Abstract
Arthropod-borne flaviviruses (FVs) are a growing world-wide health threat whose incidence and range are increasing. The pathogenicity and cytopathicity of these single-stranded RNA viruses are influenced by viral subgenomic non-protein-coding RNAs (sfRNAs) that the viruses produce to high levels during infection. To generate sfRNAs the virus co-opts the action of the abundant cellular exonuclease Xrn1, which is part of the cell's normal RNA turnover machinery. This exploitation of the cellular machinery is enabled by discrete, highly structured, Xrn1-resistant RNA elements (xrRNAs) in the 3′UTR that interact with Xrn1 to halt processive 5′ to 3′ decay of the viral genomic RNA. We recently solved the crystal structure of a functional xrRNA, revealing a novel fold that provides a mechanistic model for Xrn1 resistance. Continued analysis and interpretation of the structure reveals that the tertiary contacts that knit the xrRNA fold together are shared by a wide variety of arthropod-borne FVs, conferring robust Xrn1 resistance in all tested. However, there is some variability in the structures that correlates with unexplained patterns in the viral 3′ UTRs. Finally, examination of these structures and their behavior in the context of viral infection leads to a new hypothesis linking RNA tertiary structure, overall 3′ UTR architecture, sfRNA production, and host adaptation.
Introduction
Arthropod-borne flaviviruses (FVs) are single-stranded positive-sense RNA viruses that include Dengue, Yellow Fever, Japanese Encephalitis, West Nile, Murray Valley Encephalitis, Zika, and many others. The range of their arthropod vectors continues to increase due to human trade and climate change, making them growing worldwide health threats.Citation1-5 The genomes of these FVs comprise a single-stranded RNA molecule containing one open reading frame that is flanked by structured 5′ and 3′ untranslated regions (UTRs) important for genome circularization and processes including viral replication, packaging, and translation of the viral genome.Citation6-16 Several decades ago, it was recognized that in addition to the full-length positive-sense genomic RNA, other smaller viral RNA species accumulate to high levels during FV infection.Citation17-19 These subgenomic FV RNAs (sfRNAs) were subsequently linked to viral pathogenesis in fetal mice and cytopathicity in cell culture, thus sfRNAs are directly associated with disease.Citation19 sfRNAs in diverse viruses have been reported to alter mRNA degradation patterns,Citation20 affect miRNA-dependent pathwaysCitation21, disrupt the interferon-induced antiviral response,Citation22-25 and interact with the viral replication complex.Citation26 For a more comprehensive overview of sfRNA function during infection, the reader is directed to recent reviews.Citation27-29
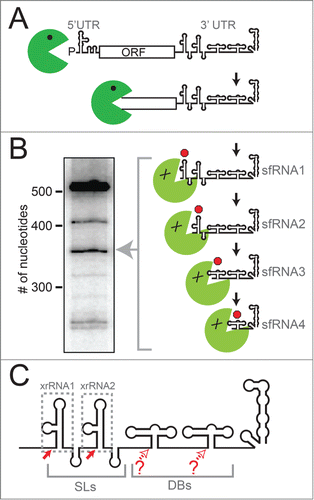
The mechanism by which sfRNAs are produced during viral infection is an example of how viruses can co-opt cellular pathways using structured RNAs. In a seminal publication, Pijlman et al. discovered that arthropod-borne FVs exploit cellular exonuclease Xrn1Citation19, an enzyme that is an important component of the cell's RNA turnover machinery.Citation30,31 During normal RNA turnover, mRNAs destined for degradation are decapped, leaving a 5′ monophosphate that is the substrate for Xrn1, which then degrades the RNA processively in a 5′ to 3′ direction.Citation30-32 During sfRNA production, Xrn1 loads on a subset of viral genomic RNAs that presumably have either been decapped or cleaved internally to leave a 5′ monophosphate, then degrades the RNA from the 5′ to the 3′ end (). When Xrn1 reaches the 3′ UTR of the viral genomic RNA it encounters structured RNA elements called Xrn1-resistant RNAs (xrRNAs).Citation33-35 Xrn1 cannot proceed through these xrRNAs, thus the RNA located 3′ of each structure is protected from degradation, resulting in sfRNA. Often more than one xrRNA element exists in the 3′ UTR of a FV, giving rise to a set of sfRNAs of different lengths ().Citation19,27,28,33-36 Recently, more divergent members of the Flaviviridae, including hepatitis C virus, have been shown to stall Xrn1 and alter the stability of the host cell's mRNA.Citation37 However, the degree of Xrn1 resistance is much less than in the arthropod-borne FVs and the RNA elements responsible do not appear to be structurally related.
