ABSTRACT
RNA polymerase II (Pol II) pausing at promoter-proximal regions is a highly regulated step in the transcription cycle. Pause release is facilitated by the P-TEFb kinase, which phosphorylates Pol II and negative elongation factors. Recent studies suggest that P-TEFb (as part of the inhibitory 7SK snRNP) is recruited to promoter-proximal regions through interaction with KAP1/TRIM28/TIF1β to facilitate ‘on-site’ kinase activation and transcription elongation. Here, I discuss features of this model and future challenges to further hone our understanding of transcriptional regulation including Pol II pausing and pause release.
Introduction
RNA polymerase II (Pol II) activity can be regulated at several steps of the transcription cycle including pre-initiation, DNA melting, initiation, promoter clearance and elongation (reviewed inCitation1,2). However, more recently, it has become well-established that Pol II pauses at promoter-proximal regions, particularly 20–60 bp downstream of the transcription start site (TSS). Conversion of paused Pol II into the elongation competent form is a key rate-limiting step for the regulation of several biological processes including response to environmental cues, cell fate choice, differentiation, and development.Citation2-5 Pol II pausing and pause release are precisely regulated by the action of 2 sets of elongation factors: negative and positive, respectively. One key positive factor is the P-TEFb kinase, which is composed of a regulatory cyclin subunit (usually CycT1 or CycT2) and the catalytic cyclin-dependent kinase 9 (Cdk9) subunit.Citation6-8 P-TEFb phosphorylates the Pol II C-terminal domain (CTD), as well as the negative factors that promote Pol II stalling (the multi-subunit Negative Elongation Factor complex (NELF) and DRB sensitivity-inducing factor (DSIF)), transforming DSIF into a positive elongation factor and evicting NELF (reviewed inCitation6) ().
Figure 1. Simplified model of the factors controlling Pol II pausing and pause release. After assembly of the transcription pre-initiation complex, transcription initiation and synthesizes of a short nascent RNA chain (> 18 nt-long, shown in red), the negative elongation factors DSIF and NELF are recruited through cooperative interactions with nascent RNA and Pol II thereby promoting pausing downstream the TSS (arrow). P-TEFb phosphorylated in the kinase activating T-loop (P-T186) is then recruited to promoter-proximal regions where it phosphorylates NELF (which is evicted from the DSIF-Pol II complex), DSIF, and Pol II to facilitate Pol II pause release. In this simplified model, it is unclear how P-TEFb is recruited to promoter-proximal regions and what occurs after substrate phosphorylation. Since P-TEFb is known to travel with elongating Pol II, it is possible that P-TEFb transitions with phosphorylated Pol II into the transcribing unit allowing the recruitment of further P-TEFb molecules to the promoter-proximal region for subsequent rounds of elongation. Although not shown in this ‘minimalistic’ model, P-TEFb also assembles into the super elongation complex (SEC) during gene activation, but it is unclear whether P-TEFb incorporation into the SEC occurs before or after DSIF/NELF phosphorylation and pause release.
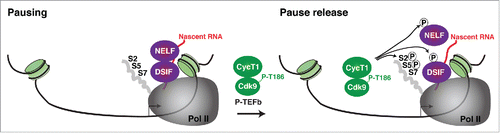
P-TEFb is both necessary and sufficient for Pol II pause release and transcription elongation because kinase inhibition produces a large increase in promoter-proximal Pol II at most regulated genes.Citation4,9,10 Although P-TEFb occupies promoter-proximal regions, one critical aspect of the activation mechanism that has not received much attention is the mode of P-TEFb recruitment to target genomic sites. Here, I discuss recent evidence for a molecular mechanism that facilitates the delivery of catalytically inactive/primed P-TEFb (as part of the 7SK small nuclear ribonucleoprotein (7SK snRNP) complex) to promoter-proximal regions to facilitate ‘on-site’ kinase activation and Pol II pause release.Citation11 Finally, I put these discoveries in the context of published literature, and discuss mechanistic insights and future challenges.
Building the 7SK snRNP complex
P-TEFb exists in at least 2 major forms that are in a reversible equilibrium in response to cell signaling to accommodate the transcriptional demands of the cell: (1) 7SK-unbound and catalytically active, and (2) 7SK-bound and catalytically inactive (reviewed inCitation12). The 7SK snRNP complex is composed of the 7SK non-coding RNA, the P-TEFb kinase, the kinase inhibitor Hexim1 and/or Hexim2 dimer (here referred as to Hexim for simplicity), the 5′-RNA methyl-capping enzyme MePCE, and the 3′-RNA stability protein Larp7Citation13-21 (). 7SK RNA is composed of 4 hairpins or stem-loops (stems I–IV) containing specific sites for the integral 7SK snRNP components.Citation16,22-25 Previous reports have shown that MePCE binds the proximal-region of stem I and Larp7 binds to the U-rich tail region of the 3′-terminal stem IV.Citation14,16,24 Then, MePCE and Larp7 form the core components on 7SK RNA (core 7SK snRNP) to provide structural stability to the 7SK complex, which does not disintegrate in response to cell signaling or transcription inhibitionCitation16,24,26,27 ().
