ABSTRACT
The eukaryotic release factor 3 (eRF3) has been involved in the control of mRNA degradation through its association with the cytoplasmic Poly(A) Binding Protein, PABP. In mammals, eRF3 N-terminal domain contains two overlapping PAM2 motifs which specifically recognize the MLLE domain of PABP. In humans, eRF3a/GSPT1 gene contains a stable GGC repeat encoding a repeat of glycine residues in eRF3a N-terminus. There are five known eRF3a/GSPT1 alleles in the human population, encoding 7, 9, 10, 11 and 12 glycines. Several studies have reported that the presence of eRF3a 12-GGC allele is correlated with an increased risk of cancer development. Using surface plasmon resonance, we have studied the interaction of the various allelic forms of eRF3a with PABP alone or poly(A)-bound PABP. We found that the N-terminal glycine repeat of eRF3a influences eRF3a-PABP interaction and that eRF3a 12-GGC allele has a decreased binding affinity for PABP. Our comparative analysis on eRF3a alleles suggests that the presence of eRF3a 12-GGC allele could modify the coupling between translation termination and mRNA deadenylation.
Introduction
The eukaryotic release factor 3 (eRF3) is a small GTPase which associates with eukaryotic release factor 1 (eRF1) to terminate translation when a stop codon enters the A site of the ribosome. In addition to its role in translation termination, eRF3 has been involved in the control of mRNA degradation through its association with the cytoplasmic poly (A) binding protein, PABP, also termed PABPC1.Citation1,2 The C-terminal domain of eRF3 which carries the translation termination activity, is well-conserved in species ranging from yeast to mammals. It contains the four canonical GTP binding motifs responsible for eRF3 GTPase activity and the eRF1 binding site at its extreme C-terminal end. In contrast, the unstructured eRF3 N-terminal domain is poorly conserved among eukaryotic species. In mammals, it carries two overlapping PAM2 (for PABP-interacting motif 2) motifs that specifically recognize the MLLE domain of PABP.Citation3
In a given cellular state, either proliferating or differentiated, the cytoplasmic degradation of mRNAs allows to maintain a constant level of mRNAs available for translation and thus plays a key role in the regulation of gene expression. The poly(A) tail of mRNAs is essential for maintaining their stability and the shortening of the poly(A) tail, referred to as deadenylation, is the first and rate-limiting step of the mRNAs degradation process.Citation4 Several molecules of PABP cover the length of the mRNA poly(A) tail (one PABP molecule interacts with ∼20 adenosine residues) and have a protective effect against mRNA degradation.Citation5 Furthermore, PABP binding to the eukaryotic initiation factor 4G (eIF4G) promotes the closed-loop mRNA structure and mediates the stimulatory effect of the poly(A) tail on translation initiation.Citation6 However, PABP also recruits the deadenylation complexes through its association with the PAM2 motif carrying proteins PAN3 and TOB,Citation7,8 that are partners of the PAN2-PAN3 and TOB-CAF1-CCR4 deadenylation complexes, respectively. In mammals, the mRNA poly(A) tail is of ∼250 adenosine residues.Citation4 It has been proposed that deadenylation proceeds in two steps: the PAN2-PAN3 complex first degrades the 3′ end of the poly(A) tail leaving ∼100 adenosine residues that are then degraded by the TOB-CAF1-CCR4 complex.Citation9
Recent studies showed that both PAN3 and TOB compete with eRF3 for their interaction with PABP. From these observations, it was proposed that upon translation termination, the PABP-poly(A)-bound termination complex, eRF1–eRF3, is recruited to the ribosome and is replaced on PABP by either PAN2-PAN3 or TOB-CAF1-CCR4 deadenylation complexes.Citation10,11 Peptide binding studies showed that the eRF3 PAM2 motifs bind PABP MLLE domain with higher affinity than PAN3 and TOB PAM2 motifs.Citation12 The competitive binding between eRF3 and deadenylation complexes suggests that eRF3 has a protective effect against deadenylation. However, the precise function of this competition in mRNA stability remains to be elucidated. Furthermore, it is not clearly understood whether deadenylation is coupled to translation and requires translation termination to proceed.Citation13 Thermodynamic analysis of human eRF3 binding to GDP, GTP and human PABP have shown that PABP associates equally with either free eRF3-GDP or eRF1-eRF3-GTP complex and that PABP-eRF3 interaction does not require GTP and does not modify eRF3 binding to GTP.Citation14 However, genetic studies in yeast have pointed out the fact that eRF3-PABP interaction, which does not involve a consensus PAM2 motif, regulates translation termination efficiency.Citation15,16
In mammals, two distinct genes, named GSPT1 and GSPT2, encode two proteins with translation termination activity, eRF3a and eRF3b, respectively.Citation17,18 eRF3a is the major form of eRF3 in the majority of the tissues and its abundance regulates the formation of translation termination complex.Citation19,20 eRF3b, which can complement eRF3a for its role in translation termination, is very poorly expressed in most tissues and likely does not play a major role in termination.Citation20 eRF3a and eRF3b proteins differ in their N-terminal domain. In human GSPT1 gene, the sequence encoding the N-terminus of eRF3a contains a repeat of GGC glycine codons,Citation21 which is not present in GSPT2 (Fig. S1). There are five known alleles of the GGC repeat, encoding 7, 9, 10, 11 and 12 glycines, in human population.Citation22 The most common allele is the 10-GGC allele. Intriguingly, in two studies on the Portugese population, the 12-GGC allele was detected exclusively in cancer patients.Citation22,23 Regardless of the genotype, Portugese patients with the 12-GGC allele had a ~20-fold increased risk for gastric cancer Citation22 and a 13-fold increased risk for breast cancer.Citation23 Consolidating these observations, a recent study on eRF3a GGC repeat polymorphism in Iranian women with breast cancer showed that the presence of the 12-GGC allele is correlated with a 3 fold increased risk of cancer development.Citation24 Complementation experiments in eRF3a-depleted cells were unable to detect differences in translation termination activity for eRF3a alleles including the 12-GGC allele.Citation23 Todate, the link between eRF3a 12-GGC allele and tumorigenesis remains unknown. However, the presence of the glycine repeat in the vicinity of PABP binding site leads to the possibility that the glycine repeat acts on eRF3a-PABP interaction and hence interferes with its role in mRNA deadenylation.
