ABSTRACT
Every ribonucleic acid begins its cellular life as a transcript. If the transcript or its processing product has a function it should be regarded an RNA. Nonfunctional transcripts, by-products from processing, degradation intermediates, even those originating from (functional) RNAs, and non-functional products of transcriptional gene regulation accomplished via the act of transcription, as well as stochastic (co)transcripts could simply be addressed as transcripts (class 0). The copious functional RNAs (class I), often maturing after one or more processing steps, already are systematized into ever expanding sub-classifications ranging from micro RNAs to rRNAs. Established sub-classifications addressing a wide functional diversity remain unaffected. mRNAs (class II) are distinct from any other RNA by virtue of their potential to be translated into (poly)peptide(s) on ribosomes. We are not proposing a novel RNA classification, but wish to add a basic concept with existing terminology (transcript, RNA, and mRNA) that should serve as an additional framework for carefully delineating RNA function from an avalanche of RNA sequencing data. At the same time, this top level hierarchical model should illuminate important principles of RNA evolution and biology thus heightening our awareness that in biology boundaries and categorizations are typically fuzzy.
Historical considerations
As RNomes continue to increase in complexity and with a new RNA category being proclaimed almost monthly,Citation1-3 it is time to contemplate the basic question as to what an RNA is. About a century ago, the veil on the structure of a substance initially known as yeast nucleic acid or zymonucleic acid, later as pentose nucleic acid (PNA), and finally as ribonucleic acid, began to lift largely due to the contributions of Phoebus A.T. Levene and Jean L.A. Brachet.Citation4-6 Since the late fifties to early sixties, we learned that most if not all cellular ribonucleic acid is copied from DNA templates in an enzymatic process.Citation7 Around the same time, knowledge concerning RNA's participation in protein biosynthesis emerged, including microsomal RNA (rRNA) and soluble RNA (tRNA) as well as mRNA.Citation8 In the sixties and seventies other RNAs were detected and characterized, including members of RNA families that we classify today as small nuclear and nucleolar RNAs.Citation9-11. In those days, RNAs other than mRNAs were termed structural RNAs, presumably owing to their anticipated structural tasks, following rRNA that was simply regarded as a rack or backbone for the “only” functional components, the ribosomal proteins.Citation8 An additional motivation for this term might have been the presumption that these RNA species could form higher order structures (secondary and tertiary structures) akin to tRNACitation12 - at that time in alleged contrast to mRNAs. Remarkably, in the early 60s, isolated investigators predicted the roles of RNA not only in the evolution of life but also as functional and regulatory entities in extant organisms.Citation13
Apart from the few aforementioned RNAs as well as others thought to be fossils from the RNA world, the contributions of RNAs in cellular function and evolution were largely underappreciated. Only now, after 3-4 decades, does RNA finally receive the recognition it truly merits and in some instances, this attention has led to an exaggeration of its functions. All DNA-templated RNAs are transcripts, but the question is whether every transcript, albeit clearly a ribonucleic acid in the chemical sense, deserves to be classified as an RNA. In the biological context, we ought to be more discriminating.
