Abstract
YciH is a bacterial protein, homologous to eukaryotic translation initiation factor eIF1. Preceding evidence obtained with the aid of in vitro translation initiation system suggested that it may play a role of a translation initiation factor, ensuring selection against suboptimal initiation complexes. Here we studied the effect of Escherichia coli yciH gene inactivation on translation of model mRNAs. Neither the translation efficiency of leaderless mRNAs, nor mRNAs with non AUG start codons, was found to be affected by YciH in vivo. Comparative proteome analysis revealed that yciH gene knockout leads to a more than fold2- increase in expression of 66 genes and a more than fold2- decrease in the expression of 20 genes. Analysis of these gene sets allowed us to suggest a role of YciH as an inhibitor of translation in a stress response rather than the role of a translation initiation factor.
Introduction
The ribosome, a machine for protein biosynthesis, is one of the most conserved macromolecular complexes in the cell. While the elongation phase of protein synthesis is organized in a similar way in bacteria and eukaryotes, the mechanisms of translation initiation are drastically different.Citation1,2 In bacteria, initiation factors 1, 2 and 3 (IF1, IF2, IF3) are sufficient to facilitate initiator fMet-tRNAfMet binding to the mRNA-programmed 30S ribosomal subunit and subsequent 50S subunit joining.Citation3,4 In a eukaryotic cell, a wider set of translation initiation factors is employed.
Several initiation factors reveal significant sequence and structure similarities between bacterial and eukaryal translation machineries. Eukaryal eIF1a (SUI1) and eIF5B are homologues of bacterial IF1 and IF2.Citation5,6 No sequence homology between bacterial IF3 and any eukaryal initiation factors could be found; however, experimental evidence is available that eIF1 fulfills the same function in eukarya.Citation7-9 Genomes of some, but not all, bacterial species such as enterobacteria and cyanobacteria encode a homolog of eIF1.Citation5 While the significant sequence,Citation5 and structuralCitation10 similarity, between eIF1 and its bacterial homolog YciH is undisputable, the functional similarity is less clear. YciH was previously demonstrated to prevent initiation complex formation on mismatched initiation codons and serve as a functional, albeit less efficient, substitute of eIF1 in vitro,Citation9 although its in vivo significance was unknown. In this work we analyzed an influence which yciH gene inactivation has on translation in bacteria.
Results
Influence of YciH on translation efficiency of model mRNAs in vivo
The YciH gene is present only in a subset of bacteriaCitation5 and is not essential,Citation11 at least at the 37°C in the rich media.Citation11,12 We used an Escherichia coli JW1274 strain from Keio knockout collectionCitation11 with an inactivated yciH gene to investigate functional significance of this putative translation initiation factor. Previous in vitro data suggested that the function of YciH is similar to that of IF3 (albeit less efficiently) in that it performs disassembly of initiation complexes formed at AUG codons located at 5′ mRNA terminus or at internal non-AUG initiation codons. In contrast to IF3, as was demonstrated in previous experiments in vitro, YciH could not dissociate 70S ribosomes into ribosomal subunits.Citation9
To test the influence of YciH on translation initiation from leaderless mRNA and non-AUG initiation codons in vivo, we used a set of reporter plasmids, encoding 2 fluorescent proteins, RFP (531/595 nm) and CER (430/486 nm) with readily distinguishable spectral properties.Citation13 In each plasmid, the RFP gene is identical and used as an internal control, while the CER fluorescent protein gene has variable translation initiation sites. In the first set of constructs () RFP and CER fluorescent proteins are encoded on a separate mRNAs transcribed from identical promoters. CER mRNA is leaderless or contain 5′UTRs with variable Shine-Dalgarno sequence or variable start codons (, left panel; Table S1).Citation14 All these plasmids were transformed into the strain with an inactivated yciH gene and in the isogenic parental strain. The translation efficiencies of leaderless mRNA and mRNAs containing non-AUG codons were demonstrated not to depend on YciH presence (, right panel). Similarly, we transformed a ΔyciH strain and the wild-type strain with a set of plasmids that contained variable lengths of the Shine-Dalgarno sequence, starting with the 6 nucleotide and ending by a complete lack of complementarity to the 16S rRNA 3′-end region (, left panel).Citation14 Again, no significant difference was detected between the translation efficiency in either the presence or absence of the functional yciH gene (, right panel). Introduction of a hairpinCitation14 into variable positions of mRNA (, left panel) affected translation efficiency dramatically (, right panel), but the presence of YciH had no influence on translation efficiency of any tested model mRNA.