Here we study the interaction of the various allelic forms of human eRF3a with PABP using surface plasmon resonance (SPR). This is a powerful approach that provides high-quality data describing the kinetics of protein-protein interactions as shown for Paip1 and Paip2 interacting with PABP.Citation25,26 Whereas SPR was also used to measure the kinetics of RNA–protein interactions,Citation27-29 it has never been applied to the study of protein complexes in which one of the partner is bound to RNA. In this report, we present a kinetic analysis of eRF3-PABP interaction when PABP is bound to a synthetic poly(A) oligoribonucleotide of ∼120 adenosine residues. This synthetic oligo(A) is able to carry several PABP molecules and therefore mimics a functional mRNA poly(A) tail. Whatever the model used, PABP alone or poly(A)-bound PABP, our comparative analysis of the different forms of human eRF3, eRF3a alleles and eRF3b, reveals that the glycine repeat at the N-terminal end of eRF3a plays a role in the interaction of eRF3 with PABP. Moreover, we show that the binding affinity of eRF3a 12-GGC allele for PABP is ten fold lower than that of the other eRF3a alleles. These results bring new insights into the mechanism by which eRF3a 12-GGC allele could promote carcinogenesis.
Results
The glycine repeat at the N-terminal extremity of human eRF3 influences eRF3-PABP interaction
Our goal was to compare the binding affinities of the different human eRF3a alleles for PABP. We included in this study eRF3b, which is devoid of the N-terminal glycine stretch encoded by the GGC repeat in GSPT1 gene (Fig. S1). To obtain quantitative kinetic measurements of eRF3-PABP interaction, experiments were conducted on an SPR-based biosensor (Biacore 3000). For the first series of experiments, purified His-tagged full-length PABP (1,000–2,500 RU) was immobilized on a CM5 sensor chip. In order to substract any background noise from each dataset, the same coupling procedure, but in the absence of protein, was used to prepare a mock surface (see Materials and Methods). A range of analyte (either eRF3a 7-GGC, 10-GGC, 11-GGC, 12-GGC alleles or eRF3b) concentrations was injected for 5 min, followed by a dissociation phase of 5–8 min over ligand (PABP) surface or mock surface. The resulting set of curves was then analyzed by curve fitting using numerical integration methods (see Materials and Methods). Removal of the noise was achieved by subtracting the signal generated from a “blank” (running buffer) injection (, 0 nM curves) over the entire data set ; this is known as “double referencing.”Citation28 After blank subtraction, the analysis of the sensorgrams yielded good fits when a simple 1:1 Langmuir interaction model was applied. The fitting to this model was judged by the quality of the residual (typically not exceeding +/− 2 RU). The sensorgrams and residual plots of typical SPR experiments are shown in , and the kinetic parameters are summarized in . The eRF3a 10-GGC allele which is the most frequent allele in the human population (∼70% in all samples of populations studied so far, see ) was chosen as a reference for the interactions, and eRF3a in the following text will refer to the 10-GGC allele.
Figure 1. SPR analysis of the interaction between eRF3 and PABP (2000–2200 RU) immobilized on a CM5 sensor chip surface. Typical experimental sensorgrams (top panels) and residual plots (difference between calculated and experimental data points, bottom panels) are shown for eRF3a 10-GGC allele (A), eRF3a 12-GGC allele (B), eRF3a 7-GGC allele (C), eRF3a 11-GGC allele (D), and eRF3b (E). The different concentrations of the injected analytes are indicated on the right of the sensorgrams (for experimental details see Materials and Methods). Data were treated and integrated using a simple Langmuir 1:1 model. For each analyte concentration the fitted curve is shown in black. (F) Comparison of eRF3a alleles and eRF3b equilibrium dissociation constants (KD) listed in . Error bars indicate the standard error (SEM). Statistical significance (P-values from unpaired Student's t-test) of differences between eRF3a 12-GGC and other eRF3a alleles are indicated.
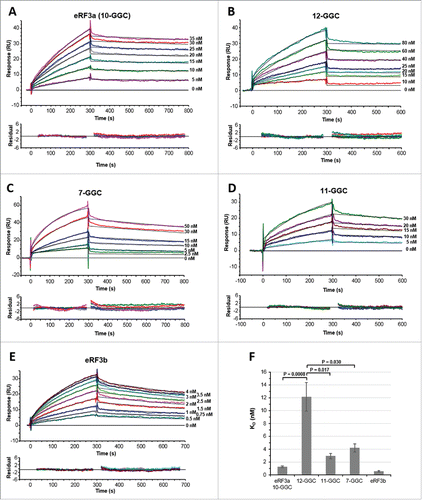
Table 1. Kinetic parameters for eRF3a alleles and eRF3b interaction with PABP immobilized on a CM5 sensor chip surface.aFootnotea
The results of kinetic determinations indicate that eRF3a binds to PABP with a fast association (ka = 1.61 × 105 M−1 s−1) and a slow dissociation (kd = 2.12 × 10−4 s−1). The dissociation equilibrium constant (KD) indicates a high binding affinity between eRF3a and PABP (KD = 1.5 nM). The fitting results reveal that 7-GGC and 11-GGC alleles bind to PABP with association and dissociation rate constants comparable to those measured for eRF3a (), resulting in KD values of the same order of magnitude (4.2 nM and 2.9 nM, respectively). In contrast, the equilibrium constant of PABP interaction with the eRF3a 12-GGC allele differed by one order of magnitude (KD = 12.0 nM, P = 0.0008) with that observed for PABP-eRF3a interaction ( and ). This difference was mainly due to a loss in the association rate while the dissociation rate was of the same magnitude as that observed for eRF3a (). Conversely, with the fastest association rate (ka = 9.66 × 105 M−1 s−1), the affinity of the PABP-eRF3b interaction (KD = 0.5 nM) was 3-fold higher than that of the PABP-eRF3a interaction.
Remembering that (i) the eRF3a alleles only differ by the length of their N-terminal glycine stretch, (ii) the glycine stretch is lacking in eRF3b, and (iii) the glycine stretch is located close to the PABP binding site in the unstructured N-terminal domain of eRF3, our results support the idea that the variations of the equilibrium constant could be attributed to the variations of the glycine stretch encoded by the GGC repeat. This is particularly clear for the eRF3a 12-GGC allele for which the presence of an additional glycine residue, compared to the 11-GGC allele, seems to play a critical role in its association with PABP.
