Abstract
Pre-mRNA splicing has been considered one of the hallmarks of eukaryotes, yet its diversity is astonishing: the number of substrate introns for splicing ranges from hundreds of thousands in humans to a mere handful in certain parasites. The catalytic machinery that carries out splicing, the spliceosome, is similarly diverse, with over 300 associated proteins in humans to a few tens in other organisms. In this Point of View, we discuss recent work characterizing the reduced spliceosome of the acidophilic red alga Cyanidioschyzon merolae, which further highlights the diversity of splicing in that it does not possess the U1 snRNP that is characteristically responsible for 5′ splice site recognition. Comparisons to other organisms with reduced spliceosomes, such as microsporidia, trypanosomes, and Giardia, help to identify the most highly conserved splicing factors, pointing to the essential core of this complex machine. These observations argue for increased exploration of important biochemical processes through study of a wider ranger of organisms.
Abbreviations
| RNA | = | ribonucleic acid |
| RNP | = | ribonucleoprotein |
| snRNA | = | small, nuclear RNA |
| mRNA | = | messenger RNA |
| pre-mRNA | = | precursor mRNA |
| intron | = | interrupting sequence in pre-mRNA |
| exon | = | protein-coding portion of pre-mRNA. |
Pre-mRNA Splicing is Catalyzed by the Spliceosome
Spliceosomal introns are intervening sequences present in eukaryotic precursor mRNAs (pre-mRNAs) that interrupt protein-coding regions and must be excised prior to mRNA export to the cytoplasm. The accurate removal of introns and ligation of exons is catalyzed by a large RNA-protein (RNP) complex termed the spliceosome. (; reviewed in Will.Citation1) The spliceosome typically consists of five small nuclear RNAs (snRNAs) that associate with dozens to hundreds of proteins, forming multi-snRNP complexes equalling or exceeding the complexity of the ribosome.Citation2,3 Currently, two spliceosomes of partially distinct composition have been identified in eukaryotes: the U2-dependent (major) and U12-dependent (minor) spliceosomes.(reviewed in ref. Citation4 and ref. Citation5) The U2-dependent spliceosome is responsible for the removal of the majority of spliceosomal introns, designated as U2-type, while the U12-dependent spliceosome removes a separate, rarer class of spliceosomal introns called U12-type. Notably, the U2-dependent spliceosome can catalyze both cis splicing (in which the exons to be ligated are in the same transcript) and trans splicing (a rarer process in which exons are in separate transcripts).Citation6 While most of what we know about splicing comes from studies in humans and the yeast Saccharomyces cerevisiae (), it is now clear that these well-characterized spliceosomes are not representative of those found in diverse organisms.
Figure 1. Schematic diagram of the pre-mRNA splicing reaction. Splicing factors and snRNPs are shown as ellipses and the pre-mRNA branchpoint adenosine is indicated as a circled “A.” Base pairing interactions between snRNAs and the pre-mRNA are denoted by short, vertical lines. Biochemically identified steps are indicated as “complex A,” “complex B,” etc.
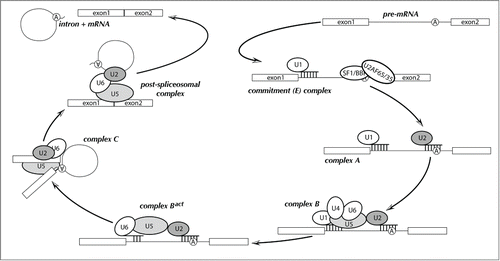
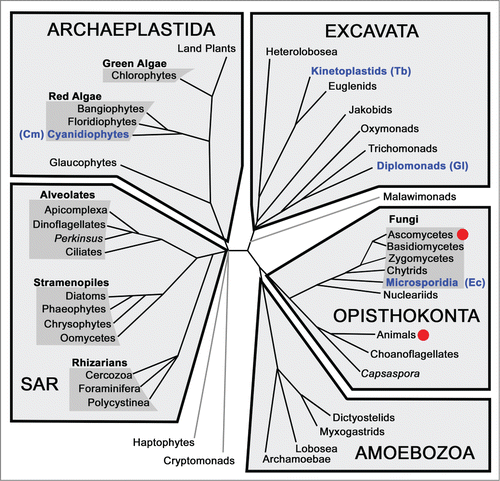
Figure 2. Schematic of eukaryotic relationships highlighting studies of splicing. Red dots indicate a lineage in which there has been extensive biochemical characterization of splicing factors and mechanisms. Clades containing organisms with reduced spliceosomes discussed in this review are in blue text with species names in parentheses: Cm - C. merolae, Ec - E. cuniculi, Gl - G. lamblia, Tb - T. brucei.