Figure 1. Efficiency of model monocistronic (A) CER mRNA translation relative to the control RFP mRNA translation efficiency or second cistron CER relative to control first cistron RFP in CER-RFP bicistronic (B) mRNA in the wild type strain (white bars) and ΔyciH strain (black bars). Schematic representation of mRNA CER is presented on the left panel, while translation efficiencies are shown on the right panel. All constructs' designations are indicated next to the schematic representations. Translation efficiencies of the CER reporter were normalized to the reference RFP construct. Exact values of relative translation efficiencies are shown next to the corresponding bars.
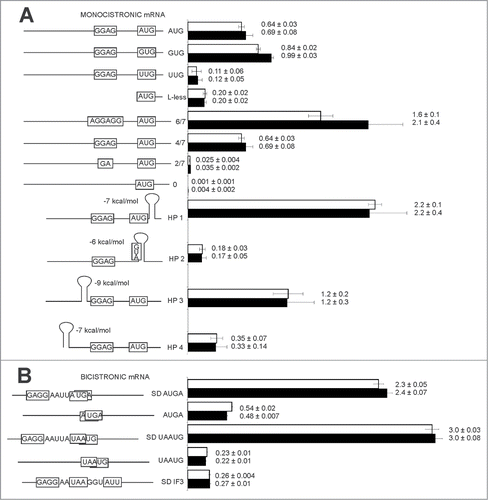
In bacteria, translation initiation could follow translation termination at the preceding open reading frame of an operon. Frequently, open reading frames can even overlap by 1 or 4 nucleotides.Citation15 We tested whether YciH could play a role in translation reinitiation on polycistronic mRNAs which have both RFP as a first cistron and CER as a second cistron on the same transcript (, left panel; Table S1).Citation14 No influence of YciH on the efficiency of re-initiation was observed (, right panel). A model of natural translation initiation site of the infC gene, starting from a non-canonical AUU codon,Citation16 was also utilized with nearly the same efficiency in the wild type strain, and the strain lacking a yciH gene (, right panel).
Influence of YciH on the proteome of the cell
We did not find any significant influence of YciH on translation efficiency of model monocistronic mRNA that differ by 5′UTR length, secondary structure, length of the Shine-Dalgarno sequence, initiation codons, as well as a set of model bicistronic mRNA. To investigate the influence of YciH on the protein composition of the cell we compared total proteomes of the ΔyciH strain and the parental strain, possessing an intact yciH gene. A label free quantitative proteome analysis revealed a set of proteins that were up- and down-regulated in the ΔyciH strain (Table S2). While a quantity of only 20 proteins was diminished more than twice upon inactivation of the yciH gene, as much as 66 proteins were found to increase their abundance upon yciH inactivation. Gene ontology (GO) term analysisCitation17 of genes downregulated in the ΔyciH strain revealed a response to heat (p-value 3·10−3), a response to stimulus (p-value 3·10−3), a response to stress (p-value 4·10−3), and a single organism catabolic process (p-value 6·10−3). No common transcription or translation factor known to control the genes downregulated in the ΔyciH strain could be revealed. Analysis of the genes significantly, more than twice, upregulated in the ΔyciH strain, revealed with high confidence relation to translation (p-value 1.3·10−6) and single organism metabolic process (p-value 4·10−6). Common transcription factors affecting a substantial number of the genes upregulated in the ΔyciH strain were found to be DksA (p-value 10−4), ArcA (p-value 3·10−3) and Fnr (p-value 8·10−3). No common features in the ribosome binding sites of genes, up- or downregulated in the ΔyciH strain could be revealed.