SPR analysis of PABP interacting with poly(A) tail
In cells, cytoplasmic PABP is mainly associated to the poly(A) tail of mRNAs. Thus, eRF3-PABP complex is likely formed mainly when PABP is bound to poly(A). In addition, PABP multimerizes on the mRNA poly(A) tail. Both elements, poly(A) tail and PABP multimerization could potentially modify eRF3-PABP interaction. We therefore decided to study the kinetic parameters of the eRF3a-PABP interaction when PABP is bound to an oligoribonucleotide carrying a poly(A) tail. To this end, we enzymatically lengthened a 5′ biotinylated 18-mer oligoribonucleotide with 120–140 adenosine residues using E. coli polyA-polymerase (Fig. S2). The resulting oligoribonucleotide (hereafter named OligoA120) could theoretically carry up to 7 PABP molecules, considering that a single PABP covers ∼20 adenosine residues of the poly(A) tail.Citation30,31
To confirm the specificity of PABP binding toward the poly(A) sequence, we immobilized either the biotinylated 18-mer synthetic oligoribonucleotide (68 RU) used for the polyA-polymerase reaction or the biotinylated OligoA120 (72 RU) through streptavidin onto a SA sensor chip. Small amounts of RNA were used to coat the surface in order to minimize mass transport effects. Purified full-length PABP was used as the analyte in the running buffer. As expected, we did not observe any mass accumulation on the SA sensor chip when the 18-mer oligoribonucleotide was immobilized, showing that PABP has no binding affinity for the 18-mer synthetic oligoribonucleotide (data not shown). Because we wished to perform multiple-binding (PABP plus increasing concentrations of eRF3a) SPR experiments with the immobilized OligoA120, we first checked whether eRF3a had by itself binding affinity for the biotinylated OligoA120. eRF3a was injected at increasing concentrations (10, 50 and 100 nM) over 220 RU of OligoA120 immobilized on the SA sensor chip surface. As a control 5 nM of PABP was injected on the same surface. The resulting set of curves unambiguously showed that, in contrast to PABP, eRF3a had no binding affinity for the OligoA120 (Fig. S3). We next determined the binding capacity of the OligoA120 for PABP. In a first experiment, the biotinylated OligoA120 (22 RU) was immobilized on the sensor chip surface and multiple injections were performed with increasing concentrations of PABP (25–200 nM) without removing bound proteins (20 min injection of PABP followed by a dissociation phase of 20 min with running buffer alone). In a second experiment, PABP at 100 nM concentration was injected for 20 min over the same surface (22 RU of biotinylated OligoA120). These experiments allow (i) to assess the stability of the OligoA120 surface over the time of the experiments and (ii) to estimate the number of PABP molecules bound per OligoA120 molecule. Both recorded sensorgrams reached a plateau during the injection of PABP at high concentration, consistent with steady-state binding (Fig. S4). This result allowed us to calculate (see Materials and Methods) that the OligoA120 molecule could bind 5 to 7 PABP molecules, in good agreement with a binding of one PABP for ∼20 adenosine residues as previously described.Citation31 This result showed that PABP binds in a linear manner along the poly(A) of the OligoA120 molecule as described for natural mRNA poly(A) tail. In addition to these results which demonstrated the high stability of the OligoA120 surface over experimental time, we also verified the stability of the immobized OligoA120 when several cycles of PABP injection (5 min) at various concentrations (10 to 100 nM), dissociation (8 min) with running buffer, and regeneration (1 min) with 0.1% SDS, were applied (in our hands regeneration with 0.1% SDS solution was the most efficient method to remove all bound proteins without damaging the RNA surface). As shown in Fig. S5, the base line corresponding to immobilized OligoA120 was no modified by repeated cycles of PABP binding, dissosciation and regeneration. The sensorgram also showed that, at the end of the association phase, bound PABP dissociated slowly from the immobilized OligoA120 suggesting that multimerized PABP formed a stable complex with the poly(A) tail of the OligoA120.
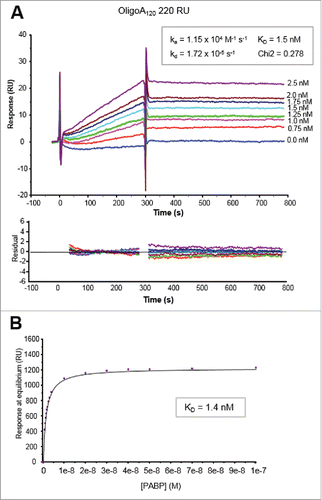
We next determined the kinetic parameters of PABP-OligoA120 interaction (). A range of PABP (0.75 to 2.5 nM) concentrations was injected for 5 min, followed by a dissociation phase of 8 min over the immobilized OligoA120 (220 RU). Disruption of any complex that remained bound after a 8 min dissociation was achieved using a 1 min injection of 0.1% SDS at 30 µl/min. After blank subtraction (no target RNA was captured on the reference flow cell surface), the analysis of the sensorgrams yielded good fits when a single site interaction model was applied. The fitting to this model was also judged by the low value of the Chi2 and the good quality of the residuals (). The results from the fittings of the PABP-OligoA120 interaction indicated a moderatly fast association rate (ka) of 1.15 × 104 M−1 s−1, a very slow dissociation rate (kd) of 1.72 × 10−5 s−1, and a resulting KD of 1.5 nM. The values of response at equilibrium obtained for PABP-OligoA120 interaction in the experiments shown in and Fig. S5, which were performed on the same immobilized OligoA120 surface, were also employed to calculate the equilibrium constant (KD) of PABP-OligoA120 interaction. These values were processed using the BIAevalution software for the steady-state affinity fitting. The curve of the response at equilibrium versus PABP concentration confirmed the KD value of 1.5 nM ().
Figure 2. Kinetic analyses of PABP binding to 5′ biotinylated OligoA120 RNA (220 RU) immobilized onto a SA sensor chip surface. (A) Sensorgrams of the binding profiles using a Langmuir 1:1 binding model (top panel) and residuals plots (bottom panel) are shown. Concentrations of PABP injected are indicated on the right of the sensorgrams. Kinetic parameters (ka, kd and KD) and the Chi2 value of the fitting are also indicated. (B) Plot of the response vs. PABP concentration used for the steady-state affinity fitting with the BIAevaluation software (see text for details).
Altogether these results validated our synthetic model of poly(A) tail that can be used for SPR studies of proteins that associate with poly(A)-bound PABP.
