (F). Cartoon view of thecatalytic sites of AfCsx3. Five residues, H57, K58, H60, H80 and E85, are involved in the coordination of 2 manganese ions. Catalytic residues are shown as sticks and colored by element. Manganese ions (Mn A and Mn B) are indicated and colored in wheat. (G). Ribonuclease activity of wild-type and mutant AfCsx3 with mutations on the key residues participating in the coordination of manganese ions. (H). Ribonuclease activity of wild-type and mutant AfCsx3 with mutations on the key residues participating in the dimerization. Reaction with no protein added serves as control.
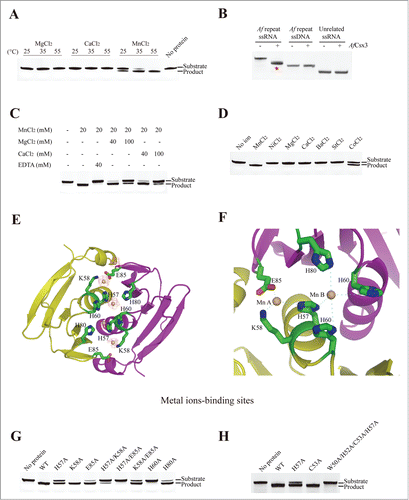
Moreover, the addition of 40 mM EDTA to chelate manganese ions dramatically reduced the cleavage activity of AfCsx3 (). Although the addition of excess calcium (40 mM or 100 mM) or magnesium ions (40 mM or 100 mM) had negative impact on AfCsx3-mediated ssRNA cleavage, AfCsx3 still showed obvious ribonuclease activity, presumably due to the high binding affinity between AfCsx3 and manganese ions (). Thus, these results demonstrated that AfCsx3 is a manganese-dependent ribonuclease, in contrast to Cas6 homologues that function in an ion-independent manner.Citation27,30
To explore the importance of divalent metal ions in AfCsx3-mediated ssRNA cleavage, more divalent metal ions were tested. Interestingly, the repeat ssRNA was partially cleaved in the presence of 20 mM cobalt ions, in contrast to other divalent cations, such as nickel, barium and strontium ions (). Notably, although magnesium, calcium, manganese and cobalt ions have the coordination number of 6, the radius of the cobalt ion is closest to that of the manganese ion. We speculate that cobalt ion may mimic the structural and functional role of the manganese ion during AfCsx3-mediated catalysis. Hence, having the right coordination property and suitable size of the divalent ion is important for AfCsx3-mediated ssRNA cleavage activity.
Structure of AfCsx3 bound to manganese ions
To investigate the structural role of manganese ions in the catalytic mechanisms of AfCsx3, the structure of AfCsx3 in complex with the manganese ion was determined by soaking crystals of AfCsx3 with 1 mM manganese (II) chloride solution. The manganese-soaked AfCsx3 crystals were screened and the diffraction data set was collected (Table 1). The crystal structure of AfCsx3 in complex with manganese ions was determined and refined at 3.0 Å. The Fo-Fc Fourier difference map showed 2 pairs of strong ball-shaped peaks along the dimer interface, which were assigned as manganese ions (). In the crystal, each pair of manganese ions is coordinated by critical residues contributed by 2 molecules. Manganese ion A is coordinated by 1-H57, 1-K58 and 2-E85 (The prefix number refers to the ordering of the molecule), whereas manganese ion B is coordinated by 1-H57, 1-H60, 2-H60 and 2-H80 ().
To validate whether the residues involved in manganese coordination are the catalytic residues for ssRNA cleavage, alanine mutations were introduced to these critical residues and ssRNA cleavage assays were performed. Consistent with the structural observation that 1-H60 and 2-H60 residues are involved in manganese ion Mn B coordination and H57 residue is involved in the coordination of both Mn A and Mn B, the introduction of alanine mutation at residue H60 or H57 abolished or significantly decreased RNA cleavage activity (). Similarly, consistent with the structural observation that residues E85 and H80 are involved in Mn A and Mn B coordination, respectively, the introduction of alanine mutation at either H80 or E85 decreased the ssRNA cleavage activity (). Surprisingly, although K58 is observed to be located near manganese ion Mn A in the crystal structure suggesting that K58 may also be involved in the coordination of manganese ions, the introduction of alanine mutation at residue K58 did not show any negative impact on ssRNA cleavage (). Consistent to our observation, the structure of Cmr2 protein, another Cas protein, in complex with manganese ions showed that the side chain of lysine residue directly coordinated with 2 manganese ions.Citation23 Nevertheless, our cleavage results showed that the introduction of double alanine mutations at H57/K58 or K58/E85 significantly decreased RNA cleavage activity (). Consistent with the structural observation, lysine residues have been proposed to participate in coordinating metal ions, such as Ca2+, Mg2+, Fe2+ and Mn2+ in previous studies.Citation31 These results suggested that K58 should also contribute to the manganese ion coordination, yet not as significantly as the other residues. Therefore, we conclude that the active sites of AfCsx3 comprise 2 manganese ions and the surrounding residues, including H57, K58, H60, H80 and E85.
