ABSTRACT
Coenzyme Q (CoQ) is a key component of the mitochondrial respiratory chain carrying electrons from complexes I and II to complex III and it is an intrinsic component of the respirasome. CoQ concentration is highly regulated in cells in order to adapt the metabolism of the cell to challenges of nutrient availability and stress stimuli. At least 10 proteins have been shown to be required for CoQ biosynthesis in a multi-peptide complex and COQ7 is a central regulatory factor of this pathway. We found that the first 765 bp of the 3′-untranslated region (UTR) of COQ7 mRNA contains cis-acting elements of interaction with RNA-binding proteins (RBPs) HuR and hnRNP C1/C2. Binding of hnRNP C1/C2 to COQ7 mRNA was found to require the presence of HuR, and hnRNP C1/C2 silencing appeared to stabilize COQ7 mRNA modestly. By contrast, lowering HuR levels by silencing or depriving cells of serum destabilized and reduced the half-life of COQ7 mRNA, thereby reducing COQ7 protein and CoQ biosynthesis rate. Accordingly, HuR knockdown decreased oxygen consumption rate and mitochondrial production of ATP, and increased lactate levels. Taken together, our results indicate that a reduction in COQ7 mRNA levels by HuR depletion causes mitochondrial dysfunction and a switch toward an enhanced aerobic glycolysis, the characteristic phenotype exhibited by primary deficiency of CoQ10. Thus HuR contributes to efficient oxidative phosphorylation by regulating of CoQ10 biosynthesis.
Introduction
Coenzyme Q (CoQ) or ubiquinone is a lipid-soluble molecule synthesized and mainly concentrated in the mitochondrial inner membrane of all eukaryotic cells. CoQ is an electron carrier from Complexes I and II to Complex III of the mitochondrial respiratory chain, it has antioxidant properties and regulates permeability transition pore, pyrimidine biosynthesis and β–oxidation of fatty acids.Citation1 CoQ is not only a component of complex III in which involves the CoQ cycle,Citation2 but it also contributes to its stability.Citation3 Recently, we demonstrated that CoQ is a key component of the respiratory chain superassembly complex,Citation4 which contributes to selecting internal electrons routes and to regulating the production of reactive oxygen species in the oxidative phosphorylation pathway.Citation5
CoQ is composed by a benzoquinone ring synthesized from tyrosine, attached to a poly-isoprenoid side chain derived from the mevalonate pathway. Benzoquinone ring final modifications take place in mitochondria by a pathway that involves at least 8 genes (COQ3-COQ9, ADCK4), whose products appear to form a multi-enzyme complex.Citation1,6 CoQ homeostasis is essential for mitochondria efficiency, and thus its biosynthesis is highly regulated. Disruption of CoQ homeostasis can induce its deficiency caused by either mutations in any of the genes involved in its biosynthesis (primary deficiency), which leads to a rare disorder with variable clinical phenotypes exhibiting major effects on the neuromuscular axis and nephrotic syndrome,Citation7-Citation9 or as an adaptive response to dysfunctional mitochondria.Citation10,11 On the contrary, healthy aging interventions such as calorie restriction and treatment with resveratrol induce mitochondrial efficiency, activating diverse bioenergetics pathways including CoQ biosynthesis.Citation12,13 Additionally, it has been demonstrated that the levels of CoQ are essential for regulating longevity in invertebrate models.Citation14,15 Therefore, CoQ levels in mitochondria must be tightly regulated to balance oxidative phosphorylation and to adapt to changes in nutritional and/or physiological conditions.
Regulatory mechanisms of CoQ biosynthesis include the transcriptional regulation of enzymes in the mevalonate pathway via the transcription factor PPARα,Citation16,17 and the transcriptional induction of COQ7 expression by NF-κB in response to oxidative stress.Citation18 Furthermore, CoQ biosynthesis is also regulated by post-translational modification of COQ7 by either a nutrient-dependent phosphorylation-dephosphorylation cycle in yeastCitation19,20 or by binding to COQ9.Citation21
Post-transcriptional control of gene expression includes the formation of ribonucleoprotein (RNP) complexes that can regulate mRNA translation.Citation22,23 COQ7 mRNA was identified as a putative target of HuR by ribonucleoprotein immunoprecipitation (RIP) followed by microarray detection of target mRNAs using arrays.Citation24 HuR is a ubiquitously expressed RNA-binding protein (RBP) member of ELAVL protein familyCitation25 that can regulate the stability and translation of hundreds mRNA involved in cell division, carcinogenesis, muscle cell differentiation, replicative senescence, immune cell activation, and stress response.Citation26-Citation30 HuR and several members of the hnRNP family as hnRNP D (also known as AUF1) are able to bind specifically to AU-rich elements (AREs) in the 3′UTR of target mRNAs,Citation30-Citation32 in cooperative or competitive manners to regulate the translation of shared target mRNAs.Citation31,33
We show here that HuR and hnRNP C1/C2 proteins, both of which bind to the COQ7 3′UTR, determine the rates of CoQ10 biosynthesis. On the one hand, HuR enhances COQ7 mRNA stability and helps maintain physiological COQ7 mRNA, COQ7 protein levels, and CoQ10 biosynthesis, while hnRNP C1/C2 binds to COQ7 mRNA by interacting with HuR and triggers its degradation. The stability of COQ7 mRNA is decreased by serum withdrawal leading to an enhanced aerobic glycolysis. We propose that these 2 proteins are required to maintain the aerobic respiratory metabolism through the post-transcriptional regulation of CoQ10 level.
Results
3′UTR of COQ7 mRNA is involved in regulating its translation
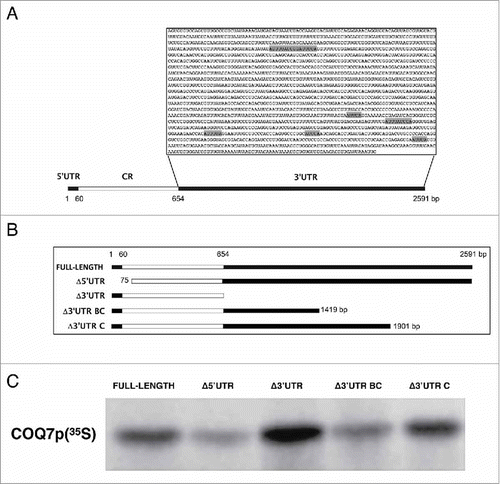
The COQ7 3′UTR (1.9 kb) contains a U/AU-rich segment that includes 3 AUUUUA repeats in the proximal 765 base pairs of the 3′UTR (). We assessed the involvement of COQ7 3′UTR in the translational regulation of coding region (CR), by using different constructs spanning the COQ7 mRNA (). The translational efficiency of each in vitro-synthesized RNA was assayed in a rabbit reticulocyte lysate system. All COQ7 transcripts were translated into a single 25-kDa polypeptide, in agreement with the size of COQ7 (). Interestingly, deletion of the entire 3′UTR increased COQ7 protein production compared to the full-length transcript, while deletion of the 5′UTR decreased it. COQ7 production was moderately reduced by the removal of the distal 1172 bp of the 3′UTR (fragment Δ3′UTR BC, bearing the 3′UTR proximal 765 bp), but expression was restored to full-length levels after additional segments spanning all but the distal 690 bp (fragment Δ3′UTR C) was tested. These findings suggested that the first 765-bp segment of the COQ7 3′UTR contains cis-acting elements involved in controlling the stability and/or translation of COQ7 mRNA.
Figure 1. The 3′UTR of COQ7 mRNA is involved in post-transcriptional regulation of COQ7 protein levels. (A) Schematic representation of the COQ7 mRNA and the AU-rich 3′UTR sequences. SHADED BOXES, location of AUUUA or AUUUUA sequences; CR: coding region. (B) Different segments of COQ7 mRNA (COQ7 full length, Δ3′UTR, Δ5′UTR, Δ3′UTR BC and Δ3′UTR) were synthesized by in vitro translation and labeled with 35S-methionine and 35S-cysteine. (C) Translational efficiency of each mRNA construct analyzed by Rabbit Reticulocyte Lysate System.
