ABSTRACT
Pat1 is a key cytoplasmic mRNA degradation factor, the loss of which severely increases mRNA half-lives. Several recent studies have shown that Pat1 can enter the nucleus and can shuttle between the nucleus and the cytoplasm. As a result, many nuclear roles have been proposed for Pat1. In this study, we analyzed four previously suggested nuclear roles of Pat1 and show that Pat1 is not required for efficient pre-mRNA splicing or pre-mRNA decay in yeast. However, lack of Pat1 results in accumulation of pre-rRNA processing intermediates. Intriguingly, we identified a novel genetic relationship between Pat1 and the rRNA decay machinery, specifically the exosome and the TRAMP complex. While the pre-rRNA processing intermediates that accumulate in the pat1 deletion mutant are, at least to some extent, recognized as aberrant by the rRNA degradation machinery, it is unlikely that these accumulations are the cause of their synthetic sick relationship. Here, we show that the dysregulation of the levels of mRNAs related to ribosome biogenesis could be the cause of the accumulation of the pre-rRNA processing intermediates. Although our results support a role for Pat1 in transcription, they nevertheless suggest that the primary cause of the dysregulated mRNA levels is most likely due to Pat1's role in mRNA decapping and mRNA degradation.
Introduction
Modulation of transcription and mRNA degradation is a crucial aspect of eukaryotic gene expression.Citation1 The stability of mRNA is controlled by two important structures, the m7G cap at the 5′ end and poly(A) tail at the 3′ end. Shortening of the poly(A) tail is the rate limiting step in the process of mRNA degradation in yeast.Citation2 After the shortening of the poly(A) tail, mRNA can be degraded either by the 5′ to 3′ or the 3′ to 5′ decay pathways. mRNA decay from the 3′ to 5′ direction is facilitated by the cytoplasmic exosome complex. Degradation of mRNA by the 5′ to 3′ decay pathway is the major pathway of mRNA degradation in yeast.Citation2 It begins with the removal of the 5′ m7G cap by the decapping enzyme followed by the rapid degradation of the mRNA by the cytoplasmic 5′ to 3′ exonuclease Xrn1.Citation3,4
Pat1 is a key protein that has been proposed to act as a scaffold to coordinate the process of mRNA degradation in yeast. Pat1 is an evolutionarily conserved mRNA decapping activator found from yeast to mammals.Citation5,6 It has been suggested that Pat1 first represses translation, then recruits and activates the decapping enzyme Citation5,7-9. Pat1 is a part of Pat1/Lsm1-7 complex, which regulates mRNA decay in the cytoplasm.Citation10,11
While the cytoplasmic role of Pat1 as an mRNA decay factor is well studied, it has been recently shown that Pat1 can enter the nucleus and can shuttle between the nucleus and cytoplasm.Citation12-14 The nuclear role of Pat1 has been largely overlooked since its initial identification as a Top2-interacting protein, which is the source of its name: “Protein Associated with Topoisomerase” (Pat1).Citation15,16 In addition, recent studies have proposed a role for Pat1 in splicing, transcription and retrograde tRNA import.Citation12,14,17-19 Human Pat1b co-localizes with splicing speckles, dynamic structures that contain poly(A)+ mRNA, SR proteins and splicing factors, supporting a splicing related function for Pat1. Its role in transcription was documented by a genome-wide transcriptomics study and in vivo transcriptional assays.Citation12,17 Furthermore, Pat1 has also been shown to be a structural component of the yeast kinetochore. Pat1 has been implicated to be crucial for integrity of the centromeric chromatin and chromosome segregation.Citation20
Another speculation was that Pat1 could bind the nuclear Lsm2-8 complex via the Lsm2 and Lsm3 proteins.Citation21 Lsm2-8 is the nuclear counterpart to the cytoplasmic Lsm1-7 complex, which functions in splicing, pre-mRNA decay and ribosome biogenesis (RB).Citation22-24 Despite these suggestions, its function in the nucleus, beyond transcription and chromosome segregation, has been mostly speculative. In this work, we addressed these possible roles and show that while Pat1 does affect rRNA processing and transcription, the overwhelming importance of Pat1 is in regulating mRNA stability.
Results
Pat1 is not required for pre-mRNA splicing
To elucidate if Pat1 functions in pre-mRNA splicing, we first examined whether the growth behavior of the pat1Δ strain is comparable to the known non-essential splicing deficient mutants of the nuclear Lsm2-8 complex (i.e. lsm6Δ and lsm7Δ mutants). The deletion of lsm1, a component of the cytoplasmic Lsm1-7 complex that does not affect splicing was used as a negative control.Citation25,26 However, Pat1 is found in the nucleus in this mutant (Fig. S1).Citation13 At 30°C all the deletion strains grew analogously to the WT strain, but at 37°C all the mutants grew slightly slower. At 20°C, the splicing defective mutants lsm6Δ and lsm7Δ grew at least 20-fold slower than the WT, while the pat1Δ and lsm1Δ strains had no growth defect ().
Figure 1. Deletion of Pat1 does not affect splicing of pre-U3 snoRNA. (A) Strains indicated were grown overnight at 30°C. Cells were then serially diluted 1:10, spotted on YEPD plates and grown at the indicated temperatures. (B) Northern analysis of the pre-U3 snoRNA splicing on a 6% denaturing polyacrylamide gel, blotted, and hybridized with oligonucleotide probe complementary U3 RNA. The unspliced U3 RNA is indicated as “Pre-U3 Species.” The mature and truncated U3 species are also indicated. scR1 is the loading control.
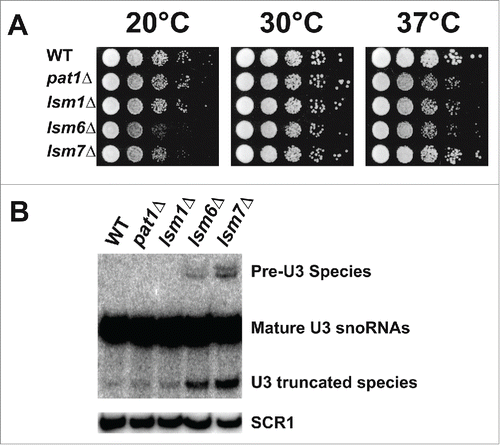
Thereafter, we examined the accumulation of the intron-containing precursor of the U3 snoRNA by northern analysis, which is a sensitive measure of splicing in yeast.Citation26,27 In the pat1Δ mutant, like the lsm1Δ strain, the pre-U3 species does not accumulate when compared to WT strain. However, as expected, the levels of the pre-U3 RNA were significantly elevated in lsm6Δ and lsm7Δ (). Since Pat1 is not required for splicing of the pre-U3 snoRNA, Pat1 is also most likely not required for efficient splicing in the nucleus in general.