Figure 1. xrRNAs and sfRNA production. (A) Diagram of the mechanism of sfRNA production by partial degradation of the viral genomic RNA by Xrn1 (green). Upon reaching the 3′ UTR, Xrn1 encounters xrRNA structures that halt enzyme progression, leading to a set of sfRNAs. (B) Northern blot analysis of the sfRNAs produced during WNVKUN infection, with sizes of RNA (number of nucleotides) shown. Diagram and blot are adapted from a previous publication.Citation35 (C) Cartoon diagram of the secondary structure of a “generic” FV 3′ UTR. The two stem-loop structures (SLs) and two dumbbell (DB) structures are shown. Different FVs have variations on this architecture, with some having only one SL or DB, and some with additional sequence or structure between these elements. Solid red arrows denote robust Xrn1 halt sites and the xrRNAs that correspond to those halt sites are boxed. Open red arrows denote possible halt sites that are less well characterized.
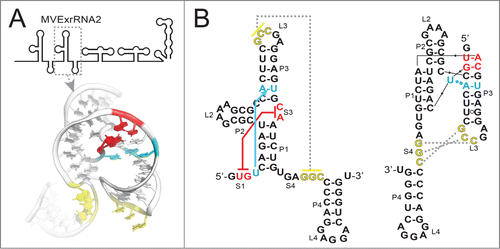
How can an RNA element less than 100 nucleotides long halt an exoribonuclease capable of degrading large structured RNAs such as rRNAs? To help answer this question, we recently solved the structure of a functional xrRNA from the Murray Valley Encephalitis Virus (MVE).Citation36 This structure revealed an RNA fold centered on a 3-way junction that forms a novel ring-like feature through which the 5′ end of the xrRNA passes (
Figure 2. Structure of the second xrRNA from MVE. (A) Top: Cartoon representation of the 3′ UTR of MVE, with the xrRNA that was crystallized indicated (MVExrRNA2). Below: structure of the xrRNA in ribbon representation. Nucleotides involved in the two important tertiary interactions are colored as per panel B. (B) Left: Secondary structure of the MVExrRNA2 drawn in a “traditional” way. Red indicates the base pairs that form between the 5′ end (S1) and nucleotides in the 3-way junction (S3), and cyan indicates the base triple that forms between a U in S1 and a base pair in P1. Yellow indicates the pseudoknot interaction between L3 and S4 that was previously predicted. Right: The secondary structure redrawn to more accurately depict the structure and these tertiary interactions.
Here, we present additional analysis and a discussion of the high-resolution crystal structure of the MVE xrRNA with the goal of gaining greater insight into how this structure relates to other FV xrRNAs and to viral infection. Our analysis reveals that certain features observed in this structure are very likely present in xrRNAs in diverse arthropod-borne FV 3′ UTRs, and these features may reveal a mechanism for how xrRNAs properly fold. Also, we find that xrRNA structures in FVs display unexpected patterns in the placement and characteristics of the xrRNAs, motivating more experiments. Together this invites a new hypothesis linking RNA tertiary structure modulation, overall 3′ UTR architecture, sfRNA production, and host adaptation.