Influence of YciH on the growth of bacterial culture in rich and poor media
Translation efficiency is of primary value for the fast growth of bacterial cultures at optimal growth conditions, while adaptation to restricted nutrition may involve various mechanisms of translation regulation. We monitored growth of the wild type and ΔyciH strains in the rich LB and minimal M9 medium as well as upon a rapid switch from rich to poor media at mid-logarithmic growth phase (). No difference between the strains' growth in the rich medium was observed, while in the poor medium the ΔyciH strain demonstrated moderate growth retardation relative to the wild type strain. A growth disadvantage of the ΔyciH strain was also observed upon rapid transition from the rich to the poor medium.
Figure 2. Influence of yciH on E. coli growth. (A) Growth curves of the WT (gray curves) and ΔyciH (black curves) strains. Squares correspond to the growth in LB rich medium at 37°C, triangles correspond to the growth in M9 poor medium at 37°C, while circles correspond to the growth in LB rich medium at 37°C for 120 minutes followed by the substitution of LB by M9. All curves are marked on the right by the medium used. A point of LB by M9 substitution is indicated by an arrow. (B) Growth competition between the wild type and yciH knockout strains in the rich LB media. The Y-axis shows the proportion of the yciH knockout strain cells in the mixture with the wild-type calls (log scale). Each point corresponds to a 24 hour growth cycle. (C) Growth curves of the WT (light gray squares), ΔyciH (black squares) and WT with plasmid-bourne yciH (pYciH) superexpression (dark gray circles) strains in LB rich medium at 37°C. Curves are marked on the right by strain designation.
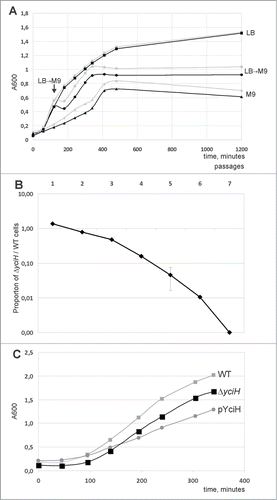
To test significance of small observed difference in growth characteristics we performed a growth competition assay. Equal numbers of cells of the kanamycin-sensitive parental strain and kanamycin resistant ΔyciH knockout strain (A600 0.001) were mixed, and grown for 24 hours at 37 °C in rich LB media. The growth cycles were repeated 7 times so that at the end of each growth cycle the mixture of the cells from preceding cycle was used to re-inoculate fresh media. After each growth cycle the cells were serially diluted and plated on LB agar plates with or without kanamycin. The kanamycin-resistant ΔyciH cell titer relative to the total cell titer is presented in . Gradual loss of ΔyciH strain from co-cultivation mixture with the parental WT strain was observed.
To check the phenotype of yciH overexpression we transformed the wild type strain with the plasmid pCA24yciH.Citation18 Expression of yciH was induced by IPTG in the rich LB media at 37°C and growth curve was monitored in comparison with untransformed strain (). Overexpression of yciH appeared to retard bacterial growth more than its inactivation.