Pat1 does not function in nuclear pre-mRNA and snoRNA decay
The major function of Pat1 in the cytoplasm is to activate the decapping enzyme complex and to promote mRNA decay.Citation5,7 Therefore, we hypothesized that Pat1 could potentially have similar functions in the nucleus. mRNAs are transcribed in the nucleus as pre-mRNA, which then undergo co-transcriptional splicing and several other post-transcriptional modifications before being exported to the cytoplasm as mature mRNAs. Pre-mRNAs whose introns are not spliced out undergo degradation in the nucleus. Inhibition of the nuclear decay machinery leads to a slight accumulation of mature mRNAs indicating that significant amount of unspliced pre-mRNAs undergo degradation even in wild-type yeast suggesting that pre-mRNA splicing is in constant competition with their decay.Citation28
To determine the role of Pat1 in nuclear pre-mRNA decay, we used an assay examines pre-mRNAs that contain a small nucleolar RNA (snoRNA) in their intronic region. snoRNAs are often located within the intronic region of pre-mRNAs of genes encoding proteins related to ribosome synthesis or function, although less so in yeast than in vertebrates. They are processed out when the pre-mRNA undergoes either splicing or degradation.Citation29,30 The presence of snoRNA serves as a steric block to exonucleases. In the deletion mutants that are defective in nuclear decapping, only 3′ to 5′ exosome mediated decay occurs. This blockage leads to an accumulation of 5′ degradation intermediates.Citation23,28 Since the Lsm2-8 complex has been shown to facilitate nuclear pre-mRNA decapping and degradation, strains lacking Lsm6 and Lsm7 were used as positive controls.Citation23
To examine if Pat1 affects nuclear pre-mRNA decay, we monitored the accumulation of 5′ unprocessed decay fragments of the TEF4 pre-mRNA that contains a snoRNA, snR38, in its intronic region, in the pat1Δ mutant by northern analysis. First, we found the pat1Δ and lsm1Δ strains did not accumulate 5′ unprocessed products of TEF4 pre-mRNA as compared to the WT strain ().Citation23 Second, as expected, the lsm6Δ and lsm7Δ mutants accumulated 5′-unprocessed fragments.
Figure 2. Deletion of Pat1 does not accumulate 5′-unprocessed fragments of pre-mRNAs having intronic snoRNAs. (A) Pictorial representation of an mRNA containing snoRNA in its intronic region, showing the relative positions of the oligonucleotide probes used in this study. (B) Left: Northern analysis of the TEF4 mRNA indicating 5′ decay product, the snR38 snoRNA and spliced full-length TEF4. To the left of each gel panel is indicated in brackets the oligonucleotide used for probing. The strains used are indicated above the figure. Total RNA was analyzed by agarose or PAGE northern analysis as appropriate. Right: The splicing products and decay intermediates are schematically depicted next to the indicated RNA species on the left. (C) As above for the EFB1 gene. (D) Northern analysis for the ACT1 gene in the strains indicated above probed with randomly primed DNA. (E) As above for the PGK1 gene. (F) As above for the SCR1 gene and probed with oTN100 (G) Quantification of the relative percentages of the mature mRNA and snoRNA of EFB1 and TEF4 normalized to scR1. The data represents averages of at least 3 independent experiments, Error = SD. Two tailed independent sample t-test with unequal variance was used. * Indicates p-values of < 0.05, ** indicates p-values of < 0.01, and *** indicates p-values of < 0.001. (H) Quantification of the relative percentages of the PGK1 and ACT1 mRNA normalized to scR1.
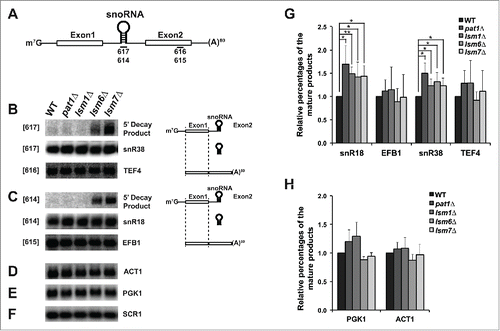
To confirm these results, we analyzed the pre-mRNA of EFB1 that contains a snoRNA, snR18 in its intronic region and observed similar results (). Despite the accumulation of the decay products in the lsm6Δ and lsm7Δ mutants, the levels of mature mRNAs of TEF4 and EFB1 did not change significantly from the WT strain, similar to the intron containing ACT1 and intron-less PGK1 mRNA ().Citation23 In contrast, the abundance of the snoRNAs, snR38, and snR18, increased slightly but significantly in pat1Δ mutant (). Both intronic and independently transcribed snoRNAs were elevated in the pat1Δ mutant and the lsm mutants tested here (Fig. S2). The cause for the increase in the levels of snoRNAs is unclear.
A possible explanation for snoRNA accumulation could be that Pat1 may be required for the destruction of mature snoRNA similar to that of strains defective in the nuclear Lsm2-8 complex.Citation31 As expected, in the strains lacking Lsm6 or Lsm7, a stable degradation product of U3 snoRNA accumulated. However, the pat1Δ and lsm1Δ strains did not have significant accumulations, indicating that they are not involved in degradation of the mature U3 snoRNA (). In conclusion, these results illustrate that Pat1 does not facilitate nuclear pre-mRNA or snoRNA decay similar to Lsm1 and unlike Lsm6 and Lsm7.
Absence of Pat1 leads to accumulation of rRNA processing intermediates
Next, we speculated that Pat1 might be required for efficient ribosome biogenesis (RB). Two main observations support this hypothesis. First, Pat1 was shown to associate with ribosomes.Citation32,33 Second, the Lsm2-8 complex to which Pat1 has been proposed to bind in the nucleus, is required for rRNA processing.Citation21,24,34 Strains deprived of proteins required for RB tend to accumulate pre-rRNA processing intermediates, produced due to inefficient or defective rRNA processing.Citation35 We hypothesized that if Pat1 is required for RB; the pat1Δ mutant should accumulate such intermediates (). The positions of the probes used in this study are diagrammed in the .
Figure 3. Pat1 is required for rRNA maturation. (A) Schematic representation of pre-35S rRNA and the relative positions of the oligonucleotide probes used for northern analysis. (B) Northern analysis of rRNA processing. The oligonucleotide probes used for probing are indicated in the brackets. RNA products analyzed are indicated to the right. The rRNA species are schematically depicted next to the indicated rRNA products. Strains examined are indicated at the top of the panels. (C) Quantification of rRNA intermediates shown in panel B normalized to scR1. n = 3, Error bar = SD and p-values as indicated in Pulse-chase analysis of WT and pat1Δ strains. Strains were pulse-labeled for 1 min and chased with unlabeled uracil for the indicated times listed above. The pre-rRNAs and rRNAs are indicated on the left and the schematic representation of rRNA species is on the right. (E) Growth assay for the WT, single, and double deletion strains indicated in the figure. Cells were grown overnight, serially diluted, spotted on YEPD plates and grown at the given temperatures.
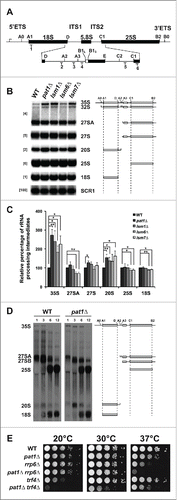
The 35S pre-rRNA transcript accumulated most prominently in the pat1Δ strain to nearly 3-fold more that of the WT strain (, ). This effect could be rescued with a centromeric plasmid containing a full-length copy of PAT1 (data not shown). Consistent with the previous studies, in the lsm1Δ, lsm6Δ and lsm7Δ mutants, there was also significant accumulation of 35S pre-rRNA precursor.Citation24,36
Table 1. Relative amounts of rRNA processing intermediates in pat1 and lsm mutants.
The 35S primary transcript undergoes initial processing by cleavages at the A0 and A1 sites to form a 32S pre-rRNA precursor, which then rapidly undergoes cleavage at A2. The A2 cleavage separates the pre-rRNA transcript into precursors of the mature 25S and 18S rRNAs. 20S pre-rRNA, the precursor of the 18S rRNA significantly accumulated in the pat1Δ mutant, which also could be rescued with a centromeric plasmid containing genomic copy of PAT1 (, and data not shown). The 20S pre-rRNA precursor clearly accumulated in lsm1Δ, lsm6Δ and lsm7Δ mutants as well at 30°C.