Spliceosomal Diversity
Despite the essential role they play in eukaryotic mRNA processing, spliceosomes show significant variation in their protein composition and individual snRNA structures when compared across divergent lineages. Much of our understanding of the proteomic composition of the spliceosome comes from biochemical analyses of U2-dependent spliceosomes from humans and yeast, which have identified at least 200 stable and transiently-interacting proteins in humans (). A much smaller subset of ‘core’ spliceosomal proteins appear to be conserved in eukaryotes. When comparing yeast and humans, there are ∼60 conserved abundant spliceosomal proteins.Citation7 Biochemical and bioinformatic characterization of U2-dependent spliceosomal components from the more distantly-related trypanosomes reveals an even smaller set of conserved spliceosomal proteins.Citation8 Detailed proteomic analyses in other diverse eukaryotes (particularly protists) remain to be performed (). Instead, our current understanding of spliceosomal protein (and snRNA) composition and conservation is derived primarily from bioinformatic surveys of the increasing genomic sequence data now available.
Table 1. Splicing protein complement of various organisms. Protein counts for the human spliceosome come from Agafonov,Citation3 those for C. merolae are from Stark,Citation1,7 and those for Giardia and for E. cuniculi come from Collins.Citation9.
In a comprehensive genomic survey of conserved core spliceosomal proteins across eukaryotic taxa, Collins and Penny identified orthologues for much of the U2-dependent core splicing machinery in numerous distantly-related eukaryotes.Citation9 Such conservation suggests a pivotal role for this core set of proteins in splicing, and may also indicate their presence in the eukaryotic ancestor. The spliceosomes of plants and animals appear significantly expanded in protein repertoire with many more transiently-associated protein factors.Citation10,11 The additional protein arsenal affords finer control over splice site selection under different cellular conditions, generating complex alternative splicing patterns and increased proteomic diversity. One such important class of accessory proteins are the serine-arginine rich (SR) proteins that have important regulatory roles in alternative splice site selection. In humans, there are at least 18 SR proteins,Citation3,10,12 but in intron-reduced organisms (such as S. cerevisiae) that rarely utilize alternative splicing, SR proteins and other types of splicing regulators are largely absent.
The biochemical characterization of spliceosomal proteins from different eukaryotes has also revealed several lineage-specific proteins. For example, examination of the composition of the fruit fly Drosophila melanogaster U2-dependent spliceosomal B and C complexes revealed several Drosophila proteins that do not have obvious homologues in humans, including many with known RNA-binding domains.Citation13 Similarly, biochemical analysis of flagellated protist Trypanosoma brucei spliceosomal complexes identified additional associated proteins without recognizable homologues outside of kinetoplastids that may represent lineage-specific spliceosomal innovations.Citation8
Although some spliceosomal proteins appear to be widely distributed, there are noteworthy instances where homologues of “core” U2-dependent spliceosomal proteins appear to be absent (). A recent bioinformatic search for conserved core proteins in another protozoan parasite, Giardia lamblia, revealed only 30 predicted homologues of abundant human spliceosomal proteins.Citation9 This analysis and other studies could not identify obvious candidates for numerous typically conserved U1, U4/U6 and U5 snRNP core proteins.Citation9,14 Similarly, the microsporidian Encephalitozoon cuniculi is predicted to have a spliceosome of only ∼35 proteins (), and apparently lacks U1 snRNA, although four associated proteins have been identified.Citation15,16 We therefore cannot rule out the possibility that a U1 homolog exists in E. cuniculi that has escaped bioinformatic detection. Nonetheless, absence or divergence of core spliceosomal proteins and RNAs further underscores the diversity evident in different eukaryotic spliceosomes. In addition, it raises the question of how much splicing diversity remains to be discovered.
A Highly Reduced Spliceosome in Cyanidioschyzon Merolae
We recently reported a biochemical and bioinformatic characterization of the spliceosomal components from C. merolae,Citation17 an acidophilic red alga, whose genome sequence revealed only 27 introns.Citation18 Consistent with the reduced number of introns, we determined that the core of the spliceosome is correspondingly reduced to only 43 proteins. We identified candidates for the U2, U4, U5, and U6 snRNAs, but were unable to find a candidate for U1. Immunoprecipitation of tri-methyl guanosine-capped RNAs confirmed that the snRNA candidates were indeed expressed, and TMG-capped, but again failed to reveal any RNAs with the characteristics we would expect of U1, including complementarity to the 5′ splice site, an Sm protein binding region, and secondary structure elements to which the highly conserved U1 proteins bind.