Two tertiary interactions form an unexpected structure and suggest a folding pathway
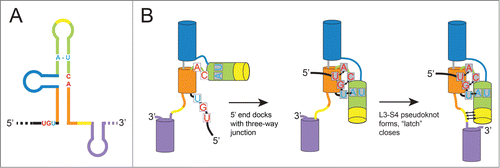
The MVE xrRNA structure reveals two long-range tertiary interactions important for stabilizing the active conformation,Citation36 which may be defining characteristics of an xrRNA. The first is a pseudoknot interaction between the L3 loop and the single-stranded S4 segment (
The xrRNA structure presents an interesting folding problem: in the context of the full viral genomic RNA, how does the 5′ end thread through the ring-like element? Consider a thought experiment in which the L3-S4 pseudoknot formed and closed the ring before the S1-S3 interaction between the 5′ end and the 3-way junction formed. In such a scenario, the entire viral genomic RNA, starting at the 5′ end, would have to thread through the closed ring (a seemingly impossible task). Fortuitously, the fact that within the crystal structure the L3-S4 pseudoknot was unpaired, yet the ring is largely formed and the 5′ end is docked, provides a hypothesis for how this scenario is avoided (
Figure 3. Hypothesized folding pathway for SL-type xrRNAs. (A) Cartoon diagram of an xrRNA, with different helical elements colored. The conserved nucleotides involved in key tertiary interactions are shown colored as per
Is there evidence for this model? In addition to the crystal structure, chemical probing experiments with an xrRNA from Dengue Virus 2 (DENV2) suggest the L3-S4 pseudoknot is conformationally dynamic or transiently formed.Citation35 The importance of this pseudoknot for Xrn1 resistance also appears to vary between xrRNAs, suggesting that the specific characteristics of this pseudoknot may modulate the robustness of Xrn1 resistance for different xrRNAs.Citation35,36 These ideas, suggested by the structure, remain largely untested, but application of modern biophysical methods linked with virology promises to give additional insight.
How conserved is the xrRNA structure and ability to robustly resist Xrn1?
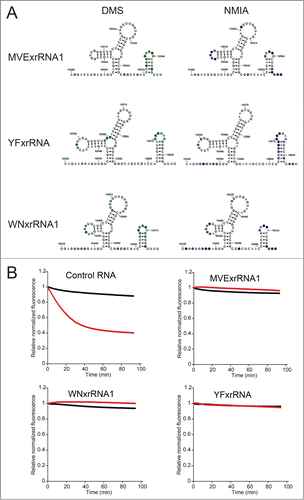
Our previously published analysis of Xrn1 resistance by elements of the DENV2 3′ UTR showed that 2 xrRNAs in this UTR resist Xrn1 degradation in a reconstituted resistance assay and also share similar secondary structures.Citation35 To extend these findings we probed three other xrRNAs. This included the first xrRNA from MVE (MVExrRNA1) (the crystal structure was of the second MVE xrRNA, MVExrRNA2), an upstream xrRNA from West Nile Virus (WNVxrRNA1) and the single identified xrRNA from Yellow Fever Virus (YFxrRNA). Chemical probing was conducted as previously described.Citation35 All displayed similar probing patterns suggesting that despite variation in primary sequence they form similar secondary structures (
Figure 4. Chemical probing and Xrn1 resistance of diverse xrRNAs. (A) Summary of chemical probing of 3 xrRNAs. All three were probed with dimethyl sulfate (DMS), left, and N-methyl isatoic anhydride (NMIA), right. Nucleotides that were modified are indicated with colored circles overlaid on the secondary structures, the degree of modification is indicated by the darkness of the color. (B) Time-course degradation assays of the 3 xrRNAs from panel A. This assay was previously describedCitation35; briefly, the loss of fluorescence (y-axis) over time (x-axis) indicates degradation of the input RNA. Black indicates no added Xrn1, red indicates reactions with Xrn1. While a control RNA is degraded by addition of Xrn1 (upper left), all 3 xrRNAs quantitatively resist Xrn1.