The amount of the 27SA pre-rRNA species, the 25S rRNA precursor products that form after A2 and A3 cleavages, did not change significantly in the pat1Δ and lsm1Δ mutants. In contrast, their levels significantly decreased in lsm6Δ and lsm7Δ mutants. We also observed a significant increase in the levels of 27S pre-rRNA species in pat1Δ but not the other strains (). Finally, the amount of mature 18S and 25S rRNA did not change significantly in the pat1Δ strain ().
Taken together, these results suggest that Pat1, like Lsm1, is also required for pre-rRNA processing of 35S rRNA, 27S pre-rRNA and 20S pre-rRNA. Therefore, Pat1 and Lsm1 might act together or in similar pathways to affect rRNA processing differently from Lsm2-8 complex.
rRNA processing is delayed in the pat1Δ mutant
The accumulated intermediates in the pat1Δ mutant could be from a delay in rRNA processing. To examine this question directly, we performed pulse-chase analyses of pre-rRNA processing in the WT and pat1Δ strains. One of the first pre-rRNA products generated is the 35S pre-rRNA transcript. However, it is rapidly processed into 27S pre-rRNA and 20S pre-rRNA species.Citation37 It is noteworthy that we observed a much longer persistence of the 35S pre-rRNA in the pat1Δ mutant consistent with the accumulation of intermediates observed at steady-state (Compare ), while the WT strain underwent rapid processing into the subsequent products.
There was also a delay in the processing of the 27SA pre-rRNA species in the pat1Δ mutant compared to the WT strain. The final maturation of the 20S pre-rRNA to 18S occurs in the cytoplasm. Therefore, the delay in processing of this pre-rRNA precursor suggests that Pat1 may either affect the export of the ribosome or the cytoplasmic processing of 18S rRNA ().Citation38,39
Pat1 mutant has synthetic growth defects with the rRNA surveillance machinery
The pat1Δ mutant accumulates abnormal levels of rRNA processing intermediates. However, additional aberrant intermediates could be eliminated rapidly by the nuclear surveillance machinery and thus not observed. To analyze if any such products are formed in the pat1Δ strain, we deleted TRF4, the poly(A) polymerase of the TRAMP (Trf4/Air2/Mtr4 Polyadenylation). The TRAMP complex facilitates the addition of 4 to 6 adenosines at the 3′ end of an abnormal rRNA intermediates. We also deleted RRP6, the nuclear-specific component of the exosome complex that degrades aberrant rRNAs in the pat1Δ and WT strain backgrounds.Citation40-42
To determine if a genetic relationship exists between Pat1 and the nuclear surveillance machinery, we examined the growth of the single (pat1Δ, trf4Δ, and rrp6Δ) and the double deletion strains (pat1Δ trf4Δ and pat1Δ rrp6Δ). As in , the pat1Δ mutant grew slightly slower than WT at 37°C. The growth of the rrp6Δ mutant was severely inhibited at 37°C, but grew comparably to the WT strain at 20°C and 30°C as reported previously () (46). When combined with the pat1Δ mutation, the 37°C growth inhibition of the rrp6Δ mutant became more pronounced.
The single deletion mutant trf4Δ was cold sensitive at 20°C, but grew like WT strain at all the other temperatures examined (). The pat1Δ trf4Δ double deletion mutant was synthetically slow growing at all temperatures, exhibiting the strongest growth defect at 20°C. Growth inhibition of the pat1Δ rrp6Δ mutant at 37°C and pat1Δ trf4Δ strain at 20°C is due to slower growth as viability was unaffected (data not shown). Altogether, these results show that Pat1 has a synthetic sick relationship with the nuclear exosome and TRAMP complex mutants. One possible source of these synthetic relationships could be presence of abnormal levels of rRNA intermediates generated in the pat1Δ strain. We therefore examined the levels of rRNA processing intermediates in the single and double mutants.
Although there are subtle rRNA processing defects in the double deletion mutants (pat1Δ rrp6Δ and pat1Δ trf4Δ) compared to the single mutants, we did not observe significant synergistic increases of rRNA intermediates in either the trf4Δ pat1Δ or the rrp6Δ pat1Δ mutant (Fig. S4). For instance, the levels of 27SA, 27S, 18S and 25S rRNA increased in the trf4Δ pat1Δ mutant at both 30°C and 37°C but not at 20°C. In rrp6Δ pat1Δ mutant, 27SA and 27S pre-rRNA intermediates accumulated at 30°C and their levels decreased at 37°C. The levels of 20S pre-rRNA stabilized at 20°C and 30°C but not at 37°C in the double mutants compared to the single mutants (rrp6Δ and trf4Δ). Stabilization of the 23S pre-rRNA was also observed in trf4Δ pat1Δ at all the temperatures. Interestingly, while the mature 25S to 18S rRNA ratio was altered in the trf4Δ pat1Δ strain at 30°C, there was only a modest growth defect. This suggests that the synthetic sickness observed in these mutants might not be linked to RB ( and S4). We envisage that the synthetic sick growth of trf4Δ pat1Δ is due to either undetected intermediates, which are not eliminated by the TRAMP complex or alternatively a TRAMP independent role of Trf4.
The nuclear exosome also promotes the degradation of abnormal rRNA processing intermediates. As shown previously, we also observed the accumulation of decay products in both the rrp6Δ and pat1Δ rrp6Δ mutants (, rrp6Δ and pat1Δ rrp6Δ lanes with products indicated with *).Citation40 In the trf4Δ single mutant, similar decay intermediates were observed at 20°C ( with products indicated with ‘). We do not know the source of these decay products. Interestingly, in the pat1Δ trf4Δ double mutant, we observed an increase of these species indicating a possible role of Pat1 in Trf4 dependent decay of the pre-rRNA processing intermediates.
Figure 4. Domains of Pat1 that are required for mRNA decapping are also required for rRNA processing. (A) Growth of different truncation mutants of Pat1. Constructs containing different domains of Pat1 were transformed into pat1Δ mutant. Strains were grown in SDC–Ura medium overnight at 30°C. Cells were serially diluted 1:10, spotted on SDC–Ura plates and grown at the temperatures indicated. (B) In vivo mRNA decapping efficiency of Pat1 truncation mutants grown. Northern blots were probed for the MFA2pG gene with oTN121. The relative percentage of pG decay fragment to full-length MFA2pG is indicated. n = 3, Error bars and p-values as in rRNA processing in the Pat1 truncation mutants. Northern analysis performed as panel B. n = 3. The oligonucleotide probes used for probing are indicated in the brackets to the left. rRNA processing intermediates are indicated to the right. The relative amounts of the rRNA intermediates is given at the bottom of the each panel, corresponding to the order of the species on the rRNA northern, with the species in parenthesis given next to it. The schematic representation of the rRNA processing intermediates is depicted to the right of the indicated rRNA species. (D) Pat1 mutants are sensitive to 6-AU. Growth of the pat1 mutants was analyzed as in panel A. Cells were spotted on SDC–Ura plates or SDC–Ura+ 6-AU (50ug/ml) plates.