The controversy
Recently, large-scale experimental approaches, such as microchip analysis and ultra-deep sequencing of the cellular RNA componentry, revealed copious transcripts including those of very low abundance. A debate is raging as to which fraction exerts a function. One camp claims that most if not all identified transcripts, including detectable degradation products, are functional.Citation14-20. The other side stipulates that although genomes of most organisms likely encode, in addition to mRNAs, as many as thousands or tens of thousands of functional RNAs, a large fraction, in particular the low abundant transcripts, merely represent transcriptional noise resulting from stochastic initiation in intergenic regions,Citation20-27 previously un-annotated (alternatively spliced) untranslated regions (UTRs),Citation28 read-throughs of bona fide gene termination sites,Citation29 and more or less stable debris or leftovers from the processing of primary/precursor transcripts. The most prominent are discarded introns, internal transcribed spacers, leaders, trailers, etc., but occasionally also sequences comprising spliced exons if, for example, the genes are merely hosts for miRNAs or snoRNAs and the corresponding exons lost (or never had) protein coding capacity.Citation30 as well Furthermore, molecules generated during the turnover and degradation of mature RNAs fall into this class.Citation31-37 Notably, transcriptional interference, which is gaining acceptance as yet another important layer of gene regulation, produces ribonucleic acids whose sequences are completely irrelevant; simply the act of transcription mediates an effect on, for example, a downstream promoter, including the blockade of transcription factor binding sitesCitation38-41 or the alteration of chromatin structure.Citation42,43
The difference between a transcript and an RNA
If we think about a ribonucleic acid chain as an RNA, it is usually with the association of a certain functional (including regulatory) or structural role – in analogy to a protein. This should not be expected from every stochastic transcript or degradation product or other “non-functional” ribonucleic acids. In keeping with the tradition of the oxymoronic term non-coding ribonucleic acids (ncRNAs, see also below) for functional non-mRNAsc , one is tempted to address the ribonucleic acids that are devoid of function as “non-RNAs.” Instead, we endorse the generic term transcripts for this class of ribonucleic acid, because some members of this category had previously been designated as stable untranslated transcripts or transcripts of unknown function (SUTs, TUFs).Citation46,47 As it is not trivial to assess whether such transcripts are never translated or otherwise functional,Citation48 and as ultra-deep RNA sequencing also identifies spurious or background transcripts of very low abundance, we prefer the simple term transcripts, adding that it includes stochastic transcripts, leftovers from primary transcript processing, or degradation products from the turnover of mature RNAs as well as products from regulatory acts of transcription. It follows then that transcripts constitute a further category, class 0d , in addition to class I (functional including structural RNA)e and class II (mRNA encoding peptide or protein) RNAs.Citation49
Gray areas
A recurring theme in biology is the frequent lack of clear boundaries or states; we are confronted instead with broad, fuzzy interphases or with chimera (). Evidently, most mRNAs (class II) not only feature an open reading frame (ORF) but also contain other codes in UTRs or even within ORFs. Those codes function as regulatory elements influencing the stability or translatability of mRNAs, and thus, mRNAs are chimeras of class I and II RNAs.Citation50-53 The borders between RNA and mRNA are also fuzzy because some bona fide class I RNAs might also encode ORFs, which are occasionally translatedCitation54 (i.e., they are bifunctionalf RNAs).Citation56,57 Likewise, an RNA might be in the process of exaptation as mRNA.Citation58 An example for a chimera is the bacterial hybrid of a transfer and a mRNA, tmRNA, formerly known as 10Sa RNA, which functions to deblock ribosomes engaged in translating truncated mRNAs devoid of an in-frame stop codon.Citation59 Furthermore, rRNA also encodes short peptides conveying antibiotic resistance.Citation60
Figure 1. A basic classification of transcripts and RNAs including the fuzziness of boundaries. Depicted is a continuum of the 3 ribonucleic acid superclasses: the first includes stochastic transcripts, other transcripts generated during gene regulation by acts of transcription, secondary products of RNA maturation such as introns etc., and RNA turnover products (transcripts, class 0) in yellow, functional RNAs (RNAs, class I) in blue, and classic mRNAs (mRNAs, class II) in red. Hybrid zones, interphases, or transitions between the 3 classes, for example, reflecting chimeras of 2 classes or those in the process of exaptation or abandonment are shown in purple, orange, and green. This figure does not represent quantitative measures of ribonucleic acid species or their abundances.