Figure 2. Composition and assembly of the 7SK snRNP complex. The 7SK snRNP complex is composed of the 7SK non-coding RNA, the P-TEFb (CycT1:Cdk9) kinase, the kinase inhibitor Hexim, the 5′-RNA methyl-capping enzyme MePCE, and the 3′-RNA stability protein Larp7. Assembly of the 7SK snRNP is promoted by 7SK RNA binding by Hexim and recruitment of the P-TEFb kinase phosphorylated on the activating T-loop (P-T186).
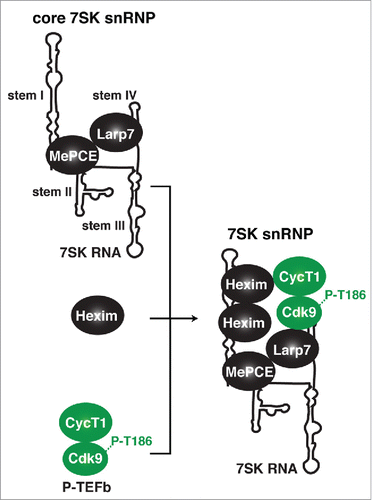
Systematic 7SK RNA mutagenesis showed that precluding the Hexim-7SK RNA interaction abolishes P-TEFb's recruitment to RNA, thereby indicating its absolute requirement for 7SK snRNP assemblyCitation23 and that Hexim-P-TEFb interacts on 7SK RNA to drive kinase inhibition.Citation18,19,28 Moreover, Larp7 also has an active role in facilitating P-TEFb recruitment to the 7SK RNA through its interaction with Cdk9.Citation24,29
Importantly, 7SK snRNP formation requires phosphorylation of the Cdk9 T-loop (at residue T186), which promotes interactions with Hexim and 7SK RNA in vitroCitation17,30 (). In support of this model, it was found that Cdk9 T-loop mutants were incapable of associating with Hexim-7SK RNA and assembling the inhibitory snRNP complex.Citation17,30 Therefore, Hexim physically inhibits the enzymatic activity of P-TEFb when bound to 7SK RNA and allows the cell to maintain gene expression equilibrium when the kinase is not transcriptionally engaged. Interestingly, T-loop phosphorylation is also what primes the kinase for activation, similar to other Cdks,Citation31 and this modification can be carried out by either P-TEFb's intrinsic auto-phosphorylation or by the TFIIH kinase (Cdk7) as first demonstrated by the Fisher lab.Citation32-34
Together, these previous studies led to the foundation for the functional characterization of the 7SK snRNP complex in controlling Pol II activity.
7SK snRNP recruitment to chromatin: ‘On-site’ kinase activation?
In rapidly growing cells, a large fraction of P-TEFb (up to 90%) exists in the catalytic inactive state bound to Hexim-7SK RNA (as part of the 7SK snRNP complex).Citation6,35 Therefore, the 7SK snRNP is a very abundant complex because it probably regulates most of both Pol II protein-coding and non-coding gene transcription. Previous studies have shown that the 7SK snRNP disassembles during activation of signaling cascades in response to environmental cues leading to an increase in free P-TEFb levels that positively correlate with gene activation (reviewed inCitation6,12). Given that the 7SK snRNP is highly abundant in the nucleoplasm, and that it appears not to stably associate with chromatin, it has been proposed that P-TEFb is globally released in this broad cellular compartment. However, it appears to be an inefficient mechanism for released P-TEFb to first diffuse around in the nucleoplasm before being attracted to promoter-proximal regions where it phosphorylates the negative elongation factors and paused Pol II to facilitate the transcriptional pause release ().
Most molecular reactions (such as enzyme-substrate recognition) are rapidly coordinated through factor compartmentalization.Citation36,37 This leads to the question of whether P-TEFb (which acts locally on chromatin): (i) is released at target genomic domains where its substrates exist, (ii) is released in the nucleoplasm before encountering chromatin, or (iii) is released by a combination of both mechanisms. While completely answering these questions will require further investigation, one previous study showed for the first time that the 7SK snRNP is recruited to the integrated HIV promoter where the kinase substrates exist.Citation38,39 Concomitantly, P-TEFb is released from the inhibitory 7SK snRNP complex during gene activation and the P-TEFb-free snRNP is repositioned or dislodged from chromatin ().