HuR and hnRNP C1/C2 bind COQ7 mRNA
Stability and translation of mRNA are governed robustly by binding of RBPs to 3′UTRs, and thus we explored the interaction of COQ7 mRNA with RBPs by synthesising several RNA probes spanning the entire COQ7 transcript by in vitro transcription (). UV-crosslinking assays after incubation with a protein extract from HeLa cells revealed the existence of protein complexes interacting with the first 765 bp of 3′UTR of the COQ7 mRNA ().
Figure 2. 3′UTR of COQ7 forms RNA-protein complexes with cellular proteins. (A) Riboprobes were synthesized by in vitro transcription and labeled with 32P (upper panel). UV crosslinking assays and electrophoresis SDS page reveal the specific interaction of COQ7 3′UTR with at least 3 human proteins (bottom panel). (B) Biotin pull-down assays were performed after incubating biotinylated transcript of COQ7 with whole cell lysates. Following pulldown, bound proteins were detected by Western blotting using the indicated antibodies. GAPDH 3′UTR was used as a negative control. Input, whole-cell lysates.
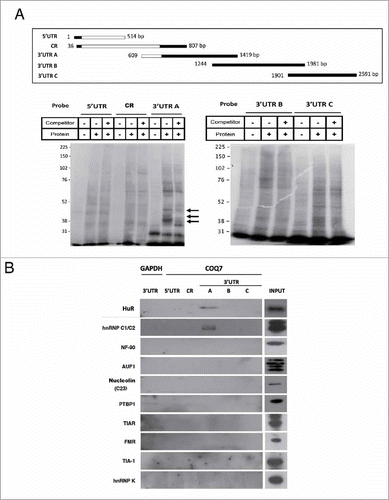
In order to identify what RBPs might associate with COQ7 mRNA, we studied the interaction between the biotinylated partial COQ7 transcripts and RBPs from HeLa cells. The COQ7 mRNA-RBP complexes (mRNPs) were further tested by biotin pulldown followed western blot analysis using antibodies against 10 different RBPs (Materials and Methods) of sizes in the range of molecular weights of the signals detected in . A biotinylated GAPDH 3′UTR RNA was used as a negative control. Two out of the 10 proteins assayed, HuR and hnRNP C1/C2, appeared to bind to a common region inside of the first 765 bp of 3′UTR (fragment A) of the COQ7 mRNA (). No other RBPs appeared to bind using this assay.
To further determine if COQ7 mRNA is a direct target of HuR and hnRNP C1/C2, we performed RIP analysis from cytoplasmic extract of HeLa cells, using anti-HuR and anti-hnRNP C1/C2 antibodies under conditions that prevented the dissociation of the RNPs. After the RNA associated with HuR or hnRNP C1/C2 was isolated, reverse transcription (RT) and real-time quantitative (q)PCR analysis was used to determine the degree of enrichment of COQ7 mRNA in each RIP sample. These results show that both anti-HuR and anti-hnRNP C1/C2 IP materials were enriched in COQ7 PCR product compared with control IgG IP samples (). Von Hippel-Lindau (VHL) mRNA and GAPDH mRNA were included as positive and negative controls, respectively.Citation24
Figure 3. HuR silencing, but not hnRNP C1/C2, reduces COQ7 expression. (A) After IP of RNA-protein complexes from HeLa cell lysates using anti-HuR, anti-hnRNP C1/C2 or control IgG antibodies, RNA was isolated and used in RT reactions. Fold differences in transcript abundance in HuR and in hnRNP C1/C2 IP samples compared with IgG IP samples were measured by RT-qPCR analysis. VHL mRNA and GAPDH mRNA were measured in the IP samples as positive and loading controls, respectively. (B) IP reactions using anti-hnRNP or anti-HuR antibodies were carried out using lysates from cells expressing normal or reduced (silenced) HuR or hnRNP, respectively, and COQ7 mRNA was quantified by RT-qPCR reactions. Data represent the means ±SD from 3 independent experiments. siRNA control vs. siRNA HuR. *P< 0.05. (C) The expression of HuR, hnRNP C1/C2, COQ7, and loading controls β-ACTIN and GAPDH, after silencing of HuR or hnRNP C1/C2, were monitored by western blot analysis.
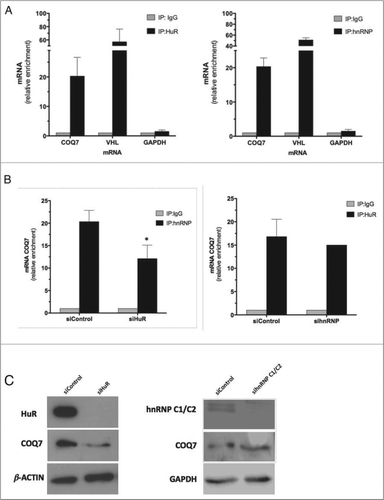
Given that the 2 RBPs associate with the same RNA region, we wondered whether the interaction between HuR and hnRNP C1/C2 with COQ7 mRNA might be competitive or cooperative. To test this possibility, we performed HuR RIP analysis under conditions of anti-hnRNP C1/C2 silencing, and conversely performed hnRNP C1/C2 RIP analysis under conditions of HuR silencing. We found that the reduced levels of hnRNP C1/C2 did not affect HuR binding. However, COQ7 mRNA enrichment in hnRNP C1/C2-IP fraction decreased significantly in HuR-silenced cells compared with control IgG IP samples (). This result suggests that HuR promotes the binding of hnRNP C1/C2 to COQ7 mRNA.
To ascertain the role of HuR and hnRNP C1/C2 on COQ7 protein production, MRC-5 cells were transfected with small interfering RNA (siRNA) to reduce the levels of these RBPs. illustrates the levels of COQ7 protein after siRNA transfection. This intervention caused a decrease in COQ7 protein levels (˜50%) in the absence of HuR but not after hnRNP C1/C2 silencing. Given that HuR promotes the stability of many target mRNAs, we set out to investigate if it also enhanced COQ7 mRNA half-life. We were unable to examine directly if COQ7 mRNA translation was affected by analyzing polysomes because the levels of COQ7 mRNA on each polysome fraction was too low (not shown).
HuR, but not hnRNP C1/C2, stabilizes the COQ7 mRNA and regulates CoQ10 biosynthesis rate
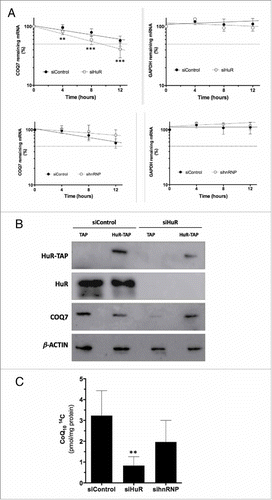
We measured the stability of COQ7 mRNA by treating cells with actinomycin D to inhibit de novo transcription. Total cellular RNA was isolated at the times indicated, and the levels of COQ7 mRNA and GAPDH mRNA (a stable mRNA encoding the housekeeping protein GAPDH) were quantified by real-time qPCR analysis. The stability of COQ7 mRNA was significantly reduced in HuR-silenced cells (), but not in hnRNP C1/C2-silenced cells (). To assess the specificity of the decrease of COQ7 in cells lacking HuR, MRC-5 cells we transfected with a siHuR targeted to HuR 3′UTR to reduce the levels of the endogenous protein, followed by overexpression of a chimeric protein (HuR-TAP) that lacks of 3′UTR region. shows that COQ7 protein levels, decreased by HuR silencing, were restored by HuR-TAP expression. After these interventions, CoQ10 biosynthesis rate was assayed by incorporation of 14C-pHB, a radiolabeled precursor of the CoQ10 biosynthesis pathway, in HuR-silenced and in hnRNP C1/C2-silenced cells. As the shows, CoQ10 biosynthesis rate was only significantly decreased in the absence of HuR, ruling out a direct involvement for hnRNP C1/C2 in regulating COQ7 expression and CoQ10 biosynthesis.