Evidence for conserved tertiary structures and unexpected patterns in diverse FV 3′ UTRs
The three-dimensional xrRNA structure revealed two critical tertiary interactions; if the hypothesis that this structural architecture is used by a wide range of xrRNAs from diverse FVs is correct, we should be able to predict these tertiary interactions in a wide range of FV 3′ UTRs. To this end, we examined a large set of mosquito-borne FV 3′ UTRs to determine if they contained sequence patterns consistent with conservation of 3-dimensional tertiary structure. We found patterns of putative xrRNA elements in a variety of viral 3′ UTRs, all with the potential tertiary interactions observed in the 3-dimensional structure of the MVE xrRNA (
Figure 5 (see previous page). Structural patterns in FV xrRNAs and 3′ UTRs. (A) Secondary structure cartoon of a generic xrRNA, with the highly conserved nucleotides shown and colored. (B) Structure of the MVExrRNA2 shown from 2 perspectives, with nucleotides and secondary structure elements colored to match panel A. (C) Alignments of xrRNAs from 23 arthropod-borne FVs, with sequences colored to match panels A and B. Underlined sequences are base-paired elements that are not as highly conserved. Gray colored nucleotides indicate bases that pair in most xrRNAs, but not in that particular xrRNA. The xrRNA sequences are grouped according to their position in the 3′ UTR (in the case of UTRs with 2 xrRNAs), or by the fact that they are the only xrRNA identified in the UTR. Numbers in parentheses indicate the length (in number of nucleotides) of the xrRNA. The first number is length excluding P4+L4; the second is the length including P4+L4. (D) Left: Graph depicting the correlation between the length of the upstream xrRNA (x-axis) with the length of the second xrRNA (y-axis) when there are 2 copies in a 3′ UTR. Right: Comparison of the lengths of xrRNAs in different contexts. P-values from a student's t-test are shown. Accession numbers for sequences: Bagaza, NC_012534.1; Bussuquara, NC_009026.2; Chaoyang, NC_017086.1; Dengue 1, NC_001477.1; Dengue 2, NC_001474.2; Dengue 3, NC_001475.2; Dengue 4, NC_002640.1; Donggang, NC_016997.1; Ilheus, NC_009028.2; Japanese encephalitis, NC_001437.1; Kedougou, NC_012533.1; Kokobera, NC_009029.2; Murray Valley encephalitis, NC_000943.1; Ntaya, NC_018705.3; Sepik, NC_008719.1; St. Louis encephalitis, NC_007580.2; Tembusu, NC_015843.2; Usutu, NC_006551.1; Wesselsbron, NC_012735.1; West Nile, NC_001563.2; Yellow fever, NC_002031.1; Yokose, NC_005039.1; Zika, NC_012532.1.
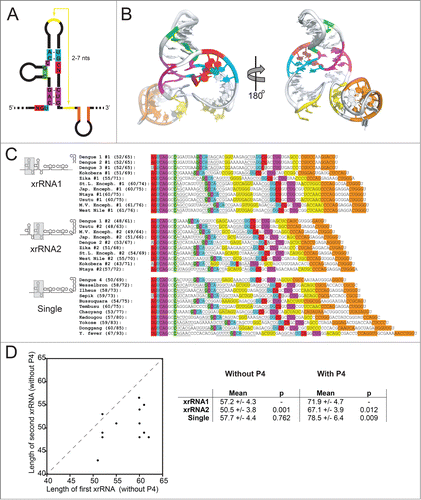
The analysis presented in
Although we cannot yet answer these questions, as a first step we compared characteristics of the sequences in
In the FV 3′UTRs that contain 2 putative xrRNAs, there is a clear trend; within each individual 3′ UTR the length of the upstream (#1) xrRNA is longer than the downstream (#2) xrRNA. This was true for all FVs except DENV2. On average, the #2 xrRNAs are shorter overall. This is true whether or not the P4 stem-loop is included in the length measurement.
The average length of putative xrRNAs in FV 3′ UTRs that are predicted to have just one copy is roughly the same as the length of the first xrRNA (#1) in those predicted to contain two copies if the P4 stem-loop is not included.
If the P4 stem-loop is included in the length measurement, the single xrRNAs were larger on average than either of the xrRNAs when two copies are present. Thus, in general, the P4/L4 stem-loop appears to have expanded in these single xrRNAs.
Is there any biological or virological significance to these patterns? The answer is unknown, in large part because the measure of length of xrRNA is not a value we can yet relate to a specific characteristic that relates to function. Does length correlate with thermodynamic stability of secondary structures or tertiary interactions, with robustness of Xrn1 resistance, or with some other undiscovered feature? Have the longer xrRNAs evolved additional functionality unrelated to Xrn1 resistance? We also note that the potential length of the L3-S4 pseudoknot varies dramatically between xrRNAs - could this correlate with stability or activity? In addition, we do not know the significance of having one xrRNA versus 2; this further complicates efforts to understand the patterns. Clearly, more experiments are needed to understand how individual xrRNA structures operate within a larger context and if, within the highly conserved xrRNAs, there are subtle variations in stability, structure, or conformational dynamics that modulate function.
