 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
A rapidly increasing number of RNA helicases are implicated in several distinct cellular processes, however, the modes of regulation of multifunctional RNA helicases and their recruitment to different target complexes have remained unknown. Here, we show that the distribution of the multifunctional DEAH-box RNA helicase Prp43 between its diverse cellular functions can be regulated by the interplay of its G-patch protein cofactors. We identify the orphan G-patch protein Cmg1 (YLR271W) as a novel cofactor of Prp43 and show that it stimulates the RNA binding and ATPase activity of the helicase. Interestingly, Cmg1 localizes to the cytoplasm and to the intermembrane space of mitochondria and its overexpression promotes apoptosis. Furthermore, our data reveal that different G-patch protein cofactors compete for interaction with Prp43. Changes in the expression levels of Prp43-interacting G-patch proteins modulate the cellular localization of Prp43 and G-patch protein overexpression causes accumulation of the helicase in the cytoplasm or nucleoplasm. Overexpression of several G-patch proteins also leads to defects in ribosome biogenesis that are consistent with withdrawal of the helicase from this pathway. Together, these findings suggest that the availability of cofactors and the sequestering of the helicase are means to regulate the activity of multifunctional RNA helicases and their distribution between different cellular processes.
Abbreviations
| ATP | = | Adenosine triphosphate |
| Cmg1 | = | Cytoplasmic and mitochondrial G-patch protein 1 |
| CRAC | = | UV crosslinking and analysis of cDNA |
| GFP | = | Green fluorescent protein |
| G-patch | = | Glycine-rich domain |
| His | = | Histidine |
| IMS | = | Intermembrane space |
| LDH | = | Lactate dehydrogenase |
| Leu | = | Leucine |
| MIA | = | Mitochondrial intermembrane space import and assembly |
| mRNA | = | mRNA |
| NADH | = | Reduced nicotinamide adenine dinucleotide |
| OB-fold | = | Oligonucleotide-binding fold |
| PEP | = | Phosphoenolpyruvate |
| PK | = | Proteinase K |
| Pre-rRNA | = | Precursor rRNA |
| RFP | = | Red fluorescent protein |
| RNA | = | Ribonucleic acid |
| RNP | = | RNA-protein complex |
| rRNA | = | rRNA |
| SDS-PAGE | = | Sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| snoRNA | = | Small nucleolar RNA |
| snRNP | = | Small nuclear RNA-protein complex |
| TAP | = | Tandem affinity purification |
| TCA | = | Trichloroacetic acid |
| Trp | = | Tryptophan |
Introduction
RNA helicases are central players in all major pathways of RNA metabolism, including pre-mRNA splicing, translation and ribosome biogenesis.Citation1-5 The ATP-dependent DExD/H-box RNA helicases are characterized by specific structural motifs for nucleotide binding and hydrolysis as well as helicase activity, which are required for their functions in the remodelling of RNAs and RNA-protein complexes (RNPs).Citation1-3 Besides the conserved helicase core domain, most RNA helicases possess additional N- or C-terminal extensions, which are thought to confer substrate specificity or mediate interactions with cofactors.Citation3 However, interacting proteins have only been identified for a small fraction of RNA helicases and few cofactors have been shown to be required for helicase recruitment or activation. These include the eukaryotic translation initiation factors eIF4G and eIF4B that synergistically stimulate the activity of the DEAD-box RNA helicase eIF4A and several proteins that interact with RNA helicases involved in splicing.Citation6-8 In ribosome biogenesis, the activity of the DEAD-box RNA helicase Dbp8 is stimulated by its cofactor Esf2, which has also been suggested to recruit Dbp8 to its target RNA in pre-ribosomal complexes.Citation9 In addition, Rrp5 has been suggested to act as a cofactor for the RNA helicase Rok1, which is required for the release of the snoRNA snR30 from pre-ribosomal complexes.Citation10-13
A family of cofactors that interact with and modulate the activity of RNA helicases are the G-patch proteins, which have been identified in multiple eukaryotes and some retroviruses and are named after their glycine-rich region that mediates their interaction with the corresponding helicase.Citation14,15 Little is known about the roles of the many G-patch proteins identified in higher eukaryotes and retroviruses. Only five G-patch proteins have been identified in the yeast Saccharomyces cerevisiae, the two pre-mRNA splicing factors Spp2 and Spp382/Ntr1, the proteins Pfa1/Sqs1 and Pxr1/Gno1 that are implicated in ribosome biogenesis, and the uncharacterised open reading frame YLR271W. The splicing factors Spp2 and Spp382 are best characterized so far. Spp2 contains a C-terminal G-patch domain with which it interacts with the DEAH-box RNA helicase Prp2.Citation16-18 The G-patch protein Spp382/Ntr1 forms a complex with Ntr2, which recruits the DEAH-box RNA helicase Prp43 to postspliceosomes containing the excised intron lariat and the U2, U5, and U6 snRNPs.Citation19,20 Here, Prp43 is required for spliceosome disassembly and release of the intron lariat.Citation21,22 The N-terminal G-patch domain of Spp382 mediates the interaction with Prp43 and is required for stimulation of the ATPase activity of the helicase.Citation18 This mode of interaction and the splicing function of the mouse orthologues of Prp43 and Spp382, mDEAH9/mDHX15 and tuftelin interacting protein 11 (mTFIP11), respectively, seem to be conserved.Citation23-25
In yeast, Pxr1 co-migrates with pre-ribosomal particles in sucrose density gradients, and deletion of Pxr1 leads to early pre-rRNA processing defects.Citation26,27 Interestingly, Pxr1 was also shown to interact with and activate the RNA helicase Prp43.Citation28,29 Indeed, as well as its role in pre-mRNA splicing, Prp43 is a key factor required for ribosome biogenesis and co-purifies with various pre-ribosomal complexes.Citation4,28,30,31 Prp43 depletion affects pre-rRNA processing in both ribosomal subunits. Using UV Crosslinking and Analysis of cDNA (CRAC) several binding sites of Prp43 have been identified on pre-rRNAs.Citation32-34 One such site mapped close to the 3′-end of the 18S rRNA in the small ribosomal subunit, where Prp43 has been linked to a late processing step mediated by the endonuclease Nob1.Citation35 Here, Prp43 is thought to bind the G-patch protein Sqs1/Pfa1, which is a component of pre-40S complexes.Citation27,28 Sqs1 has also been implicated in the Nob1-mediated 18S rRNA processing, and it can stimulate the ATPase activity of Prp43 in vitro.Citation27,35 For the large ribosomal subunit, Prp43 was shown to be required for the pre-ribosomal release of several small nucleolar RNAs (snoRNAs),Citation36 which basepair at a cluster of snoRNP-guided methylation sites.Citation34 Taken together, Prp43 is a multifunctional RNA helicase that interacts with several cofactors. It has so far remained unclear, however, whether the interaction of cofactors with Prp43 regulates the distribution of the RNA helicase between the different target complexes.