Figure 3. Model for 7SK snRNP recruitment to chromatin for ‘on site’ P-TEFb kinase activation. P-TEFb (as part of the 7SK snRNP complex) is recruited to promoter-proximal regions containing Pol II that remains paused by the action of the negative elongation factors DSIF and NELF. The 7SK snRNP complex can be disassembled on chromatin by the action of phosphatases activated in response to signaling pathways and dephosphorylate the T-loop of Cdk9 (P-T186). This step is not shown to keep the Figure simpler (seeCitation33,64 for examples). Upon 7SK snRNP disassembly, the P-TEFb-free snRNP becomes repositioned in the promoter-proximal region or dislodged from chromatin (an aspect of the model that requires further investigation).Citation33,38 Released P-TEFb can then phosphorylate the Pol II CTD on Ser5 (S5) and Ser2 (S2) residues, DSIF (which is converted into a positive elongation factor), and NELF (which becomes evicted from Pol II) thereby relieving Pol II stalling at promoter-proximal regions. It remains unclear if released P-TEFb rapidly incorporates into the SEC to phosphorylate its substrates or whether SEC assembly occurs after P-TEFb substrate phosphorylation at promoter-proximal regions.
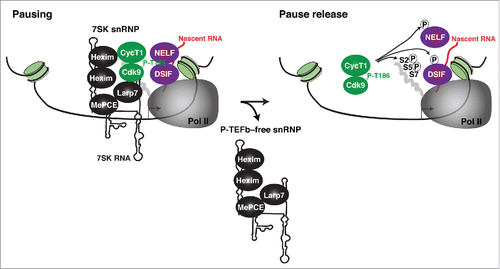
These discoveries were initially seen to be controversial in the field because of the low crosslinking efficiency of 7SK snRNP components compared to factors directly bound to chromatin. However, several subsequent studies extended this initial finding by providing compelling evidence that components of the 7SK snRNP complex are recruited to promoter-proximal regions of human and mouse genes.Citation33,40-42 This provides evidence that 7SK snRNP recruitment to chromatin is not unique to the viral promoter, but a general feature of cellular gene transcriptional control by Pol II. Because regulation of transcription occurs in the context of chromatin, it makes sense that kinase recruitment would be coupled with its activation on chromatin (‘on-site’ model) ().
To recruit or not to recruit the 7SK snRNP: Where, when and how?
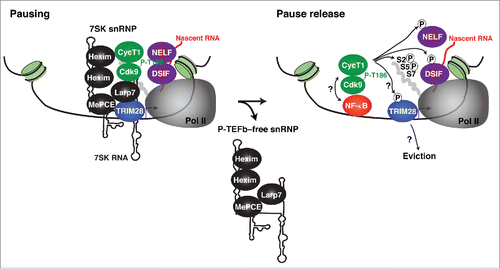
If the 7SK snRNP complex occupies promoter-proximal regions there has to be one or more factors that contribute to its recruitment. In a tour-de-force study, McNamara et al. used a biochemical approach (affinity-purification of 7SK snRNP components coupled with mass spectrometry analysis) and identified one candidate factor (the transcriptional regulator KAP1/TRIM28/TIF1βCitation11,43). TRIM28 interacted directly with the Larp7 subunit, which is constitutively bound to 7SK RNA.Citation16,26,27 Supporting the biochemical data, TRIM28 and the 7SK snRNP co-occupy the integrated HIV promoter in a manner that correlates with levels of paused Pol II and transcriptional output. Interestingly, loss of TRIM28 reduces levels of both TRIM28 (as expected) and the 7SK snRNP complex at the viral promoter at steady state. Importantly, the loss of TRIM28 did not alter levels of Pol II recruited to the promoter nor the assembly of the transcription pre-initiation complex (PIC) at promoter-proximal regions, thus suggesting that TRIM28 functions at a post-initiation step by recruiting the 7SK snRNP complex to promoter-proximal regions. The same results hold true for 2 NF-κB regulated genes, indicating that these cellular targets might be regulated in a similar manner.
Given the effect of TRIM28 knockdown in reduced Pol II elongation and gene activation at steady-state (basal conditions), McNamara et al. examined TRIM28s requirement for transcription in response to TNF stimulation (activated conditions). Consistent with the results obtained in basal conditions, loss of TRIM28 antagonized the recruitment of Cdk9 (via the 7SK snRNP complex) in response to the stimulus, but without affecting kinetics of transcription factor (NF-κB) recruitment. Reduced Cdk9 recruitment correlated with both decreased Pol II pause release and gene activation. Together, TRIM28 participates in the recruitment of inactive/primed P-TEFb to promoter-proximal regions to facilitate activation in both basal conditions and in response to stimulation ().