Figure 4. HuR silencing, but not hnRNP C1/C2 silencing, regulates COQ7 mRNA stability and CoQ10 biosynthesis. (A) The half-lives of COQ7 and GAPDH mRNAs after silencing HuR (upper panel) or hnRNP C1/C2 (bottom panel) were measured by incubating cells with actinomycin D, extracting total RNA at the times shown, and measuring mRNA levels by RT-qPCR analysis. The data were normalized to 18S rRNA levels and represented as a percentage of the mRNA levels measured at time 0, before adding actinomycin D, using a semilogarithmic scale. mRNA half-life was calculated as the time required for each mRNA decrease to 50% of its initial abundance (discontinuous horizontal line). (B) MRC-5 cells were transfected with a siRNA targeting the HuR 3′UTR (siHuR), or a control siRNA (siControl), along with either TAP- or HuR-TAP-expressing vectors. HuR-TAP is a chimeric protein expressed from a cDNA that lacks the HuR 3′UTR; the levels of endogenous (HuR) or ectopic (HuR-TAP) HuR, COQ7, and loading control β-actin were tested by Western blot analysis. (C) CoQ10 biosynthesis rate. HuR-silenced and hnRNP-silenced cells were incubated (24 h) with radiolabeled CoQ10 precursor (pHB). Cells were subjected to a lipid extraction and separation by HPLC coupled to radioactivity detector to quantify 14C CoQ10. All data represent the means ± SD of 3 independent experiments. siControl vs. siHuR ** p < 0.01, *** p < 0.001.
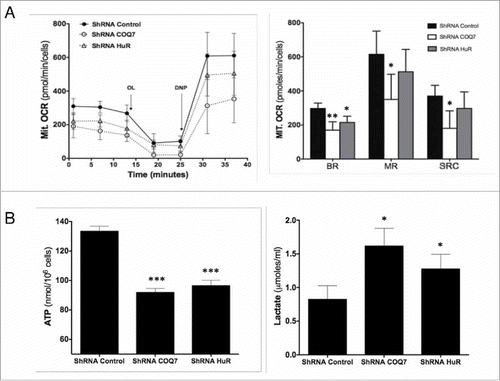
HuR affects mitochondrial respiration
As CoQ is an electron carrier of the mitochondrial respiratory chain, it is a key component of the respiratory chain superassembly complex.Citation4 Thus, the reduced CoQ10 biosynthesis capacity caused by reducing HuR might disrupt mitochondrial metabolism in a way similar to what is seen in primary CoQ10 deficiency. To test this hypothesis, we analyzed mitochondrial oxygen consumption rate (OCR) in MRC5 human cells expressing low HuR levels after transfection of HuR shRNA using a Seahorse Bioscience XF analyzer. In parallel, we performed the same assay in cells expressing reduced COQ7 mRNA levels (from transfection with COQ7 shRNA), included as a positive control. The OCR data in shows the mitochondrial respiratory response before and after sequential addition of oligomycin (OL) and 2,4 dinitrophenol (DNP) to HuR shRNA HuR and control shRNA cells. Basal mitochondrial respiration (BR) was reduced in HuR-silenced cells compared with control counterparts. However, unlike COQ7 shRNA cells, maximal mitochondrial respiration (MR) and spare mitochondrial respiration capacity (SRC) were not significantly reduced (). Furthermore, total ATP levels were decreased after HuR silencing while lactate production was higher than in control cells (). The enhanced production of lactate in both shRNA- HuR and shRNA- COQ7 silenced cells indicates a stimulation of aerobic glycolysis, likely due to reduced ATP production in mitochondria by oxidative phosphorylation.
Figure 5. HuR regulates mitochondrial respiration. Oxygen consumption rate (A) and ATP levels and lactate production (B) were measured in HuR-silenced and in COQ7-silenced cells. Mitochondrial oxygen consumption rates (Mit. OCR) following the sequential addition of oligomycin A (OL), 2,5-dinitrophenol (DNP) to MRC-5 cells stably transfected with HuR or COQ7 shRNA (B, left panel). Right panel: quantification of the mitochondrial basal respiration (BR), maximal respiration (MR), and spare respiration capacity (SRC). These parameters were calculated as described in the Materials and Methods section. Values are the means ± SD of 5 independent experiments. control shRNA vs. HuR shRNA or COQ7 shRNA. *P < 0.05, **P < 0.01, ***P < 0.001.
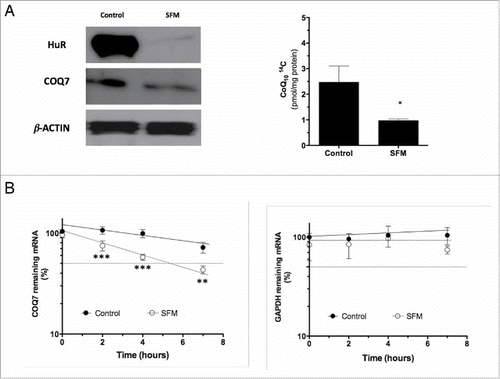
Serum withdrawal reduces levels of HuR, COQ7 protein and CoQ10 biosynthesis rate
Serum administration is required to boost mitochondrial respiratory capacity in cultured cells,Citation34 and serum withdrawal induces aerobic glycolysis in fibroblasts with increasing lactate production,Citation35 similarly to what is seen in primary CoQ10-deficient human fibroblasts.Citation36 As indicated by Atasoy et al (1998),Citation26 HuR expression decreased in NIH-3T3 cells in serum limiting condition. Thus, we hypothesized that the decreased expression of HuR in serum-deprived cells should lead to a reduction in levels of COQ7 and CoQ10 biosynthesis, playing a role in the switch of the 2 main pathways involved in energy provision, i.e., from oxidative phosphorylation to an enhanced aerobic glycolysis. To ascertain the role of serum on the functional consequences of HuR-COQ7 binding, we monitored the cytoplasmic levels of HuR and COQ7 by western blotting (). The levels of these proteins were decreased in MRC5 cells after 48 h in the absence of serum. CoQ10 biosynthesis rate assayed by incorporation of 14C-pHB, was also decreased by 50% in MRC5 grown without serum (). Furthermore, the stability of COQ7 mRNA was also significantly decreased in serum-deprived MRC5 cells ().
Figure 6. Serum deprivation decreases cytoplasmic levels of HuR and COQ7 and reduces CoQ10 biosynthesis rate. (A) Levels of cytoplasmic HuR and COQ7 proteins (left panel) and CoQ10 biosynthesis rate (right panel) in cells cultured in serum-free media (SFM). Cell culture and serum withdrawal were performed as described in the Materials and Methods section. Values are the means ± SD of 3 independent experiments. Control vs. SFM. *P< 0.05. (B) The half-lives of COQ7 and GAPDH mRNAs after serum deprivation (SFM) was measured by incubating cells with actinomycin D, extracting of total RNA at the times shown, and measuring mRNA levels by RT-qPCR analysis. The data were normalized to 18S rRNA levels and represented as a percentage of the mRNA levels measured at time 0, before adding actinomycin D, using a semilogarithmic scale. The half-lives were calculated as the time required for each mRNA decrease to 50% of its initial abundance (discontinuous horizontal line). Data represent the means ±SD of 3 independent experiments. Control vs. SFM. **P < 0.01, ***P < 0.001.