Interaction of eRF3a alleles with poly(A)-bound PABP
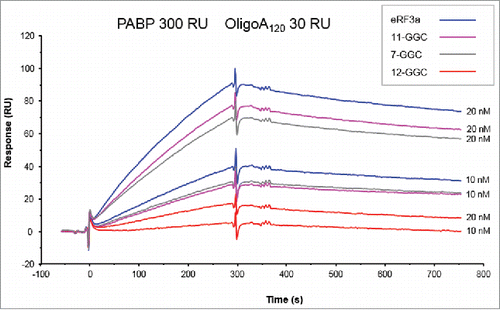
We next determined the kinetic parameters of eRF3a alleles when interacting with poly(A) bound PABP in multiple-binding SPR experiments. In a preliminary experiment, we evaluated the binding capacities of the different eRF3a alleles. To this end, the biotinylated OligoA120 (30 RU) immobilized on the SA sensor chip surface was covered with PABP by manual injection of a solution of 80 nM PABP for 5 min at 5 µl/min followed by a dissociation phase of 8 min with running buffer. This yielded approximately 300 RU of bound PABP, correponding to ∼5.4 PABP per OligoA120 molecule. Then, eRF3a or one of its allelic forms (7-GGC, 11-GGC and 12-GGC) were injected at either 10 nM or 20 nM for 5 min at 5 µl/min. Each eRF3a and allele injection was followed by a dissociation phase of 8 min and by regeneration with 0.1% SDS for 1 min. The resulting sensorgrams showed that eRF3a alleles mainly differed in their association with PABP whereas the dissociation phases were identical (). The 12-GGC allele had the slowest association rate when compared to eRF3a (10-GGC), 7-GGC and 11-GGC alleles. We also reproducibly observed that the 7-GGC and 11-GGC alleles have slightly lower association rates than that of eRF3a.
Figure 3. Sensorgrams of single-binding analysis of eRF3a and 7-GGC, 11-GGC, and 12-GGC alleles. eRF3a alleles were injected at 10 and 20 nM concentrations (indicated on the right of each sensorgram) over PABP (300 RU) bound to OligoA120 RNA (30 RU) immobilized on a SA sensor chip.
We therefore studied the kinetic parameters of the interaction of eRF3a and eRF3a 12-GGC allele with poly(A)-bound PABP using the same method as above. The immobilized OligoA120 was covered with PABP and a range of eRF3a or eRF3a 12-GGC concentrations was manually injected over the PABP-bound OligoA120 surface. For each analyte concentration, after injection and dissociation in running buffer, the immobilized OligoA120 surface was regenerated with 0.1% SDS prior a new cycle. After blank subtraction, the resulting set of curves was analyzed by curve fitting with a simple 1:1 Langmuir interaction model. The fitting to this model was judged by the quality of the residual plots. The sensorgrams and residuals plots of typical experiments are shown in , and the kinetic parameters of 4 independent experiments are summarized in . In complete agreement with the kinetic parameters obtained with PABP immobilized on a CM5 sensor chip, the fitted results indicated that eRF3a interacted with poly(A)-bound PABP with a fast association rate (ka = 1.64 × 105 M−1 s−1) and a slow dissociation rate (kd = 2.37 × 10−4 s−1), yielding an equilibrium dissociation constant of 1.3 nM (). Moreover, we also observed a ∼20-fold loss in the association rate of eRFa 12-GGC when compared to that of eRF3a, resulting in a KD value of 56 nM for eRFa 12-GGC binding to poly(A)-bound PABP (). These results confirmed that the binding affinity of eRF3a 12-GGC allele for PABP was lower than that of eRF3a (10-GGC allele) by one order of magnitude. For both, eRF3a and 12-GGC allele, the dissociation rate constant kd was in same order (2–3 × 10−4 s−1). It is interesting to note that this dissociation is ∼10-fold faster than that of PABP binding to poly(A) (). This suggests that dissociation of PABP from the immobilized OligoA120 surface only marginally affects the dissociation rate of eRF3a-PABP interaction.
Figure 4. Kinetic analyses of eRF3a 10-GGC allele (A) and eRF3a 12-GGC allele (B) interacting with PABP (300 RU) bound to OligoA120 RNA (30 RU) immobilized onto a SA sensor chip surface. Typical sensorgrams and fitting of the binding profiles to a Langmuir 1:1 binding model (top panel) and residuals plots (bottom panel) are shown. Concentrations of injected analytes are indicated on the right of the sensorgrams. For each analyte concentration the fitted curve is shown in black.
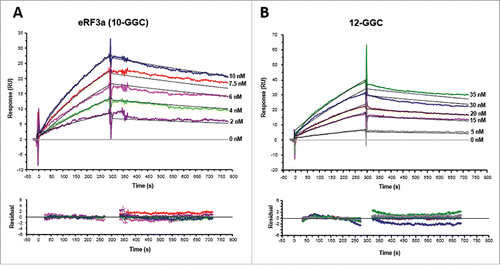
Table 2. Kinetic parameters for eRF3a and eRF3a 12-GGC allele interaction with PABP bound to OligoA120 immobilized on a SA sensor chip surface.aFootnotea
Table 3. eRF3a allele frequencies in human population.
Discussion
Surface plasmon resonance (SPR) can visualize the formation of a macromolecular complex in real time providing unique insights into the dynamics of its association and dissociation. We took advantage of this method to reevaluate the interaction between PABP and the different forms of human translation termination factor eRF3 including eRF3a alleles and eRF3b. What is new is that we show that SPR allows to determine the kinetic parameters of eRF3a-PABP interaction when PABP is multimerized onto a poly(A) tail. Our synthetic model of poly(A) tail mimics the natural situation of cytoplasmic PABP multimerized on the mRNA poly(A) tail.
Our measurements of the kinetic parameters of eRF3 revealed that eRF3a binds to PABP with a nanomolar affinity (KD = 1.5 nM). The binding affinity for PABP was also measured by SPR for two other proteins, Paip1 (KD = 5.7 nM) and Paip2 (KD = 85 nM), which carry PAM2 sites and interact with PABP MLLE domain.Citation25,26 Differences between these three PABP-interacting proteins are mainly due to the mode of binding to the C-terminus of PABP. SPR data show that there is a rearrangement step after the initial binding for both Paip1 and Paip2. This rearrangement step was attributed to a change in conformation, which occurs when Paip1 or Paip2 bind to PABP C-terminus.Citation26 Such a rearrangement step was not observed during eRF3-PABP interaction. This suggests that the different partners of PABP C-terminal region are recruted in different ways depending on their relative affinities reflecting their mechanism of action: Paip1 and Paip2 in translation regulation, and eRF3 in mRNA stability.