Understanding how the interplay of the cofactors of the DEAH-box protein Prp43 regulates the action of the helicase in different cellular pathways requires a complete inventory of its interacting G-patch proteins. Helicase interaction partners and possible functions have been proposed for Spp2, Spp382, Sqs1 and Pxr1, while the remaining G-patch protein in yeast, encoded by ylr271w, has remained uncharacterised. Here, we identify YLR271W as a direct interaction partner of Prp43 and show that the G-patch protein stimulates RNA binding and ATPase activity of the RNA helicase. YLR271W is localized in the cytoplasm and in the intermembrane space of mitochondria, and we therefore named the protein cytoplasmic and mitochondrial G-patch protein 1 (Cmg1). Interestingly, we found that the Prp43-interacting G-patch proteins compete for binding to the helicase. Modulation of their expression levels results in changes in the cellular localization of Prp43, affecting Prp43 function in the pathway from which the helicase has been withdrawn. Our data suggest that the interplay of cofactors and the sequestering of the helicase are novel means to regulate the activity of multifunctional RNA helicases and their distribution between different cellular functions.
Results
YLR271W/Cmg1 specifically interacts with and stimulates the activity of the RNA helicase Prp43
The G-patch proteins Spp382/Ntr1, Sqs1/Pfa1 and Pxr1/Gno1 have been shown to modulate the functions of the RNA helicase Prp43 in splicing and ribosome biogenesis, and Spp2 regulates the activity of Prp2 in splicing. However, the helicase interaction partner of the remaining G-patch protein YLR271W (here named Cmg1) has remained unknown. We therefore performed pulldown experiments using a yeast strain expressing TAP-tagged Cmg1 and with a wildtype control strain. Interestingly, Cmg1 significantly co-precipitated a protein of about 90 kDa, which was identified by mass spectrometry as the RNA helicase Prp43 (). The enrichment of Prp43 with TAP-tagged Cmg1 was also confirmed by Western blot analysis using antibodies recognizing Prp43 (, lower panel). To further analyze the interaction, we purified recombinant Cmg1, Spp2 and Prp43 and performed binding experiments in vitro. Prp43 was co-purified with immobilised Cmg1, but not Spp2, demonstrating a direct and specific interaction of the two proteins (). Using the yeast two-hybrid approach, we observed an in vivo interaction between Prp43 and both full length Cmg1 and the Cmg1 G-patch domain alone (amino acids 1–85; ). In contrast, neither the C-terminal part of Cmg1 (amino acids 86–274) nor Swm2 showed any interaction with Prp43, indicating that the G-patch domain of Cmg1 is required and sufficient for Prp43 interaction. The C-terminal OB-fold of Prp43 has been suggested to serve as a binding platform for other G-patch proteins.Citation27,29,37 To determine whether the same region of Prp43 mediates the interaction with Cmg1, N- or C-terminally truncated versions of Prp43 (Prp43 92–767 and Prp43 1–657) were expressed recombinantly, purified and used in binding assays with MBP-tagged versions of full length Cmg1 and the Cmg1 G-patch domain (Cmg1 1–85). Consistent with the yeast two-hybrid analysis, both full length Cmg1 and Cmg1 1–85 bound to full length Prp43, whereas only a weak interaction was observed with either of the Prp43 truncations (, Fig. S1A). This indicates that both the N- and C-terminus of Prp43 are required for efficient interaction with Cmg1, which is consistent with the close proximity of these domains as revealed by the crystal structure of Prp43,Citation38 For the full length G-patch protein this is in contrast to the interaction of Sqs1 with Prp43 where full length Sqs1 is still able to bind to Prp43 1–657.Citation38 However, the Sqs1 G-patch domain alone behaves similar to the G-patch domain of Cmg1 and hardly binds to Prp43 1–657. Importantly, these data suggest that several G-patch proteins have overlapping interaction sites on the RNA helicase.
Figure 1. Cmg1 specifically interacts with the RNA helicase Prp43. (A) Proteins were retrieved on IgG sepharose from extracts with or without tagged Cmg1, separated by SDS PAGE and stained with Coomassie. Inputs are shown on the left. Prp43 was identified in the Cmg1 eluate by mass spectrometry and Western blotting (bottom panel). The asterisk marks a background band also present in the control. (B) Recombinant Protein A-tagged Spp2 (as negative control) or Cmg1 were immobilised on IgG sepharose and incubated with purified GFP-tagged Prp43. After washing, co-purified GFP-Prp43 was eluted, then inputs and eluates were separated by SDS PAGE and analyzed by Coomassie staining. (C) Yeast two-hybrid analysis of Prp43 (fused to the GAL4 activation domain; GAL4-AD) was performed with full length (FL), the N-terminal G-patch domain (amino acids 1–85) or the C-terminus (amino acids 86–274) of Cmg1 (fused to the GAL4 binding domain; GAL4-BD), and Swm2 and Tgs1 as controls. The strains were spotted on plates not selecting (left) or selecting (right) for a yeast two-hybrid (Y2H) interaction. The domain structure of Cmg1, containing the G-patch domain and the domain of unidentified function (DUF4187) is shown at the top. (D) Protein A-tagged full length Prp43 or N- (92–767) or C- (1–657) terminally truncated versions of Prp43 were immobilised on IgG sepharose and incubated with MBP-tagged full length Cmg1 or Cmg1 1–85. After washing the beads, proteins were eluted, separated by SDS PAGE and analyzed by Commassie staining. Cmg1 and Cmg1 1–85 proteins co-precipitated with the different forms of Prp43 are indicated by the asterisk and double asterisk, respectively. For inputs of the binding experiments see Figure S1A.
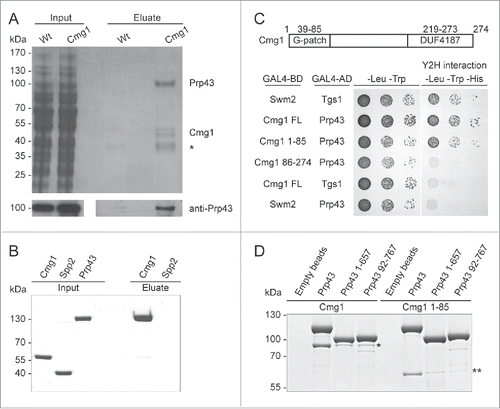
The G-patch proteins Spp382, Sqs1 and Pxr1, have previously been shown to modulate Prp43 activity. To reveal whether Cmg1 is also a bona fide cofactor of Prp43 we analyzed whether Cmg1 influences RNA binding by Prp43 using fluorescence anisotropy measurements with a fluorescein-labeled RNA and increasing concentrations of Prp43 in the presence or absence of Cmg1 or the G-patch domain of Spp382. As full-length recombinant Spp382 is difficult to obtain, we used its G-patch domain (Spp382 51–110), which had previously been shown to be sufficient for Prp43 interaction and activation.Citation39 Interestingly, Cmg1 or Spp382 51–110 stimulated RNA binding by Prp43 by about 25-fold in anisotropy experiments (; Table S2), while Cmg1 itself, similar to Spp2,Citation40 showed no interaction with the RNA (data not shown). This is in contrast to other G-patch proteins, such as Spp382 and Sqs1,Citation27,39 which have been shown to bind RNA directly and were suggested to recruit the helicase to its targets. To test whether Cmg1 activates or represses Prp43, we performed ATPase assays in vitro. Cmg1, which does not hydrolyse ATP itself (Fig. S1B), stimulated the ATPase activity of Prp43 more than 6-fold, similar to the G-patch domain of Spp382 (Spp382 51–110; and Table S2). Besides the reduction in the KM, the turnover number (kcat) for ATP hydrolysis was mildly increased by Cmg1. Furthermore, the G-patch domain of Cmg1 (Cmg1 1–85) is sufficient to stimulate the ATPase activity of full length Prp43, but although the N- and C-terminal truncations of Prp43 themselves show ATPase activity, this was hardly stimulated by the presence of either full length Cmg1 or Cmg1 1–85 (Fig. S1B), which is consistent with the strongly reduced binding of the Prp43 truncations to Cmg1 (). Taken together, we have identified the G-patch protein Cmg1 as a novel interaction partner of the RNA helicase Prp43 and show that Cmg1 stimulates both the RNA binding and ATPase activity of Prp43.