To test the possibility that U1 was present, but so divergent that we failed to recognize it, we computationally searched for homologues of all known U1 proteins, as well as all of the other known spliceosomal proteins. While we were able to identify clear homologues of many splicing proteins, including ones that associate with U2, U4, U5, and U6, we were unable to detect any proteins with similarity to known U1-associated proteins. We therefore concluded that C. merolae is in fact missing the U1 snRNP. This naturally raises the question of how the 5′ splice site is recognized (see below).
A summary of the splicing proteins present in C. merolae is shown in and . The most complex particle in this spliceosome is the U2 snRNP, which contains all three known SF3a components, and five of the known SF3b proteins, along with the U2-associated factors Msl5/BBP/SF1 and U2AF65/Mud2. The presence of so many of the elements associated with branch site recognition suggests that this is one of the most critical steps of pre-mRNA splicing, especially given how many other spliceosomal components are missing. The U5 snRNP also retains most of its associated proteins, whereas U4 appears to have lost all but two proteins, aside from the Sm ring common to U2, U4, and U5.
Figure 3. Spliceosomal protein complement from various organisms. (following ref.Citation7) Proteins are divided up according to the snRNP or sub-particle with which they are associated. Protein orthologues conserved in humans and yeast (S. cerevisiae), but not C. merolae or G. lamblia, are in light gray font, while those additionally found in C. merolae are in blue, those in G. lamblia in orange. Proteins conserved in all four organisms are in black font.
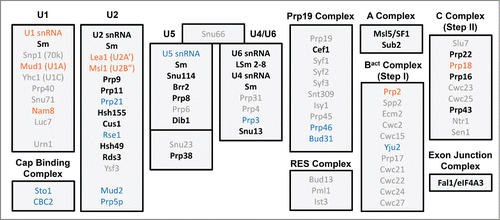
The identity of splicing proteins that appear to be missing in C. merolae is also informative. The Prp19/CDC5L complex. (NTC.Citation19) has lost all but three of its proteins - Cef1, Prp46, and Bud31 - meaning that 15 previously identified NTC proteins are absent, including the eponymous Prp19. Similarly, step-specific proteins that join the splicing machinery for only one step of splicing, such as the 7 B complex-specific proteins in humans,Citation3 are almost all absent in C. merolae.
We evaluated the set of proteins retained in C. merolae by considering their properties in other organisms. One prediction is that proteins with a more fundamental role in the assembly, organization, or catalytic function of the spliceosome would be more likely to be essential. Approximately 70 of the 97 splicing proteins from S. cerevisiae (yeast) are essential in that organism (i.e. a strain containing a deletion of the gene is inviable).Citation20 In contrast, all but three of the 43 core splicing proteins in C. merolae have orthologues that have been shown to be essential in either yeast or mice. Furthermore, all of the C. merolae proteins predicted to be present in the catalytically active C complex have orthologues associated with the salt-resistant core of the human C complex.Citation12 In other words, they correspond to those proteins that are most stably associated with the catalytic center of the spliceosome. Strikingly, the set of proteins retained in C. merolae appears to be substantially similar to those retained in G. lamblia, further supporting the central role of these proteins in splicing ( and ). Taken together, these observations suggest that the proteins missing in C. merolae perform functions that are peripheral or modulatory, for example increasing the efficiency of splicing for certain transcripts or classes of transcript, mediating interactions with regulatory factors, or coordinating with other cellular processes such as transcription and mRNA export from the nucleus.
One obvious difference between C. merolae and humans is the vast number of introns, and particularly alternatively-spliced introns, in the latter compared to the former. A large intron repertoire permits a highly flexible splicing program in which transcript-specific splicing can change in response to cell type, stage of development, or environmental factors. An extensive network of regulatory factors effects the appropriate splicing program. These include splicing enhancers and silencers in the transcripts, splicing factors such as SR proteins that bind such signals, and a variety of kinases, phosphatases and other protein and RNA-modifying enzymes that can influence the activity and substrate specificity of the spliceosome.Citation21 It appears that nearly all of this layer of regulation is absent in C. merolae. Factors that couple splicing to other processes, such as transcription and mRNA export, are also substantially reduced. The handful of regulatory and coupling proteins that have C. merolae homologues includes two SR proteins, one hnRNP protein, three proteins from the EJC/TREX complex, and a few miscellaneous proteins including Quaking, which has been implicated in alternative splicing as well as a number of other cellular processes.Citation22-24
Hybrid Spliceosomes in G. Lamblia
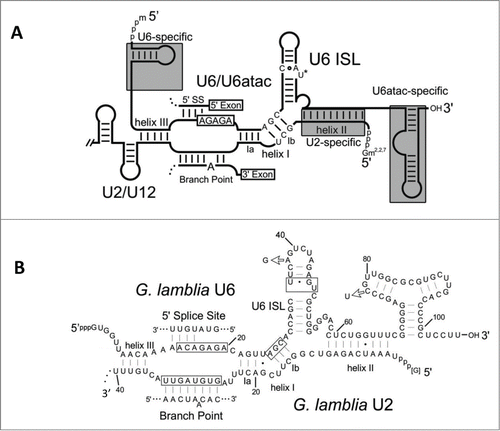
In 2012, we reported a set of novel U1, U2, U4 and U6 spliceosomal snRNAs from G. lamblia, which further exemplifies the diversity of eukaryotic spliceosomes.Citation14 Instead of being either U2 or U12-dependent snRNAs, the snRNAs identified in G. lamblia appear to be a mixture from both spliceosomes. For example, the G. lamblia U6 snRNA seems most similar to U6atac: it is truncated at its 5′ end and lacks the U6-specific 5′ stem-loop; it has an extended 3′ domain containing a stem-loop characteristic of U6atac snRNAs; and it has substantial sequence identity to U6atac (but not major U6) snRNAs (). In contrast, the 5′ half of the Giardia U2 snRNA is longer than canonical U12 snRNAs. Also, its primary sequence is more similar to U2 snRNAs from diverse eukaryotes than to U12 snRNAs ().Citation14 Thus, the G. lamblia spliceosomes appear to be a hybrid of the major and minor spliceosomes found in other organisms.