The ∼120 A-long RNA (OligoA120) molecule we used to study eRF3a-PABP interaction is a polyvalent ligand for PABP. Whereas the preferred binding polarity of multiple PABP molecules along the poly(A) tail remains unknown, in a recent report, Lin et al.Citation32 have shown a ∼50% cooperative binding conformation of two PABPs using a C50A25 RNA oligomer. In the cooperative binding conformation, the C-termini of adjacent PABPs interact directly with each other thereby stabilizing the overall complex. This cooperative binding conformation resulted in a longer dissociation time of poly(A)-PABP complex when compared with a noncooperatively bound complex. In our experiments, PABPs binding onto the poly(A) of the OligoA120 involved multiple molecular interactions with probably cooperative and noncooperative PABP conformations. This heterogeneous binding mode, and particularly its cooperative component, is likely responsible for the very slow dissociation kinetics observed. Thus, the single-step binding model that we used to measure the kinetic parameters of PABP-OligoA120 interaction, did not take into consideration the heterogeneous PABP binding mode and provided only an effective method for measuring PABP-Poly(A) binding constants. Recently, ITC experiments with peptides containing eRF3a PAM2 motif (residues 64 to 94) showed that the presence of a polyA1000 polymer during the calorimetric titration resulted in 100-fold increase in eRF3 affinity for PABP.Citation11 However, we did not observe such a considerable variation in eRF3a binding affinity when PABP was bound to poly(A). Conversely, in our experiments, PABP binding to poly(A) did not seem to affect eRF3 binding to PABP. In addition, the lower dissociation rate of poly(A)-PABP complex (kd = 1.72 × 10−5 s−1) when compared to that of eRF3-PABP (kd = 2.37 × 10–4 s−1) suggests that, in cells, eRF1-eRF3 termination complex dissociates from PABP which remains bound on the poly(A) tail. These results tend to confirm the model proposed recently by Osawa et al.Citation11: upon translational termination, recruitment of PABP-dissociated translation termination complex to the ribosomes permits the binding of the deadenylase complexes to the PABP molecule bound at the 3′ end of the poly(A) tail, thus coupling mRNA deadenylation with translation termination. However, it is not clear whether eRF3a dissociates from the PABP molecule bound to the 3′ extremity of poly(A) tail, or conversely, from the PABP molecule at the 5′ end of the poly(A), in closest proximity to the stop codon. Thus, it is not yet clearly understood whether deadenylation is coupled to translation and requires translation termination to proceed.
eRF3 is the only known example of protein with two overlapping PAM2 sites (PAM2-N and PAM2-C). However, the eRF3-PABP complex exists exclusively in one-to-one stoichiometry and it has been shown that the presence of overlapping PAM2 sites increases the affinity of eRF3-PABP interaction.Citation11,33 Structural studies also showed that the overlapping PAM2 sites bind slightly different regions of PAPB MLLE domain: PAM2-N occupies the hydrophobic pocket between MLLE helices α2 and α3 whereas PAM2-C interacts between helices α3 and α5.Citation11,12 The presence of the overlapping PAM2 sites may explain the small heterogeneity of experimental dissociation curves relative to the fitting curves and the asymetrical deviations in the dissociation phase of residual plots observed for all eRF3a forms in our SPR experiments ().
Our kinetic studies reveal that the different forms of human eRF3, i. e., eRF3 7-, 10-, 11-, 12-GGC alleles and eRF3b, which differ by the length of the glycine extension at their N-terminal extremity, have different affinities for the C-terminal region of PABP. These differences in affinity are mainly due to variations in the association rates suggesting that the glycine extension is able to change the kinetics of complex formation. This effect is particularly clear (i) for the 12-GGC allele of eRF3a which has a lower (∼5 fold with PABP alone and ∼20 fold with poly(A)-bound PABP) association rate than that of the most common eRF3a 10-GGC allele and (ii) for eRF3b which has a 6-fold higher association rate than that of eRF3a 10-GGC allele (). What could be the structural basis for these differences? Mutations in eRF3a PAM2 sites compromise binding to PABP Citation14,34 showing that these PAM2 motifs are the only interaction sites with PABP. We can then speculate that the presence of the glycine repeat at the end of eRF3a N-terminal domain leads to a significant change in the adjustment of PAM2 motif upon PABP binding. This is supported by the facts that (i) only the association rate is changed by the presence of the glycine repeat and (ii) the absence of glycine repeat in eRF3b results in an enhanced affinity for PABP (). It has been reported previously that deletions of Paip1 outside of its PAM2 motif decrease its affinity for PABP Citation26 suggesting that structural elements outside of the PAM2 motif are critical for the interaction. The crystal structure of the MLLE domain from PABP in complex with the two PAM2 regions of eRF3 reveals that the MLLE-PAM2 interactions are mostly hydrophobic. The eRF3a PAM2 peptide in an extended conformation is wrapped around the highly conserved MLLE domain and interacts with its hydrophobic pockets.Citation12 The glycine repeat at eRF3a N-terminal extremity is separated from the PAM2 sites by a 48 amino acids sequence in which 4 prolines with propensity to form β-turns are interspersed. The folding of this region could bring the glycine extension in the vicinity of the PAM2 motifs lowering its affinity for the MLLE domain. In this conformation, the length of the glycine repeat could be of great significance, the addition of a single glycine, as in eRF3a 12-GGC allele relative to the 11-GGC allele, inducing even more pronounced changes in the adjustment of PAM2 motifs upon PABP binding.
Interestingly, the eRF3a 12-GGC allele was involved in cancer susceptibility in several studies on portugese and iranian populations with gastric and breast cancer.Citation22-24 Strikingly, the comparison of allele frequencies in different population samples including the portugese and iranian populations mentioned above and the population that participates to the NHLBI Exome Sequencing Project (Exome Variant Server, NHLBI GO Exome Sequencing Project, Seattle, WA), reveals substantial variations (). The 9-GGC allele was not found in the iranian population studied by Miri et al. (2012), whereas the 12-GGC allele was completely absent in the population samples of NHLBI Exome Sequencing Project. Further studies with a worldwide distribution of human population samples is needed to accurately ascertain eRF3a allele frequencies, in particular for the 12-GGC allele. Using complementation experiments in eRF3a-depleted cells, we have previously shown that there were no obvious differences in the ability of the alternate eRF3a alleles to complement for the lack of endogenous eRF3a.Citation23 However, what could be the biological significance of the lower affinity of eRF3a 12-GGC allele for PABP? Lowering the affinity of eRF3a for PABP could be the basis of a defect in translation termination by modifying the recruitment of termination complexes to the ribosome. Alternatively, by affecting the coupling of mRNA deadenylation with translation termination, it could favor the recruitment of deadenylation complexes, and thus, enhance the degradation of the subset of mRNAs that are sensitive to PAN2-PAN3 degradation pathway. Among these mRNAs, those involved in the control of cellular proliferation would be of paramount importance. The status of mRNA degradation in cells carrying the 12-GGC allele needs to be further examined.