Discussion
CoQ10 biosynthesis is a complex process involving the products of at least 10 nuclear genes, although additional genes have been identified recently in yeastCitation37 Both CoQ levels and COQ genes expression in mammals are tissue-specific and depend on nutritional conditions and the age of the organism,Citation13,38 which suggests the existence of highly conserved regulatory mechanisms to support cellular metabolism.Citation39 To-date, transcriptional regulation has been demonstrated to involve PPARα and NF-κB transcription factors mainly in response to either nutrient or stress conditions.Citation16-Citation18 Additionally, recent literature highlights the importance of the post-transcriptional control of COQ proteins and the structure of the complex in the regulation of CoQ10 levels.Citation40 Specifically, COQ7 is regulated by phosphorylation depending on nutritional conditions and is regulated in part by the PTC7 mitochondrial phosphatase.Citation19,20 COQ7 protein also requires COQ9 encoded peptide interaction for its activity.Citation21 COQ7 enzyme activity is the only step in CoQ biosynthesis in which the non-reactive intermediate demethoxyubiquinone is accumulated by either nutritional limitations or mutant strains, and it is proposed to be a key step in this pathway.Citation41,42
Recently, it has been described that Clu1/CluA homolog (CLUH) encodes for mRNA binding protein that bound to hundreds of mRNAs encoding mitochondrial-directed proteins, including COQ3 and COQ6 mRNAs although there is not demonstration of its role in their stability and/or translation.Citation43 COQ7 mRNA was included as a putative target of HuR,Citation24 and thus we focused on COQ7 mRNA interaction with RBPs as a novel post-transcriptional regulatory mechanism of CoQ biosynthesis. Over the past decade, ARE-binding RBPs have been described to regulate the turnover and/or translation of target RNAs,Citation44-Citation46 including mitochondrial RNAs.Citation47 In this work, we show evidence that COQ7 mRNA forms an RNP complex that regulates COQ7 expression post-transcriptionally. The elements regulating the translational efficiency tend to be located at 5′UTR of the transcripts,Citation48,49 although it was also described that the 3′UTR of β-F1 ATPase mRNA promoted the translation initiation,Citation50 and 3 clustered AUUUA motifs in the 3′UTR of the interleukin-1α (IL-1α) caused the instability to the transcript.Citation51 In an in vitro assay to measure protein production, deletion of 5′UTR, a region essential to initiate translation, reduced the translation of COQ7 mRNA. However, deletion of the proximal 3′UTR segment, which included 3 repeats of the AUUUUA hexanucleotide and also contained the binding sites for HuR and hnRNP C1/C2, decreased in vitro COQ7 production. The explanation for this effect is not clear, but perhaps the regulatory elements identified in the proximal segment were not accessible to positive regulators such as HuR to form a functional RNP complex. Whether other positive or negative regulators (e.g., other RBPs or possibly microRNAs) become available to this truncated RNA remains to be examined.
Among the proteins targeting the proximal 3′UTR segment of COQ7 mRNA, we identified HuR and hnRNP C1/C2, which do not have competitive effects on mRNA stability. Interestingly, HuR promoted the formation of hnRNP C1/C2-COQ7 mRNA complex, indicating a functional association between both RBPs and a modest cooperative impact on binding. In contrast, hnRNP C1/C2 was not required for HuR binding to COQ7 mRNA. HuR induced the stability of COQ7 mRNA and consequently the rate of CoQ biosynthesis. The decrease of HuR led to CoQ deficiency and reduced oxidative phosphorylation, which depleted ATP and increased lactate levels. Similarly, primary deficiency of CoQ10 led to a decrease of mitochondrial respiratory chain activities affecting oxidative phosphorylation and forcing cells toward enhanced glycolysis.Citation52
The functional association between HuR and other RBPs, such as AUF1, has been widely described,Citation30,33,53,54 and the interactions between different RPBs may be cooperative or competitive depending on cell type, growth conditions, RBP concentration, cellular compartment and target mRNA.Citation31,55,56 Cooperative hnRNP C1 and HuR interaction to modulate alternative Fas splicing has been also described.Citation57,58
HuR and hnRNP C1/C2 are predominantly located in the nucleus, but these proteins can shuttle between the nucleus and the cytoplasm in different cellular conditions, such as oxidative stress, UV radiation exposure or genotoxic stress.Citation59-Citation62 The relation of HuR with mitochondria is described throughout its association with β-F1-ATPase mRNACitation63 and the regulation of antiapoptotic proteins such as Bcl2 or BCLxL,Citation64,65 and here we show the direct interaction of both HuR and hnRNP C1/C2 to COQ7 mRNA, the key enzyme of CoQ biosynthesis pathway.
The CoQ10is synthesized in mammals in quantities that vary widely among tissues and among pathophysiological conditions, such as aging, dietary intake and mitochondrial diseases.Citation1,38,66 The dynamic and composition of the COQ7 mRNA-RBPs complex could depend on the specific tissue but also on metabolic and stress conditions. For example, expression of target mRNAs encoding proteins implicated in cellular proliferation and replicative senescence may be linked to the decreased levels of HuR with senescence,Citation30 and HuR expression is reduced in NIH-3T3 fibroblasts in the absence of growth factors after serum withdrawal.Citation26 We observed that the levels of cytoplasmic HuR decreased in serum-deprived MRC-5 cells in parallel to the reduction of COQ7 levels and CoQ10 biosynthesis, in agreement to the increase of aerobic glycolysis described in serum-deprived cells.Citation35 Importantly, deprivation of serum of 3T6 mouse cells triggered an arrest of the cell cycle and a decrease of CoQ10 biosynthesis independent of the mevalonate pathway.Citation67 Our results suggest that serum withdrawal affects CoQ biosynthesis by reducing levels of COQ7 mRNA through a decline in HuR levels.
In summary, we have identified a new mechanism for the regulation of CoQ levels that could correspond to specific requirements between long-term transcriptional regulation and the transient modulation of biosynthesis of certain enzymes in the complex. Furthermore, an increasing number of patients display CoQ10 deficiency syndrome; for some of them, mutations in CoQ biosynthesis genes are responsible for the deficiency, but for many others the molecular pathogenic causes are unknown.Citation8 We hypothesize that alterations in regulatory sequences in the UTRs of COQ genes or in the levels or activities of specific RBPs might contribute to the CoQ10 deficiency in patients with unknown genetic defects.
Materials and methods
Cell culture
Human cervical carcinoma HeLa (ATCC) and primary human fibroblasts MRC5 cells (CCL−171, ATCC, Manassas, USA) were cultured in Dulbecco's modified essential medium (DMEM, Gibco) with 4.5 g/L glucose and 1 g/L glucose respectively, supplemented with 10% bovine serum and antibiotics (FBS, Sigma). Cells were cultured in a humidified incubator at 37°C and 5% CO2, and detached by trypsinization. Serum withdrawal condition was established by culturing cells 48 h without FBS.
Immunoprecipitation (IP) of RNP complexes (RIP analysis)
IP of endogenous RNA-protein complexes was performed using 400 μg of cytoplasmic extract (10 mM Tris-HCl pH 7.4, 100 mM NaCl, 2.5 mM MgCl2, 100 U de RNase-OUT) prepared from HeLa cells. Previously, Protein A Sepharose-beads (Sigma) were pre-coated with 30 μg (overnight at 4°C) of either anti-IgG1 (BD PharMingen), anti-HuR or anti-hnRNP C1/C2 (Santa Cruz Biotech.). Next, lysates were incubated with beads (50% v/v) for 2 h at 4°C, and washed using NT2 (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM MgCl2, 0.05% Nonidet P-40). To isolate and analyze the RNA in the IP fraction, the beads were incubated with NT2 buffer containing 20 U of DNase I (Sigma) (30 min at 37°C) and washed with NT2 buffer. Finally, complexes were incubated with 0.1% SDS and 0.5 mg/ml proteinase K (15 min at 55°C) in NT2 buffer. RNA was isolated from the supernatant by phenol-chloroform extraction and measured by real-time qPCR analysis.