Interestingly, although the introduction of W50A/H52A/C53A/H57A quadruple mutation didn't display any effects in AfCsx3 dimerization in solution (Fig. S2), the introduction of the quadruple mutations indeed disrupted the cleavage activity of AfCsx3 (). Such observation indicated that the catalytic activity change corresponding to introduction of alanine mutations, especially at residue H57, could induce significant local structural rearrangement at the active sites instead of dimeric interface.
Illustrative model of AfCsx3 bound to RNA fragment
To investigate the ssRNA binding properties of AfCsx3, the structure of AfCsx3 in complex with an ssRNA fragment was determined by molecular replacement at space group I212121 at 3.0 Å (Table 1). The refined model comprises 2 AfCsx3 molecules with residues ranging from 1 to 104 aa and one 4 nt ssRNA fragment, which is located along the dimer interface (). The observed ssRNA binding pocket has a cuboid-like shape with the dimension of 20 Å × 20 Å × 8Å (length × width × depth) (). The cuboid-like ssRNA binding pocket is formed by 2 parts. The side chains of the critical residues R46 and R71 serve as the walls lining along the bound ssRNA fragment. Meanwhile, the side chains of the conserved residues I49, H52 and P48, together with the main chain of residues G47 and R46, serve as the base holding the bound ssRNA fragment (). Particularly, the bound ssRNA fragment adopts an extended conformation with the phosphorus moieties clustering inside and the base moieties extending out and inserting into the preformed pockets within AfCsx3 ().
Figure 3. Illustrative model of AfCsx3 bound to RNA. (A). Overall cartoon representation of AfCsx3 in complex with an ssRNA fragment. (B). The cartoon and ribbon (left) and electrostatic potential surface (right) representation of detailed view of RNA binding region. Electron densities for bound ssRNAs are shown in red mesh contours. The map was calculated using Fo-Fc coefficients and phases from the refined structure, but with the RNA residues omitted from the Fc calculation. The map is contoured at 3σ. The ssRNA 5′-N1-N2-N3-N4-3′ is shown in stick model. The key residues involved in AfCsx3-RNA interaction are indicated and colored in red (serving as walls) and cyan (serving as base), respectively.
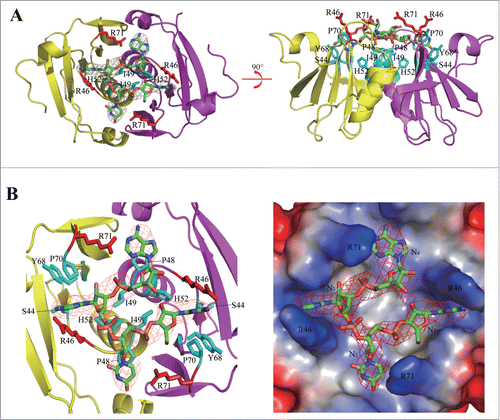
Although the 37 nt repeat ssRNA was incubated with AfCsx3 for co-crystallization, only the 4 nt ssRNA model was built in our complex. We found that the repeat ssRNA was degraded during the long-term crystallization incubation. Therefore, the extra electronic density could be assigned to any tetramer derived from repeat ssRNA. Nevertheless, to build an illustrative model showing the interactions between AfCsx3 and RNA, we assigned 4 unidentified nucleotides (5′-N1-N2-N3-N4-3′) for refinement. The bases of N1 and N3 are buried inside the narrow pockets formed by residues P70, Y68, S44 and G45, whereas the bases of N2 and N4 are located at the top of the peptide backbones of residues G47 and P48 (). In addition, the bound ssRNAs, including the moieties of ribose, phosphorus backbone and base, are located at the top of a specific AfCsx3 structural motif (45GRGPI50) adopting a sharp “U-turn” shape linking the well-structured β3 and α2 (). Hence, the strong hydrophobic interactions between the “U-turn” structure and the ssRNA fragment, within 5 Å distance, stabilize ssRNA binding. Notably, residue R71 and the “45GRGPI50” structural motif are well conserved among Csx3 proteins, suggesting the unique ssRNA binding pocket within the AfCsx3 dimer should be conserved among Csx3 proteins.
AfCsx3 is a deadenylation exonuclease
To investigate the molecular mechanism of AfCsx3-mediated ssRNA cleavage, cleavage assays were performed by AfCsx3 in parallel with AfCas6, a homolog to Cas6 in Pyrococcus furiosus.Citation27 Notably, both AfCsx3 and AfCas6 were able to cleave the repeat ssRNA, yet the cleavage sites recognized by AfCsx3 and AfCas6 are apparently different (). Additionally, the cleavage product produced by AfCsx3 is cleavable by AfCas6, whereas the cleavage product produced by AfCas6 is resistant to AfCsx3 cleavage (). Since Cas6 specifically recognizes a 3′- hairpin loop sequence embedded within the repeat ssRNA and cleaves the 3′ terminus 8 nt off the repeat ssRNA (), we speculate that AfCsx3 may recognize the sequence at 3′-terminus of the repeat ssRNA.