Figure 4. Model for the assembly of the TRIM28-7SK snRNP complex on chromatin: current knowledge and future challenges. TRIM28 recruits the 7SK snRNP complex to promoter-proximal regions containing the negative elongation factors (NELF and DSIF) that induce Pol II to pausing. In response to TNF stimulation, NF-κB is recruited to target promoters and P-TEFb is simultaneously released from the 7SK snRNP by the action of Cdk9 T-loop phosphatases (seeCitation33,64 for examples). Then, the precise molecular steps and ordered recruitment remain poorly understood. It is unclear whether NF-κB binds P-TEFb directly to stimulate Pol II pause release,Citation44 or via BRD4,Citation65,66 which could also help release P-TEFb from the 7SK snRNP in combination with the action of the Cdk9 T-loop phosphatases. In addition, the P-TEFb-free snRNP and TRIM28 appear to be dislodged from chromatin. It would be interesting to determine whether P-TEFb phosphorylates TRIM28 and whether this molecular event promotes its eviction from chromatin (similarly to NELF ejection from Pol II). The eviction of TRIM28 and the P-TEFb-free snRNP from chromatin could promote that released P-TEFb is ‘hands off’ to NF-κB,Citation44 or another subunit of the transcriptional apparatus that brings P-TEFb in proximity to its substrates to promote molecular crowding and kinase activity or promote SEC assembly. The question mark (?) is to indicate a lack of understanding of the highlighted steps in the transcription cycle. Further studies are required to precisely define these uncertainties.
During gene activation TRIM28 appears to cycle on and off promoters, indicating that TRIM28 may continuously tether the 7SK snRNP complex to deliver several kinase molecules. Now, if TRIM28 cycles on and off from chromatin, then the question is how does it stimulate gene activity? One possibility is that TRIM28 delivers P-TEFb to promoter-proximal regions and then is evicted from chromatin (probably similarly to NELF) to allow for further cycles of TRIM28-mediated P-TEFb recruitment to facilitate multiple rounds of transcription elongation. Another alternative scenario would be that TRIM28 leaves the promoter-proximal region along with Pol II (like DSIF) and that both TRIM28 and the 7SK snRNP complex have additional functions in transcription and/or pre-mRNA processing. Although the first evidence of TRIM28 cycling on/off promoters is tempting, additional experiments are needed to validate the proposed model by McNamara et al.Citation11
These discoveries suggest an interesting scenario. Given that P-TEFb associates with several transcription factors (such as NF-κB and c-Myc) it has been proposed that the kinase is recruited to promoters via the activator.Citation6,10,44 However, although NF-κB normally binds target gene promoters in response to stimulation, the P-TEFb kinase is not properly recruited in the absence of TRIM28. Therefore, future studies are needed to test a possible TRIM28 role as co-activator to ‘hand off’ the kinase to the transcription factor and/or another subunit in the transcription machinery. This possibility could provide evidence of an ordered recruitment mechanism as a paradigm for precisely controlling gene activation ().
Whether TRIM28 is the only or the dominant scaffold that mediates P-TEFb/7SK snRNP recruitment to promoter-proximal regions remains to be shown. Even though the results by McNamara et al. clearly favor a model in which TRIM28 is a critical factor in the recruitment of 7SK-bound P-TEFb to promoter-proximal regions to facilitate gene induction, at this point is not possible to formally exclude the involvement of additional mechanisms that regulate 7SK snRNP recruitment. In fact, Mizutani et al. proposed that the 7SK snRNP complex is recruited to the HIV promoter via newly-synthesized nascent RNAs arrested at +50–70 bp from the TSS,Citation45 but it remains unclear what makes the bridge between the nascent transcript and the inhibitory snRNP. Thus, the possibility that TRIM28 and/or additional factors contribute to placing the 7SK snRNP at these genomic sites certainly warrants further examination.
TRIM28-7SK snRNP: General or gene-specific co-activator?
Genome-wide studies revealed that the majority of human genes containing promoter-proximal paused Pol II also show evidence of TRIM28-7SK snRNP recruitment.Citation11 This suggests that the requirement of TRIM28 for 7SK snRNP recruitment to these genomic sites might be a widespread regulatory mechanism to deliver P-TEFb to facilitate Pol II pause release. Moreover, the presence of P-TEFb as part of the 7SK snRNP complex at gene promoters correlates with the degree to which Pol II pauses and transcription levels (as revealed by GRO-seq), probably indicating a direct role in positively controlling transcription elongation.Citation11,46 In this context, the 7SK snRNP complex appears not to be simply an inactive reservoir of P-TEFb but to play a functional role in delivering primed kinase to promoter-proximal regions containing paused Pol II.
McNamara et al. showed that loss of TRIM28 reduces RNA steady-state levels for only a subset of the genes examined. Since some of them, such as ACTB and GAPDH, produce highly abundant and long-lived transcripts, loss of TRIM28 does not appear to have an effect. However, examination of RNA levels through qPCR does not permit us to distinguish between effects at the transcriptional and/or post-transcriptional levels. Therefore, techniques that directly measure transcription in cells (such as metabolic labeling of newly transcribed RNAsCitation47) and run-on with isolated nuclei (GRO-seqCitation48) would be useful tools to determine if TRIM28-7SK snRNP function in a gene-specific or condition-specific manner rather than as a general transcription elongation factor.
In addition, TRIM28 belongs to a family of transcriptional regulators known as transcription intermediary factor 1 (TIF1) composed of 3 members: TIF1α/TRIM24, TIF1β/TRIM28, and TIF1γ/TRIM22.Citation49-51 Since TRIM22 controls transcription elongation of erythroid specific genes in a P-TEFb-dependent manner it would be interesting to determine whether these family members play unique roles at selected group of genes or fulfill similar roles at different genes classes to control pause release through 7SK snRNP recruitment and/or alternative mechanisms.