Figure 2. RNA binding and the ATPase activity of Prp43 are stimulated by Cmg1. (A) Fluorescence anisotropy titrations of fluorescein-labeled 11 nt RNA with Prp43 were performed with or without Cmg1 or the G-patch domain of Spp382 (Spp382 51–110). The Kd values are 60 ± 5 nM (Prp43 alone), 2.3 ± 1.6 nM (+Cmg1) and 2.5 ± 1.3 nM (+Spp382 51–110). (B) RNA-stimulated ATPase activity of Prp43 was measured in the absence or presence of Spp382 51–110 or Cmg1. The kcat values were 3.61 ± 0.14 s (Prp43 alone), 5.09 ± 0.26 s (+Cmg1) and 3.44 ± 0.13 s (+Spp382 51–110), the KM values 1.41 ± 0.14 μM (Prp43 alone), 0.23 ± 0.05 µM (+Cmg1) and 0.22 ± 0.04 µM (+Spp382 51–110).
Cmg1 is localized in the cytoplasm and in the intermembrane space of mitochondria
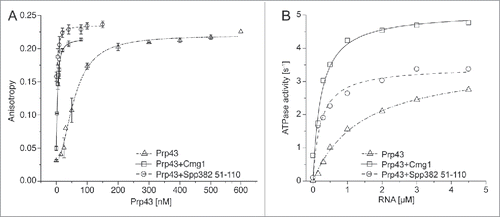
Prp43 is largely localized in the nucleolus/nucleus but also functions in the cytoplasm, and it interacts with different G-patch proteins in each of these compartments. To elucidate the cellular localization of Cmg1 we analyzed a yeast strain that expressed GFP-tagged Cmg1 under the control of its own promoter using fluorescence microscopy. This showed largely cytoplasmic staining, which overlapped with a cytoplasmic marker protein, Ssa2-RFP, implying that Cmg1 likely interacts with Prp43 in the cytoplasm (). Interestingly, cytoplasmic foci were also observed and Cmg1 was found to co-localize with a Mitotracker (). To determine whether this fraction of Cmg1 is bound to or imported into mitochondria and inaccessible to cytoplasmic Prp43, we analyzed the sub-organellar localization of Cmg1 using isolated mitochondria and protease protection assays.Citation41 Incubation of whole mitochondria with proteinase K left Cmg1 intact, suggesting that it is indeed imported into the organelle. Mitochondrial Cmg1 only became susceptible to protease digestion after rupture of the outer mitochondrial membrane by osmotic swelling (), indicating that the mitochondrial fraction of Cmg1 is localized in the intermembrane space (IMS). We have therefore named the previously uncharacterised YLR271W as cytoplasmic and mitochondrial G-patch protein 1 (Cmg1).
Figure 3. Cmg1 is localized in the cytoplasm and in the intermembrane space of mitochondria. (A) The localization of Cmg1-GFP (green in overlays) and Ssa2-RFP (panels on the left; red in overlay) or Mitotracker (panels on the right; red in overlay) and overlays are shown. Scale bar represents 5 µm for the main images. A 2-fold zoom of a representative cell is shown in each panel. (B) To analyze submitochondrial localization, yeast mitochondria were left untreated, swollen (Mitoplasts), or solubilised with 1% Triton X-100 (TX-100), then treated with Proteinase K (PK) where indicated, and subjected to SDS PAGE and Western blotting using antibodies against tagged Cmg1, the outer membrane protein Tom70, inner membrane proteins Mic10, Tim21 and Tim44. Note, Tim44 extrudes into the matrix, while a domain of Tim21 and the majority of Mic10 extends into the IMS. (C) Serial dilutions of Cmg1 overexpression (oex), overexpression of Cmg1 without its G-patch domain (86–274 oex), cmg1 deletion (Δcmg1) and wildtype strains were spotted for growth analysis with and without induction of apoptosis by treatment with acetic acid. Growth was documented after 2 days.
Since Cmg1 localizes to mitochondria we analyzed whether its deletion or overexpression affects mitochondrial function. Mitochondria play a central role in the generation of metabolic energy and perturbation of mitochondrial metabolism or translation leads to impaired growth on carbon sources such as glycerol or lactate, which require functional mitochondria and the electron transport chain for energy metabolism. The growth of yeast strains in which cmg1 had been genomically deleted or was overexpressed was therefore analyzed on each of these media. No significant differences were observed however (Fig. S2A), implying that Cmg1 is not required for mitochondrial energy production. Another major function of mitochondria is in apoptosis and interestingly, cell survival assays revealed that overexpression of Cmg1 promotes apoptosis while deletion of cmg1 increased survival following acetic acid treatment (increased number of colonies; ), indicating that Cmg1 is a pro-apoptotic factor. Overexpression of Cmg1 without its G-patch domain (Cmg1 86–274 oex) had a mild effect on growth under normal conditions (), which likely reflects the slightly higher expression levels of the truncated protein compared to the full length protein (Fig. S2B). However, like overexpression of the full length protein, overexpression of Cmg1 without its G-patch domain promoted apoptosis after acetic acid treatment (), suggesting that this function is independent of an interaction with the RNA helicase.
G-patch protein cofactors compete for Prp43 binding
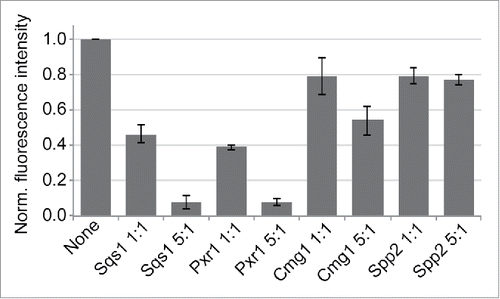
Our data identify the G-patch protein Cmg1 as a novel Prp43 cofactor that, like Sqs1, Pxr1 and Spp382,Citation27,29,37 can interact with the C-terminal OB-fold of the multifunctional RNA helicase. How Prp43 is distributed between its roles in different cellular pathways remained unknown and we hypothesized that its interaction partners might compete for interaction with Prp43 and thereby regulate the targeting of the helicase to its diverse functions. To test this model, we expressed and purified recombinant G-patch domains of the five G-patch proteins from yeast and performed competition assays in vitro. The Protein A-tagged G-patch domain of Spp382 was immobilised on IgG sepharose and binding with GFP-Prp43 performed in the absence or in the presence of the other G-patch domains. As expected, the presence of the Spp2 G-patch domain had little effect on Prp43 binding to immobilised Spp382 (). However, the presence of the G-patch domains of Sqs1 and Pxr1 strongly reduced Prp43 retrieval by Spp382, while Cmg1 had a weaker effect (), indicating that Cmg1 might have a lower affinity for Prp43 than Spp382. Our data therefore indicate that the different cofactors can compete for interaction with Prp43.