Figure 4. AfCsx3 demonstrates deadenylation exonuclease activity. (A). Subsequent-cleavage assay showing different target sites in repeat ssRNA by AfCsx3 and AfCas6. Solo cleavage by AfCsx3 or AfCas6 and reaction with no protein added serve as controls. The RNA substrates and products cleaved by AfCsx3 and AfCas6 are indicated. (B). Statistical analysis of small RNA library derived from cleaved repeat ssRNA by AfCsx3. Red arrows under and above the sequence indicate the cleavage sites of repeat ssRNA by AfCas6 and AfCsx3, respectively. In consistence with sequencing results, urea-denaturing PAGE showed 4 bands corresponding to the ssRNA substrate and 3 dominant cleavage products (arrows on the gel). The secondary structure of repeat ssRNA was predicted by using the Mfold web server (http://mfold.rna.albany.edu/?q=mfold). (C). Ribonuclease activity of AfCsx3 on different ssRNA substrates derived from CRISPR sequence of Clostridium thermocellum (Ct), Pyrococcus furiosus (Pf) and Thermobifida fusca (Tf), respectively. The asterisks indicate the products of cleavable repeat ssRNAs. (D). Time-course deadenylation assays toward an in vitro transcribed RNA substrate ending with 3′-oligo (A) tail. A7 indicates the intact RNA substrates containing 7 3′ adenylate residues and A0 indicates the fully deadenylated RNA product. The cleavage sites are indicated with arrows above the sequence. (E). Deadenylation assays toward an in vitro transcribed RNA substrate ending with 10 nt 3′-oligo (A) tail. The ladder is shown with a size range from 4˜14 nt. (F). Ribonuclease activity of AfCsx3 on in vitro transcribed ssRNA substrates ending with various types of terminal nucleotides at 3′-oligo (A) tail. (G). Ribonuclease activity of AfCsx3 on ssRNA substrates with various types of terminal 3′-oligo tails.
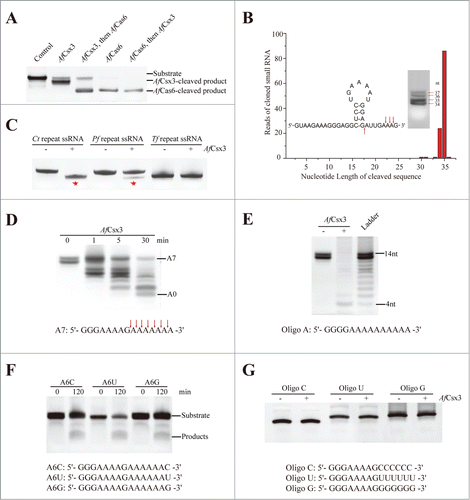
To identify the cleavage sites of the repeat ssRNA catalyzed by AfCsx3, we constructed a small RNA library by cloning the cleaved ssRNA fragments into PCR TOPO-2.1 vector for sequencing. The RNA products derived by AfCsx3 catalysis showed 4 consecutive bands by decreasing the incubation time and fine-tuning the protein/ion concentration when the cleavage assays were performed (). To avoid any false positive hits during the following selection and sequencing process, we excised the 2 product bands far away from the substrate band (). Remarkably, the sequencing results showed that 75% of repeat ssRNA was cleaved between 35th nt and 36th nt, whereas 21% of repeat ssRNA was cleaved between 34th nt and 35th nt by AfCsx3 (). In associated with the cleavage results in , all 3 cleavage sites were located adjunct to adenines at the 3′ ending sequence of the repeat ssRNA, which suggests that AfCsx3 may specifically target ssRNA substrates with the “AA” stretch at 3′ terminus ().
To validate our speculation, 3 more repeat ssRNAs derived from the CRISPR sequences of Clostridium thermocellum, Pyrococcus furiosus and Thermobifida fusca Citation24 were tested for cleavage by AfCsx3, respectively (Table S1). Notably, 2 ssRNAs were cleavable by AfCsx3 while the third ssRNA was resistant to AfCsx3 cleavage (). Surprisingly, all the cleavable ssRNAs tested so far contain “(A)AAG/C” sequences at 3′-terminus. By contrast, the 2 non-cleavable ssRNAs end with 5′-GACCG-3′ and 5′-CAUUU-3′, respectively, which lack the “AA” stretch (Table S1). These results suggest that the “AA” stretch located near the 3′ terminus might be targeted by AfCsx3. In general, AfCsx3 is able to cleave the ssRNA with the “AA” stretch at 3′ terminus and produce 3 consecutive dominant cleavage products derived from A. fulgidus repeat ssRNA ending with the 5′-GAAAG-3′ sequence. These results prompted us to speculate that AfCsx3 may function as a deadenylation exonuclease specifically targeting the poly (A) tail of the RNA in nature.