The dilemma: How does TRIM28 bind promoter-proximal regions?
The study by McNamara et al.Citation11 has found that TRIM28 is recruited to promoter-proximal regions genome-wide, consistent with previous evidence from the Farnham lab.Citation43 However, it is still unclear whether TRIM28 binds to the template DNA directly, through a bridging interaction (Pol II or other), by contacting post-translationally modified histone tails as part of nucleosome +1, or any non-mutually exclusive combination of these mechanisms.
One recent study linking TRIM28 to pausing and pause release identified TRIM28 as a factor that binds the non-template single-stranded DNA formed in the elongation bubble,Citation52 and contains a motif that was previously derived from genome-wide studies.Citation53 This study would argue that TRIM28 is recruited directly to the DNA. However, proving that this model is correct would require an identification of the TRIM28 DNA-binding domain and evidence of direct in vivo interaction. In addition, TRIM28 contains a bromodomain, a unique protein module that recognizes acetylated Lysines on histone tails, suggesting that TRIM28 could be recruited to promoter-proximal regions by directly contacting DNA and/or chromatin.
Historically, TRIM28 is known to interact with a family of Kruppel-associated box (KRAB)-containing Cys2-His2 zinc finger (C2H2-ZF) proteins to silence both exogenous retroviruses and specific endogenous retroelemens.Citation54 The majority of C2H2-ZF proteins, including KRAB proteins, show widespread binding to regulatory regions in the human genome, indicating that through the C2H2-ZF regulatory network the TRIM28-7SK snRNP complex could target a diverse range of genes and pathways. Certainly, while the possibility of direct DNA recognition first described by Bunch et al.Citation52 cannot be excluded, TRIM28 could use multiple surfaces to interact with DNA (alone or combinatorially), histone tails and/or the transcription machinery. Dissecting out these possible models will require a combination of biochemical, genetic and genomics approaches.
Nature repeats itself: The negative to positive transition?
It has been previously shown that TRIM28 is phosphorylated at the C-terminal domain (S824) to relieve negative interactions and to induce chromatin relaxation.Citation55 These previous observations probably prompted Bunch et al. to examine the role of TRIM28 site-specific phosphorylation on Pol II pausing and pause release. They found that S824 phosphorylation (possibly through the ATM or ATR PI3-kinases) appears to positively control Pol II pause release in vitro and in vivo, but only under specific conditions.Citation52
Much recently, Sanso et al. also provided preliminary evidence that the mechanism of TRIM28 site-specific phosphorylation could still hold true.Citation56 Using Shokat's ‘chemical genetic’ approach they identified TRIM28 as one substrate of the P-TEFb kinase. However, Ser residues in the N-terminal of TRIM28 rather than S824 in the C-terminus were identified as phospho-acceptor sites. In addition to the study by McNamara et al., these exciting discoveries would suggest that TRIM28 regulates Pol II pausing and pause release by controlling the recruitment of primed P-TEFb to promoter-proximal regions and then by acting as a substrate of the activated kinase ().Citation11,56
Together, these observations open the possibility that TRIM28 can also function as a negative elongation factor that promotes pausing probably by repressing P-TEFb through the 7SK snRNP at promoter-proximal regions. TRIM28 could then either be converted into a positive elongation factor through site-specific phosphorylation (like DSIF) or evicted from chromatin (like NELF) (). The preliminary evidence that TRIM28 cycles on and off from chromatin would imply that it can recruit the 7SK snRNP to promoters, become a substrate of the P-TEFb kinase (as soon as it is released from the 7SK snRNP) and then either travel with Pol II (like DSIF) or is dislodged from chromatin with or without the inhibitory 7SK snRNP subunits (like NELF).Citation33,38 Either mechanism would allow the subsequent recruitment of further TRIM28-7SK snRNP molecules aiding in the control of multiple rounds of elongation (necessary for robust gene induction), as previously suggested by McNamara et al.Citation11 Whether upon P-TEFb release from the 7SK snRNP, the core 7SK snRNP is dislodged from chromatin or repositioned at the promoter-proximal region is still unclear. One other possible scenario is that TRIM28 keeps the core 7SK snRNP at promoters and upon resolution of a signaling/activating event the elongation program is shut down by capturing active P-TEFb into the TRIM28-7SK snRNP complex assembled at promoters thereby contributing to Pol II pausing. Using an artificial gene reporter system, Prasanth et al. found that 7SK RNA transiently associates with the promoter-proximal region upon transcriptional down-regulation and its presence correlates with efficient displacement of P-TEFb from the locus and relocation into speckles.Citation57 It would be really interesting to define whether there are groups of genes regulated through the TRIM28-7SK snRNP ‘on site’ or through recruitment of the inactive complex to speckles or both. Testing these hypotheses would be critical for ascertaining the precise basis of the molecular mechanism through which TRIM28 and the 7SK snRNP control Pol II pausing and pause release.