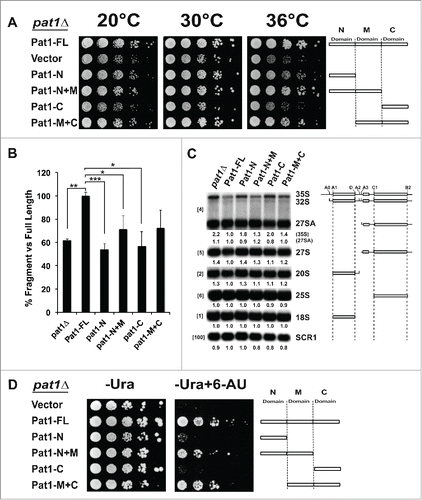
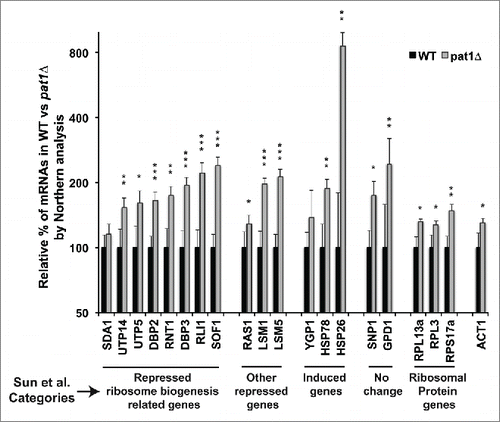
Figure 5. RB-related mRNAs are upregulated in pat1 deletion mutant. Quantification of the northern analysis of the mRNAs belonging to different classes of differentially expressed mRNAs in pat1Δ mutant from Sun et al. Citation17 normalized to SCR1 RNA. Total RNA from WT and pat1Δ strains grown at 30°C to 0.5 O.D was extracted and separated on a 1.2 % agarose gel. mRNAs indicated in the X-axis were probed with randomly primed probes made with primers listed in Table S3. Y-axis is plotted in log2 scale. Different categories of mRNAs from Sun et al. Citation17 examined in this study are indicated at the bottom of the graph. n = 3, Error bar = SD and p-values as given in .
We also considered the possibility that the abnormal rRNA precursors containing pre-ribosomes generated in the pat1Δ mutant might be exported to the cytoplasm where they could be detected by the nonfunctional rRNA decay pathway that recognizes ribosomes with aberrant rRNA.Citation43 This pathway relies on the Dom34 and Hbs1 proteins to recognize faulty 40S ribosomes and target them for destruction by Xrn1 and the cytoplasmic exosome. To determine whether a genetic relationship exists between PAT1 and DOM34 or HBS1, we monitored the growth of the individual deletion mutants, and those combined with the pat1Δ mutation at various temperatures. The growth assays demonstrated no synthetic genetic relationship between Pat1 and nonfunctional rRNA decay pathway proteins Dom34 and Hbs1 (Fig. S3).
Pat1 mutants that are defective in decapping also exhibit rRNA processing defects
Next, we tested if the mRNA decapping function of Pat1 and RB are linked. Pat1 affects multiple processes of gene expression. Pat1 for example, not only strongly affects mRNA decapping but can also alter translation and transcription.Citation5,7,12 Previous studies have examined the role of different truncation mutants of Pat1 in mRNA decapping, translation repression and protein-protein interaction.Citation5,9
The Middle or M-domain of Pat1, consisting of amino acids 254–422, was shown to be required but not sufficient for decapping.Citation9 When expressed alone, the M-domain behaved like a pat1 null mutant in decapping ability (unpublished data). The N-terminal or N-domain (1–254) and C-terminal or C-domain (422–796) were proposed to enhance the ability of M-domain to promote decapping. However, the M and C-domains together were demonstrated to be the most important for translational repression in vitro.Citation5,9
First, we tested the growth behavior of Pat1 truncation mutants. We wished to examined them in the background used in our study (BY4741) to confirm that they perform the same as in the different strain background previously used.Citation9,44 As shown in Pilkington et al. Citation9, at 36°C, the pat1Δ mutant demonstrates growth inhibition ().Citation9 Consistent with the previous results, all the mutants lacking the M-domain showed growth defects, although to differing extents. While our data qualitatively agrees with Pilkington et al., some strain specific differences may account for the pat1Δ strain and the mutants lacking the M-domain being unable to grow at 35°C and the growth inhibition in other mutants being much more pronounced than in BY4741 than in the strain used by Pilkington et al.Citation9
Since the pat1Δ mutant in our strain background exhibited growth differences compared to the one previously examined, we wished to confirm the mutants linkage to mRNA decapping in BY4741 strain. mRNA decapping in vivo can be assessed using a reporter mRNA containing a poly(G) tract in its 3′ UTR, which serves as a steric block to 5′ to 3′ degradation and results in the generation of an mRNA decay intermediate or decay fragment (Fig. S5). As the generation of the fragment is analogous to the efficiency of decapping, the levels of this fragment compared to the full-length mRNA represents an accurate measure of the ability of mRNA decapping.Citation9 Using the MFA2 mRNA, we found that the M-domain is required for decapping, and that the N and M promote decapping to differing extents (), similar to published results.Citation9
Since we have determined the role of the truncations in decapping, we then investigated how different domains of Pat1 affect pre-rRNA processing. We observed 4 main results from these analyses (). First, 35S pre-rRNA, 27S pre-rRNA and 20S pre-rRNA clearly accumulated in the pat1Δ strain with no significant decrease in the mature 25S and 18S rRNAs (). Second, the truncation mutants lacking the M-domain accumulated 35S pre-rRNA (). Third, deletion of both the M and C-domain together showed similar defects as deletion of the entire PAT1 gene (). Finally, the Pat1 N+M or M+C domains did not fully rescue the rRNA processing defects, suggesting that the N-domain and the C-domain are also needed for efficient rRNA processing. In conclusion, these results show that the mutants of Pat1 that are deficient in mRNA decapping are also defective in rRNA processing, indicating that these 2 processes are linked.
Pat1 is linked to transcription
One possibility for the delay in ribosomal processing when Pat1 is absent could be due to RNA polymerase transcription. Pat1 has been shown to facilitate transcription elongation in yeast.Citation12 To evaluate how the different truncation mutants of Pat1 affect transcription elongation, we expressed the Pat1 mutants in pat1Δ strain and examined their growth on media containing 6-azauracil (6-AU). 6-AU is a transcriptional elongation inhibitor that depletes the intracellular GTP pools. Mutants that are deficient in Pol I or Pol II transcriptional elongation are hypersensitive to 6-AU.Citation45,46
The Pat1 domains required for efficient decapping ability are also required for transcriptional elongation as assessed by 6-AU sensitivity. Loss of Pat1 leads to complete inhibition of growth in 50µg/ml 6-AU. The Pat1 mutants’ growth inhibition was correlated with the amount of decapping defect observed, with the M-domain being most important (Compare ). Similarly, it was previously found that Pat1 lacking the M-domain was also defective in chromosome segregation.Citation20 These results indicate that, as suggested by previous studies, regulation of mRNA decay by Pat1 is linked to its role in transcriptional elongation.
Lack of Pat1 leads to increase in mRNA abundance
To further examine if transcription could be the source of the rRNA processing defects, we analyzed transcriptomics data from Sun et al.Citation17 This study calculated the total abundance, transcription rate and decay rate of around 4000 mRNAs in 46 deletion mutants that affect mRNA decay, including pat1Δ. We analyzed the mRNAs that had been significantly reduced in abundance in the pat1Δ mutant, and found a significant enrichment in GO categories involved in RB (Fig. S6). This decrease in abundance is due to reduced transcription in the pat1Δ mutant in the study. These results suggest that the rRNA processing defects in pat1Δ could be attributable to decrease in the levels of these RB mRNAs and that the M-domain of Pat1 is important for to transcription of these genes.