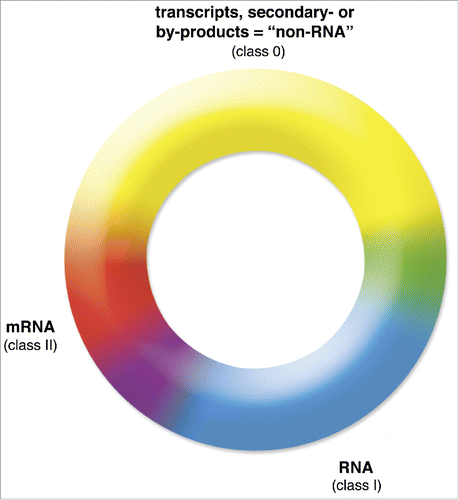
Interphases between transcripts on one hand and RNAs or mRNAs on the other, are also of interest. A broad range is expected between transcripts and RNAs, with some transcripts on their way to exaptation and a significant fraction on their way to oblivion.Citation21,61-63 In contrast to the interphase between transcripts and RNAs, the range between transcripts and mRNA is presumably somewhat condensed by mRNA's requirement of at least temporal ribosome association, which can be assessed experimentally.Citation64 While this does not necessarily assure translatability, its absence would not clearly exclude it, as this association might not have been investigated in the appropriate cell types or developmental times.
Ribonucleic acids that serve as binding partners for one or more proteins to bring them into close proximity for a combined function or to shuttle them to a specific subcellular location exert a task and hence are RNAs (class I). Then again, any transcript or degradation product can become or remain decorated with protein, which does not automatically imply a function; hence, they would constitute transcripts (class 0). In contrast, ribonucleic acids that act as decoys or sinks for other RNAs, proteins, or other molecules should be categorized as RNAs (class I).Citation65-68 This includes some of the circular RNAs (circRNAs) and non-translatable transcripts generated from duplicated genes, such as retroposed pseudogenes.Citation69 However, the majority of circRNAs, often generated by aberrant splicing, is expected to be devoid of function and consequently should be considered transcripts (class 0).Citation70 Likewise, a ribonucleic acid generated by a regulatory act of transcription also is not an RNA in the biological sense. As argued above, this does not rule out a fortuitous future exaptation of any class 0 transcript as a functional RNA or mRNA.Citation21 Notably, a role for extra transcripts as evolutionary raw material was proposed by Henry Harris half a century ago.Citation71,72
Concluding remarks
If we address a functional ribonucleic acid as RNA (class I), a translated or messenger ribonucleic acid as mRNA (class II) and everything else as a transcript (class 0), we do not need terms such as non-protein coding RNA (npcRNA), which sometimes might be subject to revision if a templated translation product is subsequently revealed. Neither do we need the unfortunate term non-coding RNA (ncRNA), because it reduces the RNA to something that it is not, obscures the fact that there is a gene coding for it (“a gene encoding a non-coding RNA”), and ignores the fact that RNAs carry many codes other than the one translated at ribosomes.Citation44,45 In any event, there is absolutely no need for the “nc” qualifier for RNA, as the term mRNA (mRNA) already provides the necessary qualifying differentiator. We do not propose to abandon clearly defined categories of RNAs, such as, for example, tRNAs, rRNAs, snRNAs, snoRNAs, miRNAs, or piRNAs, Instead, we simply add a top-level hierarchical layer to RNA classification using established categories of ribonucleic acids. Importantly, with this basic framework in mind, it should be easier to comprehend that defining a (sub)class of RNA does not necessarily imply that all its members are functional, such as any circular RNA by-product from splicing or any RNA snippet that happens to be in a size range of miRNAs or piRNAs.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Marsha Bundman and Stephanie Klco-Brosius for editing.