Figure 4. Secondary structures of major and minor spliceosomal snRNAs. (A) Consensus secondary structures and base pairing for the interaction between major U2/U6 and minor U12/U6atac spliceosomal snRNAs. Features distinctive of U2- and U12-dependent spliceosomes are indicated in gray boxes.(adapted from Hudson.Citation14) (B) Secondary structural predictions for the G. lamblia U2 and U6 snRNA interaction.Citation14 Conserved U2/U6 snRNA-snRNA intermolecular helices I to III are indicated, and important snRNA regions that bind intron elements or catalytic metal ions are boxed. ISL = intramolecular stem loop, SS = splice site.
Except for the shared U5 snRNA, spliceosomes containing a mix of U2-dependent and U12-dependent snRNA components have not previously been observed in vivo. This is particularly interesting because several in vivo studies have revealed that some snRNA components (or subdomains) are functionally interchangeable between the two spliceosomes.Citation25,26 For example, the highly conserved U6 snRNA intramolecular stem loop (ISL) involved in Mg2+ binding.Citation27 and essential to spliceosome catalysis may be replaced by the ISL from U6atac snRNA.Citation25 or the equivalent domain V of group II introns.Citation28 It is also intriguing that a modified U4 snRNA that can interact with U6atac can functionally replace the U4atac snRNA in U12-dependent splicing.Citation26 Taken together, these results indicate that components of U2 and U12-dependent spliceosomes are functionally interchangeable, but are segregated (at least in those eukaryotes characterized to date) through the recognition of spliceosome-specific snRNA motifs by U2 and U12 class-specific spliceosomal proteins.
It is tempting to speculate that loss of major-minor spliceosome segregating factors in eukaryotic lineages undergoing extensive genomic reduction and intron loss may permit the formation of spliceosomes containing mixtures of U2- and U12-dependent snRNAs capable of splicing novel “chimeric” U2/U12-type intron classes. Intriguingly, we found that one of the G. lamblia introns has the AT/AC termini characteristic of U12-type introns, while possessing extended 5′ splice site and branch site sequences generally found in U2-type introns.Citation14 The finding of mixed snRNA types and introns with chimeric major-minor features underscores the remarkable diversity of spliceosome and intron evolution.
Group II Introns and Reversible Complexity
Spliceosomal introns are defining features of eukaryotic nuclear genomes and have not been identified in any eukaryotic organellar or prokaryotic genome. There is compelling evidence, however, that spliceosomes share ancestry with self-splicing group II introns,Citation29-33 which are found in certain bacterial genomes and eukaryotic organelles, but are absent from all characterized nuclear genomes.Citation34 This raises a variety of questions about the timing of the split between self-splicing and spliceosomal introns, the evolutionary steps by which the latter acquired additional protein and RNA components, and the ramifications for nuclear function of having a system in which a trans-acting spliceosome can remove an arbitrary number of genomically-encoded introns. We suggest that the acquistion of complexity.Citation35,36 happened largely through a process of accretion (i.e., growth by serial addition of factors to the outside of the spliceosome), as has been elegantly demonstrated for the ribosome.Citation37 We therefore predict that in reduced genomes such as that of C. merolae, reduction of complexity would happen by the reverse process, that is removing the most peripheral splicing factors first.