Analysis of binding of RBPs to biotinylated RNA
The MAXIscript T7 kit (Ambion) was used to synthesize the biotinylated transcripts, including the modified nucleotide biotin-CPT (Enzo Life Sciences AG), following the manufacturer's instructions. Biotinylated transcripts were purified with NucAway Spin Columns (Ambion). DNA templates for the production of biotinylated transcripts were synthesized using the following oligonucleotides (sense and antisense): for coding region: 5′-(T7)ACGAAGTGGTTGCTTTTTTTAG-3′ and 5′-(T7)ACGAAGTGGTTGCTTTTTTTAG-3′; for 5′UTR: 5′-(T7)GTCCGAGCCAAGGGCACTA-3′ and 5′-TGCTCCATATTCGCCTGCA-3′; for 3′UTR A: 5′-(T7)GCTTGAGCACCATGACATAGG-3′ and 5′-TAGGAGAAAATGGGCCTGG-3′; for 3′UTR B: 5′-(T7)GGCTCAGTGATCCTCCCG-3′ and 5′-TTGGGGGATTTTTTTGGG-3′: for 3′UTR C: 5′-(T7)AGTTGTGGATTATTTGTGAAATTG-3′ and 5′-CCTTAAGAAAACCTTGTTGTGC-3′; for 3′UTR GAPDH: 5′-(T7)CCTCAACGACCACTTTGTCA-3′ and 5′-GGTTGAGCACAGGGTACTTTATT-3′. Biotin pulldown assays were carried out by incubating either whole-cell lysates (HeLa) with purified biotinylated transcripts (40 μg lysate, 1 μg RNA) for 2 h at room temperature. Complexes were isolated with paramagnetic streptavidin-conjugated Dynabeads (Dynal, Oslo, Norway) as previously described.Citation24,68 Proteins bound in the pulldown material were analyzed by Western blotting using specific antibodies against HuR, hnRNP C1/C2, Nucleolin, TIAR, FMR, TIA-1, TTP (Santa Cruz Biotech.), NF90 (BD-biosciences), AUF1 (Millipore), PTBP1 and hnRNP K (Abcam).
UV cross-linking assays
The MAXIscript T7 kit (Ambion) was used to synthesize the radiolabeled riboprobes, including 20 μCi of the nucleotide α-(P32)-UTP (GE Healthcare), following the manufacturer's instructions. DNA templates for the production of biotinylated transcripts were synthesize using the following oligonucleotides (sense and antisense): for the coding region: 5′-(T7)ACGAAGTGGTTGCTTTTTTTAG-3′ and 5′-(T7)ACGAAGTGGTTGCTTTTTTTAG-3′; for 5′UTR: 5′-(T7)GTCCGAGCCAAGGGCACTA-3′ and 5′-TGCTCCATATTCGCCTGCA-3′; for 3′UTR A: 5′-(T7)GCTTGAGCACCATGACATAGG-3′ and 5′-TAGGAGAAAATGGGCCTGG-3′; for 3′UTR B: 5′-(T7)GGCTCAGTGATCCTCCCG-3′ and 5′-TTGGGGGATTTTTTTGGG-3′: for 3′UTR C: 5′-(T7)AGTTGTGGATTATTTGTGAAATTG-3′ and 5′-CCTTAAGAAAACCTTGTTGTGC-3′. Whole-cell lysate of HeLa cells (30 μg) was incubated with radiolabeled probes (1×105 c.p.m.), 10 min at 30°C, in RLP buffer (NaPHO4- Na2PO4 50 mM pH 7.4, NaCl 140 mM, MgCl2 1.5 mM, DTT 1 mM), containing 4 μg of Escherichia coli ARNt. Next, 20 U of RNase T1 (Boehringer Mannheim) was added (30 min a 37°C). The UV cross-linking assay was carried out by exposition of the reaction mixtures to 254 nm of UV light (Stratalinker 1800; Stratagene) for 6 min. in ice. For competition studies, an excess of unlabeled RNA was added 10 min before the addition of the radiolabeled RNA to liver extracts. The RNA-protein complexes were resolved by SDS–12% PAGE. After electrophoresis, the gels were vacuum dried and the 32P-labeled complexes were visualized by exposure of the gels to X-ray films.
In vitro translation of mRNAs
In vitro-synthesized mRNAs (100 ng), derived from the corresponding plasmids, were utilized as templates for protein synthesis in a nuclease-treated rabbit reticulocyte lysate (Amersham). DNA templates for the production of the transcripts were synthesized using the following oligonucleotides (sense and antisense): for COQ7 full length: 5′-(T7)GTCCGAGCCAAGGGCACTA-3′ and 5′-CCTTAAGAAAACCTTGTTGTGC-3′; for Δ3′UTR: 5′-(T7)GTCCGAGCCAAGGGCACTA-3′ and 5′-AACGATAACTGTACACCTGTTTCTC-3′; for ΔBC: 5′-(T7)GTCCGAGCCAAGGGCACTA-3′ and 5′-TAGGAGAAAATGGGCCTGG-3′: for ΔC: 5′-(T7)GTCCGAGCCAAGGGCACTA-3′ and 5′-TTGGGGGATTTTTTTGGG-3′. The riboprobes were quantified by spectrophotometry at 260 nm and the quality was monitored by A260/A280 and A260/A230 ratios. The reactions were performed in the presence of 40 mCi of L-[35S]methionine (1,000 Ci/mM) and 40 U of RNase A. At different reaction times (up to 1 h), the products were analyzed by SDS-PAGE.
Determination of mitochondrial oxygen consumption rate
Cellular oxygen consumption rates (OCR) were determined in an XF24 Extracellular Flux Analyzer (Seahorse Bioscience). Cells were plated on XF24 microplates at 15,000 cells/well in supplemented medium and incubated at 37°C and 5% CO2 for 24 h. After measuring of basal respiration rate, 6 μM oligomycin was added to inhibit complex V, next 0.75 mM 2,4-dinitrophenol was added to uncouple respiration. Finally, complex I and III were inhibited by addition of 1 μM rotenone and 1 μM antimycin A, respectively. OCR was determined by subtracting the ‘non mitochondrial OCR’ after treatment with rotenone+actinomycin A. Mitochondrial basal respiration was determined from mitochondrial OCR before administration of oligomycin. Mitochondrial maximal respiration was mitochondrial OCR after administration of 2,4-dinitrophenol. Spare respiration capacity was mitochondrial maximal respiration minus mitochondrial basal respiration.
Determination of lactate and ATP levels
Cells were plated on 24-well microplates and cultured for 24 h. Medium was collected and lactate determined by using colorimetric lactate oxidase-peroxidase assay lactate kit (Spinreact, Spain) as instructed by the manufacturer. The concentration of lactate in the medium was calculated by comparison with the signal obtained from standard with known amount of lactate provided by the manufacturer. Signals determined in medium without cells were also analyzed and subtracted from the amount determined in the presence of cells. ATP levels in cells were determined by using the CellTiter-Glo Luminiscent assay (Promega, USA), following the instructions of the manufacturer. Briefly, the same amount of reactive solution was added to culture medium to produce cell lysis with gentle movement. The mixture was homogenized and transferred to a white polystyrene 96-well assay plate. A standard with known amounts of ATP were also added in the same plate and mixed with reactive solution in a 1:1 ratio. A POLAR Star Omega fluorimeter and Omega Data Analysis Software (BMG Labtech) was used to analyze luminiscence. ATP amount was referred to the total of cells counted by hemocytometer after trypsin detachment seeded in a 24 well plate seeded in parallel.