To validate our hypothesis, we selected 8 RB-related mRNAs that were especially decreased in the Sun et al. dataset in the pat1Δ mutant and analyzed them by northern analyses. Surprisingly, the levels of these mRNAs increased significantly in the pat1Δ mutant as compared to WT strain (). We then selected an additional set of mRNAs that are non-RB-related. Again, we saw a significant increase in mRNA abundance of these genes. We next examined 3 mRNAs increased in abundance in the Sun et al. data set. These mRNA increased as well, although more modestly than reported except for the HSP26 mRNA. Finally, we wished to examine whether the mRNA encoding for ribosomal protein are also increased in the pat1Δ mutant. We selected 3 genes, and found only a slight increase of their levels. When normalized to ACT1 mRNA instead of SCR1 RNA, no significant difference in their levels between the pat1Δ and WT strains was observed (Fig. S7). We could not compare the mRNA encoding for ribosomal protein levels to the published transcriptomics dataset, as they were largely absent in the study.
To eliminate the possibility of strain specific differences being the cause of the discrepancy, some of the above tested mRNAs were analyzed in the same pat1Δ strain used to obtain the Sun et al. data set. We observed a similar increase in the levels of RB genes related mRNAs and no change in the levels of ribosomal protein gene mRNAs consistent with the results with our strain (data not shown). To test if this increase in the levels of mRNA is due to lack of efficient mRNA decapping, we analyzed the amounts of full-length mRNAs in the decapping assay in Fig. S5. The strain lacking Pat1 accumulated at least 4-fold more full-length mRNA compared to the strain having a genomic copy of PAT1 (Figs. S5A and S5B).
These results suggest that the mRNA levels are dysregulated in the pat1Δ mutant, presumably due to the effects of mRNA decapping and mRNA decay. We found that ribosomal biogenesis mRNAs tend toward significant increases in abundance, while the ribosomal protein mRNAs are not increased. This discordance at the mRNA level may be the source of the biogenesis defect.
To further confirm our hypothesis that the increase in mRNA levels is due to the lack of efficient decapping in the pat1Δ mutant, we examined and compared other mutants in 5′-to-3′ decay pathway to that of the pat1Δ deletion strain by northern analysis. Similar to the pat1Δ deletion mutant, the levels of the RB mRNAs were raised in xrn1Δ and lsm1Δ mutants (Fig. S8). Taken together, these results are consistent with Pat1 affecting the levels of RB mRNAs through decapping.
Discussion
Since its discovery as a topoisomerase II interacting protein, Pat1 has been implicated to have a role in the nucleus (Fig. S1) Citation15,16. Although, subsequent studies have primarily concentrated on its cytoplasmic role as a translational repressor and mRNA decapping activator, several recent studies have provided evidence and proposed that Pat1 could have a function in the nucleus.Citation5-7,12,14 One line of evidence for Pat1 entering the nucleus is that Pat1 accumulates there when the Lsm1 protein is absent.Citation13 The nuclear accumulation of Pat1 in the lsm1Δ mutant could be due to loss of cytoplasmic anchoring, perhaps via P bodies.Citation13
In this work, we investigated these roles and show that the absence of Pat1 does not affect splicing or nuclear pre-mRNA degradation. Interestingly, it affects rRNA processing resulting in abnormal intermediates, generated from multiple steps, especially the 35S, 27S and 20S pre-rRNAs ().
The ribosome biogenesis defects of the pat1Δ and other mutants we examined are largely consistent with the previously published data on the lsm1Δ, lsm6Δ and lsm7Δ mutants.Citation24 Namely, the defects in processing of the 35S rRNA was delayed, which is a commonly observed defect in ribosome biogenesis mutants.Citation36 We found an increase in the 20S pre-rRNA levels at 30°C for the pat1Δ mutant and lsm1Δ mutant. As lsm1Δ mutant was shown to have decreased levels of 20S at 37°C, we observed that the amount of the 20S pre-rRNA decreased in the pat1 Δ mutant at 37°C (Fig. S4).Citation24 Similarly, the lsm6 and lsm7 mutants have 20S accumulation at 20°C by northern blot analyses in another study.Citation36 Taken together, the lsm1Δ has broadly similar defects in rRNA processing to the pat1Δ mutant.
These results suggest that Pat1 does not affect a single step in RB, but rather affects the process of rRNA processing globally. Nevertheless, absence of Pat1 could allow generation of products that are recognized by the ribosome quality control machinery and not be revealed through our steady-state and pulse-chase analyses as has been previously described.Citation47 In this study, we showed that Pat1 has a synthetic sick relationship with Rrp6 and Trf4. Despite these synthetic relationships, we did not uncover any clear rRNA processing defects that could account for the synthetic sickness.
Pat1 has been shown to shuttle between the nucleus and the cytoplasm, and can recognize oligoadenylated (oligo(A)) RNAs. Therefore, it is possible that Pat1 together with Lsm1 facilitates the recognition of the short oligo(A) tails that are added by the TRAMP complex in the nucleus. This interaction may facilitate the processing of the TRAMP dependent oligoadenylated RNAs such as non-coding RNA, snoRNAs etc. or other alternate functions of the TRAMP complex.Citation12,14,48 Strengthening our hypothesis, we also observed an increase in the levels of snoRNAs in pat1Δ mutant (, Fig. S2).
Other roles proposed for the TRAMP complex and the exosome include termination of snoRNA transcription, determination of mRNA poly(A) length and control of pervasive transcription of noncoding RNA in general as well as rDNA copy number in particular.Citation49-52 The most likely linkage of Pat1 to the TRAMP complex may be in control of rDNA copy number. Evidence supporting this is that the pat1Δ mutant mimics the rDNA recombination phenotype of top1Δ and top2Δ mutants. Furthermore, this is consistent with several studies reporting that top1Δ is either synthetically sick or lethal with trf4Δ.Citation52-55
A possibility exists that the Pat1/Lsm1-7 complex could directly interact with rRNA processing intermediates, suggested by its ribosomal association and precipitation of 20S pre-rRNA with Lsm1.Citation24 However, based on the observations that Pat1 affects multiple steps in rRNA processing, we theorized that lack of Pat1 might lead to down-regulation of transcription of RB genes as a class. Such a model was suggested by our analysis of a recent study that had examined the effect of a pat1 deletion on mRNA abundance, decay and transcription.Citation17 In their data, we found that the transcription of mRNAs related to RB was down regulated leading to a decrease in the total abundance of these mRNAs in pat1Δ mutant (Fig. S6).
Surprisingly, when we validated these results by northern analyses, the total abundance of most RB mRNAs that we examined was instead increased in the pat1Δ mutant. This increase was seen both with normalization against the non-coding RNA component of the signal recognition particle, scR1 and with ACT1 mRNA (). However, the study observed that the absence of Pat1 reduced transcription genome-wide.Citation17 Similarly, GAL1 transcription assays in another study suggested that Pat1 promotes transcription elongation.Citation12 Taking together, these data suggest that in the absence of Pat1, there should be reduced transcription, while we find the opposite result for the ribosomal biogenesis genes. Although previous studies and our results demonstrated that Pat1s role in transcriptional elongation is linked to its role in mRNA degradation, the increase in total mRNA abundance of RB mRNAs tested here suggest that the dysregulation of mRNA levels in the pat1Δ mutant is due to poor mRNA decapping and as result reduced decay. Increase in RB mRNA levels in other decapping pathway mutants, xrn1Δ and lsm1Δ, strongly support our assumption that the ribosome biogenesis defects observed in pat1Δ strain are due to defects in mRNA decay (Fig. S8). However, it should be noted that both Xrn1 and Lsm1 have been previously reported as ribosome biogenesis factors with defects in rRNA proccesing.Citation35,56,57 Specifically, the deletion of xrn1 affects processing at the A0, A1, A2 and A3 sites as well as the 5′ end processing of the 27SA3 and 5.8S rRNAs. For its part, the absence of Lsm1 delays A0 and A1 cleavages and affects 20S rRNA levels.