Group II intron RNAs are organized into 6 conserved structural domains (I – VI) with domains I, V and VI showing similarity to the U5, U6 and U2 snRNAs from the spliceosome, respectively. Consequently, one theory posits that the spliceosomal U2, U5 and U6 snRNAs were the first to emerge from the common ancestor, and were derived from fragments of group II introns.Citation38 The U1 and U4 snRNAs (which have no obvious counterparts in group II introns) evolved later as key components of the splicing reaction. In light of this, the case of a missing U1 snRNA in C. merolae is intriguing and suggests that this organism possesses a less complex mechanism of 5′ splice site recognition, possibly via more extensive interaction of U6 snRNA with the 5′ splice site. The lack of U1 in C. merolae mimics the proposed ancestral state in which primordial spliceosomes utilized fewer snRNA components. However, it is important to note that algal relatives of C. merolae possess a U1 snRNP, hence it is clear that the U1 absence in C. merolae is a derived situation, not ancestral. Indeed, we propose that C. merolae is an exemplary case of reversible spliceosomal complexity, where an entire snRNP particle may have been lost. Assuming that complexity is lost roughly in the reverse order by which it is gained, every characterized spliceosome, and particularly the most reduced ones, would yield a snapshot of a possible step in the evolutionary divergence of spliceosomes from group II introns.
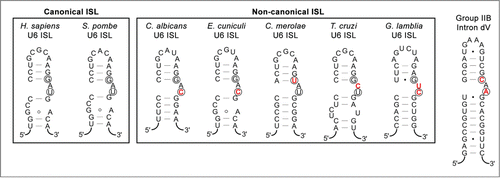
While loss of U1 provides a dramatic example of the loss of complexity, the U6 ISL provides an example of the opposite, namely a central catalytic feature that has apparently been retained largely intact since spliceosomes diverged from group II introns. Even in the organisms with highly reduced spliceosomes discussed above, the ISL maintains many of the same features as in highly elaborate spliceosomes (). The recent EM structure of the S. pombe spliceosome reveals some of the structural constraints that have ensured the U6 ISL's endurance.Citation33 Notably, the second bulged nucleotide in the U6 ISL () forms a base-triple with U2/U6 stem Ib, an interaction that is also seen in the core of the group II intron. This nucleotide has been shown to coordinate a catalytic magnesium atom,Citation31 suggesting that the base triple with the “C” of the AGC catalytic triad anchors the magnesium in the correct location. While the “canonical” (i.e. well-studied) human and S. pombe spliceosomes happen to have highly similar U6 ISLs, the reduced splicesomes discussed above demonstrate a substantial amount of flexibility in the composition of the bulged nucleotides (). While we predict that all of these ISLs will turn out to form a similar 3-dimensional structure in the respective cores of their spliceosomes, it would be interesting to determine whether these less canonical U6 ISL motifs reduce catalytic efficiency during splicing and are therefore better tolerated in organisms that are extremely intron poor.
Figure 5. U6 snRNA intramolecular stem-loops (ISLs) compared to domain V of group II introns. Secondary structure predictions for canonical and non-canonical U6 ISLs from Homo sapiens,Citation50 Schizosaccharomyces pombe,Citation33 Candida albicans,Citation51 Encephalitozoon cuniculi,Citation15 Cyanidioschyzon merolae,Citation17 Trypanosoma cruzi,Citation52 and Giardia lamblia.Citation14 are compared to the P.li.LSUI2 group IIB intron from Pylaiella littoralis.Citation53 Nucleotides involved in Mg2+ binding are circled and nucleotides in the U6 ISL bulge region that differ from the eukaryotic consensus are indicated in red text.
Intron Loss and Spliceosome Evolution
In contrast to the notion that spliceosome reduction can only follow one path, changes in intron and spliceosome structure show markedly different patterns in eukaryotes assumed to be in the process of progressive intron loss and possibly even spliceosome elimination. C. merolae, G. lamblia, T. brucei, and E. cuniculi are examples of very intron-poor eukaryotes that display unique differences in spliceosome evolution. In C. merolae, there is no evidence for a U1 snRNP, and its function may be replaced by U5 or U6.Citation17 The parasitic protist T. brucei has only two documented cis-spliced spliceosomal introns.Citation39,40 and undergoes spliced leader (SL) trans-splicing,Citation41 a process that utilizes the SL RNA to put common 5′ end leader sequences on a subset of mRNAs.Citation6 The U1 snRNA is highly structurally divergent in this organism and in other trypanosomatids.Citation42,43 In G. lamblia, where only 12 introns have been identified to date, a significant number of those undergo genic trans-splicing, i.e., joining exons from different transcripts with no SL RNA.Citation44-46 Further spliceosome variability is seen in the Microsporidia, a group of unicellular, parasitic, early-diverging fungi. E. cuniculi is predicted to have a small spliceosome of ∼35 proteins.Citation15,16 The E. cuniculi genome contains only 37 introns, which we found to be spliced at very low levels, presumably as a direct result of spliceosome reduction.Citation47 Indeed, a number of species of microsporidia have taken spliceosome and intron reduction to the extreme, where no evidence for spliceosomal introns or spliceosome machinery can be identified.Citation48,49
The presence of spliceosomal introns and their removal from messages has long been considered a hallmark of the eukaryotic form, yet the process has only been carefully examined in a small number of organisms (). When more diverse lineages are assessed, this glimpse into splicing landscapes reveals highly varied genome architectures and spliceosomal complexity, indicating a surprising level of plasticity for this large RNP complex. The full extent of this plasticity is unknown, and will await investigation of a larger collection of eukaryotes. Indeed, each example of a missing spliceosomal component previously deemed universal helps us to further refine our view of what defines the essential functional core of this enigmatic macromolecular machine.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank the members of our labs for thoughtful comments on the manuscript.