Transfection and plasmids
Cells were seeded in DMEM supplemented with 5% serum without antibiotics 24 h after transfection with lipofectamine 2000 reagent (Invitrogen) and 2 mg of TAP and HuR-TAP plasmids were diluted in Opti-MEN media (Gibco) and incubated 10 min at room temperature, separately. After 30 min of incubation of plasmid and lipofectamine at room temperature, the transfection complex was added to the cells and incubated for 6 h. Then, the medium was replaced with fresh medium for the ensuing 48 h.
siRNA transfection and shRNA infection
siRNAs targeting HuR or hnRNP C1/C2 and control siRNA were acquired from Qiagen. Cells were transfected with oligofectamine (Invitrogen) using the manufacturer's conditions. Briefly, 24 h before transfection cells were incubated in DMEM 5% serum without antibiotics. Then, for each single transfection, siRNA (10 nM) and oligofectamine were diluted in Opti-MEM media (Gibco). After 30 min of incubation at room temperature the transfection complexes were added to the cells. Lentiviral particles and shRNA of HuR, COQ7 and control incorporated into lentivirus were acquired from Santa Cruz Biotechnology. Cells were plated 24 h before viral infection in supplemented medium. Medium was removed from the plate and replaced with supplemental medium with Polybrene (sc-134220) at final concentration of 10 mg/ml. Cells were infected by adding of shRNA lentiviral particles to the culture.
Quantitative real-time PCR
Total RNA was isolated with the TriPure Isolation Reagent (Roche, Mannheim, Germany), treated for DNA removal with DNase I (Sigma-Aldrich) and reverse transcribed by using the iScript cDNA Synthesis Kit (Bio-Rad). Real-time qPCR was performed to measure the expression of select target genes. The reactions were carried out with iQ SYBR Green Supermix (Bio-Rad) and MyiQTM Single-Color Real-Time PCR Detection System (Bio-Rad) on a Bio-Rad conventional thermocycler. The primers were designed with the Beacon Designer software, and the primer pair sequences used in this study were as followed: human forward and reverse primers were GGACGCTGATGGAGGAGGAC and AGGACGGCATAGGCTGGAC for COQ7, and TGCACCACCAACTGCTTAGC and GGCATGGACTGTGGTCATGAG for GAPDH. The quality of the PCR reaction products was determined by melting curve analysis and visualization on agarose gels. Negative controls containing water only confirmed the absence of cross-contamination. Efficiency (E) of the PCR reaction was determined using a dilution series, and the data analyzed using the formula (ETARGET-CT/EHOUSEKEEPING-CT) with GAPDH mRNA as loading control. Each PCR reaction was carried out with 3 or more biological replicates in each group.
Statistical analysis
Significant differences between the data groups were evaluated using a paired t test (2-tailed P values) for comparing 2 groups and by 2-way ANOVA for >2 groups using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA, USA). Multiple comparisons of ANOVA were followed with post hoc Bonferroni. Results are expressed as the means ± SD and were considered significant at P ≤ 0 .05.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by grants from the Spanish Ministry of Health, Instituto de Salud Carlos III (ISCIII), FIS PI14–01962 to PN, the Spanish Ministry of Economy and Competitively, SAF2013–41945-R to JMC, and the Intramural Research Program of the NIA, NIH (KA, MG).
References
- Bentinger M, Tekle M, Dallner G. Coenzyme Q – Biosynthesis and functions. Biochem Biophys Res Commun 2010; 396:74-9; PMID:20494114; http://doi.dx.org/10.1016/j.bbrc.2010.02.147
- Cramer WA, Hasan SS, Yamashita E. The Q cycle of cytochrome bc complexes: A structure perspective. Biochim Biophys Acta - Bioenerg 2011; 1807:788-802; http://doi.dx.org/10.1016/j.bbabio.2011.02.006
- Padilla S, Jonassen T, Jimenez-Hidalgo MA, Fernandez-Ayala DJM, Lopez-Lluch G, Marbois B, Navas P, Clarke CF, Santos-Ocana C. Demethoxy-Q, an intermediate of coenzyme Q biosynthesis, fails to support respiration in saccharomyces cerevisiae and lacks antioxidant activity. J Biol Chem 2004; 279:25995-6004; PMID:15078893; http://doi.dx.org/10.1074/jbc.M400001200
- Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, et al. Supercomplex Assembly Determines Electron Flux in the Mitochondrial Electron Transport Chain. Science (80-) 2013; 340:1567-70; PMID:23812712; http://doi.dx.org/10.1126/science.1230381
- Genova ML, Lenaz G. The Interplay Between Respiratory Supercomplexes and ROS in Aging. Antioxid Redox Signal 2015; 23:208-38; PMID:25711676; http://doi.dx.org/10.1089/ars.2014.6214
- Doimo M, Desbats MA, Cerqua C, Cassina M, Trevisson E, Salviati L. Genetics of coenzyme q10 deficiency. Mol Syndromol 2014; 5:156-62; PMID:25126048; http://doi.dx.org/10.1159/000362826
- Laredj LN, Licitra F, Puccio HM. The molecular genetics of coenzyme Q biosynthesis in health and disease. Biochimie 2014; 100:78-87; PMID:24355204; http://doi.dx.org/10.1016/j.biochi.2013.12.006
- Desbats MA, Lunardi G, Doimo M, Trevisson E, Salviati L. Genetic bases and clinical manifestations of coenzyme Q10 (CoQ 10) deficiency. J Inherit Metab Dis 2014; 38:145-56; PMID:25091424; http://doi.dx.org/10.1007/s10545-014-9749-9
- Desbats MA, Vetro A, Limongelli I, Lunardi G, Casarin A, Doimo M, Spinazzi M, Angelini C, Cenacchi G, Burlina A, et al. Primary coenzyme Q10 deficiency presenting as fatal neonatal multiorgan failure. Eur J Hum Genet 2015; 23:1254-8; PMID:25564041; http://doi.dx.org/10.1038/ejhg.2014.277
- Cornelius N, Byron C, Hargreaves I, Guerra PF, Furdek AK, Land J, Radford WW, Frerman F, Corydon TJ, Gregersen N, et al. Secondary coenzyme Q10 deficiency and oxidative stress in cultured fibroblasts from patients with riboflavin responsive multiple Acyl-CoA dehydrogenation deficiency. Hum Mol Genet 2013; 22:3819-27; PMID:23727839; http://doi.dx.org/10.1093/hmg/ddt232
- Asencio C, Rodríguez-Hernandez MA, Briones P, Montoya J, Cortés A, Emperador S, Gavilán A, Ruiz-Pesini E, Yubero D, Montero R, et al. Severe encephalopathy associated to pyruvate dehydrogenase mutations and unbalanced coenzyme Q10 content. Eur J Hum Genet 2015; PMID:26014431; http://doi.dx.org/10.1038/ejhg.2015.112.