Figure 4. G-patch proteins compete for binding to Prp43. Recombinant Protein A-tagged G-patch domain of Spp382 was immobilised and incubated with GFP-Prp43 in the presence or absence of the indicated G-patch domains of Sqs1, Pxr1, Cmg1 or Spp2. Competitors were used in ratios of 1:1 and 5:1 compared to the immobilised G-patch domain of Spp382. Eluted GFP-Prp43 was quantified by fluorescence measurements. Data from three independent experiments are presented as mean +/− SEM.
Cofactor competition regulates the distribution of Prp43 between target pathways
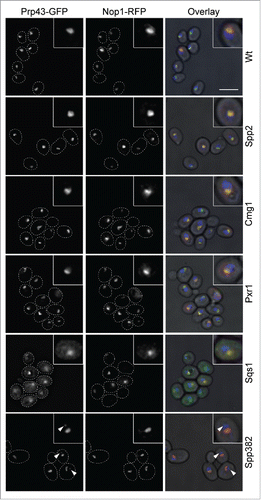
To analyze the effect of such competition in vivo we individually overexpressed all five yeast G-patch proteins (Fig. S3A) and analyzed the cellular localization of GFP-tagged Prp43. The nuclear/nucleolar localization of Prp43 observed in wildtype cells was unaffected by overexpression of Spp2, the G-patch protein that does not interact with Prp43 (). As expected, increased levels of the Pxr1, which itself localizes to the nucleolus, did not affect the nucleolar Prp43 localization. In contrast, increasing the expression of Sqs1, which leads to accumulation of the G-patch protein in the cytoplasm (Fig. S3B), also resulted in cytoplasmic accumulation of Prp43 (). Furthermore, overexpression of the nuclear splicing protein Spp382 lead to partial redistribution of Prp43 from the nucleolus to the nucleoplasm/nucleoplasmic foci (marked by arrowheads) and a change in nucleolar morphology in some cells (). In the case of Cmg1 overexpression, only few cells showed a slight increase in cytoplasmic Prp43 levels, but the major pool of the RNA helicase remained in the nucleolus. These data suggest that the Prp43-interacting cofactors also compete for interaction with the RNA helicase in vivo.
Figure 5. Overexpression of G-patch proteins leads to cellular relocalisation of Prp43. The localization of Prp43-GFP was analyzed by fluorescence microscopy in yeast cells overexpressing individual G-patch proteins or in control cells (Wt). The localization of the Prp43-GFP is shown on the left, the nucleolar marker Nop1-RFP in the middle, and the overlay with DAPI staining (blue) and a brightfield image is presented on the right. In the left and middle panels, cells are outlined with a white, dotted line. Nuclear foci containing GFP-Prp43 after overexpression of Spp382 are marked with arrowheads. The scale bar represents 5 µm.
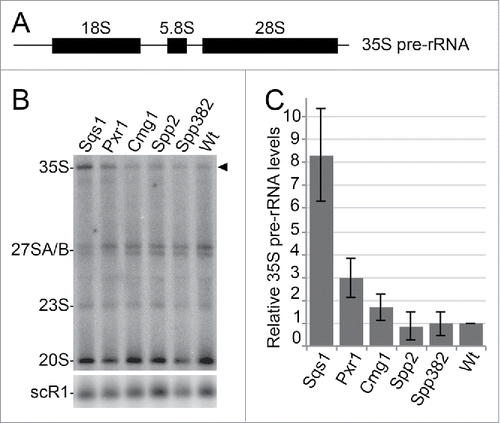
Based on the competition of the cofactors and the relocalisation of Prp43 upon G-patch protein overexpression, we investigated whether the removal of Prp43 from target complexes leads to defects in pathways from which the RNA helicase is withdrawn. We analyzed the effects of G-patch protein overexpression on the processing of pre-rRNA intermediates (pre-rRNAs). Genetic depletion of Prp43 affects the biogenesis of both ribosomal subunits and impedes pre-rRNA processing, leading to a strong accumulation of the 35S pre-rRNA primary transcript ().Citation28,34 Here, we observed that overexpression of Spp2 or Spp382 did not significantly affect pre-rRNA processing (), which is consistent with the absence or only minimal effects on Prp43 localization, respectively (). Conversely, overexpression of Sqs1 or Pxr1 caused accumulation of the 35S pre-rRNA transcript (). The increase in 35S pre-rRNA levels resembles the effect of Prp43 depletion and is especially strong for overexpression of Sqs1. Sqs1 overexpression had also led to a major redistribution of Prp43 to the cytoplasm and therefore to a withdrawal of Prp43 from its interactions and functions in the nucleolus (). Although Prp43 is still localized in the nucleolus upon overexpression of Pxr1, a significant accumulation of the 35S pre-rRNA was also observed, suggesting that Prp43 is sequestered by the additional Pxr1 and therefore not able to fulfil its functions in the early steps of ribosome biogenesis. An increase in the level of the 35S pre-rRNA was also observed upon Cmg1 overexpression, but this effect is much milder, likely because a significant fraction of Cmg1 is sequestered in mitochondria and not accessible to Prp43. Taken together, our findings imply that Prp43-interacting proteins compete for the binding and recruitment of the RNA helicase and that the distribution of Prp43 can be regulated by changes in the availability of interaction partners.
Figure 6. G-patch protein overexpression leads to defects in ribosome biogenesis. (A) Scheme of the 35S pre-rRNA transcript, which contains the sequences of the mature 18S, 5.8S and 25S rRNAs. (B) Total RNA was isolated from yeast cells with or without overexpression of the individual G-patch proteins indicated, RNA was separated by denaturing agarose gel electrophoresis and analyzed by Northern blotting using probes for the detection of rRNA precursors (indicated on the left). The 35S pre-rRNA is marked by an arrowhead. (C) Levels of the 35S rRNA precursor transcript in three independent experiments were quantified, normalized to the scR1 RNA (loading control) and the wildtype, and are presented as mean +/− SEM.
Discussion
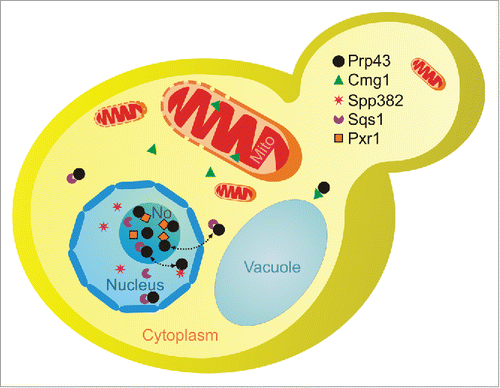
A growing number of RNA helicases have recently been shown to act in several different pathways, indicating that the “one enzyme – one function” model does not apply for these proteins. However, little is known about their interaction partners and it has remained unclear how the distribution of such multifunctional RNA helicases between individual target pathways and RNA-protein complexes is regulated. Here, we show that multiple G-patch protein cofactors compete for binding of the multifunctional RNA helicase Prp43. Prp43 is mainly localized in nucleoli, where it interacts with the G-patch proteins Sqs1/Pfa1 and Pxr1/Gno1 in ribosome biogenesis (). In addition, Prp43 acts in pre-mRNA splicing together with Spp382/Ntr1 in the nucleoplasm and has been suggested to shuttle to the cytoplasm with Sqs1 on pre-40S complexes. Interestingly, changes in the levels of its G-patch protein cofactors modulate the cellular localization of Prp43, suggesting a model in which cofactor competition regulates the distribution of the multifunctional RNA helicase between different pathways. This competition can even occur between different functions in the same pathway, as illustrated here in the case of ribosome biogenesis, where Sqs1 and Pxr1 compete for Prp43 interaction. The overexpression of either Sqs1 or Pxr1 results in redirection of Prp43 to complexes containing the overexpressed cofactor, likely leading to withdrawal of Prp43 from other pre-ribosomal intermediates, preventing its function in their maturation. Indeed, redistribution of Prp43 affects its activity in ribosome biogenesis and leads to defects in the processing of rRNA precursors. Interestingly, these results are supported by an independent report that also demonstrates defects in splicing upon overexpression of the G-patch protein Sqs1,Citation42 indicating that sequestering of Prp43 to the cytoplasm by Sqs1 causes defects in mRNA maturation as well as in ribosome biogenesis. The strong defects observed upon overexpression of Sqs1 also further support the suggestion that Prp43 and Sqs1 shuttle to the cytoplasm, where they stimulate the final processing of the 18S rRNA by the endonuclease Nob1Citation35 and that this might involve a larger fraction of Prp43 than previously anticipated.