To further confirm that AfCsx3 displays deadenylase activity, we performed the RNA cleavage assay by using an in vitro transcribed ssRNA containing a 3′- 7 nt oligo (A) tail as the substrate. Strikingly, time-course deadenylation assay showed that the ssRNA fragments derived from AfCsx3 catalysis displayed obvious 8 consecutive RNA bands (). Similarly, in vitro transcribed ssRNA with a 3′- 10 nt oligo (A) tail was also cleaved into 11 consecutive RNA bands (Fig. S4). These results strongly suggest that AfCsx3 is a deadenylation exonuclease specifically targeting ssRNA substrates with poly (A) tails. Consistent with this hypothesis, the 2 ssRNAs tested above also contain internally embedded oligo (A) stretches, which are not recognizable by AfCsx3, demonstrating that the internal oligo (A) sequence is a poor substrate for AfCsx3-mediated cleavage. Furthermore, to test the maximal length of the poly (A) stretch that can be removed, we generated an in vitro transcribed RNA substrate Oligo A (14 nt) with 10 consecutive adenines at 3′ terminus. As expected, the Oligo A substrate was fully deadenylated to produce a 4 nt product ().
Moreover, in order to identify whether the 3′-terminal nucleotide other than A could be tolerated, RNA oligos with an additional G/C/U were generated by in vitro transcription and subjected to cleavage assay. The results showed that all 3 RNA oligos were cleavable by AfCsx3, yet the cleavage rates were decreased compared to the RNA oligo substrate with homogenous oligo (A) tail (). In addition, AfCsx3 showed no cleavage activity toward oligos with G, C or U stretches at the 3′-terminus (). Taken together, AfCsx3 displays robust deadenylation exonuclease activity in vitro.
The robust deadenylation exonuclease activity displayed by AfCsx3 prompted us to investigate the putative deadenylation mechanism of AfCsx3. Notably, Csx3 family proteins are a group of proteins possessing great variation in the number and sequence of amino acids. Sequence alignment showed that only the central Csx3_III-U homologous domain, approximately containing the residues of AfCsx3 40GVVI—VVQS,80 was relatively conserved. No classical DEDDH-like motifs, typical for deadenylation exonuclease, were observed among the aligned Csx3 proteins (). However, in the AfCsx3 structure, the observation of 2 pairs of well-refined manganese ions coordinated by H57, K58, H60, H80 and E85 within each catalytic core prompted us to speculate that AfCsx3 may adopt the two-metal-ion-dependent catalysis mechanism for RNA deadenylation.
Discussion
Csx3 family proteins are mostly found in archaea and bacteria and are considered as putative CRISPR/Cas proteins. However, the exact function of Csx3 is largely unknown. In this study, we have determined the crystal structures of AfCsx3 in free form, in complex with manganese ions and in complex with an ssRNA fragment. Our structures have shown that AfCsx3 forms a homodimer comprising tandem ferredoxin domains and functions as a manganese-dependent deadenylation exonuclease. Surprisingly, our biochemical and structural data indicate that the recognition and cleavage of ssRNA by AfCsx3 occurs at physically distant sites, which are located at opposite surfaces. The crystal structure of AfCsx3 in complex with an ssRNA fragment shows that ssRNA binding sites are located at the shallow cuboid lining of residues R46 and R71, which is opposite to the active sites revealed by the crystal structure of AfCsx3 in complex with manganese ions (). Moreover, structure-based mutagenesis analysis further confirmed that residues H57, K58, H60, H80 and E85 comprise the catalytic active sites, since the introduction of alanine mutation at either one or 2 of them significantly decreased catalytic activity ().
Figure 5. Catalytic mechanisms of AfCsx3. (A). Structural superimposition of AfCsx3 bound to ssRNA fragment (green) and AfCsx3 bound to manganese (cyan). The key residues involved in interaction with ssRNA and in coordination of manganese are indicated and colored in yellow and magenta, respectively. Manganese ions and ssRNA fragment are colored in red. (B). Electrostatic potential surface views of the structural superimposition. The views are the same as those in (a). The surface views show the separated RNA binding site and manganese ions binding site. The positive-charged residues which may contribute to the hypothetical path are indicated. (C). Alternative-syringe model of RNA recognition and cleavage by AfCsx3. The RNA substrate is colored in red. Incoming RNA substrate is recognized by AfCsx3 with the 5′-terminus bound to binding sites (B sites) while the 3′-polyA tail is captured and tethered to active sites (A sites, scissors) for cleavage.