Discussion
Several lines of evidence published in a scientific literature suggest a common origin and similar binding site on a ribosome for bacterial YciH protein and the translation initiation factor eIF1 of eukaryotes.Citation5,9,10 Previous in vitro experiments revealed that YciH could partially substitute IF3 or eIF1 in selection against erroneously formed translation initiation complexes on leaderless mRNA or non-AUG initiation codons.Citation9 Testing the significance of YciH encoded in the E. coli genome for translation initiation on leaderless mRNA and non-AUG start codons performed in this work failed to reveal any influence of YciH on the fidelity of translation initiation. No influence of YciH on translation of model mRNA carrying other specific ribosome binding site features could be found either. It might be concluded that YciH does not fulfill a role in maintaining the fidelity of translation initiation in bacteria. As was suggested in the previously published reports, this function in bacteria might be taken over by IF3.Citation9
Recent data on quantitative proteome analysis of E. coliCitation19 revealed that an average E. coli cell in a stationary phase contains 6.5 thousand ribosomes, 1 thousand IF1 molecules, 650 IF2 molecules and 2 thousand IF3 molecules. The same analysis revealed that only 15 molcules of YciH could be found in an average bacterial cell, obviously not sufficient to play a role in translation initiation. Analysis of the ribosome profile data gives a rough estimation of one order of magnitude difference between ribosome density on translation initiation factors and yciH mRNA.Citation20,21 YciH is encoded as a second gene in an operon after the pyrF gene,Citation22 encoding orotidine-5′-phosphate decarboxylase involved in pyrimidine nucleotide biosynthesis and expressed at a level 8 times exceeding that of yciH according to the quantitative proteome analysisCitation19 or comparable to that of yciH as judged from ribosome profile analysis.Citation20,21 The lack of a separate promoter and the absence of any known regulatory features, such as attenuators separating pyrF and yciH genes, make it unlikely that yciH could be significantly upregulated at any specific conditions to reach an expression level comparable with translation initiation factors.
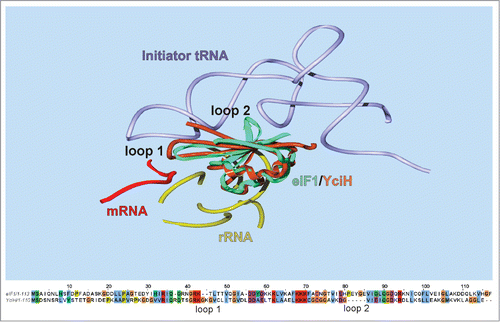
Despite an obvious structural resemblance between YciHCitation10 and eIF1Citation23 and the undoubtedly similar binding site on the small subunit of a ribosome,Citation9 YciH and eIF1 structures differ in several important regions (). YciH lacks a loop 2 corresponding to aminoacids 77–81 of eIF1, intercalating between the D- and anticodon helices of initiator tRNA (). A loop 1 of eIF1 interacting with the anticodon loop of the initiation tRNACitation24-26 is appended in YciH by 2 aminoacids. These very parts of eIF1 structure were proposed to fulfill its major function of competition with initiator tRNA fitting to the P-site thus ensuring the fidelity of start codon recognition. Both loops undergo conformational rearrangements upon start codon recognition,Citation25 leading to ejection of eIF1 from the ribosome.Citation27 In contrast, the eIF1 surface that interacts with the ribosome is conserved in YciH. On the basis of this structural comparison it might be suggested that the YciH binding site on a ribosome is the same for YciH and eIF1, while a functional interaction with initiator tRNA is specific to eIF1, but not to YciH. The fact that a bulkier loop of YciH is inserted to a place where the P-site bound tRNA anticodon is located speaks in favor of YciH possibly being an inhibitor of translation rather than an initiation factor.
Figure 3. Comparison of eIF1 and YciH primary (lower panel) and tertiary (upper panel) structure. Upper panel presents a superimposition of (I) 48S initiation complex containing 40S subunit, mRNA, initiator tRNA and eIF1A (not included into the presentation) (4KZZ), (II) PIC2 complex containing 40S subunit, eIF1A (not included into the presentation) and eIF1 (4KZY) and (III) the structure of E. coli YciH (1D1R). Only parts proximal to the position of eIF1 are shown. All components are designated on the picture. Position of loops 1 and 2 discussed in the text are indicated. Lower panel contains an alignment of mammalian (rabbit) eIF1 and bacterial (E. coli) YciH proteins.