Interestingly, while we found most RB genes were up regulated in the pat1Δ mutant, the ribosomal protein encoding mRNAs were not elevated in the pat1Δ mutant when normalized to ACT1 mRNA (Fig. S7). These data suggest a possibility that Pat1 specifically targets the RB genes for degradation and thus these genes are elevated in its absence. Similarly, mRNA encoding ribosomal biogenesis factors and ribosomal proteins have been shown previously to have different decay profiles.Citation58 It is tempting to speculate that Pat1 could control this differential decay, which was observed most significantly upon glucose deprivation, where Pat1 plays a role in translational repression.Citation7 Such a regulation of the decay of a class of mRNA was first suggested from a genome-wide study of mRNA decay.Citation59 By specifically looking at mRNA degradation genome-wide, ribosome biogenesis factors were suggested to be regulated at the level of mRNA stability,Citation60 which is supported by our results. Further analysis of the dataset Citation17 combined with analysis of UTR lengths revealed a motif specific for rapid degradation of RB factors.Citation61 This sequence is similar to that recognized by the Pat1/Lsm1-7 complex, suggesting a possibility that Pat1 might play a role in the differential decay of RB and ribosomal proteins' mRNAs.Citation62
The increase in the levels of mRNA for RB factors may alter the composition of the RB machinery to assemble and process ribosomes. These results suggest a model in which ribosomal biogenesis is a sensitive sensor for correct stoichiometry between ribosomal components, since this process has more than 200 trans-acting factors.Citation63 Consistent with this proposition, in a genome-wide study, overexpression of 4 of the top 5 most up-regulated mRNAs involved in ribosome biogenesis that we examined were either lethal or sick.Citation64 While many studies have examined the use of conditional promoters to deplete ribosomal biogenesis factors’ mRNAs, few have examined the effect of over-expression. However, overexpression of RB transcription factors such as Sfp1 causes toxicity concomitant with the induction of RB mRNA.Citation65
In conclusion, our results suggest that Pat1s role in the cell is predominantly in mRNA degradation rather than in transcription. Pat1 appears to affect the levels of mRNA encoding ribosomal proteins and those encoding RB biogenesis factors. These data support a model in which Pat1 regulates mRNA mRNA stability differentially among different classes of mRNAs in a coordinated fashion to modulate gene expression.
Materials and methods
Yeast strains, plasmids, and cell culture
The strains and plasmids used in this work are listed in Tables S1 and S2 respectively. Double deletion mutants were constructed by crossing two respective isogenic single mutants of opposing mating type. Strains used in this study were cultured using rich YEPD or synthetic complete medium lacking the appropriate amino acids. Unless otherwise mentioned cells were inoculated from overnight cultures to 0.05-0.08 OD in fresh medium, grown at 30°C and harvested at ∼0.5 O.D. For decapping assay, cells were grown in SC -Ura –Leu with 2% sucrose overnight. Cells were washed twice and inoculated to 0.05 O.D in SC -Ura –Leu with 2% galactose to induce the expression of pGAL-MFA2pG and were harvested at ∼0.3 O.D.Citation66
Live cell imaging
Cells were grown in SDC-His liquid medium to exponential phase at 30°C. Fluorescence images were captured using Nikon eclipse 90i microscope with Hamamatsu ORCA-ER digital camera. Pictures were then processed using ImageJ and Adobe Illustrator CS6.Citation67
Yeast growth assay
Strains were grown overnight at 30°C in the appropriate medium. 10ml of overnight grown cultures were harvested and re-suspended to1.0 O.D. Cells were 10-fold serially diluted and ∼3µl of each dilution was spotted with a pin tool (V&P Scientific) onto appropriate plates. Plates were incubated at the indicated temperatures for 2 d (30°C or 37°C) or 4 d (20°C).
Northern analysis
Total RNA extractions were performed as described in Citation68 with minor modifications. Unless mentioned otherwise, 10 µg of total RNA was analyzed by either 1.2% formaldehyde agarose gel or 6% polyacrylamide/7.5 M urea gel. RNA was either capillary transferred or electro blotted onto a nylon membrane.Citation69 Northern analysis was performed with oligonucleotide probes or random labeled probes. The oligonucleotides and primers used for the preparation of probes in this work are listed in Table S3.
Pulse–chase analysis
The pulse-chase analysis was done as described Citation37 with modifications. WT and pat1Δ strains were grown to 0.4–0.5 O.D at 30°C, harvested, and re-suspended to 1.0 O.D in SDC-Ura liquid medium. Cells were further grown for 30 minutes before they were pulse-labeled for 1 min with 3H uracil and then chased with excess of nonradioactive uracil. At the indicated times, 800µl sample was transferred to a 2ml Eppendorf half filled with crushed ice placed in ice. Samples were spun down quickly and frozen in dry ice.Citation70 Total RNA was isolated by the hot phenol method as described in.Citation71 20,000 cpm/sample of RNA was separated on a 1.2% formaldehyde agarose gel and transferred to a Hybond-N membrane (GE) by capillary transfer overnight. The membrane was UV-cross-linked, sprayed with En3HanceTM (PerkinElmer) according to the manufacturer's instructions and exposed to a film for 5 d at −80°C.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Data.pdf
Download PDF (64.9 MB)Acknowledgements
We would like to thank Marcus Johansson for helpful comments on the manuscript. We thank Firoj Mohmud for his suggestions for the data analysis. We also thank Roy Parker, Paul Herman, Patrick Cramer and Anders Byström for strains and plasmids.
Funding
This work was supported by Vetenskapsrådet - The Swedish Research Council [621-2010-4602 to T.N.]; Umeå University and the Carl Tryggers Stiftelse [CTS 11:330 to T.N.].