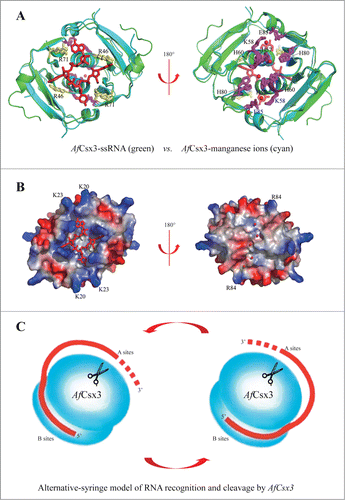
Notably, superimposition of the structures of AfCsx3 in free form and in complex with ssRNA did not show structural rearrangement after ssRNA binding (r.m.s.d. 1.1Å, 97Cα), demonstrating that the ssRNA-binding cuboid is pre-formed. Such observation indicates that the ssRNA binding sites and the catalytic sites should be constantly separated before and after ssRNA binding. The physical separation of active sites from ssRNA binding sites observed in our AfCsx3 structures prompts us to speculate that there should be a path leading from the ssRNA recognition face of AfCsx3 to the catalytic active sites at the other face of AfCsx3. Consistently, it is observed that patches of positive electrostatic potential regions (K20, K23 and R84) are lined between the active sites and the binding sites (). In the crystal structure, the half-perimeter of AfCsx3, connecting the binding and active sites, is around 45 Å, suggesting a minimal linker length of around 10 nucleotides is required to bridge the binding and cleavage sites. However, AfCsx3 was capable of cleaving RNA substrate and generated a final product with the length of 4 nt (), indicating that AfCsx3 protein moved along the RNA substrate while catalyzing consecutive deadenylation reactions.
Hence, we suggest that the AfCsx3 dimer provides 2 separate sites at opposite faces for RNA binding and cleavage, respectively. AfCsx3 binds to the incoming RNA near its 5′-terminus by binding sites on one surface and binds to the 3′-poly (A) tail by active sites on the opposite surface (). Thus, the RNA target can anchor its 5′-terminus at binding sites of AfCsx3, wrap around AfCsx3 and tether the downstream cleavage sites to AfCsx3 active sites on the opposite surface (). The 2-fold-axis dimerization arrangement of AfCsx3 can provide an alternating-syringe model to bind, tether and cleave the incoming RNA using 2 different surfaces of AfCsx3 by turns (). Therefore, our results strongly suggest that AfCsx3 is a deadenylation exonuclease employing a two-metal-ion catalytic mechanism.
Notably, the AfCsx3-encoded gene is located within a cluster of cmr genes in the genome, which suggests that the deadenylase activity of AfCsx3 may be associated with CMR-mediated RNA cleavage. RNA polyadenylation is a general post-transcriptional modification occurring in almost all organisms. In addition to functioning as the stabilizing element in eukaryotes, polyadenylation is also involved in the RNA degradation process throughout all life dom-ains.Citation32-36 During the decay process, RNA is sequentially cleaved by endonuclease, added with poly (A) or poly (A)-rich tail, and degraded by hydrolytic exoribonucleases. In archaea, exosome-like complexes are known to accomplish the process of RNA polyadenylation and exonucleolytic degradation.Citation37,38 Since CMR-mediated RNA cleavage only occurs in the region complementary to crRNA, little is known about the RNA degradation mechanisms after being cleaved by the CMR complex.Citation16,23 We speculate that the Type III-B CRISPR/Cas defense process might be accomplished by CMR-mediated RNA cleavage and polyadenylation-stimulated RNA degradation by exosome-like complexes. AfCsx3 could be employed by A. fulgidus to cooperate with an exosome-like complex to facilitate the degradation of RNA products cleaved by CMR machinery. However, the role of AfCsx3 in the defense process is yet to be determined and more data are required to further investigate the function of AfCsx3 in vivo. Nevertheless, our current work has demonstrated the deadenylase activity of AfCsx3 and should provide important insights revealing the functions of Csx3 related to CRISPR/CMR machinery for future studies.
Materials and Methods
Sequence alignment
Based on the blast-search against UniProt Knowledgebase (http://www.uniprot.org), AfCsx3, together with other 49 proteins, is classified as the Csx3_III-U homolog protein. These proteins share relatively low sequence similarities, which are varied from 27% to 58%. The sequences of the top 14 Csx3 homologues were aligned by DNAMAN (http://www.lynnon.com/dnaman.html). The aligned sequences (SWISS-PROT ID) are as follows: Y1864_ARCFU (Archaeoglobus fulgidus), Y377_AQUAE (Aquifex aeolicus), F8AJ05_PYRYC (Pyrococcus yayanosii), I3ZWS0_9EURY (Thermococcus sp. CL1), B5YBJ8_DICT6 (Dictyoglomus thermophilum), B8DZG8_DICTD (Dictyoglomus turgidum), D9S1M9_THEOJ (Thermosediminibacter oceani), D3S350_FERPA (Ferroglobus placidus), K8GEC4_9CYAN (Oscillatoriales cyanobacterium JSC-12), W6SD50_9CYAN (Arthrospira sp), Q2JVY5_SYNJA (Synechococcus sp.), K1W731_ARTPT (Arthrospira platensis C1), A0ZJA1_NODSP (Nodularia spumigena CCY9414), A0YT15_LYNSP (Lyngbya sp) and I0JXU5_9BACT (Methylacidiphilum fumariolicum SolV).