Figure 7. Scheme of the localisations of Prp43 and its G-patch protein cofactors. Prp43 is largely localized in nucleoli (No) where it is involved in ribosome synthesis and likely interacts with Pxr1/Gno1 and Sqs1/Pfa1. Prp43 is also found in nucleoplasm, where it acts in splicing with Spp382/Ntr1, and it is present in the cytoplasm, possibly binding Sqs1 or Cmg1. Dotted arrows indicate the exchange of Prp43 between its different cofactors.
Our findings also raise questions about the function of Prp43 together with Cmg1. Our data suggest that this complex is formed in the cytoplasm, but since deletion of Cmg1 does not affect pre-rRNA processing (data not shown), it is unlikely that Cmg1 participates in the described cytoplasmic role of Prp43 in the final steps of 18S rRNA maturation. It is likely that Cmg1 and Prp43 act together on other cytoplasmic substrates, which remain to be identified. However, since Cmg1, in contrast to other G-patch proteins, does not directly contact RNA, the identification of such targets is challenging. Furthermore, as Cmg1 is normally expressed at much lower levels than Prp43 (less than 10%),Citation43 putative cytoplasmic targets of the Prp43-Cmg1 complex likely evade detection among the multitude of nuclear Prp43-interacting RNAs. Prp43 has been shown to mediate the dissociation of aberrant and late spliceosomes and to release small nucleolar RNPs from pre-ribosomal complexes, suggesting a general role of this RNA helicase in the disassembly of RNP complexes.Citation19,34,44,45 It is possible, therefore, that Cmg1 and Prp43 function together in other RNP restructuring events in the cytoplasm.
Interestingly, our data show that in addition to the cytoplasmic pool, a fraction of Cmg1 is localized in the mitochondrial IMS (). Cmg1 does not contain a classical N-terminal target sequence and N-terminal tagging did not abolish import into the IMS, suggesting that Cmg1 might be transported across the outer membrane via the MIA-dependent pathway.Citation46 Our data show that overexpression of Cmg1 decreases cell survival after induction of apoptosis, implying that Cmg1 functions in promoting apoptosis. Since overexpression of a truncated form of Cmg1, from which the G-patch domain is absent, also decreased survival of yeast cells after acetic acid treatment, it is likely that this function of Cmg1 is independent of an interaction with an RNA helicase. Similar to our findings for Cmg1, a helicase-independent function has also been described for the human homolog of Pxr1, PINX1.Citation47 While the roles of the other G-patch proteins in pre-mRNA splicing and ribosome biogenesis are thought to be mediated via their helicase interaction partners, PINX1 was shown to independently act in telomerase regulation by sequestering the catalytic subunit Est2 in an inactive complex.Citation48
Together, our findings suggest that cofactor levels and availability can regulate the distribution of a multifunctional RNA helicase between different pathways and thereby modulate its activity in target complexes (). We also observed that the G-patch domains of Pxr1 and Sqs1 showed stronger competition than the domain of Cmg1 for the Prp43-Spp382 interaction. Although regions of the proteins other than the G-patch domain are suggested to also contribute to interactions with Prp43,Citation38,49 our data suggest different affinities of the G-patch proteins for the RNA helicase. Interestingly, the ability of the G-patch domains of Spp382, Sqs1 and Pxr1 to outcompete that of Cmg1 correlates with the lower conservation of the G-patch domains of Cmg1 and especially Spp2 (Fig. S4), which does not stably bind to Prp43. Future structural analysis of G-patch-RNA helicase complexes will reveal which residues in the G-patch domain mediate the interaction and how specificity for the cofactor is conferred. In addition, there might be further levels of regulation, such as cofactor multimerisation as proposed for Sqs1 and Pxr1Citation27 and post-translational modification of G-patch proteins (see, for example, extensive phosphorylation of Spp382; ref.Citation50), which might modulate their interaction with the target helicases.
Our findings highlight the importance of cofactor identification for understanding the cellular functions of RNA helicases. G-patch proteins in particular are found in many eukaryotes and likely represent the largest family of RNA helicase cofactors.Citation14,15 The more than 20 human G-patch proteins include factors involved in angiogenesis, splicing, transcription regulation, and tumor suppression (see, for example, refs.Citation51,52). Interestingly, G-patch proteins are also encoded by various retroviruses.Citation14 As retroviruses rely on cellular RNA helicases for their replication,Citation53 it seems likely that they make use of their own G-patch proteins and other factors to recruit and redirect cellular RNA helicases to the viral replication machinery. Understanding the modulation of cellular RNA helicases by viral binding partners might allow the development of anti-viral drugs that specifically interrupt such interactions. Our findings now provide a mechanism for the regulation of multifunctional RNA helicases and thereby a foundation for the analysis of their modulation by cofactors on a cellular level and in disease.
Materials and methods
Expression and purification of recombinant proteins
His10-ZZ-TEV-taggedCitation54 Prp43, Prp43 1–657, Prp43 92–767, Cmg1, Spp2, Spp382 51–110, His10-MBP-TEV-tagged Sqs1 705–767, Cmg1 1–85, Pxr1 1–82, Spp2 92–157 and His-GFP-tagged Prp43 were expressed in Escherichia coli BL21 (DE3), bacteria lysed in a buffer containing 50 mM Tris/HCl pH 7.4, 600 mM NaCl, 10 mM imidazole, 0.1 mM PMSF, 1 mM MgCl2 and fusion proteins retrieved from soluble extracts on cOmplete His-tag Purification Resin (Roche). Wash steps initially using 50 mM Tris/HCl pH 7.4, 1 M NaCl, 10 mM imidazole, 0.1 mM PMSF, 1 mM MgCl2 followed by 50 mM Tris/HCl pH 7.4, 150 mM KCl, 5 mM MgCl2, 1 mM ATP and 50 mM GTP were performed to remove contaminating proteins and chaperones. Elution by 500 mM imidazole in a buffer containing 50 mM Tris/HCl pH 7.4, 150 mM NaCl, 1 mM MgCl2 was followed by size exclusion chromatography and purified proteins were stored in buffer containing 20 % (v/v) glycerol.