References
- Pérez-Ortín JE, de Miguel-Jiménez L, Chávez S. Genome-wide studies of mRNA synthesis and degradation in eukaryotes. Biochim Biophys Acta 2012; 1819:604–15; PMID:22182827; http://dx.doi.org/10.1016/j.bbagrm.2011.12.002
- Balagopal V, Fluch L, Nissan T. Ways and means of eukaryotic mRNA decay. Biochim Biophys Acta 2012; 1819:593–603; PMID:22266130; http://dx.doi.org/10.1016/j.bbagrm.2012.01.001
- Parker R, Song H. The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 2004; 11:121–7; PMID:14749774; http://dx.doi.org/10.1038/nsmb724
- Franks TM, Lykke-Andersen J. The Control of mRNA Decapping and P-Body Formation. Mol Cell 2008; 32:605–15; PMID:19061636; http://dx.doi.org/10.1016/j.molcel.2008.11.001
- Nissan T, Rajyaguru P, She M, Song H, Parker R. Decapping Activators in Saccharomyces cerevisiae Act by Multiple Mechanisms. Molecular Cell 2010; 39:773–83; PMID:20832728; http://dx.doi.org/10.1016/j.molcel.2010.08.025
- Marnef A, Standart N. Pat1 proteins: a life in translation, translation repression and mRNA decay. Biochem Soc Trans 2010; 38:1602; PMID:21118134; http://dx.doi.org/10.1042/BST0381602
- Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell 2005; 122:875–86; PMID:16179257; http://dx.doi.org/10.1016/j.cell.2005.07.012
- Holmes LEA, Campbell SG, De Long SK, Sachs AB, Ashe MP. Loss of Translational Control in Yeast Compromised for the Major mRNA Decay Pathway. Mol Cell Biol 2004; 24:2998–3010; PMID:15024087; http://dx.doi.org/10.1128/MCB.24.7.2998-3010.2004
- Pilkington GR, Parker R. Pat1 Contains Distinct Functional Domains That Promote P-Body Assembly and Activation of Decapping. Mol Cell Biol 2008; 28:1298–312; PMID:18086885; http://dx.doi.org/10.1128/MCB.00936-07
- Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature 2000; 404:515–8; PMID:10761922; http://dx.doi.org/10.1038/35006-676
- Bouveret E, Rigaut G, Shevchenko A, Wilm M, Séraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J 2000; 19:1661–71; PMID:10747033; http://dx.doi.org/10.1093/emboj/19.7.1661
- Haimovich G, Medina DA, Causse SZ, Garber M, Millán-Zambrano G, Barkai O, Chávez S, Pérez-Ortín JE, Darzacq X, Choder M. Gene Expression Is Circular: Factors for mRNA Degradation Also Foster mRNA Synthesis. Cell 2013; 153:1000–11; PMID:23706738; http://dx.doi.org/10.1016/j.cell.2013.05.012
- Teixeira D, Parker R. Analysis of P-body assembly in Saccharomyces cerevisiae. Mol Biol Cell 2007; 18:2274–87; PMID:17429074; http://dx.doi.org/10.1091/mbc.E07-03-0199
- Marnef A, Weil D, Standart N. RNA-related nuclear functions of human Pat1b, the P-body mRNA decay factor. Mol Biol Cell 2011; 23:213–24; PMID:22090346; http://dx.doi.org/10.1091/mbc.E11-05-0415
- Wang X, Watt PM, Louis EJ, Borts RH, Hickson ID. Pat1: a topoisomerase II-associated protein required for faithful chromosome transmission in Saccharomyces cerevisiae. Nucleic Acids Res 1996; 24:4791–7; PMID:8972867; http://dx.doi.org/10.1093/nar/24.23.4791
- Wang X, Watt PM, Borts RH, Louis EJ, Hickson ID. The topoisomerase II-associated protein, Pat1p, is required for maintenance of rDNA locus stability in Saccharomyces cerevisiae. Mol Gen Genet 1999; 261:831–40; PMID:10394921; http://dx.doi.org/10.1007/s004380050027
- Sun M, Schwalb B, Pirkl N, Maier KC, Schenk A, Failmezger H, Tresch A, Cramer P. Global Analysis of Eukaryotic mRNA Degradation Reveals Xrn1-Dependent Buffering of Transcript Levels. Mol Cell 2013; 52:52–62; PMID:24119399; http://dx.doi.org/10.1016/j.molcel.2013.09.010
- Hurto RL, Hopper AK. P-body components, Dhh1 and Pat1, are involved in tRNA nuclear-cytoplasmic dynamics. RNA 2011; 17:912–24; PMID:21398402; http://dx.doi.org/10.1261/rna.2558511
- Kramer EB, Hopper AK. Retrograde transfer RNA nuclear import provides a new level of tRNA quality control in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 2013; 110:21042–21047; PMID:24297920; http://dx.doi.org/10.1073/pnas.1316579110
- Mishra PK, Ottmann AR, Basrai MA. Structural integrity of centromeric chromatin and faithful chromosome segregation requires Pat1. Genetics 2013; 195:369–79; PMID:23893485; http://dx.doi.org/10.1534/genetics.113.155291
- Sharif H, Conti E. Architecture of the lsm1-7-pat1 complex: a conserved assembly in eukaryotic mRNA turnover. Cell Rep 2013; 5:283–91; PMID:24139796; http://dx.doi.org/10.1016/j.celrep.2013.10.004
- He W, Parker R. Functions of Lsm proteins in mRNA degradation and splicing. Curr Opin Cell Biol 2000; 12:346–50; PMID:10801455; http://dx.doi.org/10.1016/S0955-0674(00)00098-3
- Kufel J, Bousquet-Antonelli C, Beggs JD, Tollervey D. Nuclear pre-mRNA decapping and 5' degradation in yeast require the Lsm2-8p complex. Mol Cell Biol 2004; 24:9646–57; PMID:15485930; http://dx.doi.org/10.1128/MCB.24.21.9646-9657.2004
- Kufel J. Lsm Proteins Are Required for Normal Processing and Stability of Ribosomal RNAs. J Biol Chem 2003; 278:2147–56; PMID:12438310; http://dx.doi.org/10.1074/jbc.M208856200
- Séraphin B. Sm and Sm-like proteins belong to a large family: identification of proteins of the U6 as well as the U1, U2, U4 and U5 snRNPs. EMBO J 1995; 14:2089–98; PMID:7744014
- Mayes AE. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J 1999; 18:4321–31; PMID:10428970; http://dx.doi.org/10.1093/emboj/18.15.4321
- Luhtala N, Parker R. LSM1 over-expression in Saccharomyces cerevisiae depletes U6 snRNA levels. Nucleic Acids Res 2009; 37:5529–36; PMID:19596813; http://dx.doi.org/10.1093/nar/gkp572
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell 2000; 102:765–75; PMID:11030620; http://dx.doi.org/10.1016/S0092-8674(00)00065-9
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 1999; 18:5399–410; PMID:10508172; http://dx.doi.org/10.1093/emboj/18.19.5399
- Tollervey D, Kiss T. Function and synthesis of small nucleolar RNAs. Curr Opin Cell Biol 1997; 9:337–42; PMID:9159079; http://dx.doi.org/10.1016/S0955-0674(97)80005-1
- Kufel J. A complex pathway for 3' processing of the yeast U3 snoRNA. Nucleic Acids Res 2003; 31:6788–97; PMID:14627812; http://dx.doi.org/10.1093/nar/gkg904
- Antic S, Wolfinger MT, Skucha A, Hosiner S, Dorner S. General and MicroRNA-Mediated mRNA Degradation Occurs on Ribosome Complexes in Drosophila Cells. Mol Cell Biol 2015; 35:2309–20; PMID:25918245; http://dx.doi.org/10.1128/MCB.01346-14
- Wyers F, Minet M, Dufour ME, Vo LT, Lacroute F. Deletion of the PAT1 gene affects translation initiation and suppresses a PAB1 gene deletion in yeast. Mol Cell Biol 2000; 20:3538–49; PMID:10779343; http://dx.doi.org/10.1128/MCB.20.10.3538-3549.2000
- Wu D, Muhlrad D, Bowler MW, Jiang S, Liu Z, Parker R, Song H. Lsm2 and Lsm3 bridge the interaction of the Lsm1-7 complex with Pat1 for decapping activation. Cell Res 2014; 24:233–46; PMID:24247251; http://dx.doi.org/10.1038/cr.2013.152
- Woolford JL, Baserga SJ. Ribosome Biogenesis in the Yeast Saccharomyces cerevisiae. Genetics 2013; 195:643–81; PMID:24190922; http://dx.doi.org/10.1534/genetics.113.153197