References
- Will CL, Lührmann R. Spliceosome Structure and Function. Cold Spring Harbor Perspectives Biol 2011; 3:1-23; http://dx.doi.org/10.1101/cshperspect.a003707
- Nilsen TW. The spliceosome: the most complex macromolecular machine in the cell? Bioessays 2003; 25:1147-9; PMID:14635248; http://dx.doi.org/10.1002/bies.10394
- Agafonov DE, Deckert J, Wolf E, Odenwälder P, Bessonov S, Will CL, Urlaub H, Lührmann R. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol Cell Biol 2011; 31:2667-82; PMID:21536652; http://dx.doi.org/10.1128/MCB.05266-11
- Patel AA, Steitz JA. Splicing double: insights from the second spliceosome. Nat Rev Mol Cell Biol 2003; 4:960-70; PMID:14685174; http://dx.doi.org/10.1038/nrm1259
- Will CL, Lührmann R. Splicing of a rare class of introns by the U12-dependent spliceosome. Biol Chem 2005; 386:713-24; PMID:16201866; http://dx.doi.org/10.1515/BC.2005.084
- Lasda EL, Blumenthal T. Trans-splicing. Wiley Interdiscip Rev RNA 2011; 2:417-34; PMID:21957027; http://dx.doi.org/10.1002/wrna.71
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 2009; 36:593-608; PMID:19941820; http://dx.doi.org/10.1016/j.molcel.2009.09.040
- Luz Ambrósio D, Lee JH, Panigrahi AK, Nguyen TN, Cicarelli RMB, Günzl A. Spliceosomal proteomics in Trypanosoma brucei reveal new RNA splicing factors. Eukaryotic Cell 2009; 8:990-1000; http://dx.doi.org/10.1128/EC.00075-09
- Collins L, Penny D. Complex spliceosomal organization ancestral to extant eukaryotes. Mol Biol Evol 2005; 22:1053-66; PMID:15659557; http://dx.doi.org/10.1093/molbev/msi091
- Barbosa-Morais NL, Carmo-Fonseca M, Aparício S. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res 2006; 16:66-77; PMID:16344558; http://dx.doi.org/10.1101/gr.3936206
- Hegele A, Kamburov A, Grossmann A, Sourlis C, Wowro S, Weimann M, Will CL, Pena V, Lührmann R, Stelzl U. Dynamic protein-protein interaction wiring of the human spliceosome. Mol Cell 2012; 45:567-80; PMID:22365833; http://dx.doi.org/10.1016/j.molcel.2011.12.034
- Bessonov S, Anokhina M, Krasauskas A, Golas MM, Sander B, Will CL, Urlaub H, Stark H, Lührmann R. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 2010; 16:2384-403; PMID:20980672; http://dx.doi.org/10.1261/rna.2456210
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Lührmann R. Conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosomal complexes. Mol Cell Biol 2009; 29:281-301; PMID:18981222; http://dx.doi.org/10.1128/MCB.01415-08
- Hudson AJ, Moore AN, Elniski D, Joseph J, Yee J, Russell AG. Evolutionarily divergent spliceosomal snRNAs and a conserved non-coding RNA processing motif in Giardia lamblia. Nucleic Acids Res 2012; 40:10995-1008; PMID:23019220; http://dx.doi.org/10.1093/nar/gks887
- Katinka MD, Duprat S, Cornillot E, Méténier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature 2001; 414:450-3; PMID:11719806; http://dx.doi.org/10.1038/35106579
- Lopez MD, Alm Rosenblad M, Samuelsson T. Computational screen for spliceosomal RNA genes aids in defining the phylogenetic distribution of major and minor spliceosomal components. Nucleic Acids Res 2008; 36:3001-10; PMID:18390578; http://dx.doi.org/10.1093/nar/gkn142
- Stark MR, Dunn EA, Dunn WSC, Grisdale CJ, Daniele AR, Halstead MRG, Fast NM, Rader SD. Dramatically reduced spliceosome in Cyanidioschyzon merolae. Proc Natl Acad Sci USA 2015; 112:E1191-200; PMID:25733880; http://dx.doi.org/10.1073/pnas.1416879112
- Matsuzaki M, Misumi O, Shin-I T, Maruyama S, Takahara M, Miyagishima S-Y, Mori T, Nishida K, Yagisawa F, Nishida K, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 2004; 428:653-7; PMID:15071595; http://dx.doi.org/10.1038/nature02398