Protein interaction analysis
In vitro binding experiments with recombinant proteins were performed essentially as previously described.Citation55 Briefly, His10-ZZ-TEV-tagged Cmg1/Spp2 () were immobilised on IgG sepharose (GE Healthcare) and incubated with GFP-tagged Prp43 in a buffer containing 50 mM Tris/HCl pH 7.4, 100 mM NaCl, 1.5 mM MgCl2 and 2 mM ß-mercaptoethanol. Prey proteins were eluted in buffer containing 50 mM Tris/HCl pH 7.4 and 2 M MgCl2 and both inputs and eluates separated by SDS PAGE and analyzed by Coomassie staining (). Alternatively, Prp43/Prp43 1–657/Prp43 92–767 () were immobilised on IgG sepharose and incubated with MBP-tagged Cmg1 or Cmg1 1–85 in a buffer containing 50 mM Tris/HCl pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 1 mM ß-mercaptoethanol and 2 mg/mL BSA. After washing with the buffer lacking BSA, both bait and prey proteins were eluted using 0.5 M HAc pH 3.4 and inputs and eluates were analyzed as above (Fig. S1A and ). In the case of the competition experiments (), a binding experiment using His10-ZZ-TEV-tagged Spp382 G-patch domain (amino acids 51–110) and GFP-tagged Prp43 was performed as described above, but MBP-Sqs1 705–767, MBP-Pxr1 1–82, MBP-Cmg1 1–85 or MBP-Spp2 92–157 were used as competitors in ratios of 1:1 and 5:1 compared to the immobilised His10-ZZ-TEV-tagged Spp382 G-patch domain. Eluted GFP-Prp43 was quantified by fluorescence quantification. Pulldown assays from soluble yeast extracts () were performed as previously described.Citation13
Anisotropy measurements
Fluorescence anisotropy titrations of fluorescein-labeled 11 nt RNA were performed in a Jobin Yvon Fluoromax3 spectrometer at 37°C in 45 mM Tris/HCl pH 7.4, 25 mM NaCl, 2 mM MgCl2. 20 nM, 10 nM and 5 nM RNA, respectively, were titrated with Prp43 in the absence of cofactors, or in presence of 5 µM Spp382 51–110 or Cmg1. Data were analyzed using the Hill equation for Prp43 alone (n = 2), or using a 1:1 binding model for Prp43 in presence of cofactors (eq. 1).where r0 is the anisotropy of the free RNA, Δrmax is the amplitude, and [Prp43]tot and [RNA]tot are the total concentrations of Prp43 and RNA.
Steady-state ATP hydrolysis
The steady-state ATPase activity of Prp43 was monitored as a decrease of absorption at λ = 340 nm in a coupled enzymatic assay that couples ATP hydrolysis to the oxidation of NADH to NAD+.Citation56 Assays were performed using 400 nM Prp43, 250 nM Prp43 with 1 µM cofactor (Spp382 51–110 or Cmg1) in 45 mM Tris/HCl pH 7.4, 25 mM NaCl, 2 mM MgCl2, and 4 mM ATP and indicated concentrations of 11 nt RNA. Reactions were supplemented with 1 mM PEP, 20 U/ml pyruvate kinase, 20 U/ml LDH and 300 µM NADH and absorbance at 340 nm measured using a BioTEK Synergy HT microplate spectrophotometer equipped with Gene5 software. ATPase rates were determined from the decrease in A340, and kcat and KM values were obtained using Michaelis-Menten equation.
Yeast two-hybrid analysis
The bait plasmid pGBKT7 for expression of a fusion to the GAL4 DNA binding domain (GAL4-BD) and the prey plasmid PGADT7 for expression of a fusion to the GAL4 activation domain (GAL4-AD) were co-transformed into the reporter yeast strain PJ69-4A.Citation57 The transformants were spotted in a 10-fold serial dilution on SDC-Trp-Leu, and SDC-Trp-Leu-His and grown at 30°C. As a positive control, the combination of plasmids GAL4-BD-Swm2 and GAL4-AD-Tgs1 was used.Citation58
Yeast strains and protein expression/localization analysis
Yeast strains expressing fusion proteins and deletion strains were generated as previously described.Citation59,60 For fluorescence microscopy, yeast cells were grown in minimal medium to exponential phase and the cellular localization of fluorescent fusion proteins was analyzed using a DeltaVision microscope and the softWoRx (AppliedPrecision) software and 10 cycles of deconvolution. The Nop1-mRFP nucleolar marker protein was expressed from a pRS415 vector derivative. To monitor protein expression levels, yeast cells were grown in exponential phase before harvesting, lysis and precipitation of proteins using 15% trichloroacetic acid. Proteins were then separated by SDS PAGE and analyzed by Western blotting using the indicated antibodies. Yeast strains used are listed in Table S1.
Cell survival assays
Cell survival assays were performed as described.Citation61
Isolation of mitochondria and suborganellar protein localization
Mitochondrial isolation and sublocalisation assays were performed following the protocol previously described.Citation41 For the analysis of suborganellar protein localization, isolated mitochondria were untreated or converted to mitoplasts by hypotonic swelling in EM buffer (1 mM EDTA, 10 mM MOPS pH 7.2) or lysed in 1% Triton X-100 before treatment with different concentration of Proteinase K (0, 20 and 100 μg/ml) for 15 min at 4°C. Samples were precipitated with TCA and analyzed by SDS PAGE and Western blotting.
RNA analysis
RNA isolation and Northern transfer and hybridization were performed as previously described.Citation11,34 Probes used for pre-rRNA detection hybridize to sequences in the internal transcribed spacer 1 (probe 004: CGGTTTTAATTGTCCTA) and 2 (probe 020: TGAGAAGGAAATGACGCT). A probe against scR1 RNA (ATCCCGGCCGCCTCCATCAC) was used as loading control and band intensities were quantified using the ImageQuant software.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_datas.docx
Download MS Word (2.2 MB)Acknowledgments
We thank Yves Henry for providing the anti-Prp43 antibody, Mirjam Wissel, Monika Raabe, Uwe Pleßmann and Olaf Bernhard for technical assistance, and Johannes Heininger for help preparing .
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (SFB860 to H.U., R.L., J.E., P.R. and M.T.B.; SFB902 to E.S.; KL1153/7-1 to D.K.); Max-Planck Society (to R.L. and P.R.); Alexander von Humboldt foundation (to K.E.S. and M.T.B.); University Medical Center Göttingen (to B.S., P.R. and M.T.B.). Funding for open access charge: Open access publication fund of the Georg-August University, Göttingen.