- Li Z, Lee I, Moradi E, Hung N-J, Johnson AW, Marcotte EM. Rational Extension of the Ribosome Biogenesis Pathway Using Network-Guided Genetics. PLoS Biol 2009; 7:e1000213; PMID:19806183; http://dx.doi.org/10.1371/journal.pbio.1000213
- KoS M, Koš M, Tollervey D. Yeast Pre-rRNA Processing and Modification Occur Cotranscriptionally. Molecular Cell 2010; 37:809–20; PMID:20347423; http://dx.doi.org/10.1016/j.molcel.2010.02.024
- Venema J, Tollervey D. Ribosome synthesis in Saccharomyces cerevisiae. Annu Rev Genet 1999; 33:261–311; PMID:10690410; http://dx.doi.org/10.1146/annurev.genet.33.1.261
- Kressler D, Linder P, la Cruz de J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol 1999; 19:7897–912; PMID:10567516; http://dx.doi.org/10.1128/MCB.19.12.7897
- Allmang C, Mitchell P, Petfalski E, Tollervey D. Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res 2000; 28:1684–91; PMID:10734186; http://dx.doi.org/10.1093/nar/28.8.1684
- Kuai L, Fang F, Butler JS, Sherman F. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 2004; 101:8581–6; PMID:15173578; http://dx.doi.org/10.1073/pnas.0402888101
- Wery M, Ruidant S, Schillewaert S, Lepore N, Lafontaine DL. The nuclear poly(A) polymerase and Exosome cofactor Trf5 is recruited cotranscriptionally to nucleolar surveillance. RNA 2009; 15:406–19; PMID:19141608; http://dx.doi.org/10.1261/rna.1402709
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell 2009; 34:440–50; PMID:19481524; http://dx.doi.org/10.1016/j.molcel.2009.04.017
- He W, Parker R. The yeast cytoplasmic LsmI/Pat1p complex protects mRNA 3′ termini from partial degradation. Genetics 2001; 158:1445–55; PMID:11514438
- Tansey WP. 6-azauracil sensitivity assay for yeast. CSH Protoc 2006; 2006; PMID:22484669
- Laribee RN, Hosni-Ahmed A, Workman JJ, Chen H. Ccr4-Not Regulates RNA Polymerase I Transcription and Couples Nutrient Signaling to the Control of Ribosomal RNA Biogenesis; PLoS Genetics; 11(3); e1005113; PMID:25815716; http://dx.doi.org/10.1371/journal.pgen.1005113
- Hage el A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev 2010; 24:1546–58; PMID:20634320; http://dx.doi.org/10.1101/gad.573310
- Chowdhury A, Kalurupalle S, Tharun S. Pat1 contributes to the RNA binding activity of the Lsm1-7-Pat1 complex. RNA 2014; 20:1465–75; PMID:25035297; http://dx.doi.org/10.1261/rna.045252.114
- Grzechnik P, Kufel J. Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Mol Cell 2008; 32:247–58; PMID:18951092; http://dx.doi.org/10.1016/j.molcel.2008.10.003
- Schmid M, Poulsen M, Olszewski P, Pelechano V, Saguez C, Gupta I, Steinmetz L, Moore C, Jensen T, Poulsen M, et al. Rrp6p Controls mRNA Poly(A) Tail Length and Its Decoration with Poly(A) Binding Proteins. Molecular Cell 2012; 47:267–80; PMID:22683267; http://dx.doi.org/10.1016/j.molcel.2012.05.005
- Tudek A, Porrua O, Kabzinski T, Lidschreiber M, Kubicek K, Fortova A, Lacroute F, Vanacova S, Cramer P, Stefl R, et al. Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol Cell 2014; 55:467–81; PMID:25066235; http://dx.doi.org/10.1016/j.molcel.2014.05.031
- Houseley J, Kotovic K, Hage El A, Tollervey D. Trf4 targets ncRNAs from telomeric and rDNA spacer regions and functions in rDNA copy number control. EMBO J 2007; 26:4996–5006; PMID:18007593; http://dx.doi.org/10.1038/sj.emboj.7601921
- Sadoff BU, Heath-Pagliuso S, Castaño IB, Zhu Y, Kieff FS, Christman MF. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics 1995; 141:465–79; PMID:8647385
- Castaño IB, Brzoska PM, Sadoff BU, Chen H, Christman MF. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae.
- Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 2006; 124:1069–81; PMID:16487579; http://dx.doi.org/10.1016/j.cell.2005.12.036
- Petfalski E, Dandekar T, Henry Y, Tollervey D. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol Cell Biol 1998; 18:1181–9; PMID:9488433; http://dx.doi.org/10.1128/MCB.18.3.1181
- Lindahl L, Bommankanti A, Li X, Hayden L, Jones A, Khan M, Oni T, Zengel JM. RNase MRP is required for entry of 35S precursor rRNA into the canonical processing pathway. RNA 2009; 15:1407–16; PMID:19465684; http://dx.doi.org/10.1261/rna.1302909
- Arribere J, Doudna J, Gilbert W. Reconsidering Movement of Eukaryotic mRNAs between Polysomes and P Bodies. Mol Cell 2011; 44:745–58; PMID:22152478; http://dx.doi.org/10.1016/j.molcel.2011.09.019
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO. Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA 2002; 99:5860–5; PMID:11972065; http://dx.doi.org/10.1073/pnas.092538799
- Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-Wide Analysis of mRNA Stability Using Transcription Inhibitors and Microarrays Reveals Posttranscriptional Control of Ribosome Biogenesis Factors. Mol Cell Biol 2004; 24:5534–47; PMID:15169913; http://dx.doi.org/10.1128/MCB.24.12.5534-5547.2004
- Shalgi R, Lapidot M, Shamir R, Pilpel Y. A catalog of stability-associated sequence elements in 3' UTRs of yeast mRNAs. Genome Biol 2005; 6:R86; PMID:16207357; http://dx.doi.org/10.1186/gb-2005-6-10-r86
- Chowdhury A, Tharun S. lsm1 mutations impairing the ability of the Lsm1p-7p-Pat1p complex to preferentially bind to oligoadenylated RNA affect mRNA decay in vivo. RNA 2008; 14:2149–58; PMID:18719247; http://dx.doi.org/10.1261/rna.1094208
- Kressler D, Hurt E, Baßler J. Driving ribosome assembly. Biochim Biophys Acta 2010; 1803:673–83; PMID:19879902; http://dx.doi.org/10.1016/j.bbamcr.2009.10.009
- Sopko R, Huang D, Preston N, Chua G, Papp B, Kafadar K, Snyder M, Oliver SG, Cyert M, Hughes TR, et al. Mapping Pathways and Phenotypes by Systematic Gene Overexpression. Molecular Cell 2006; 21:319–30; PMID:16455487; http://dx.doi.org/10.1016/j.molcel.2005.12.011
- Jorgensen P. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev 2004; 18:2491–505; PMID:15466158; http://dx.doi.org/10.1101/gad.1228804
- Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature 1994; 370:578–81; PMID:8052314; http://dx.doi.org/10.1038/370578a0
- Nissan T, Parker R. Analyzing P-bodies in Saccharomyces cerevisiae. Meth Enzymol 2008; 448:507–20; PMID:19111192; http://dx.doi.org/10.1016/S0076-6879(08)02625-6
- Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA 2001; 7:1717–27; PMID:11780629; http://dx.doi.org/10.1017/S135583820101994X
- Sambrook J, Russell DW. Sambrook: Molecular cloning. A laboratory manual. Third - Google Scholar. Cold pring Harbor Laboratory Press; 2001.
- Tollervey D. A yeast small nuclear RNA is required for normal processing of pre-ribosomal RNA. EMBO J 1987; 6:4169–75; PMID:3327689
- Zhou Y, Chen C, Johansson MJO. The pre-mRNA retention and splicing complex controls tRNA maturation by promoting TAN1 expression. Nucleic Acids Research 2013; 41:5669–78; PMID:23605039; http://dx.doi.org/10.1093/nar/gkt269