Protein expression and purification
The AfCsx3-encoding gene AF1864 was amplified by polymerase chain reaction (PCR) using the genomic DNA of A. fulgidus DSM 4304 as template. The PCR fragment was cloned into expression vector pET28-b (Novagen), which provides a hexahistidine tag at the N-terminus of the recombinant protein. The constructed plasmid was transformed into Escherichia coli (BL21/DE3) strain using a heat-shock method. Recombinant protein was induced by 0.4 mM isopropyl β-d-thiogalactoside (IPTG) and was expressed at 20°C overnight. Selenium-substituted AfCsx3 was prepared as described previously.Citation39 Cells were harvested by centrifugation and the pellets were re-suspended and passed through a cell disruptor (www.avestin.com) 3 times. The suspension was centrifuged at 35,000 rpm for 1 h to remove insoluble cell debris. The supernatant was applied onto Ni–NTA affinity column and washed sequentially with elution buffer containing increasing concentrations of imidazole. The pure protein was eluted at the concentration of 500 mM imidazole. The eluted protein was again passed through the Ni-NTA column and eluted as previously mentioned, so that the protein was more than 95% pure. The protein was resolved in a buffer solution containing 20 mM Tris-HCl (pH 7.5), 125 mM NaCl, 5% (v/v) Glycerol and 500 mM imidazole and then concentrated to ∼15 mg/ml for subsequent crystallization and biochemical analysis.
Crystallization and structure determination
Due to the lack of methionine residues, the mutant I49M/L51M, named as AfCsx3 thereafter, was generated for selenium-substituted AfCsx3 production in E. coli. Crystals of both native AfCsx3 and selenium-substituted AfCsx3 were grown at 20 ˚C by mixture of 1.0 μl of protein with 1.0 μl of reservoir containing 100 mM Citrate/Phosphate (pH 4.0) and 2 M Ammonium sulfate. These crystals grew to a maximal size of 0.1 mm × 0.1 mm × 0.3 mm over the course of 10 days.
The crystals of AfCsx3 in complex of manganese were prepared by soaking selenium-substituted AfCsx3 crystals with 0.1 M manganese chloride overnight. AfCsx3 was mixed with ssRNA target at a ratio of 1:1.1 and subjected for crystallization. The crystals of AfCsx3-ssRNA complex were grown at 20°C by mixture of 1.0 μl of AfCsx3-ssRNA with 1.0 μl of reservoir containing 20 mM magnesium chloride, 100 mM Hepes (pH 7.5) and 1.5 M lithium sulfate. The crystals were flash frozen (100 K) in the reservoir solution supplemented with 30% (v/v) glycerol.
One-wavelength data set (total 360˚ with 1˚ oscillation) for AfCsx3 in free form was collected at the wavelength of 0.9792 Å, whereas the data sets for AfCsx3 in complex with manganese and ssRNA were collected at the wavelength of 1.5418 Å at home. All data sets were processed by HKL2000.Citation40 The structure of AfCsx3 in free form was determined by single wavelength anomalous diffraction (SAD) method using SHARP/autoSHARP, whereas the structures of AfCsx3 in complex with manganese and ssRNA were determined by difference fourier electron density map and molecular replacement (Molrep/CCP4, www.ccp4.ac.uk), respectively. The models were built using the program O41 and refined using REFMAC/CCP4.Citation42 The crystallographic statistic details of the structures are listed in Table 1.
Preparation of ssRNAs
RNA oligos (A7, A10, A6C, A6U, A6G and Oligo A, were generated by in vitro transcription using T7 RiboMAX™ Express Large Scale RNA Production System (Promega). Transcription templates were prepared by annealing 2 complementary oligonucleotides (Table S1) with a T7 promoter at 5′-terminus of RNA-encoded sequence. Transcription reactions were incubated at 37°C for 4 hours and the RNA products were size-fractioned on a 20% polyacrylamide gel containing 8 M urea. Subsequently, the target bands of RNA products were excised from the gel and soaked in the elution buffer (0.3 M NaAc and 0.1% SDS) at 42°C overnight. The RNAs were extracted by TRIZOL and precipitated by isopropanol. After washed twice by 75% ice-cooled ethanol, the RNAs were dissolved in DEPC-treated water. Other RNA oligos were purchased from Research Dharmacon (http://dharmacon.gelifesciences.com).
Cleavage assay
The nucleic acid and AfCsx3 protein were incubated in buffer solution containing 200 mM KCl, 10% (v/v) Glycerol, 20 mM Hepes (pH7.0) with or without 20 mM divalent cations. Cleavage assays were performed using 20 μM protein and 10 μM substrate. All reactions were conducted at 55°C for 30 min, unless otherwise stated, and terminated by addition of formamide RNA loading buffer (90% formamide, 0.5mM EDTA pH 8.0, 0.025% SDS, 0.025% Xylene Cyanol and 0.025% Bromophenol Blue). The RNA ladder was generated by limited alkaline hydrolysis of a 14 nt RNA oligo. RNA oligo was incubated in 50 mM NaHCO3 (pH 8.0) at 100°C for 1hr and neutralized with 300 mM NaAc (pH 5.0). The RNA samples were resolved by 20% polyacrylamide gel containing 8 M urea, visualized by SYBR Gold staining and analyzed by Genesnap software.
Alanine scanning mutagenesis
Alanine scanning mutagenesis was performed against the critical residues involved in RNA recognition and manganese ion coordination, respectively. The mutant constructs were generated using PCR-driven overlapping mutagenesis method and verified by sequencing. The mutant proteins were purified by the same protocols used for wild-type AfCsx3 purification.