A model linking xrRNA tertiary structure, sfRNA production, and viral host adaptation
We do not yet understand how an individual xrRNA tertiary structure relates to the full architecture of a FV 3′ UTR and to patterns of sfRNA production during infection, but recent observations from several papers lead to an interesting model. First, when we infected human cells with the Kunjin strain of WNV (WNVKUN), the pattern of sfRNA production demonstrated that the first xrRNA (WNVxrRNA1) was very efficient in halting Xrn1, with most of the sfRNAs produced by this upstream xrRNA (
Figure 6. Hypothesis linking xrRNA tertiary structure, sfRNA formation, and host adaptation. (A) Northern blot analysis of RNA produced during WNVKUN infection in human cells. Wild-type (WT) virus and 2 mutants in which the S1-S3 tertiary interaction was abrogated in either the first xrRNA or second xrRNA are shown. These data were previously published.Citation35 (B) Secondary structure of the MVExrRNA2 as in
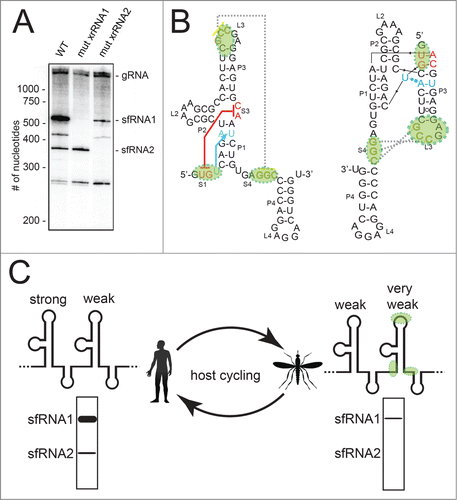
The second observation comes from the recent work of Villordo et al..Citation38 They showed that when Dengue infects mosquitos, the virus acquires mutations in the second of its 2 xrRNAs (DVxrRNA2) that revert when the mutant viruses are moved back to a mammalian host. We note that the mutations which accumulate during infection of mosquitos are specific to the parts of the xrRNA that form the two functionally critical tertiary interactions (
Conclusions
Analysis of the recent crystal structure of an xrRNA from MVE not only suggests a mechanism by which these xrRNAs halt the progression of a powerful cellular exonuclease, but also invites new hypotheses. This includes a model for how xrRNAs properly fold into their unusual structure within the context of the full viral RNA. The key tertiary structural elements appear to be well conserved across diverse mosquito-borne flaviviruses, but slight differences in these xrRNAs and their relationship to one another in the full viral 3′ UTR may alter xrRNA function in unexplored ways. Modulation of tertiary structure during infection may regulate sfRNA production and we speculate that this is related to the viruses' ability to adapt to different hosts. Understanding these phenomena may be important in continuing efforts to create new vaccines and anti-viral drugs.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors thank the members of the Kieft Lab for helpful discussions, and Catherine Musselman, Ben Akiyama, and David Costantino for critical reading of this manuscript.
Funding
This research was partially funded by NIH grant 1F32GM108257 to EGC. JSK is an Early Career Scientist of the Howard Hughes Medical Institute.
References
- Guzman MG, Harris E. Dengue. Lancet 2015; 385:453-65; PMID:25230594; http://dx.doi.org/10.1016/S0140-6736(14)60572-9
- Gray TJ, Webb CE. A review of the epidemiological and clinical aspects of West Nile virus. Int J Gen Med 2014; 7:193-203; PMID:24748813; http://dx.doi.org/10.2147/IJGM.S59902
- Hayes EB. Zika virus outside Africa. Emerg Infect Dis 2009; 15:1347-50; PMID:19788800; http://dx.doi.org/10.3201/eid1509.090442
- Weissenbock H, Hubalek Z, Bakonyi T, Nowotny N. Zoonotic mosquito-borne flaviviruses: worldwide presence of agents with proven pathogenicity and potential candidates of future emerging diseases. Vet Microbiol 2010; 140:271-80; PMID:19762169; http://dx.doi.org/10.1016/j.vetmic.2009.08.025
- Mackenzie JS, Gubler DJ, Petersen LR. Emerging flaviviruses: the spread and resurgence of Japanese encephalitis, West Nile and dengue viruses. Nat Med 2004; 10:S98-109; PMID:15577938; http://dx.doi.org/10.1038/nm1144