- Cheng SC, Tarn WY, Tsao TY, Abelson J. PRP19: a novel spliceosomal component. Mol Cell Biol 1993; 13:1876-82; PMID:8441419
- Zhang R, Ou H-Y, Zhang C-T. DEG: a database of essential genes. Nucleic Acids Res 2004; 32:D271-2; PMID:14681410; http://dx.doi.org/10.1093/nar/gkh024
- Stoltzfus CM, Madsen JM. Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing. Curr HIV Res 2006; 4:43-55; PMID:16454710; http://dx.doi.org/10.2174/157016206775197655
- Hall MP, Nagel RJ, Fagg WS, Shiue L, Cline MS, Perriman RJ, Donohue JP, Ares M. Quaking and PTB control overlapping splicing regulatory networks during muscle cell differentiation. RNA 2013; 19:627-38; PMID:23525800; http://dx.doi.org/10.1261/rna.038422.113
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015; 160:1125-34; PMID:25768908; http://dx.doi.org/10.1016/j.cell.2015.02.014
- van der Veer EP, de Bruin RG, Kraaijeveld AO, de Vries MR, Bot I, Pera T, Segers FM, Trompet S, van Gils JM, Roeten MK, et al. Quaking, an RNA-binding protein, is a critical regulator of vascular smooth muscle cell phenotype. Circ Res 2013; 113:1065-75; PMID:23963726; http://dx.doi.org/10.1161/CIRCRESAHA.113.301302
- Shukla GC, Padgett RA. The intramolecular stem-loop structure of U6 snRNA can functionally replace the U6atac snRNA stem-loop. RNA 2001; 7:94-105; PMID:11214185; http://dx.doi.org/10.1017/S1355838201000218
- Shukla GC, Padgett RA. U4 small nuclear RNA can function in both the major and minor spliceosomes. Proc Natl Acad Sci USA 2004; 101:93-8; PMID:14691257; http://dx.doi.org/10.1073/pnas.0304919101
- Yean SL, Wuenschell G, Termini J, Lin RJ. Metal-ion coordination by U6 small nuclear RNA contributes to catalysis in the spliceosome. Nature 2000; 408:881-4; PMID:11130730; http://dx.doi.org/10.1038/35048617
- Shukla GC, Padgett RA. A catalytically active group II intron domain 5 can function in the U12-dependent spliceosome. Mol Cell 2002; 9:1145-50; PMID:12049749; http://dx.doi.org/10.1016/S1097-2765(02)00505-1
- Seetharaman M, Eldho NV, Padgett RA, Dayie KT. Structure of a self-splicing group II intron catalytic effector domain 5: parallels with spliceosomal U6 RNA. RNA 2006; 12:235-47; PMID:16428604; http://dx.doi.org/10.1261/rna.2237806
- Toor N, Keating KS, Taylor SD, Pyle AM. Crystal structure of a self-spliced group II intron. Science 2008; 320:77-82; PMID:18388288; http://dx.doi.org/10.1126/science.1153803
- Fica SM, Mefford MA, Piccirilli JA, Staley JP. Evidence for a group II intron-like catalytic triplex in the spliceosome. Nat Struct Mol Biol 2014; 21:464-71; PMID:24747940; http://dx.doi.org/10.1038/nsmb.2815
- Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature 2013; 493:638-43; PMID:23354046; http://dx.doi.org/10.1038/nature11843
- Hang J, Wan R, Yan C, Shi Y. Structural basis of pre-mRNA splicing. Science 2015; 349(6253):1191-8
- Candales MA, Duong A, Hood KS, Li T, Neufeld RAE, Sun R, McNeil BA, Wu L, Jarding AM, Zimmerly S. Database for bacterial group II introns. Nucleic Acids Res 2012; 40:D187-90; PMID:22080509; http://dx.doi.org/10.1093/nar/gkr1043
- Gray MW, Lukes J, Archibald JM, Keeling PJ, Doolittle WF. Irremediable complexity? Science 2010; 330:920-1; PMID:21071654; http://dx.doi.org/10.1126/science.1198594
- Stoltzfus A. On the possibility of constructive neutral evolution. J Mol Evol 1999; 49:169-81; PMID:10441669; http://dx.doi.org/10.1007/PL00006540