Materials and methods
Plasmid constructions
Plasmids pCMV-eRF3a,Citation35 pCMV-3a-7GGC, 3a-11GGC and 3a-12GGC Citation23 contain the entire coding sequence of the human GSPT1 gene encoding eRF3a (10-GGC allele) and the other allelic forms of eRF3a containing 7 GGC, 11 GGC and 12 GGC, respectively. These plasmids were BamHI-SalI doubly digested and cloned into the similarly digested pET28b plasmid (Invitrogen). In the resulting plasmids, a 6His tag is preceding the eRF3a coding sequence and was used for affinity purification. Plasmid pET28-His-eRF3b expressing the entire human isoform eRF3b was constructed using the pBK-heRFb plasmid Citation20 as a template for PCR amplification with the oligonucleotides (eRF3b Fwd: GGACGCTAGCATGGATTCGGGCAGCAGCAGCA, and eRF3b Rev: TAATACGACTCACTATAGGGCGAATTGG) containing NheI and NotI restriction sites for cloning within the NheI and NotI sites of pET28b. The plasmid pET-His-eRF3bΔN in wich the first 129 codon of eRF3b were deleted was obtained by ligating NcoI-klenow-filled-NotI fragment of pBK-heRF3b in NheI-klenow-filled-NotI fragment of pET28b-His-eRF3a vector. The plasmid pTrcHisBPABP Citation36 encoding for the human full-length His-tagged PABP protein, was generously donated by Bertrand Cosson.
Poly (A) tail synthesis
A synthetic Poly(A) tail was synthesized by selectively adding adenosine residues to the 3′ end of a chemically synthesized 18-mer oligoribonucleotide 5′-CUCAUGUUACCUACCAUA-3′ carrying a 5′-biotin tag (Cogenics). The 5′ biotin allows immobilization of the RNA oligonucleotide onto streptavidin-coated SA sensor chips in SPR experiments (see below). In a typical 20 µl reaction, 5 units of Escherichia coli poly(A) polymerase (Biolabs) were incubated with 200 pmoles of the 18-mer oligoribonucleotide and 1 mM ATP in the reaction buffer supplied by the manufacturer at 37°C for 30 minutes. The reaction was stopped by ethanol precipitation in the presence of 0.3 M sodium acetate. The RNA precipitate was recovered by centrifugation and resuspended in 20 µl of water. The resulting poly(A)-elongated oligoribonucleotide (hereafter named OligoA120) was analyzed by electrophoresis onto a 10% polyacrylamide gel containing 7 M urea followed by Northern blotting onto a Hybond-N membrane (GE Healthcare). The membrane was then U.V. cross-linked and blocked for 1 h in Tris-buffered saline (TBS)–Tween solution (20 mM Tris-HCl, pH 7.6, 140 mM NaCl, 0.2% Tween 20) containing 3% bovine serum albumine. The membrane was then incubated for 1 h with HRP-conjugated streptavidin reagent (GE Healthcare) in TBS-Tween, washed five times for 10 min in TBS-Tween, and the biotinylated oligoribonucleotides were visualized by chemiluminescence and exposure to X-ray film (Fig. S2). The size of the elongated oligoribonucleotide was estimated by ethidium bromide staining and U. V. exposure of oligonucleotides of known size loaded onto the same gel.
Protein expression and purification
For expression and purification of PABP and of the different forms of mammalian eRF3, Escherichia coli BL21(λDE3) (eRF3) and DH5α (PABP) cells were transformed with the bacterial expression vectors described above. After induction with 1 mM IPTG (isopropyl-β-D-thiogalactopyranoside) and incubation either at 37°C for 4 h (PABP) or 22 h at 25°C (eRF3), bacteria were harvested by centrifugation. The bacterial pellets were resuspended in lysis buffer (20 mM Tris-HCl, pH 7.8, 10% glycerol, 500 mM NaCl, 1 mg/ml lysozyme and 10 mM imidazole) and cells were lysed by sonication. The lysate was clarified by centrifugation at 13,000 × g for 30 min and incubated with Ni2+ beads (Qiagen) for 20 min at room temperature. After binding, lysate and beads were loaded on a column and washed with 10 volumes of washing buffer (20 mM Tris-HCl, pH 7.4, 10% glycerol, 500 mM NaCl) containing 20 mM imidazole. The proteins were eluted with the washing buffer containing increasing concentrations of imidazole (50 mM to 250 mM). The fractions (250 µl) were collected and analyzed by electrophoresis on sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE) and Coomassie blue staining. The purest fractions were dialyzed separately against HSBN buffer (10 mM Hepes, pH7.4, 150 mM NaCl). Protein concentration of the dialysed fractions was carefully determined using a Micro BCA protein assay reagent kit (Pierce), bovine serum albumin being used as standard. The level of purity was assessed by analysis on Experion Pro260 (Bio-Rad) and SDS-PAGE (Fig. S6).
SPR method and analysis
To obtain quantitative kinetic measurements of the eRF3-PABP interactions, experiments were conducted on an SPR-based Biacore 3000 biosensor (GE Healthcare). Two different sensor chips were used: a CM5 sensor chip for the experiments with immobilized PABP and a SA sensor chip for the experiments with immobilized RNA oligonucleotides.
SPR experiments on CM5 sensor chip: purified His-tagged full-length PABP was immobilized through primary amino groups to the carboxy-methylated dextran matrix of a CM5 sensor chip. A solution of 1 µg/ml PABP diluted in immobilization buffer (10 mM sodium acetate, pH 4.5) was manually injected in order to obtain 1,000–2,500 RU of coupled PABP. The immobilization was followed by injection of 20 μl 1 M ethanolamine hydrochloride, pH 8.5, to saturate the free activated sites of the matrix. To minimize artifacts such as mass transport and rebinding effects, we coupled the minimum amount of PABP required to obtain an optimal signal-to-noise ratio when injecting eRF3a alleles or eRF3b. The absence of a mass transport step was verified by injecting the same eRF3a solution at flow rates ranging from 5 to 50 µl/min over the PABP surface. After data treatment, the interaction curves (sensorgrams) at the different flow rates were superimposable, indicating the absence of mass transport (data not shown). To subtract any background noise from each dataset, the same coupling procedure, but in the absence of PABP, was used to prepare a mock surface. Kinetic experiments were carried out at 25°C at a flow rate of 5 µl/min. Buffer HBS-P (20 mM HEPES, pH 7.4, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P20) was used as the running buffer and for diluting all the injected proteins. Different concentrations of analyte (eRF3a alleles or eRF3b) were injected over the PABP surface and mock surface for 300 s, followed by a 500 s-long buffer injection unless otherwise indicated. The surface was regenerated by 5 µl injection of a 1 M Nacl, 50 mM NaOH solution for 30 sec.