- Markoff L. 5′- and 3′-noncoding regions in flavivirus RNA. Adv Virus Res 2003; 59:177-228; PMID:14696330; http://dx.doi.org/10.1016/S0065-3527(03)59006-6
- de Borba L, Villordo SM, Iglesias NG, Filomatori CV, Gebhard LG, Gamarnik AV. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J Virol 2015; 89:3430-7; PMID:25589642; http://dx.doi.org/10.1128/JVI.02677-14
- Polacek C, Foley JE, Harris E. Conformational changes in the solution structure of the dengue virus 5′ end in the presence and absence of the 3′ untranslated region. J Virol 2009; 83:1161-6; PMID:19004957; http://dx.doi.org/10.1128/JVI.01362-08
- Zhang B, Dong H, Stein DA, Iversen PL, Shi PY. West Nile virus genome cyclization and RNA replication require two pairs of long-distance RNA interactions. Virology 2008; 373:1-13; PMID:18258275; http://dx.doi.org/10.1016/j.virol.2008.01.016
- Hahn CS, Hahn YS, Rice CM, Lee E, Dalgarno L, Strauss EG, Strauss JH. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J Mol Biol 1987; 198:33-41; PMID:2828633; http://dx.doi.org/10.1016/0022-2836(87)90455-4
- Khromykh AA, Meka H, Guyatt KJ, Westaway EG. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 2001; 75:6719-28; PMID:11413342; http://dx.doi.org/10.1128/JVI.75.14.6719-6728.2001
- Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 2005; 79:6631-43; PMID:15890901; http://dx.doi.org/10.1128/JVI.79.11.6631-6643.2005
- Villordo SM, Gamarnik AV. Genome cyclization as strategy for flavivirus RNA replication. Virus Res 2009; 139:230-9; PMID:18703097; http://dx.doi.org/10.1016/j.virusres.2008.07.016
- Villordo SM, Alvarez DE, Gamarnik AV. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 2010; 16:2325-35; PMID:20980673; http://dx.doi.org/10.1261/rna.2120410
- Alvarez DE, De Lella Ezcurra AL, Fucito S, Gamarnik AV. Role of RNA structures present at the 3′UTR of dengue virus on translation, RNA synthesis, and viral replication. Virology 2005; 339:200-12; PMID:16002117; http://dx.doi.org/10.1016/j.virol.2005.06.009
- Lindenbach BD, Thiel HJ, Rice CM. Flaviridae: The Viruses and Their Replication. In: Knipe DM, Howley PM, eds. Fields Virology, 5th Edition; 2007.
- Urosevic N, van Maanen M, Mansfield JP, Mackenzie JS, Shellam GR. Molecular characterization of virus-specific RNA produced in the brains of flavivirus-susceptible and -resistant mice after challenge with Murray Valley encephalitis virus. J Gen Virol 1997; 78(Pt 1):23-9; PMID:9010281; http://dx.doi.org/10.1099/0022-1317-78-1-23
- Lin KC, Chang HL, Chang RY. Accumulation of a 3′-terminal genome fragment in Japanese encephalitis virus-infected mammalian and mosquito cells. J Virol 2004; 78:5133-8; PMID:15113895; http://dx.doi.org/10.1128/JVI.78.10.5133-5138.2004
- Pijlman GP, Funk A, Kondratieva N, Leung J, Torres S, van der Aa L, Liu WJ, Palmenberg AC, Shi PY, Hall RA, et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008; 4:579-91; PMID:19064258; http://dx.doi.org/10.1016/j.chom.2008.10.007
- Moon SL, Anderson JR, Kumagai Y, Wilusz CJ, Akira S, Khromykh AA, Wilusz J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease XRN1 and alters host mRNA stability. RNA 2012; 18:2029-40; PMID:23006624; http://dx.doi.org/10.1261/rna.034330.112
- Schnettler E, Sterken MG, Leung JY, Metz SW, Geertsema C, Goldbach RW, Vlak JM, Kohl A, Khromykh AA, Pijlman GP. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and Mammalian cells. J Virol 2012; 86:13486-500; PMID:23035235; http://dx.doi.org/10.1128/JVI.01104-12
- Schuessler A, Funk A, Lazear HM, Cooper DA, Torres S, Daffis S, Jha BK, Kumagai Y, Takeuchi O, Hertzog P, et al. West nile virus noncoding subgenomic RNA contributes to viral evasion of the type I interferon-mediated antiviral response. J Virol 2012; 86:5708-18; PMID:22379089; http://dx.doi.org/10.1128/JVI.00207-12