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu V V, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature 2006; 444:337-42; PMID:17086191; http://doi.dx.org/10.1038/nature05354
- L pez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo M V, Allard J, Ingram DK, Navas P, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci 2006; 103:1768-73; PMID:16446459; http://doi.dx.org/10.1073/pnas.0510452103
- Asencio C, Rodriguez-Aguilera JC, Ruiz-Ferrer M, Vela J, Navas P. Silencing of ubiquinone biosynthesis genes extends life span in Caenorhabditis elegans. FASEB J 2003; 17:1135-7; PMID:12709403; http://doi.dx.org/10.1096/fj.02-1022fje
- Wang Y, Oxer D, Hekimi S. Mitochondrial function and lifespan of mice with controlled ubiquinone biosynthesis. Nat Commun 2015; 6:6393; PMID:25744659; http://doi.dx.org/10.1038/ncomms7393
- Turunen M, Peters JM, Gonzalez FJ, Schedin S, Dallner G. Influence of peroxisome proliferator-activated receptor α on ubiquinone biosynthesis. J Mol Biol 2000; 297:607-14; PMID:10731415; http://doi.dx.org/10.1006/jmbi.2000.3596
- Bentinger M, Tekle M, Brismar K, Chojnacki T, Swiezewska E, Dallner G. Stimulation of coenzyme Q synthesis. BioFactors 2008; 32:99-111; PMID:19096105; http://doi.dx.org/10.1002/biof.5520320112
- Brea-Calvo G, Siendones E, Sánchez-Alcázar JA, de Cabo R, Navas P. Cell survival from chemotherapy depends on NF-kappaB transcriptional up-regulation of coenzyme Q biosynthesis. PLoS One 2009; 4:e5301; PMID:19390650; http://doi.dx.org/10.1371/journal.pone.0005301
- Martín-Montalvo A, González-Mariscal I, Padilla S, Ballesteros M, Brautigan DL, Navas P, Santos-Ocaña C. Respiratory-induced coenzyme Q biosynthesis is regulated by a phosphorylation cycle of Cat5p/Coq7p. Biochem J 2011; 440:107-14; PMID:21812761; http://doi.dx.org/10.1042/BJ20101422
- Martin-Montalvo A, Gonzalez-Mariscal I, Pomares-Viciana T, Padilla-Lopez S, Ballesteros M, Vazquez-Fonseca L, Gandolfo P, Brautigan DL, Navas P, Santos-Ocana C. The Phosphatase Ptc7 Induces Coenzyme Q Biosynthesis by Activating the Hydroxylase Coq7 in Yeast. J Biol Chem 2013; 288:28126-37; PMID:23940037; http://doi.dx.org/10.1074/jbc.M113.474494
- Lohman DC, Forouhar F, Beebe ET, Stefely MS, Minogue CE, Ulbrich A, Stefely JA, Sukumar S, Luna-Sánchez M, Jochem A, et al. Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc Natl Acad Sci U S A 2014; 111:E4697-705; PMID:25339443; http://doi.dx.org/10.1073/pnas.1413128111
- Mata J, Marguerat S, Bähler J. Post-transcriptional control of gene expression: A genome-wide perspective. Trends Biochem Sci 2005; 30:506-14; PMID:16054366; http://doi.dx.org/10.1016/j.tibs.2005.07.005
- Pascale A, Govoni S. The complex world of post-transcriptional mechanisms: is their deregulation a common link for diseases? Focus on ELAV-like RNA-binding proteins. Cell Mol life Sci C 2011; 69:501-17; PMID:21909784; http://doi.dx.org/10.1007/s00018-011-0810-7
- López de Silanes I, Zhan M, Lal A, Yang X, Gorospe M. Identification of a target RNA motif for RNA-binding protein HuR. Proc Natl Acad Sci U S A 2004; 101:2987-92; PMID:14981256; http://doi.dx.org/10.1073/pnas.0306453101
- Hinman MN, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci 2008; 65:3168-81; PMID:18581050; http://doi.dx.org/10.1007/s00018-008-8252-6
- Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci 1998; 111 ( Pt 2:3145-56; PMID:9763509.
- Figueroa A, Cuadrado A, Fan J, Atasoy U, Muscat GE, Munoz-Canoves P, Gorospe M, Munoz A. Role of HuR in Skeletal Myogenesis through Coordinate Regulation of Muscle Differentiation Genes. Mol Cell Biol 2003; 23:4991-5004; PMID:12832484; http://doi.dx.org/10.1128/MCB.23.14.4991-5004.2003
- Kullmann M. ELAV/Hu proteins inhibit p27 translation via an IRES element in the p27 5′UTR. Genes Dev 2002; 16:3087-99; PMID:12464637; http://doi.dx.org/10.1101/gad.248902
- Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J 2000; 19:2340-50; PMID:10811625; http://doi.dx.org/10.1093/emboj/19.10.2340
- Chang N, Yi J, Guo G, Liu X, Shang Y, Tong T, Cui Q, Zhan M, Gorospe M, Wang W. HuR uses AUF1 as a cofactor to promote p16INK4 mRNA decay. Mol Cell Biol 2010; 30:3875-86; PMID:20498276; http://doi.dx.org/10.1128/MCB.00169-10
- Lal A, Mazan-Mamczarz K, Kawai T, Yang X, Martindale JL, Gorospe M. Concurrent versus individual binding of HuR and AUF1 to common labile target mRNAs. EMBO J 2004; 23:3092-102; PMID:15257295; http://doi.dx.org/10.1038/sj.emboj.7600305
- Pan Y-X, Chen H, Kilberg MS. Interaction of RNA-binding Proteins HuR and AUF1 with the Human ATF3 mRNA 3′-Untranslated Region Regulates Its Amino Acid Limitation-induced Stabilization. J Biol Chem 2005; 280:34609-16; PMID:16109718; http://doi.dx.org/10.1074/jbc.M507802200
- Zou T, Rao JN, Liu L, Xiao L, Yu T-X, Jiang P, Gorospe M, Wang J-Y. Polyamines Regulate the Stability of JunD mRNA by Modulating the Competitive Binding of Its 3′ Untranslated Region to HuR and AUF1. Mol Cell Biol 2010; 30:5021-32; PMID:20805360; http://doi.dx.org/10.1128/MCB.00807-10
- Herzig RP. Sequential Serum-dependent Activation of CREB and NRF-1 Leads to Enhanced Mitochondrial Respiration through the Induction of Cytochrome c. J Biol Chem 2000; 275:13134-41; PMID:10777619; http://doi.dx.org/10.1074/jbc.275.17.13134
- Golpour M, Akhavan Niaki H, Khorasani HR, Hajian A, Mehrasa R, Mostafazadeh A. Human fibroblast switches to anaerobic metabolic pathway in response to serum starvation: a mimic of warburg effect. Int J Mol Cell Med 2014; 3:74-80; PMID:25035856.