SPR experiments on SA sensor chip: 5′ biotinylated RNA oligonucleotide with Poly(A) tail (hereafter named OligoA120) diluted to a final concentration of 2 nM in SA running buffer (10 mM Tris-HCl, pH 7.8, 150 mM NaCl, 1 mM DTT, 5% glycerol and 0.005% Surfactant P20) was allowed to interact with streptavidin until a response of 20 to 220 RU was obtained. We prepared either low density (20–40 RU) or intermediate density (200–250 RU) OligoA120 surfaces as indicated in the figure legends. These small amounts of immobilized OligoA120 minimized mass transport effects and yielded sufficient signal. No target RNA was captured on a parallel flow cell, which was used as a reference surface. To measure the kinetic parameters of eRF3-PABP interaction, His-tagged full-length PABP (80 nM in SA running buffer) was manually injected at a flow rate of 5 µl/min for 5 min and allowed to dissociate for 2 min at 5 µl/min flow rate in SA running buffer. In these conditions, a stable signal was obtained due to the very low dissociation rate (kd = 1.72 × 10−5 s−1) of PABP from OligoA120 surface (see Results). For kinetic parameter measurements on PABP-bound OligoA120 surface, RNasin (1.5 units/µl) was added to the proteins which were serially diluted in SA running buffer and injected over the sensor chip surface at 5 µl/min for 5 min at 25°C. The surface was regenerated with a 0.1% SDS solution. In order to subtract any background noise from each data set, all samples were run over an unmodified sensor chip surface, used as a reference.
Biacore data analysis
Sensorgrams obtained with the Biacore 3000 system were prepared and each set was subjected to curve fitting with numerical integration methods using the BIAevaluation software, Version 4.1 (GE Healthcare). The data were processed by fitting the binding profiles to a 1:1 Langmuir interaction model. The quality of the fit was assessed by the statistical chi2 value provided by the software (chi2 values < 10 were considered as acceptable) and by visual inspection of the shape of residual plots (deviation between experimental and fitted data) that is a good indication of the goodness of fit (BIAevaluation Software Handbook, GE Healthcare). Residual should be minimal (residuals of +/− 2 RU are considered as good) and randomly distributed around a zero value.Citation37 The fitting of each dataset yielded rates for association (ka) and dissociation (kd), from which the equilibrium dissociation constant KD was calculated (KD = kd / ka). The ka , kd and KD from 2 to 5 experiments (number of experiments is indicated in ) were used to calculate the mean values and SEM of these variables.
The valency of the OligoA120 for PABP was calculated with the formula Valency = Mr ligand × R max / Mr analyte × R ligand.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Figure1-6.doc
Download MS Word (842.5 KB)Acknowledgments
S. J. held fellowships from the French Ministère de la Recherche et de l’Enseignement Supérieur (MENESR). We thank Bertrand Cosson for helpful advices in the purification of PABP.
Funding
The Université Pierre et Marie Curie (UPMC) and the Center National de la Recherche Scientifique (CNRS).
References
- Cosson B, Berkova N, Couturier A, Chabelskaya S, Philippe M, Zhouravleva G. Poly(A)-binding protein and eRF3 are associated in vivo in human and Xenopus cells. Biol Cell 2002; 94:205-16; PMID:12489690; http://dx.doi.org/10.1016/S0248-4900(02)01194-2
- Hoshino S, Imai M, Kobayashi T, Uchida N, Katada T. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-Poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J Biol Chem 1999; 274:16677-80; PMID:10358005; http://dx,doi.org/10.1074/jbc.274.24.16677
- Hoshino S. Mechanism of the initiation of mRNA decay: role of eRF3 family G proteins. Wiley Interdiscip Rev RNA 2012; 3:743-57; PMID:22965901; http://dx.doi.org/10.1002/wrna.1133
- Burgess HM, Gray NK. mRNA-specific regulation of translation by poly(A)-binding proteins. Biochem Soc Trans 2010; 38:1517-22; PMID:21118118; http://dx.doi.org/10.1042/BST0381517
- Bernstein P, Peltz SW, Ross J. The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol Cell Biol 1989; 9:659-70; PMID:2565532; http://dx.doi.org/10.1128/MCB.9.2.659
- Kahvejian A, Roy G, Sonenberg N. The mRNA closed-loop model: the function of PABP and PABP-interacting proteins in mRNA translation. Cold Spring Harb Symp Quant Biol 2001; 66:293-300; PMID:12762031
- Ezzeddine N, Chang TC, Zhu W, Yamashita A, Chen C-YA, Zhong Z, Yamashita Y, Zheng D, Shyu AB. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol Cell Biol 2007; 27:7791-801; PMID:17785442; http://dx.doi.org/10.1128/MCB.01254-07
- Siddiqui N, Mangus DA, Chang TC, Palermino JM, Shyu AB, Gehring K. Poly(A) nuclease interacts with the C-terminal domain of polyadenylate-binding protein domain from poly(A)-binding protein. J Biol Chem 2007; 282:25067-75; PMID:17595167; http//:dx.doi.org/10.1074/jbc.M701256200
- Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen C-YA, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat Struct Mol Biol 2005; 12:1054-63; PMID:16284618; http://dx.doi.org/10.1038/nsmb1016
- Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev 2007; 21:3135-48; PMID:18056425; http://dx.doi.org/10.1101/gad.1597707
- Osawa M, Hosoda N, Nakanishi T, Uchida N, Kimura T, Imai S, Machiyama A, Katada T, Hoshino S, Shimada I. Biological role of the two overlapping poly(A)-binding protein interacting motifs 2 (PAM2) of eukaryotic releasing factor eRF3 in mRNA decay. RNA 2012; 18:1957-67; PMID:23019593; http://dx.doi.org/10.1261/rna.035311.112
- Kozlov G, Gehring K. Molecular basis of eRF3 recognition by the MLLE domain of poly(A)-binding protein. PloS One 2010; 5:e10169; PMID:20418951; http://dx.doi.org/10.1371/journal.pone.0010169
- Brook M, Gray NK. The role of mammalian poly(A)-binding proteins in co-ordinating mRNA turnover. Biochem Soc Trans 2012; 40:856-64; PMID:22817748; http://dx.doi.org/10.1371/journal.pone.0010169