Notes
c Possibly, the sole function of miRNAs is to act as guides complementary to regions of mRNA 3'UTRs (usually), placing repressive proteins for translational regulation or stability onto the targeted mRNAs. Likewise, snoRNAs act as guides complementary to RNAs (mostly rRNAs), exactly determining the nucleosides to be modified by enzymes bound to snoRNAs. These RNA classes clearly carry (anti)codes according to Barbieri43 and Trifonov44. and thus, should not be addressed as non-coding RNAs.
d Of course, any RNA begins life as a transcript and a few of the functional RNAs remain unprocessed. Thus the primary transcript constitutes the functional RNA, but it is the function that usually sets RNAs apart from nonfunctional transcripts or parts thereof: Many processed parts of primary transcripts (e.g., introns, leaders, trailers) are nonfunctional, perhaps performing a temporary function such as providing higher order structure, e.g., necessary for processing. Yet, these ribonucleic acids should not be regarded as RNAs in the considerations presented here, due to their lack of function (biology over chemistry). Usually, a transcript without being processed does not function despite the fact that it harbors functional RNAs (e.g., pre-mRNAs, pre-rRNA, pre-miRNAs, etc.).
e It does not matter how tiny (e.g., miRNA, piRNA), or large (e.g., Xist) an RNA is or whether an RNA was chemically synthesized. The latter is an RNA since most, if not all, molecules are being designed to exert a function, in vivo or in vitro. Viral RNA genomes are RNAs where one of the functions is the mobility from cell to cell and host to host.
f The term bifunctional RNA is also used for other molecules with two domains. For example, an antisense RNA sequence specific to a target hnRNA and an untethered RNA segment that serves as a binding platform for splicing factors to guide certain desired splice variants as potential therapeutic agents. ADDIN EN.CITE.DATA 55.
References
- St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet 2015; 31:239-51; PMID:25869999; http://dx.doi.org/10.1016/j.tig.2015.03.007.
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014; 157:77-94; PMID:24679528; http://dx.doi.org/10.1016/j.cell.2014.03.008.
- Vickers KC, Roteta LA, Hucheson-Dilks H, Han L, Guo Y. Mining diverse small RNA species in the deep transcriptome. Trends Biochem Sci 2015; 40:4-7; PMID:25435401; http://dx.doi.org/10.1016/j.tibs.2014.10.009.
- Levene PA. The structure of yeast nucleic acid. IV Ammonia hydrolysis. J Biol Chem 1919; 40:415-24.
- Brachet J. La localisation des acides pentose-nucléiques dans les tissue animaux et les oeufs d'amphibiens en voie de développment. Arch Biol (Liege) 1941; 53:207-57.
- Brachet J. La détection histochimique et le microdosage des acides pentosenucléiques. Enzymologia 1941; 10:87-96.
- Hurwitz J. The discovery of RNA polymerase. J Biol Chem 2005; 280:42477-85; PMID:16230341; http://dx.doi.org/10.1074/jbc.X500006200.
- Rheinberger HJ. A history of protein biosynthesis and ribosome research. In: Nierhaus KH, Wilson DN, eds. Protein Synthesis and Ribosome Structure Translating the Genome. Weinheim: Wiley, 2004:1-71.
- Hadjiolov AA, Venkov PV, Tsanev RG. Ribonucleic acids fractionation by density-gradient centrifugation and by agar gel electrophoresis: a comparison. Anal Biochem 1966; 17:263-7; PMID:5339429; http://dx.doi.org/10.1016/0003-2697(66)90204-1.
- Nakamura T, Prestayko AW, Busch H. Studies on nucleolar 4 to 6 S ribonucleic acid of Novikoff hepatoma cells. J Biol Chem 1968; 243:1368-75; PMID:4296684.
- Weinberg RA, Penman S. Small molecular weight monodisperse nuclear RNA. J Mol Biol 1968; 38:289-304; PMID:5718554; http://dx.doi.org/10.1016/0022-2836(68)90387-2.
- Holley RW, Apgar J, Everett GA, Madison JT, Marquisee M, Merrill SH, Penswick JR, Zamir A. Structure of a Ribonucleic Acid. Science 1965; 147:1462-5; PMID:14263761; http://dx.doi.org/10.1126/science.147.3664.1462.
- Rich A. On the problems of evolution and biochemical information transfer. In: Kasha M, Pullman B, eds. Horizons in Biochemistry New York, London: Academic Press, 1962:103-26.