References
- Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene 2006; 367:17-37; PMID:16337753; http://dx.doi.org/10.1016/j.gene.2005.10.019
- Bleichert F, Baserga SJ. The long unwinding road of RNA helicases. Mol. Cell 2007; 27:339-52; PMID: 17679086; http://dx.doi.org/10.1016/j.molcel.2007.07.014
- Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nature Rev Mol Cell Biol 2011; 12:505-516;PMID: 21779027; http://dx.doi.org/10.1038/nrm3154
- Martin R, Straub AU, Doebele C, Bohnsack MT. DExD/H-box RNA helicases in ribosome biogenesis. RNA Biol 2013; 10:4-18; PMID:22922795; http://dx.doi.org/10.4161/rna.21879
- Rodriguez-Galan O, Garcia-Gomez JJ, de la Cruz J. Yeast and human RNA helicases involved in ribosome biogenesis: current status and perspectives. Biochim Biophys Acta 2013; 1829:775-90; PMID:23357782; http://dx.doi.org/10.1016/j.bbagrm.2013.01.007
- Hilbert M, Kebbel F, Gubaev A, Klostermeier D. eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res 2011; 39:2260-70; PMID:21062831; http://dx.doi.org/10.1093/nar/gkq1127
- Nielsen KH, Behrens MA, He Y, Oliveira CL, Jensen LS, Hoffmann SV, Pedersen JS, Andersen GR. Synergistic activation of eIF4A by eIF4B and eIF4G. Nucleic Acids Res 2011; 39:2678-89; PMID:21113024; http://dx.doi.org/10.1093/nar/gkq1206
- Cordin O, Hahn D, Beggs JD. Structure, function and regulation of spliceosomal RNA helicases. Curr Opin Cell Biol 2012; 24:431-8; PMID:22464735; http://dx.doi.org/10.1016/j.ceb.2012.03.004
- Granneman S, Lin C, Champion EA, Nandineni MR, Zorca C, Baserga SJ. The nucleolar protein Esf2 interacts directly with the DExD/H box RNA helicase, Dbp8, to stimulate ATP hydrolysis. Nucleic Acids Res 2006; 34:3189-99; PMID:16772403; http://dx.doi.org/10.1093/nar/gkl419
- Vos HR, Bax R, Faber AW, Vos JC, Raué HA. U3 snoRNP and Rrp5p associate independently with Saccharomyces cerevisiae 35S pre-rRNA, but Rrp5p is essential for association of Rok1p. Nucleic Acids Res 2004; 32:5827-33; PMID:15523097; http://dx.doi.org/10.1093/nar/gkh904
- Bohnsack MT, Kos M, Tollervey D. Quantitative analysis of snoRNA association with pre-ribosomes and release of snR30 by Rok1 helicase. EMBO Rep 2008; 9:1230-6; PMID:18833290; http://dx.doi.org/10.1038/embor.2008.184
- Young CL, Khoshnevis S, Karbstein K. Cofactor-dependent specificity of a DEAD-box protein. Proc Natl Acad Sci 2013; 110:2668-76; PMID: 23630256; http://dx.doi.org/10.1073/pnas.1302577110
- Martin R, Hackert P, Ruprecht M, Simm S, Brüning L, Mirus O, Sloan KE, Kudla G, Schleiff E, Bohnsack MT. A pre-ribosomal RNA interaction network involving snoRNAs and the Rok1 helicase. RNA 2014; 20:1173-82; PMID:24947498; http://dx.doi.org/10.1261/rna.044669.114
- Aravind L, Koonin EV. G-patch: a new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem Sci 1999; 24:342-4; PMID:10470032; http://dx.doi.org/10.1016/S0968-0004(99)01437-1
- Robert-Paganin J, Réty S, Leulliot N. Regulation of DEAH/RHA helicases by G-patch proteins. Biomed Res Int 2015; 2015:931857; PMID:25692149; http://dx.doi.org/10.1155/2015/931857
- Roy J, Kim K, Maddock JR, Anthony JG, Woolford JL. Jr The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1995; 1:375-90; PMID:7493316
- Edwalds-Gilbert G, Kim DH, Silverman E, Lin RJ. Definition of a spliceosome interaction domain in yeast Prp2 ATPase. RNA 2004; 10:210-20; PMID:14730020; http://dx.doi.org/10.1261/rna.5151404
- Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol 2004; 24:10101-10; PMID:15542821; http://dx.doi.org/10.1128/MCB.24.23.10101-10110.2004
- Tsai R T, Fu RH, Yeh FL, Tseng CK, Lin YC, Huang YH, Cheng SC. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev 2005; 19:2991-3003; PMID:16357217; http://dx.doi.org/10.1101/gad.1377405
- Boon KL, Auchynnikava T, Edwalds-Gilbert G, Barrass JD, Droop AP, Dez C, Beggs JD. Yeast ntr1/spp382 mediates prp43 function in postspliceosomes. Mol Cell Biol 2006; 26:6016-23; PMID:16880513; http://dx.doi.org/10.1128/MCB.02347-05
- Arenas JE, Abelson JN. Prp43: An RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. 1997; 94:11798-802; PMID:9342317; http://dx.doi.org/10.1073/pnas.94.22.11798
- Martin A, Schneider S, Schwer B. Prp43 is an essential RNA-dependent ATPase required for release of lariat-intron from the spliceosome. J Biol Chem 2002; 277:17743-50; PMID:11886864; http://dx.doi.org/10.1074/jbc.M200762200
- Ebersberger I, Simm S, Leisegang MS, Schmitzberger P, Mirus O, von Haeseler A, Bohnsack MT, Schleiff E. The evolution of the ribosome biogenesis pathway from a yeast perspective. Nucleic Acids Res 2014; 42:1509-23; PMID:24234440; http://dx.doi.org/10.1093/nar/gkt1137
- Gee S, Krauss SW, Miller E, Aoyagi K, Arenas J, Conboy J G. Cloning of mDEAH9, a putative RNA helicase and mammalian homologue of Saccharomyces cerevisiae splicing factor Prp43. Proc Natl Acad Sci 1997; 94:11803-7; PMID:9342318; http://dx.doi.org/10.1073/pnas.94.22.11803
- Wen X, Lei YP, Zhou YL, Okamoto CT, Snead ML, Paine, M.L. Structural organization and cellular localization of tuftelin-interacting protein 11 (TFIP11). Cell Mol Life Sci 2005; 62:1038-46; PMID:15868102; http://dx.doi.org/10.1007/s00018-005-4547-z
- Guglielmi B, Werner M. The yeast homolog of human PinX1 is involved in rRNA and small nucleolar RNA maturation, not in telomere elongation inhibition. J Biol Chem 2002; 277:35712-9; PMID:12107183; http://dx.doi.org/10.1074/jbc.M205526200
- Lebaron S, Papin C, Capeyrou R, Chen YL, Froment C, Monsarrat B, Caizergues-Ferrer M, Grigoriev M, Henry Y. The ATPase and helicase activities of Prp43p are stimulated by the G-patch protein Pfa1p during yeast ribosome biogenesis. EMBO J 2009; 28:3808-19; PMID:19927118; http://dx.doi.org/10.1038/emboj.2009.335
- Lebaron S, Froment C, Fromont-Racine M, Rain JC, Monsarrat B, Caizergues-Ferrer M, Henry Y. The splicing ATPase prp43p is a component of multiple preribosomal particles. Mol Cell Biol 2005; 25:9269-82; PMID:16227579; http://dx.doi.org/10.1128/MCB.25.21.9269-9282.2005
- Chen YL, Capeyrou R, Humbert O, Mouffok S, Kadri YA, Lebaron S, Henras AK, Henry Y. The telomerase inhibitor Gno1p/PINX1 activates the helicase Prp43p during ribosome biogenesis. Nucleic Acids Res 2014; 42:7330-45; PMID:24823796; http://dx.doi.org/10.1093/nar/gku357
- Combs DJ, Nagel RJ, Ares M, Stevens SW. Prp43p is a DEAH-box spliceosome disassembly factor essential for ribosome biogenesis. Mol Cell Biol 2006; 26:523-34; PMID:16382144; http://dx.doi.org/10.1128/MCB.26.2.523-534.2006