- Kononenko AV, Mitkevich VA, Atkinson GC, Tenson T, Dubovaya VI, Frolova LY, Makarov AA, Hauryliuk V. GTP-dependent structural rearrangement of the eRF1:eRF3 complex and eRF3 sequence motifs essential for PABP binding. Nucleic Acids Res 2010; 38:548-58; PMID:19906736; http://dx.doi.org/10.1093/nar/gkp908
- Cosson B, Couturier A, Chabelskaya S, Kiktev D, Inge-Vechtomov S, Philippe M, Zhouravleva G. Poly(A)-binding protein acts in translation termination via eukaryotic release factor 3 interaction and does not influence [PSI(+)] propagation. Mol Cell Biol 2002; 22:3301-15; http;//dx.doi.org/10.1128/MCB.22.10.3301-3315.2002
- Roque S, Cerciat M, Gaugué I, Mora L, Floch AG, de Zamaroczy M, Heurgué-Hamard V, Kervestin S. Interaction between the poly(A)-binding protein Pab1 and the eukaryotic release factor eRF3 regulates translation termination but not mRNA decay in Saccharomyces cerevisiae. RNA 2015; 21:124-34; PMID:25411355; http://dx.doi.org/10.1261/rna.047282.114
- Hoshino S, Imai M, Mizutani M, Kikuchi Y, Hanaoka F, Ui M, Katada T. Molecular cloning of a novel member of the eukaryotic polypeptide chain-releasing factors (eRF). Its identification as eRF3 interacting with eRF1. J Biol Chem 1998; 273:22254-9; PMID:9712840; http://dx.doi.org/10.1074/jbc.273.35.22254
- Jakobsen CG, Segaard TM, Jean-Jean O, Frolova L, Justesen J. Identification of a novel termination release factor eRF3b expressing the eRF3 activity in vitro and in vivo. Mol Biol (Mosk) 2001; 35:672-81; PMID:11524954
- Chauvin C, Jean-Jean O. Proteasomal degradation of human release factor eRF3a regulates translation termination complex formation. RNA 2008; 14:240-5; PMID:18083835; http://dx.doi.org/10.1261/rna.728608
- Chauvin C, Salhi S, Le Goff C, Viranaicken W, Diop D, Jean-Jean O. Involvement of human release factors eRF3a and eRF3b in translation termination and regulation of the termination complex formation. Mol Cell Biol 2005; 25:5801-11; PMID:15987998; http://dx.doi.org/10.1128/MCB.25.14.5801-5811.2005
- Jean-Jean O, Le Goff X, Philippe M. Is there a human [psi]? Comptes Rendus Académie Sci Sér III 1996; 319:487-92; PMID:8881282
- Brito M, Malta-Vacas J, Carmona B, Aires C, Costa P, Martins AP, Ramos S, Conde AR, Monteiro C. Polyglycine expansions in eRF3/GSPT1 are associated with gastric cancer susceptibility. Carcinogenesis 2005; 26:2046-9; PMID:15987717; http://dx.doi/10.1093/carcin/bgi168
- Malta-Vacas J, Chauvin C, Gonçalves L, Nazaré A, Carvalho C, Monteiro C, Bagrel D, Jean-Jean O, Brito M. eRF3a/GSPT1 12-GGC allele increases the susceptibility for breast cancer development. Oncol Rep 2009; 21:1551-8; PMID:19424636; http://dx.doi.org/10.3892/or_00000387
- Miri M, Hemati S, Safari F, Tavassoli M. GGCn polymorphism of eRF3a/GSPT1 gene and breast cancer susceptibility. Med Oncol 2012; 29:1581-5; PMID:22101789; http://dx.doi.org/10.1007/s12032-011-0111-x
- Khaleghpour K, Kahvejian A, De Crescenzo G, Roy G, Svitkin YV, Imataka H, O’Connor-McCourt M, Sonenberg N. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol 2001; 21:5200-13; PMID:11438674; http://dx.doi.org/10.1128/MCB.21.15.5200-5213.2001
- Roy G, De Crescenzo G, Khaleghpour K, Kahvejian A, O’Connor-McCourt M, Sonenberg N. Paip1 interacts with poly(A) binding protein through two independent binding motifs. Mol Cell Biol 2002; 22:3769-82; PMID:11997512; http://dx.doi.org/10.1128/MCB.22.11.3769-3782.2002
- Chaloin O, Peter JC, Briand JP, Masquida B, Desgranges C, Muller S, Hoebeke J. The N-terminus of HIV-1 Tat protein is essential for Tat-TAR RNA interaction. Cell Mol Life Sci 2005; 62:355-61; PMID:15723170; http://dx.doi.org/10.1007/s00018-004-4477-1
- Law MJ, Chambers EJ, Katsamba PS, Haworth IS, Laird-Offringa IA. Kinetic analysis of the role of the tyrosine 13, phenylalanine 56 and glutamine 54 network in the U1A/U1 hairpin II interaction. Nucleic Acids Res 2005; 33:2917-28; PMID:15914668; http://dx.doi.org/10.1093/nar/gki602
- Park S, Myszka DG, Yu M, Littler SJ, Laird-Offringa IA. HuD RNA recognition motifs play distinct roles in the formation of a stable complex with AU-rich RNA. Mol Cell Biol 2000; 20:4765-72; PMID:10848602; http://dx.doi.org/10.1128/MCB.20.13.4765-4772.2000
- Kühn U, Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J Mol Biol 1996; 256:20-30; PMID:8609610; http://dx.doi.org/10.1006/jmbi.1996.0065
- Sachs AB, Davis RW, Kornberg RD. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol Cell Biol 1987; 7:3268-76; PMID:3313012; http://dx.doi.org/10.1128/MCB.7.9.3268
- Lin J, Fabian M, Sonenberg N, Meller A. Nanopore detachment kinetics of poly(A) binding proteins from RNA molecules reveals the critical role of C-terminus interactions. Biophys J 2012; 102:1427-34; PMID:22455926; http://dx.doi.org/10.1016/j.bpj.2012.02.025
- Kozlov G, De Crescenzo G, Lim NS, Siddiqui N, Fantus D, Kahvejian A, Trempe JF, Elias D, Ekiel I, Sonenberg N, et al. Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase. EMBO J 2004; 23:272-81; PMID:14685257; http://dx.doi.org/10.1038/sj.emboj.7600048
- Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol 2008; 6:e111; PMID:18447585; http://dx.doi.org/10.1371/journal.pbio.0060111
- Le Goff X, Philippe M, Jean-Jean O. Overexpression of human release factor 1 alone has an antisuppressor effect in human cells. Mol Cell Biol 1997; 17:3164-72; PMID:9154815; http://dx.doi.org/10.1128/MCB.17.6.3164
- Sagliocco F, Laloo B, Cosson B, Laborde L, Castroviejo M, Rosenbaum J, Ripoche J, Grosset C. The ARE-associated factor AUF1 binds poly(A) in vitro in competition with PABP. Biochem J 2006; 400:337-47; PMID:16834569; http://dx.doi.org/10.1042/BJ20060328
- Drescher DG, Ramakrishnan NA, Drescher MJ. Surface plasmon resonance (SPR) analysis of binding interactions of proteins in inner-ear sensory epithelia. Methods Mol Biol 2009; 493:323-43; PMID:18839357; http://dx.doi.org/10.1007/978-1-59745-523-7_20