- Carninci P. RNA dust: where are the genes? DNA Res 2010; 17:51-9; PMID:20211845; http://dx.doi.org/10.1093/dnares/dsq006.
- Clark MB, Amaral PP, Schlesinger FJ, Dinger ME, Taft RJ, Rinn JL, Ponting CP, Stadler PF, Morris KV, Morillon A, et al. The reality of pervasive transcription. PLoS Biol 2011; 9:e1000625; discussion e1102; PMID:21765801; http://dx.doi.org/10.1371/journal.pbio.1000625.
- Mattick JS. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 2003; 25:930-9; PMID:14505360; http://dx.doi.org/10.1002/bies.10332.
- Mattick JS, Dinger ME. The extent of functionality in the human genome. HUGO J 2013, 7:2. doi: 10.1186/1877-6566-7-2
- Kapranov P, St Laurent G. Dark Matter RNA: Existence, Function, and Controversy. Front Genet 2012; 3:60; PMID:22536205; http://dx.doi.org/10.3389/fgene.2012.00060
- Bernstein BE, Birney E, Dunham I, Green ED, Gunter C, Snyder M. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57-74; PMID:22955616; http://dx.doi.org/10.1038/nature11247.
- Kowalczyk MS, Higgs DR, Gingeras TR. Molecular biology: RNA discrimination. Nature 2012; 482:310-1; PMID:22337043; http://dx.doi.org/10.1038/482310a.
- Brosius J. Waste not, want not – transcript excess in multicellular Eukaryotes. Trends Genet 2005; 21:287-8; PMID:15851065; http://dx.doi.org/10.1016/j.tig.2005.02.014.
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol 2007; 14:103-5; PMID:17277804; http://dx.doi.org/10.1038/nsmb0207-103.
- Raj A, van Oudenaarden A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 2008; 135:216-26; PMID:18957198; http://dx.doi.org/10.1016/j.cell.2008.09.050.
- Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell 2013; 154:26-46; PMID:23827673; http://dx.doi.org/10.1016/j.cell.2013.06.020.
- Graur D, Zheng Y, Price N, Azevedo RB, Zufall RA, Elhaik E. On the immortality of television sets: “function” in the human genome according to the evolution-free gospel of ENCODE. Genome Biol Evol 2013; 5:578-90; PMID:23431001; http://dx.doi.org/10.1093/gbe/evt028.
- Jensen TH, Jacquier A, Libri D. Dealing with pervasive transcription. Mol Cell 2013; 52:473-84; PMID:24267449; http://dx.doi.org/10.1016/j.molcel.2013.10.032.
- Mudge JM, Frankish A, Harrow J. Functional transcriptomics in the post-ENCODE era. Genome Res 2013; 23:1961-73; PMID:24172201; http://dx.doi.org/10.1101/gr.161315.113.
- van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most “dark matter” transcripts are associated with known genes. PLoS Biol 2010; 8:e1000371; PMID:20502517; http://dx.doi.org/10.1371/journal.pbio.1000371.
- Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev 2009; 23:1247-69; PMID:19487567; http://dx.doi.org/10.1101/gad.1792809.
- Tycowski KT, Shu MD, Steitz JA. A mammalian gene with introns instead of exons generating stable RNA products. Nature 1996; 379:464-6; PMID:8559254; http://dx.doi.org/10.1038/379464a0
- Moore MJ. Nuclear RNA turnover. Cell 2002; 108:431-4; PMID:11909514; http://dx.doi.org/10.1016/S0092-8674(02)00645-1.
- Cole C, Sobala A, Lu C, Thatcher SR, Bowman A, Brown JW, Green PJ, Barton GJ, Hutvagner G. Filtering of deep sequencing data reveals the existence of abundant Dicer-dependent small RNAs derived from tRNAs. RNA 2009; 15:2147-60; PMID:19850906; http://dx.doi.org/10.1261/rna.1738409.