- Leeds NB, Small EC, Hiley SL, Hughes TR, Staley JP. The splicing factor Prp43p, a DEAH box ATPase, functions in ribosome biogenesis. Mol Cell Biol 2006; 26:513-22; PMID:16382143; http://dx.doi.org/10.1128/MCB.26.2.513-522.2006
- Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci 2009; 106:9613-8; http://dx.doi.org/10.1073/pnas.0901997106
- Bohnsack MT, Tollervey D, Granneman S. Identification of RNA helicase target sites by UV cross-linking and analysis of cDNA. Methods Enzymol. 2012; 511:275-88; PMID:22713325; http://dx.doi.org/10.1016/B978-0-12-396546-2.00013-9
- Bohnsack MT, Martin R, Granneman S, Ruprecht M, Schleiff E, Tollervey D. Prp43 bound at different sites on the pre-rRNA performs distinct functions in ribosome synthesis. Mol Cell 2009; 36:583-92; PMID: 19941819; http://dx.doi.org/10.1016/j.molcel.2009.09.039
- Pertschy B, Schneider C, Gnädig M, Schäfer T, Tollervey D, Hurt E. RNA helicase Prp43 and its co-factor Pfa1 promote 20 to 18 S rRNA processing catalyzed by the endonuclease Nob1. J Biol Chem 2009; 284:35079-91; PMID:19801658; http://dx.doi.org/10.1074/jbc.M109.040774
- Watkins NJ, Bohnsack MT. The box C/D and H/ACA snoRNPs: key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 2012; 3:397-414; PMID:22065625; http://dx.doi.org/10.1002/wrna.117
- Tanaka N, Aronova A, Schwer B. Ntr1 activates the Prp43 helicase to trigger release of intron-lariat from the spliceosome. Genes Dev 2007; 21:2312-25; PMID:17875666; http://dx.doi.org/10.1101/gad.1580507
- Walbott H, Mouffok S, Capeyrou R, Lebaron S, Humbert O, van Tilbeurgh H, Henry Y, Leulliot N. Prp43p contains a processive helicase structural architecture with a specific regulatory domain. EMBO J 2010; 29:2194-204; http://dx.doi.org/10.1038/emboj.2010.102
- Christian H, Hofele RV, Urlaub H, Ficner R. Insights into the activation of the helicase Prp43 by biochemical studies and structural mass spectrometry. Nucleic Acids Res. 2014; 42:1162-79; PMID:24165877; http://dx.doi.org/10.1093/nar/gkt985
- Warkocki Z, Schneider C, Mozaffari-Jovin S, Schmitzová J, Höbartner C, Fabrizio P, Lührmann R. The G-patch protein Spp2 couples the spliceosome-stimulated ATPase activity of the DEAH-box protein Prp2 to catalytic activation of the spliceosome. Genes Dev. 2015; 29:94-107; PMID:25561498; http://dx.doi.org/10.1101/gad.253070.114
- Mick DU, Wagner K, van der Laan M, Frazier AE, Perschil I, Pawlas M, Meyer HE, Warscheid B, Rehling P. Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J 2007; 26:4347-58; PMID:17882259; http://dx.doi.org/10.1038/sj.emboj.7601862
- Pandit S, Paul S, Zhang L, Chen M, Durbin N, Harrison SM, Rymond BC. Spp382p interacts with multiple yeast splicing factors, including possible regulators of Prp43 DExD/H-Box protein function. Genetics 2009; 183:195-206; PMID:19581443; http://dx.doi.org/10.1534/genetics.109.106955
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature 2003; 425:737-41; PMID:14562106; http://dx.doi.org/10.1038/nature02046
- Mayas RM, Maita H, Semlow DR, Staley JP. Spliceosome discards intermediates via the DEAH box ATPase Prp43p. Proc. Natl. Acad. Sci. 2010; 107:10020-5; PMID:20463285; http://dx.doi.org/10.1073/pnas.0906022107
- Fourmann JB, Schmitzová J, Christian H, Urlaub H, Ficner R, Boon KL, Fabrizio P, Lührmann R. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev. 2013; 27:413-28; PMID:23431055; http://dx.doi.org/10.1101/gad.207779.112
- Gornicka A, Bragoszewski P, Chroscicki P, Wenz LS, Schulz C, Rehling P, Chacinska A A discrete pathway for the transfer of intermembrane space proteins across the outer membrane of mitochondria. Mol. Biol. Cell 2014; 25:3999-4009; PMID:25318675; http://dx.doi.org/10.1091/mbc.E14-06-1155
- Zhou XZ, Lu KP. The Pin2/TRF1-interacting protein PinX1 is a potent telomerase inhibitor. Cell 2001; 107:347-59; PMID:11701125; http://dx.doi.org/10.1016/S0092-8674(01)00538-4
- Lin J, Blackburn EH. Nucleolar protein PinX1p regulates telomerase by sequestering its protein catalytic subunit in an inactive complex lacking telomerase RNA. Genes Dev. 2004; 18:387-96; PMID:14977919; http://dx.doi.org/10.1101/gad.1171804
- Banerjee D, McDaniel PM, Rymond BC. Limited Portability of G-Patch Domains in Regulators of the Prp43 RNA Helicase Required for Pre-mRNA Splicing and Ribosomal RNA Maturation in Saccharomyces cerevisiae. Genetics 2015; 200:135-417; PMID:25808954; http://dx.doi.org/10.1534/genetics.115.176461
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villen J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods 2013; 10:676-82; PMID: 23749301; http://dx.doi.org/10.1038/nmeth.2519
- Niu Z, Jin W, Zhang L, Li X. Tumor suppressor RBM5 directly interacts with the DExD/H-box protein DHX15 and stimulates its helicase activity. FEBS Lett. 2012; 586:977-83; PMID:22569250; http://dx.doi.org/10.1016/j.febslet.2012.02.052
- Hu FY, Wu C, Li Y, Xu K, Wang WJ, Cao H, Tian XL. AGGF1 is a novel anti-inflammatory factor associated with TNF-α-induced endothelial activation. Cell Signal. 2013; 25:1645-53; PMID:23628701; http://dx.doi.org/10.1016/j.cellsig.2013.04.007
- Steimer L, Klostermeier D. RNA helicases in infection and disease. RNA Biol. 2012; 9:751-71; PMID:22699555; http://dx.doi.org/10.4161/rna.20090
- Leulliot N, Bohnsack MT, Graille M, Tollervey D, Van Tilbeurgh H. The yeast ribosome synthesis factor Emg1 is a novel member of the superfamily of α/β knot fold methyltransferases. Nucleic Acids Res. 2008; 36:629-39; PMID:18063569; http://dx.doi.org/10.1093/nar/gkm1074
- Lehmann P, Bohnsack MT, Schleiff E. The functional domains of the unusual chloroplast positioning protein 1. Plant Science 2011; 180:650-4; PMID:21421414; http://dx.doi.org/10.1016/j.plantsci.2011.01.006
- Adam H. Adenosin-5′diphosphat und Adenosin-5′monophosphat. In Methoden der enzymatischen Analyse Bergmeyer HU (ed.) 1962; Weinheim: Verlag Chemie:573-7
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 1996; 144:1425-36; PMID:8978031
- Boon KL, Kos M. Deletion of Swm2p selectively impairs trimethylation of snRNAs by trimethylguanosine synthase (Tgs1p). FEBS Lett 2010; 584:3299-304; PMID:20621096; http://dx.doi.org/10.1016/j.febslet.2010.07.001
- Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953-61; PMID:9717241; http://dx.doi.org/10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassetes. Yeast 2004; 21:947-62; PMID:15334558; http://dx.doi.org/10.1002/yea.1142
- Teng X, Hardwick JM. Reliable method for detection of programmed cell death in yeast. Methods Mol Biol 2009; 559:335-42; PMID:19609767; http://dx.doi.org/10.1007/978-1-60327-017-5_23