Evolutionary analyses
A previous analyses of the evolutionary conservation of P-TEFb subunits as well as factors that control P-TEFb activity (DSIF, NELF and Hexim) by Peterlin and Price provided some insights into the development of the transcriptional control system.Citation6 In order to examine whether 7SK snRNP components and TRIM28 potentially co-evolved, I searched and analyzed their orthologs in model organisms such as zebrafish, fruit fly, sea urchin, worm and yeast (both budding and fission). Protein sequences of the canonical P-TEFb subunits (Cdk9 and CycT1), Hexim, core 7SK snRNP subunits (Larp7, MePCE, and 7SK RNA), and TRIM28 were identified in model organisms from different evolutionary branches of the phylogenetic tree and aligned. The percentages of amino acids that were identical and similar to their human counterpart were calculated (). Conservation varies for different 7SK snRNP components and TRIM28. CycT1 and Cdk9 orthologs were identified through budding and fission yeast, but Cdk9 orthologs were more conserved than the cyclin subunit, consistent with previous observations.Citation6 The kinase inhibitor Hexim is clearly present in sea urchins and higher organisms but is absent in worms and yeast, probably indicating a gain in the kinase inactivation control system in higher organisms. The three core 7SK snRNP components (Larp7, MePCE, and 7SK RNA) are present in zebrafish but absent in worms and yeast like Hexim. However, only Larp7 and MePCE (but not 7SK RNA) are present in fruit fly and sea urchin potentially suggesting they are not part of an evolutionary conserved Pol II elongation control mechanism. The presence of the core 7SK snRNP protein components Larp7 and MePCE suggest that they might be playing other roles or working through other non-coding RNAs not related to 7SK function. The sharp difference in conservation between P-TEFb and the rest of the 7SK snRNP complex subunits and TRIM28 also suggest that the kinase control mechanisms arose as complex organisms evolved. Therefore, the control of P-TEFb may be different in less complex organisms (yeast and worm), consistent with previous proposals by Peterlin and Price.Citation6
Figure 5. Phylogenetic analyses of the 7SK snRNP complex and TRIM28 in model organisms. Sequences from the indicated proteins and species were compared to human sequences. Human sequences for CycT1 (726 residues), Cdk9 (372 residues), Hexim (359 residues), Larp7 (582 residues), MePCE (689 residues), 7SK RNA (331 nucleotides), and TRIM28 (835 residues) were retrieved from the UniProt Knowledgebase (UniProtKB), blasted and aligned using the specified reference genome at the National Center for Biotechnology Information (NCBI). Only Hexim1, which is the most studied Hexim isoform, but not Hexim2, was analyzed. Zebrafish, Danio rerio; fruit fly, Drosophila melanogaster; sea urchin, Strongylocentrotus purpuratus; worm, Caenorhabditis elegans; budding yeast, Saccharomyces cerevisiae; fission yeast, Schizosaccharomyces pombe. The first and second (in bracket) numbers indicate the percentage (%) of identical and similar residues between the human and model organism sequences, respectively. Budding and fission yeast are known to have 2 kinases with similarity to Cdk9 (Ctk1 and Bur1). For simplicity, only analysis with the Cdk9 ortholog Ctk1 is shown. For 7SK RNA, only identical nucleotides were used. Abbreviations: NF, not found. The asterisk (*) indicates predicted orthologs since no functional characterization has yet been done.
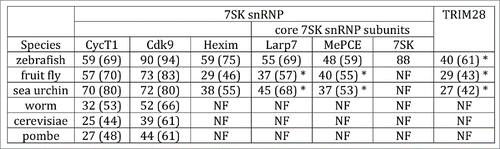
Similarly to Hexim and the core 7SK snRNP components (Larp7 and MePCE), TRIM28 predicted orthologs were found in zebrafish, fruit fly and sea urchin, potentially indicating co-evolution. However, the absence of 7SK RNA in fruit fly and sea urchin poses an interesting paradigm to this evolutionary conundrum. One possibility could be that Hexim, the core 7SK snRNP components, and TRIM28 function through a yet unknown non-coding RNA in fruit fly and sea urchin to regulate P-TEFb kinase activity. Alternatively, the control of P-TEFb may be different in those organisms. Given that the regulatory axis of P-TEFb kinase has not been found to date in worm and yeast, it is possible that inhibitory components of the 7SK snRNP and TRIM28 arose to regulate kinase activity, leading to a precise control of gene activation mechanisms. This description will surely be the beginning of more refined phylogenetic analyses to promote the discovery of alternative modes of P-TEFb regulation in other species as well as to characterize the TRIM28-7SK snRNP complex and their target genes in other model organisms.