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 2009; 23:2639-49; PMID:19933153; http://dx.doi.org/10.1101/gad.1837609.
- Keene JD. Minireview: global regulation and dynamics of ribonucleic Acid. Endocrinology 2010; 151:1391-7; PMID:20332203; http://dx.doi.org/10.1210/en.2009-1250.
- Khayrullina GA, Raabe CA, Hoe CH, Becker K, Reinhardt R, Tang TH, Rozhdestvensky TS, Kopylov AM. Transcription analysis and small non-protein coding RNAs associated with bacterial ribosomal protein operons. Curr Med Chem 2012; 19:5187-98; PMID:22680642; http://dx.doi.org/10.2174/092986712803530485.
- Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, Beach C, Nicholls RD, Zavolan M, Stamm S. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet 2010; 19:1153-64; PMID:20053671; http://dx.doi.org/10.1093/hmg/ddp585.
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 2013; 10:1798-806; PMID:24351723; http://dx.doi.org/10.4161/rna.27177.
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 2004; 429:571-4; PMID:15175754; http://dx.doi.org/10.1038/nature02538.
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference–a crash course. Trends Genet 2005; 21:339-45; PMID:15922833; http://dx.doi.org/10.1016/j.tig.2005.04.009.
- Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol Cell 2009; 34:545-55; PMID:19524535; http://dx.doi.org/10.1016/j.molcel.2009.04.018.
- Kornienko AE, Guenzl PM, Barlow DP, Pauler FM. Gene regulation by the act of long non-coding RNA transcription. BMC Biol 2013; 11:59; PMID:23721193; http://dx.doi.org/10.1186/1741-7007-11-59.
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell 2006; 127:1209-21; PMID:17174895; http://dx.doi.org/10.1016/j.cell.2006.10.039.
- Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012; 338:1469-72; PMID:23239737; http://dx.doi.org/10.1126/science.1228110.
- Barbieri M. The Organic Codes. An Introduction to semantic Biology. Cambridge: Cambridge University Press, 2003.
- Trifonov EN. The multiple codes of nucleotide sequences. Bull Math Biol 1989; 51:417-32; PMID:2673451; http://dx.doi.org/10.1007/BF02460081.
- Gingeras TR. Origin of phenotypes: genes and transcripts. Genome Res 2007; 17:682-90; PMID:17567989; http://dx.doi.org/10.1101/gr.6525007.
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature 2009; 457:1033-7; PMID:19169243; http://dx.doi.org/10.1038/nature07728.
- Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, et al. Considerations when investigating lncRNA function in vivo. eLife 2014; 3:e03058; PMID:25124674; http://dx.doi.org/10.7554/eLife.03058.
- Brosius J, Tiedge H. RNomenclature. RNA Biol 2004; 1:81-3; PMID:17179746; http://dx.doi.org/10.4161/rna.1.2.1228.
- Chen H, Blanchette M. Detecting non-coding selective pressure in coding regions. BMC Evol Biol 2007; 7 Suppl 1:S9; PMID:17288582; http://dx.doi.org/10.1186/1471-2148-7-S1-S9.
- Dinger ME, Pang KC, Mercer TR, Mattick JS. Differentiating protein-coding and noncoding RNA: challenges and ambiguities. PLoS Comput Biol 2008; 4:e1000176; PMID:19043537; http://dx.doi.org/10.1371/journal.pcbi.1000176.
- Kageyama Y, Kondo T, Hashimoto Y. Coding vs non-coding: Translatability of short ORFs found in putative non-coding transcripts. Biochimie 2011; 93:1981-6; PMID:21729735; http://dx.doi.org/10.1016/j.biochi.2011.06.024.
- Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol 2002; 3:REVIEWS0004; PMID:11897027; http://dx.doi.org/10.1186/gb-2002-3-3-reviews0004.