Small RNA library construction, sequencing and analysis
The protocols for RNA cleavage and purification are mentioned above. The cleaved RNAs were dephosphorylated by alkaline phosphatase (CIAP) before completely phosphorylated by T4 polynucleotide kinases (T4 PNK). Then 3′ RNA adapter and 5′ RNA adapter were ligated to the purified RNA sequentially by T4 ssRNA Ligase (NEB), followed by further purification. Finally, the purified RNA fragments with both 3′ and 5′ adapters were reverse transcribed using primer SBS3 and amplified by primers SBS5 and SBS3 (Table S1). The PCR products were cloned into pCR2.1-TOPO vector for small RNA library construction. After transformation, the colonies were screened and 114 positive clones were sent for sequencing. After trimming the adapter sequences, all identified sequences were compared against the original sequence to identify the cleavage site.
Data Deposition
The coordinate has been deposited in the Protein Data Bank under the accession codes 3WZG (AfCsx3 in free form), 3WZH (AfCsx3 in complex with manganese) and 3WZI (AfCsx3 in complex with ssRNA).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
X.Y. and Y.A.Y. contributed to project planning and design; X.Y. performed protein purification, crystallization and biochemical assays; Y.A.Y. performed data collection and structure determination; G.W. performed small RNA library construction; Y.A.Y. and X.Y. analyzed the data and wrote the manuscript.
Supplementary_Figures.zip
Download Zip (6.8 MB)Acknowledgments
We would like to thank J. He at Shanghai Synchrotron Radiation Facility (U17, SSRF) and H. Robinson at National Synchrotron Light Source of Brookhaven National Laboratory (X29, BNL) for assistance in data collection; S. Li at Xiamen University for analytical ultracentrifugation experiments; Y. Wang for the preliminary cloning and protein expression screening experiments.
Funding
This work was supported by research grants from Singapore Ministry of Education (MOE2014-T2-1-103); Natural Science Foundation in Jiangsu Province (BK20131189); and National University of Singapore (Suzhou) Research Institute (R-2012-N-007).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website
References
- Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature 2012; 482:331-8; PMID:22337052; http://dx.doi.org/10.1038/nature10886
- Terns MP, Terns RM. CRISPR-based adaptive immune systems. Curr Opin Microbiol 2011; 14:321-7; PMID:21531607; http://dx.doi.org/10.1016/j.mib.2011.03.005
- Yoshizumi Ishino, Hideo Shinagawa, Kozo Makino, Mitsuko Amemura, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol 1987; 169:5429-33; PMID:3316184
- Ruud Jansen, Jan DA. van Embden, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 2002; 43:1565-75; PMID:11952905; http://dx.doi.org/10.1046/j.1365-2958.2002.02839.x
- Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans 2009; 37:23-8; PMID:19143596; http://dx.doi.org/10.1042/BST0370023
- Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol 2005; 60:174-82; PMID:15791728; http://dx.doi.org/10.1007/s00239-004-0046-3
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005; 151:2551-61; PMID:16079334; http://dx.doi.org/10.1099/mic.0.28048-0
- Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005; 151:653-63; PMID:15758212; http://dx.doi.org/10.1099/mic.0.27437-0
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A Guild of 45 CRISPR-Associated (Cas) Protein Families and Multiple CRISPR-Cas Subtypes Exist in Prokaryotic Genomes. PLoS Comput Biol 2005; 1:e60; PMID:16292354; http://dx.doi.org/10.1371/journal.pcbi.0010060
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct 2011; 6:38; PMID:21756346; http://dx.doi.org/10.1186/1745-6150-6-38
- Bhaya D, Davison M, Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Ann Rev Gen 2011; 45:273-97; PMID:22060043; http://dx.doi.org/10.1146/annurev-genet-110410-132430
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 2011; 9:467-77; PMID:21552286; http://dx.doi.org/10.1038/nrmicro2577
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J 2011; 30:1335-42; PMID:21343909; http://dx.doi.org/10.1038/emboj.2011.41
- Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res 2011; 39:9275-82; PMID:21813460; http://dx.doi.org/10.1093/nar/gkr606
- Garneau JE, Dupuis ME, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadan AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010; 468:67-71; PMID:21048762; http://dx.doi.org/10.1038/nature09523
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell 2009; 139:945-56; PMID:19945378; http://dx.doi.org/10.1016/j.cell.2009.07.040
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 2008; 322:1843-5; PMID:19095942; http://dx.doi.org/10.1126/science.1165771
- Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell 2010; 37:7-19; PMID:20129051; http://dx.doi.org/10.1016/j.molcel.2009.12.033
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 2008; 321:960-4; PMID:18703739; http://dx.doi.org/10.1126/science.1159689
- Deveau H, Garneau JE, Moineau S. CRISPR/Cas system and its role in phage-bacteria interactions. Ann Rev Microbiol 2010; 64:475-93; PMID:20528693; http://dx.doi.org/10.1146/annurev.micro.112408.134123
- Huo Y, Nam KH, Ding F, Lee H, Wu L, Xiao Y, Farchione MD, Jr., Zhou S, Rajashankar K, Kurinov I, et al. Structures of CRISPR Cas3 offer mechanistic insights into Cascade-activated DNA unwinding and degradation. Nat Struct Mol Biol 2014; 21:771-7; PMID:25132177; http://dx.doi.org/10.1038/nsmb.2875
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-Guided RNA Cleavage by a CRISPR RNA-Cas Protein Complex. Cell 2009; 139:945-56; PMID:19945378; http://dx.doi.org/10.1016/j.cell.2009.07.040
- Benda C, Ebert J, Scheltema RA, Schiller HB, Baumgartner M, Bonneau F, Mann M, Conti E. Structural model of a CRISPR RNA-silencing complex reveals the RNA-target cleavage activity in Cmr4. Mol Cell 2014; 56:43-54; PMID:25280103; http://dx.doi.org/10.1016/j.molcel.2014.09.002
- Rousseau C, Gonnet M, Le Romancer M, Nicolas J. CRISPI: a CRISPR interactive database. Bioinformatics 2009; 25:3317-8; PMID:19846435; http://dx.doi.org/10.1093/bioinformatics/btp586
- van der Oost J, Jore MM, Westra ER, Lundgren M, Brouns SJ. CRISPR-based adaptive and heritable immunity in prokaryotes. Trends Biochem Sci 2009; 34:401-7; PMID:19646880; http://dx.doi.org/10.1016/j.tibs.2009.05.002
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res 2010; 38:W545-9; PMID:20457744; http://dx.doi.org/10.1093/nar/gkq366
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Gen Dev 2008; 22:3489-96; PMID:19141480; http://dx.doi.org/10.1101/gad.1742908
- Sashital DG, Jinek M, Doudna JA. An RNA-induced conformational change required for CRISPR RNA cleavage by the endoribonuclease Cse3. Nat Struct Mol Biol 2011; 18:680-7; PMID:21572442; http://dx.doi.org/10.1038/nsmb.2043
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science 2010; 329:1355-8; PMID:20829488; http://dx.doi.org/10.1126/science.1192272
- Wang R, Preamplume G, Terns MP, Terns RM, Li H. Interaction of the Cas6 riboendonuclease with CRISPR RNAs: recognition and cleavage. Structure 2011; 19:257-64; PMID:21300293; http://dx.doi.org/10.1016/j.str.2010.11.014
- Dokmanic I, Sikic M, Tomic S. Metals in proteins: correlation between the metal-ion type, coordination number and the amino-acid residues involved in the coordination. Acta crystallographica Section D, Biological crystallography 2008; 64:257-63; PMID:18323620; http://dx.doi.org/10.1107/S090744490706595X
- Portnoy V, Schuster G. The poly(A) tail of mRNAs: bodyguard in eukaryotes, scavenger in bacteria. Nucleic Acids Res 2006; 34:5923-31; PMID:17065466; http://dx.doi.org/10.1093/nar/gkl763
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 2005; 121:725-37; PMID:15935759; http://dx.doi.org/10.1016/j.cell.2005.04.030
- Portnoy V, Schuster G. RNA polyadenylation and degradation in different Archaea; roles of the exosome and RNase R. Nucleic Acids Res 2006; 34:5923-31; PMID:17065466; http://dx.doi.org/10.1093/nar/gkl763
- Slomovic S, Fremder E, Staals RH, Pruijn GJ, Schuster G. Addition of poly(A) and poly(A)-rich tails during RNA degradation in the cytoplasm of human cells. Proc Natl Acad Sci U S A 2010; 107:7407-12; PMID:20368444; http://dx.doi.org/10.1073/pnas.0910621107
- Lange H, Sement FM, Canaday J, Gagliardi D. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci 2009; 14:497-504; PMID:19716749; http://dx.doi.org/10.1016/j.tplants.2009.06.007
- Portnoy V, Evguenieva-Hackenberg E, Klein F, Walter P, Lorentzen E, Klug G, Schuster G. RNA polyadenylation in Archaea: not observed in Haloferax while the exosome polynucleotidylates RNA in Sulfolobus. EMBO Rep 2005; 6:1188-93; PMID:16282984; http://dx.doi.org/10.1038/sj.embor.7400571
- Makino DL, Halbach F, Conti E. The RNA exosome and proteasome: common principles of degradation control. Nat Rev Mol Cell Biol 2013; 14:654-60; PMID:23989960; http://dx.doi.org/10.1038/nrm3657
- Doublié S. Preparation of selenomethionyl proteins for phase determination. Methods Enzymol 1997; 276:523-30.
- Otwinowski Z, Minor W. Processing of X-Ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 1997; 276:307-26.
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved Methods for Building Protein Models in Electron Density Maps and the Location of Errors in these Models. Acta Crystallogr A 1991; 1:110-9; http://dx.doi.org/10.1107/S0108767390010224
- Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 1994; 1:760-3.