Conclusions and challenges: The ephemeral nature of ideas
The current model poses that: (i) TRIM28 and the 7SK snRNP are present at most promoters where paused Pol II is located, (ii) recruitment of the 7SK snRNP complex to promoter-proximal regions allows for the localized release of P-TEFb for ‘on site’ kinase activation, and (iii) 7SK snRNP recruitment is continuously stimulated by TRIM28 to control transcriptional homeostasis and rapidly respond to environmental cues (). Despite these previous discoveries, the study by McNamara et al. spurs new questions. Given that only a subset of genes was examined to determine effects of TRIM28 loss on gene activation, genome-wide data would be critical to determine the role of TRIM28 in transcription elongation in both basal conditions and in response to various stimuli. In addition, despite the frequent co-occupancy of TRIM28 and the 7SK snRNP at promoter-proximal regions containing paused Pol II, it still remains unknown whether TRIM28 recruits the snRNP to most promoter-proximal regions and whether the TRIM28-7SK snRNP complex regulates pause release genome-wide. Does TRIM28-7SK snRNP play a widespread regulatory role at all paused Pol II target genes or does it regulate different gene classes with various degrees of dependencies? Furthermore, the mechanism through which TRIM28 is recruited to active promoters is still not clear, nor is the model for how this might stimulate transcription activation.
Genetic evidence of TRIM28-mediated 7SK snRNP recruitment to target genes and loss of gene activation only derives from TRIM28 knockdown experiments. Therefore, in order to prove that the sole (or at least the main) function of TRIM28 in gene activation is through the recruitment of primed P-TEFb kinase, more thorough genetic/biochemical evidence is needed. For example, it would be informative to engineer a TRIM28 mutant protein that loses binding to Larp7 (and thus compromises 7SK snRNP recruitment to promoter-proximal regions) without affecting TRIM28 recruitment to chromatin nor interaction with other key players. Importantly, this mutant could be used to pinpoint 7SK-dependent and -independent TRIM28 functions in the transcriptional cycle.
One other important point that remains to be addressed is how recruitment of an inactive/primed kinase (as part of the 7SK snRNP) is faster (or more efficient) than just recruiting the active form of the kinase itself or as part of the highly active super elongation complex (SEC).Citation58-60 One explanation might be that P-TEFb is not recruited faster but in an inactive state until encountering its substrates bound at promoter-proximal regions, probably to avoid spurious kinase activation and substrate phosphorylation. Approaches to measure dynamics of P-TEFb/7SK snRNP recruitment to target gene promoters will provide answers to these conundrums and illuminate possible unexplored facets in the control of Pol II pausing and pause release.
Although TRIM28 is an elongation factor, it was not previously identified as part of the SEC by the Shilatifard lab.Citation58-60 This result can be explained by the fact that TRIM28 does not bind P-TEFb directly, but through the 7SK snRNP subunit Larp7, and that only a small fraction (10–15%) of 7SK snRNP appears to be bound to TRIM28 in cells.Citation11 Thus, even though both 7SK snRNP and SEC contain P-TEFb, the selectivity of TRIM28 for the 7SK snRNP can be dictated by the presence of the inhibitory snRNP subunits. Together, this may also help explain why a previous Cdk9 immunoprecipitation-mass spectrometry analysis did not detect TRIM28 but 7SK snRNP subunits only.Citation59
Our studies have closely examined annotated transcripts but not divergent transcripts or non-coding RNAs. Therefore, it remains to be investigated whether these unique non-coding RNA species are also regulated by TRIM28 and the 7SK snRNP or if they have unique regulatory strategies. Interestingly, a study by the Sharp lab revealed that antisense Pol II divergent transcripts are P-TEFb dependent,Citation61 thus opening the possibility that TRIM28-7SK snRNP also control divergent transcription.
In addition to 7SK snRNP recruitment to promoter-proximal regions, the Rosenfeld lab proposed that the snRNP complex occupies transcriptional enhancers,Citation62 which are genomic elements that stimulate gene activation from promoter-distal sites (reviewed inCitation63). Through gene looping between enhancer and promoters, and a specialized set of chromatin-modifying enzymes, P-TEFb appears to be released from the 7SK snRNP complex. Notably, given that enhancers play an important role in controlling gene activation from promoters and because the 7SK snRNP appears to be recruited to these genomic sites,Citation62 studies are needed to further define the role of TRIM28-7SK snRNP in controlling transcription from enhancers during development, differentiation and cell fate choice. We believe that with these recent discoveries we lay a foundation for future studies.
With my efforts to discuss these recent discoveries and put them in the context of previously published literature I hope to provoke future work that will advance our understanding of this fascinating scientific problem. New data supporting or refuting the models discussed herein will undoubtedly increase our knowledge and generate discussions rather than engendering unproductive controversy. Even the more skeptical researchers should be attracted to challenge these models and bring their own points of view to fuel the progress in the field.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
I thank Leslie Spector, Matjaz Barboric and members of the D'Orso lab for critical reading of the manuscript. I am also indebted with the reviewers for providing constructive criticisms prior to publication.