- Bokov K, Steinberg SV. A hierarchical model for evolution of 23S ribosomal RNA. Nature 2009; 457:977-80; PMID:19225518; http://dx.doi.org/10.1038/nature07749
- Zimmerly S, Semper C. Evolution of group II introns. Mob DNA 2015; 6:7; PMID:25960782; http://dx.doi.org/10.1186/s13100-015-0037-5
- Kolev NG, Franklin JB, Carmi S, Shi H, Michaeli S, Tschudi C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog 2010; 6:e1001090; PMID:20838601; http://dx.doi.org/10.1371/journal.ppat.1001090
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GAM. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res 2010; 38:4946-57; PMID:20385579; http://dx.doi.org/10.1093/nar/gkq237
- Perry KL, Watkins KP, Agabian N. Trypanosome mRNAs have unusual “cap 4” structures acquired by addition of a spliced leader. Proc Natl Acad Sci USA 1987; 84:8190-4; PMID:3120186; http://dx.doi.org/10.1073/pnas.84.23.8190
- Schnare MN, Gray MW. A candidate U1 small nuclear RNA for trypanosomatid protozoa. J Biol Chem 1999; 274:23691-4; PMID:10446125; http://dx.doi.org/10.1074/jbc.274.34.23691
- Djikeng A, Ferreira L, D'Angelo M, Dolezal P, Lamb T, Murta S, Triggs V, Ulbert S, Villarino A, Renzi S, et al. Characterization of a candidate Trypanosoma brucei U1 small nuclear RNA gene. Mol Biochem Parasitol 2001; 113:109-15; PMID:11254959; http://dx.doi.org/10.1016/S0166-6851(00)00384-4
- Roy SW, Hudson AJ, Joseph J, Yee J, Russell AG. Numerous fragmented spliceosomal introns, AT-AC splicing, and an unusual dynein gene expression pathway in Giardia lamblia. Mol Biol Evol 2012; 29:43-9; PMID:21482665; http://dx.doi.org/10.1093/molbev/msr063
- Kamikawa R, Inagaki Y, Tokoro M, Roger AJ, Hashimoto T. Split introns in the genome of Giardia intestinalis are excised by spliceosome-mediated trans-splicing. Curr Biol 2011; 21:311-5; PMID:21315596; http://dx.doi.org/10.1016/j.cub.2011.01.025
- Nageshan RK, Roy N, Hehl AB, Tatu U. Post-transcriptional repair of a split heat shock protein 90 gene by mRNA trans-splicing. J Biol Chem 2011; 286:7116-22; PMID:21209094; http://dx.doi.org/10.1074/jbc.C110.208389
- Grisdale CJ, Bowers LC, Didier ES, Fast NM. Transcriptome analysis of the parasite Encephalitozoon cuniculi: an in-depth examination of pre-mRNA splicing in a reduced eukaryote. BMC Genomics 2013; 14:207; PMID:23537046; http://dx.doi.org/10.1186/1471-2164-14-207
- Akiyoshi DE, Morrison HG, Lei S, Feng X, Zhang Q, Corradi N, Mayanja H, Tumwine JK, Keeling PJ, Weiss LM, et al. Genomic survey of the non-cultivatable opportunistic human pathogen, Enterocytozoon bieneusi. PLoS Pathog 2009; 5:e1000261; PMID:19132089; http://dx.doi.org/10.1371/journal.ppat.1000261
- Cuomo CA, Desjardins CA, Bakowski MA, Goldberg J, Ma AT, Becnel JJ, Didier ES, Fan L, Heiman DI, Levin JZ, et al. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res 2012; 22:2478-88; PMID:22813931; http://dx.doi.org/10.1101/gr.142802.112
- Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 1992; 71:803-17; PMID:1423631; http://dx.doi.org/10.1016/0092-8674(92)90556-R
- Mitrovich QM, Guthrie C. Evolution of small nuclear RNAs in S. cerevisiae, C. albicans, and other hemiascomycetous yeasts. RNA 2007; 13:2066-80; PMID:17956975; http://dx.doi.org/10.1261/rna.766607
- Ambrósio DL, Silva MTA, Cicarelli RMB. Cloning and molecular characterization of Trypanosoma cruzi U2, U4, U5, and U6 small nuclear RNAs. Mem Inst Oswaldo Cruz 2007; 102:97-105; http://dx.doi.org/10.1590/S0074-02762007000100017
- Robart AR, Chan RT, Peters JK, Rajashankar KR, Toor N. Crystal structure of a eukaryotic group II intron lariat. Nature 2014; 514(7521):193-7; PMID:25252982