- López-Martín JM, Salviati L, Trevisson E, Montini G, DiMauro S, Quinzii C, Hirano M, Rodriguez-Hernandez A, Cordero MD, Sánchez-Alcázar JA, et al. Missense mutation of the COQ2 gene causes defects of bioenergetics and de novo pyrimidine synthesis. Hum Mol Genet 2007; 16:1091-7; PMID:17374725; http://doi.dx.org/10.1093/hmg/ddm058
- Allan CM, Awad AM, Johnson JS, Shirasaki DI, Wang C, Blaby-Haas CE, Merchant SS, Loo JA, Clarke CF. Identification of Coq11, a New Coenzyme Q Biosynthetic Protein in the CoQ-Synthome in Saccharomyces cerevisiae. J Biol Chem 2015; 290:7517-34; PMID:25631044; http://doi.dx.org/10.1074/jbc.M114.633131
- Parrado-Fernández C, López-Lluch G, Rodríguez-Bies E, Santa-Cruz S, Navas P, Ramsey JJ, Villalba JM. Calorie restriction modifies ubiquinone and COQ transcript levels in mouse tissues. Free Radic Biol Med 2011; 50:1728-36; PMID:21447381; http://doi.dx.org/10.1016/j.freeradbiomed.2011.03.024
- González-Mariscal I, García-Testón E, Padilla S, Martín-Montalvo A, Pomares Viciana T, Vazquez-Fonseca L, Gandolfo Domínguez P, Santos-Ocaña C. The Regulation of Coenzyme Q Biosynthesis in Eukaryotic Cells: All That Yeast Can Tell Us. Mol Syndromol 2014; 5:107-18; PMID:25126044; http://doi.dx.org/10.1159/000362897
- He CH, Xie LX, Allan CM, Tran UC, Clarke CF. Coenzyme Q supplementation or over-expression of the yeast Coq8 putative kinase stabilizes multi-subunit Coq polypeptide complexes in yeast coq null mutants. Biochim Biophys Acta - Mol Cell Biol Lipids 2014; 1841:630-44; PMID:24406904; http://doi.dx.org/10.1016/j.bbalip.2013.12.017
- Padilla S, Tran UC, Jiménez-Hidalgo M, López-Martín JM, Martín-Montalvo A, Clarke CF, Navas P, Santos-Ocaña C. Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell Mol Life Sci 2009; 66:173-86; PMID:19002377; http://doi.dx.org/10.1007/s00018-008-8547-7
- Miyadera H, Amino H, Hiraishi A, Taka H, Murayama K, Miyoshi H, Sakamoto K, Ishii N, Hekimi S, Kita K. Altered Quinone Biosynthesis in the Long-lived clk-1Mutants of Caenorhabditis elegans. J Biol Chem 2001; 276:7713-6; PMID:11244089; http://doi.dx.org/10.1074/jbc.C000889200
- Gao J, Schatton D, Martinelli P, Hansen H, Pla-Martin D, Barth E, Becker C, Altmueller J, Frommolt P, Sardiello M, et al. CLUH regulates mitochondrial biogenesis by binding mRNAs of nuclear-encoded mitochondrial proteins. J Cell Biol 2014; 207:213-23; PMID:25349259; http://doi.dx.org/10.1083/jcb.201403129
- Pullmann R, Kim HH, Abdelmohsen K, Lal A, Martindale JL, Yang X, Gorospe M. Analysis of Turnover and Translation Regulatory RNA-Binding Protein Expression through Binding to Cognate mRNAs. Mol Cell Biol 2007; 27:6265-78; PMID:17620417; http://doi.dx.org/10.1128/MCB.00500-07
- Romano M, Buratti E. Targeting RNA Binding Proteins Involved in Neurodegeneration. J Biomol Screen 2013; 18:967-83; PMID:23954928; http://doi.dx.org/10.1177/1087057113497256
- Szostak E, Gebauer F. Translational control by 3′-UTR-binding proteins. Brief Funct Genomics 2013; 12:58-65; PMID:23196851; http://doi.dx.org/10.1093/bfgp/els056
- Richman TR, Davies SMK, Shearwood A-MJ, Ermer JA, Scott LH, Hibbs ME, Rackham O, Filipovska A. A bifunctional protein regulates mitochondrial protein synthesis. Nucleic Acids Res 2014; 42:5483-94; PMID:24598254; http://doi.dx.org/10.1093/nar/gku179
- Svoboda P, Cara AD. Hairpin RNA: a secondary structure of primary importance. Cell Mol Life Sci 2006; 63:901-8; PMID:16568238; http://doi.dx.org/10.1007/s00018-005-5558-5
- Mignone F, Gissi C, Liuni S, Pesole G. No Title. Genome Biol 2002; 3:reviews0004.1; PMID:11897027; http://doi.dx.org/10.1186/gb-2002-3-3-reviews0004
- Izquierdo JM, Cuezva JM. Control of the translational efficiency of β-F1-ATPase mRNA depends on the regulation of a protein that binds the 3′ untranslated region of the mRNA. Mol Cell Biol 1997; 17:5255-68; PMID:9271403.
- Gorospe M, Baglioni C. Degradation of unstable interleukin-1α mRNA in a rabbit reticulocyte cell-free system: Localization of an instability determinant to a cluster of AUUUA motifs. J Biol Chem 1994; 269:11845-51; PMID:8163483.
- Trevisson E, Dimauro S, Navas P, Salviati L. Coenzyme Q deficiency in muscle. Curr Opin Neurol 2011; 24:449-56; PMID:21844807; http://doi.dx.org/10.1097/WCO.0b013e32834ab528
- Blaxall BC, Pende A, Wu SC, Port JD. No Title. Mol Cell Biochem 2002; 232:1-11; PMID:12030365; http://doi.dx.org/10.1023/A:1014819016552
- David PS, Tanveer R, Port JD. FRET-detectable interactions between the ARE binding proteins, HuR and p37AUF1. RNA 2007; 13:1453-68; PMID:17626845; http://doi.dx.org/10.1261/rna.501707
- Barker A, Epis MR, Porter CJ, Hopkins BR, Wilce MCJ, Wilce JA, Giles KM, Leedman PJ. Sequence requirements for RNA binding by HuR and AUF1. J Biochem 2012; 151:423-37; PMID:22368252; http://doi.dx.org/10.1093/jb/mvs010
- Masuda K, Marasa B, Martindale JL, Halushka MK, Gorospe M. Tissue- and age-dependent expression of RNA-binding proteins that influence mRNA turnover and translation. Aging (Albany NY) 2009; 1:681-98; PMID:20157551.
- Izquierdo JM. Heterogeneous ribonucleoprotein C displays a repressor activity mediated by T-cell intracellular antigen-1-related/like protein to modulate Fas exon 6 splicing through a mechanism involving Hu antigen R. Nucleic Acids Res 2010; 38:8001-14; PMID:20699271; http://doi.dx.org/10.1093/nar/gkq698
- Papadopoulou C, Patrinou-Georgoula M, Guialis A. Extensive association of HuR with hnRNP proteins within immunoselected hnRNP and mRNP complexes. Biochim Biophys Acta - Proteins Proteomics 2010; 1804:692-703; PMID:19931428; http://doi.dx.org/10.1016/j.bbapap.2009.11.007
- Fan XC, Steitz JA. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc Natl Acad Sci U S A 1998; 95:15293-8; PMID:9860962; http://doi.dx.org/10.1073/pnas.95.26.15293
- Keene JD. Why is Hu where? Shuttling of early-response-gene messenger RNA subsets. Proc Natl Acad Sci 1999; 96:5-7; PMID:9874760; http://doi.dx.org/10.1073/pnas.96.1.5
- Lal A, Abdelmohsen K, Pullmann R, Kawai T, Galban S, Yang X, Brewer G, Gorospe M. Posttranscriptional Derepression of GADD45α by Genotoxic Stress. Mol Cell 2006; 22:117-28; PMID:16600875; http://doi.dx.org/10.1016/j.molcel.2006.03.016
- Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR Regulates p21 mRNA Stabilization by UV Light. Mol Cell Biol 2000; 20:760-9; PMID:10629032; http://doi.dx.org/10.1128/MCB.20.3.760-769.2000
- Ortega AD, Sala S, Espinosa E, Gonzalez-Baron M, Cuezva JM. HuR and the bioenergetic signature of breast cancer: a low tumor expression of the RNA-binding protein predicts a higher risk of disease recurrence. Carcinogenesis 2008; 29:2053-61; PMID:18687667; http://doi.dx.org/10.1093/carcin/bgn185
- Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional Orchestration of an Anti-Apoptotic Program by HuR. Cell Cycle 2007; 6:1288-92; PMID:17534146; http://doi.dx.org/10.4161/cc.6.11.4299
- Durie D, Hatzoglou M, Chakraborty P, Holcik M. HuR controls mitochondrial morphology through the regulation of BclxL translation. Transl (Austin, Tex) 2013; 1:e23980; PMID:25328858; http://doi.dx.org/10.4161/trla.23980
- Montero R, Grazina M, López-Gallardo E, Montoya J, Briones P, Navarro-Sastre A, Land JM, Hargreaves IP, Artuch R, del Mar O'Callaghan M, et al. Coenzyme Q10 deficiency in mitochondrial DNA depletion syndromes. Mitochondrion 2013; 13:337-41; PMID:23583954; http://doi.dx.org/10.1016/j.mito.2013.04.001
- Larsson O. Role of biosynthesis of cholesterol and isoprenoid derivatives in regulation of G1 progression and cell proliferation of 3T6 cells. J Cell Physiol 1987; 133:163-8; PMID:3667703; http://doi.dx.org/10.1002/jcp.1041330121
- Abdelmohsen K, Pullmann R, Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, et al. Phosphorylation of HuR by Chk2 Regulates SIRT1 Expression. Mol Cell 2007; 25:543-57; PMID:17317627; http://doi.dx.org/10.1016/j.molcel.2007.01.011