A comparative analysis of proteomes of the wild type and ΔyciH strains also suggests an inhibitory role of YciH on translation. Three times more genes were upregulated then downregulated upon yciH inactivation. Genes downregulated upon yciH inactivation take part in a stress response, while upregulated genes are associated with translation and metabolism. It may also evidence that YciH is unlikely to play a role in enhancing translation initiation at normal growth conditions but rather inhibits translation in stress conditions. Reduced ΔyciH strain fitness in a state of limited nutrition and lack of growth retardation in a rich medium also speaks in favor of a function of YciH in stress response rather than being a translation initiation factor.
Materials and Methods
BW25141 parental strainCitation28 and JW1274 strain, carrying kanR cassette, inserted at the place of yciH geneCitation11 and pCA24yciH plasmid were kindly provided by Dr. H. Mori.
A set of reporter constructs was used in this study and a procedure for measuring translation efficiency of model mRNAs was described.Citation14 A plasmid encoding a leaderless CER gene was created in this study by PCR from pRFPCER plasmid using oligonucleotides: F 5′-ATGAAAGAGACGGACGAGAGCGGC-3′ and R 5′-GCGGCTATTATACAGAAAAATTTTCC-3′.
Expression of RFP and CER reporter genes was measured by Victor X5 plate reader (Perkin-Elmer) at wavelengths 531/595 nm and 430/486 nm after overnight growth in LB media at 37°C at vigorous shaking in a 2 ml 96-well plate.
Growth curves were monitored by automatic A600 measurement of the cultures growing in the flat bottom 96 well plate each 1 hour by Janus workstation (Perkin-Elmer).
Proteomic analysis procedure is described in supplementary methods file.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary_Material.zip
Download Zip (242.2 KB)Acknowledgments
We thank Dr. H. Mori, NIG, Japan for providing us with the knockout strain JW1274 and yciH expression plasmid. Authors are thankful to Alex Lebedeff for his help in improvement of the manuscript.
Funding
This work was supported by grants from the Russian Foundation for Basic Research 13-04-40211-Н, 14-04-01061, 13-04-00836, 15-34-20139, from the Russian Ministry of Science 14.607.21.0086, Russian Science Foundation grant 14-14-00072 and Moscow State University Development Program PNR 5.13.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci U S A 2001; 98:7029-36; PMID:11416183; http://dx.doi.org/10.1073/pnas.111145798
- Hinnebusch AG. The scanning mechanism of eukaryotic translation initiation. Ann Rev Biochem 2014; 83:779-812; PMID:24499181; http://dx.doi.org/10.1146/annurev-biochem-060713-035802
- Milon P, Rodnina MV. Kinetic control of translation initiation in bacteria. Crit Rev Biochem Mol Biol 2012; 47:334-48; PMID:22515367; http://dx.doi.org/10.3109/10409238.2012.678284
- Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochemistry 1990; 29:5881-9; PMID:2200518; http://dx.doi.org/10.1021/bi00477a001
- Kyrpides NC, Woese CR. Universally conserved translation initiation factors. Proc Natl Acad Sci U S A 1998; 95:224-8; PMID:9419357; http://dx.doi.org/10.1073/pnas.95.1.224
- Atkinson GC. The evolutionary and functional diversity of classical and lesser-known cytoplasmic and organellar translational GTPases across the tree of life. BMC Genomics 2015; 16:78; PMID:25756599; http://dx.doi.org/10.1186/s12864-015-1289-7
- Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell 2001; 8:855-64; PMID:11684020; http://dx.doi.org/10.1016/S1097-2765(01)00356-2
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 2003; 17:2786-97; PMID:14600024; http://dx.doi.org/10.1101/gad.1141803
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J 2006; 25:196-210; PMID:16362046; http://dx.doi.org/10.1038/sj.emboj.7600904