- Ingolia NT, Brar GA, Stern-Ginossar N, Harris MS, Talhouarne GJ, Jackson SE, Wills MR, Weissman JS. Ribosome profiling reveals pervasive translation outside of annotated protein-coding genes. Cell Rep 2014; 8:1365-79; PMID:25159147; http://dx.doi.org/10.1016/j.celrep.2014.07.045.
- Baughan T, Shababi M, Coady TH, Dickson AM, Tullis GE, Lorson CL. Stimulating full-length SMN2 expression by delivering bifunctional RNAs via a viral vector. Mol Ther 2006; 14:54-62; PMID:16580882; http://dx.doi.org/10.1016/j.ymthe.2006.01.012.
- Ulveling D, Francastel C, Hube F. When one is better than two: RNA with dual functions. Biochimie 2011; 93:633-44; PMID:21111023; http://dx.doi.org/10.1016/j.biochi.2010.11.004.
- Wadler CS, Vanderpool CK. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc Natl Acad Sci U S A 2007; 104:20454-9; PMID:18042713; http://dx.doi.org/10.1073/pnas.0708102104.
- Neme R, Tautz D. Entire genome transcription across evolutionary time exposes non-coding DNA to de novo gene emergence. 2015: in press.
- Hayes CS, Keiler KC. Beyond ribosome rescue: tmRNA and co-translational processes. FEBS Lett 2010; 584:413-9; PMID:19914241; http://dx.doi.org/10.1016/j.febslet.2009.11.023.
- Tenson T, DeBlasio A, Mankin A. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc Natl Acad Sci U S A 1996; 93:5641-6; PMID:8643630; http://dx.doi.org/10.1073/pnas.93.11.5641.
- Brosius J, Gould SJ. On “genomenclature:” a comprehensive (and respectful) taxonomy for pseudogenes and other “junk DNA.” Proc Natl Acad Sci U S A 1992; 89:10706-10; PMID:1279691; http://dx.doi.org/10.1073/pnas.89.22.10706.
- Schmitz J, Brosius J. Exonization of transposed elements: A challenge and opportunity for evolution. Biochimie 2011; 93:1928-34; PMID:21787833; http://dx.doi.org/10.1016/j.biochi.2011.07.014.
- Polev D. Transcriptional noise as a driver of gene evolution. J Theor Biol 2012; 293:27-33; PMID:22001319; http://dx.doi.org/10.1016/j.jtbi.2011.10.001.
- Wilson BA, Masel J. Putatively noncoding transcripts show extensive association with ribosomes. Genome Biol Evol 2011; 3:1245-52; PMID:21948395; http://dx.doi.org/10.1093/gbe/evr099.
- Babitzke P, Romeo T. CsrB sRNA family: sequestration of RNA-binding regulatory proteins. Curr Opin Microbiol 2007; 10:156-63; PMID:17383221; http://dx.doi.org/10.1016/j.mib.2007.03.007.
- Banks IR, Zhang Y, Wiggins BE, Heck GR, Ivashuta S. RNA decoys: an emerging component of plant regulatory networks? Plant Signal Behav 2012; 7:1188-93; PMID:22899065; http://dx.doi.org/10.4161/psb.21299.
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 2007; 39:1033-7; PMID:17643101; http://dx.doi.org/10.1038/ng2079.
- Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010; 465:1033-8; PMID:20577206; http://dx.doi.org/10.1038/nature09144.
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453-61; PMID:24811520; http://dx.doi.org/10.1038/nbt.2890.
- Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science 2013; 340:440-1; PMID:23620042; http://dx.doi.org/10.1126/science.1238522.
- Harris H. The short-lived RNA in the cell nucleus and its possible role in evolution. In: Bryson V, Vogel HJ, eds. Evolving genes and proteins. New York, London: Academic Press, 1965:469-500.
- Harris H. History: Non-coding RNA foreseen 48 years ago. Nature 2013; 497:188; PMID:23657339; http://dx.doi.org/10.1038/497188d.