- Chang RY, Hsu TW, Chen YL, Liu SF, Tsai YJ, Lin YT, Chen YS, Fan YH. Japanese encephalitis virus non-coding RNA inhibits activation of interferon by blocking nuclear translocation of interferon regulatory factor 3. Vet Microbiol 2013; 166:11-21; PMID:23755934; http://dx.doi.org/10.1016/j.vetmic.2013.04.026
- Bidet K, Dadlani D, Garcia-Blanco MA. G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog 2014; 10:e1004242; PMID:24992036; http://dx.doi.org/10.1371/journal.ppat.1004242
- Manokaran G, Finol E, Wang C, Gunaratne J, Bahl J, Ong EZ, Tan HC, Sessions OM, Ward AM, Gubler DJ, et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 2015; PMID:26138103
- Fan YH, Nadar M, Chen CC, Weng CC, Lin YT, Chang RY. Small noncoding RNA modulates Japanese encephalitis virus replication and translation in trans. Virol J 2011; 8:492; PMID:22040380; http://dx.doi.org/10.1186/1743-422X-8-492
- Roby JA, Pijlman GP, Wilusz J, Khromykh AA. Noncoding subgenomic flavivirus RNA: Multiple functions in west nile virus pathogenesis and modulation of host responses. Viruses-Basel 2014; 6:404-27; PMID:24473339; http://dx.doi.org/10.3390/v6020404
- Clarke BD, Roby JA, Slonchak A, Khromykh AA. Functional non-coding RNAs derived from the flavivirus 3′ untranslated region. Virus Res 2015; 206:53-61; PMID:25660582; http://dx.doi.org/10.1016/j.virusres.2015.01.026
- Moon SL, Wilusz J. Cytoplasmic viruses: rage against the (cellular RNA decay) machine. PLoS Pathog 2013; 9:e1003762; PMID:24339774; http://dx.doi.org/10.1371/journal.ppat.1003762
- Jones CI, Zabolotskaya MV, Newbury SF. The 5′ -> 3′ exoribonuclease XRN1/Pacman and its functions in cellular processes and development. Wiley Interdiscip Rev RNA 2012; 3:455-68; PMID:22383165; http://dx.doi.org/10.1002/wrna.1109
- Nagarajan VK, Jones CI, Newbury SF, Green PJ. XRN 5′ -> 3′ exoribonucleases: Structure, mechanisms and functions. Bba-Gene Regul Mech 2013; 1829:590-603; PMID:23517755
- Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem mol Biol 2004; 39:197-216; PMID:15596551; http://dx.doi.org/10.1080/10409230490513991
- Funk A, Truong K, Nagasaki T, Torres S, Floden N, Balmori Melian E, Edmonds J, Dong H, Shi PY, Khromykh AA. RNA structures required for production of subgenomic flavivirus RNA. J Virol 2010; 84:11407-17; PMID:20719943; http://dx.doi.org/10.1128/JVI.01159-10
- Silva PA, Pereira CF, Dalebout TJ, Spaan WJ, Bredenbeek PJ. An RNA pseudoknot is required for production of yellow fever virus subgenomic RNA by the host nuclease XRN1. J Virol 2010; 84:11395-406; PMID:20739539; http://dx.doi.org/10.1128/JVI.01047-10
- Chapman EG, Moon SL, Wilusz J, Kieft JS. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. eLife 2014; 3:e01892; PMID:24692447; http://dx.doi.org/10.7554/eLife.01892
- Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 2014; 344:307-10; PMID:24744377; http://dx.doi.org/10.1126/science.1250897
- Moon SL, Blackinton JG, Anderson JR, Dozier MK, Dodd BJT, Keene JD, Wilusz CJ, Bradrick SS, Wilusz J. XRN1 stalling in the 5 ' UTR of hepatitis C virus and bovine viral diarrhea virus is associated with dysregulated host mRNA stability. PLoS Pathog 2015; 11:e1004708; PMID:25747802; http://dx.doi.org/10.1371/journal.ppat.1004708
- Villordo SM, Filomatori CV, Sanchez-Vargas I, Blair CD, Gamarnik AV. Dengue virus RNA structure specialization facilitates host adaptation. PLoS Patho 2015; 11:e1004604; PMID:25635835; http://dx.doi.org/10.1371/journal.ppat.1004604
- Olsthoorn RC, Bol JF. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA 2001; 7:1370-7; PMID:11680841