- Cort JR, Koonin EV, Bash PA, Kennedy MA. A phylogenetic approach to target selection for structural genomics: solution structure of YciH. Nucleic Acids Res 1999; 27:4018-27; PMID:10497266; http://dx.doi.org/10.1093/nar/27.20.4018
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006; 2:2006 0008; http://dx.doi.org/10.1038/msb4100050
- Gerdes SY, Scholle MD, Campbell JW, Balazsi G, Ravasz E, Daugherty MD, Somera AL, Kyrpides NC, Anderson I, Gelfand MS, et al. Experimental determination and system level analysis of essential genes in Escherichia coli MG1655. J Bacteriol 2003; 185:5673-84; PMID:13129938; http://dx.doi.org/10.1128/JB.185.19.5673-5684.2003
- Osterman IA, Prokhorova IV, Sysoev VO, Boykova YV, Efremenkova OV, Svetlov MS, Kolb VA, Bogdanov AA, Sergiev PV, Dontsova OA. Attenuation-based dual-fluorescent-protein reporter for screening translation inhibitors. Antimicrob Agents Chemother 2012; 56:1774-83; PMID:22252829; http://dx.doi.org/10.1128/AAC.05395-11
- Osterman IA, Evfratov SA, Sergiev PV, Dontsova OA. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res 2013; 41:474-86; PMID:23093605; http://dx.doi.org/10.1093/nar/gks989
- Salgado H, Moreno-Hagelsieb G, Smith TF, Collado-Vides J. Operons in Escherichia coli: genomic analyses and predictions. Proc Natl Acad Sci U S A 2000; 97:6652-7; PMID:10823905; http://dx.doi.org/10.1073/pnas.110147297
- Butler JS, Springer M, Dondon J, Graffe M, Grunberg-Manago M. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J Mol Biol 1986; 192:767-80; PMID:2438418; http://dx.doi.org/10.1016/0022-2836(86)90027-6
- Keseler IM, Mackie A, Peralta-Gil M, Santos-Zavaleta A, Gama-Castro S, Bonavides-Martinez C, Fulcher C, Huerta AM, Kothari A, Krummenacker M, et al. EcoCyc: fusing model organism databases with systems biology. Nucleic Acids Res 2013; 41:D605-12; PMID:23143106; http://dx.doi.org/10.1093/nar/gks1027
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res 2005; 12:291-9; PMID:16769691; http://dx.doi.org/10.1093/dnares/dsi012
- Wisniewski JR, Rakus D. Multi-enzyme digestion FASP and the ‘Total Protein Approach’-based absolute quantification of the Escherichia coli proteome. J Proteomics 2014; 109:322-31; PMID:25063446; http://dx.doi.org/10.1016/j.jprot.2014.07.012
- Li GW, Oh E, Weissman JS. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature 2012; 484:538-41; PMID:22456704; http://dx.doi.org/10.1038/nature10965
- Michel AM, Fox G, A MK, De Bo C, O'Connor PB, Heaphy SM, Mullan JP, Donohue CA, Higgins DG, Baranov PV. GWIPS-viz: development of a ribo-seq genome browser. Nucleic Acids Res 2014; 42:D859-64; PMID:24185699; http://dx.doi.org/10.1093/nar/gkt1035
- Turnbough CL, Jr., Kerr KH, Funderburg WR, Donahue JP, Powell FE. Nucleotide sequence and characterization of the pyrF operon of Escherichia coli K12. J Biol Chem 1987; 262:10239-45; PMID:2956254
- Fletcher CM, Pestova TV, Hellen CU, Wagner G. Structure and interactions of the translation initiation factor eIF1. EMBO J 1999; 18:2631-7; PMID:10228174; http://dx.doi.org/10.1093/emboj/18.9.2631
- Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature 2013; 500:307-11; PMID:23873042; http://dx.doi.org/10.1038/nature12355
- Hussain T, Llacer JL, Fernandez IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 2014; 159:597-607; PMID:25417110; http://dx.doi.org/10.1016/j.cell.2014.10.001
- Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat Struct Mol Biol 2013; 20:1015-7; PMID:23851459; http://dx.doi.org/10.1038/nsmb.2622
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell 2005; 17:265-75; PMID:15664195; http://dx.doi.org/10.1016/j.molcel.2004.11.051
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 2000; 97:6640-5; PMID:10829079; http://dx.doi.org/10.1073/pnas.120163297