Funding
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the NIH under award number R01AI114362 and Welch Foundation grant I-1782 to Iván D'Orso.
References
- Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol 2009; 21:344-51; PMID:19411170; http://dx.doi.org/10.1016/j.ceb.2009.03.006
- Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 2012; 13:720-31; PMID:22986266; http://dx.doi.org/10.1038/nrg3293
- Danko CG, Hah N, Luo X, Martins AL, Core L, Lis JT, Siepel A, Kraus WL. Signaling pathways differentially affect RNA polymerase II initiation, pausing, and elongation rate in cells. Mol Cell 2013; 50:212-22; PMID:23523369; http://dx.doi.org/10.1016/j.molcel.2013.02.015
- Henriques T, Gilchrist DA, Nechaev S, Bern M, Muse GW, Burkholder A, Fargo DC, Adelman K. Stable pausing by RNA polymerase II provides an opportunity to target and integrate regulatory signals. Mol Cell 2013; 52:517-28; PMID:24184211; http://dx.doi.org/10.1016/j.molcel.2013.10.001
- Smith E, Shilatifard A. Transcriptional elongation checkpoint control in development and disease. Genes Dev 2013; 27:1079-88; PMID:23699407; http://dx.doi.org/10.1101/gad.215137.113
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 2006; 23:297-305; PMID:16885020; http://dx.doi.org/10.1016/j.molcel.2006.06.014
- Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev 1998; 12:755-62; PMID:9499409; http://dx.doi.org/10.1101/gad.12.5.755
- Fu TJ, Peng J, Lee G, Price DH, Flores O. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J Biol Chem 1999; 274:34527-30; PMID:10574912; http://dx.doi.org/10.1074/jbc.274.49.34527
- Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem 2001; 276:31793-9; PMID:11431468; http://dx.doi.org/10.1074/jbc.M102306200
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell 2010; 141:432-45; PMID:20434984; http://dx.doi.org/10.1016/j.cell.2010.03.030
- McNamara RP, Reeder JE, McMillan EA, Bacon CW, McCann JL, D'Orso I. KAP1 Recruitment of the 7SK snRNP Complex to Promoters Enables Transcription Elongation by RNA Polymerase II. Mol Cell 2016; 61:39-53; PMID:26725010; http://dx.doi.org/10.1016/j.molcel.2015.11.004
- Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem 2012; 81:119-43; PMID:22404626; http://dx.doi.org/10.1146/annurev-biochem-052610-095910
- Barboric M, Kohoutek J, Price JP, Blazek D, Price DH, Peterlin BM. Interplay between 7SK snRNA and oppositely charged regions in HEXIM1 direct the inhibition of P-TEFb. EMBO J 2005; 24:4291-303; PMID:16362050; http://dx.doi.org/10.1038/sj.emboj.7600883
- He N, Jahchan NS, Hong E, Li Q, Bayfield MA, Maraia RJ, Luo K, Zhou Q. A La-related protein modulates 7SK snRNP integrity to suppress P-TEFb-dependent transcriptional elongation and tumorigenesis. Mol Cell 2008; 29:588-99; PMID:18249148; http://dx.doi.org/10.1016/j.molcel.2008.01.003
- Jeronimo C, Forget D, Bouchard A, Li Q, Chua G, Poitras C, Thérien C, Bergeron D, Bourassa S, Greenblatt J, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell 2007; 27:262-74; PMID:17643375; http://dx.doi.org/10.1016/j.molcel.2007.06.027
- Krueger BJ, Jeronimo C, Roy BB, Bouchard A, Barrandon C, Byers SA, Searcey CE, Cooper JJ, Bensaude O, Cohen EA, et al. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res 2008; 36:2219-29; PMID:18281698; http://dx.doi.org/10.1093/nar/gkn061
- Li Q, Price JP, Byers SA, Cheng D, Peng J, Price DH. Analysis of the large inactive P-TEFb complex indicates that it contains one 7SK molecule, a dimer of HEXIM1 or HEXIM2, and two P-TEFb molecules containing Cdk9 phosphorylated at threonine 186. J Biol Chem 2005; 280:28819-26; PMID:15965233; http://dx.doi.org/10.1074/jbc.M502712200
- Michels AA, Fraldi A, Li Q, Adamson TE, Bonnet F, Nguyen VT, Sedore SC, Price JP, Price DH, Lania L, et al. Binding of the 7SK snRNA turns the HEXIM1 protein into a P-TEFb (CDK9/cyclin T) inhibitor. EMBO J 2004; 23:2608-19; PMID:15201869; http://dx.doi.org/10.1038/sj.emboj.7600275
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell 2003; 12:971-82; PMID:14580347; http://dx.doi.org/10.1016/S1097-2765(03)00388-5
- Yik JH, Chen R, Pezda AC, Zhou Q. Compensatory contributions of HEXIM1 and HEXIM2 in maintaining the balance of active and inactive positive transcription elongation factor b complexes for control of transcription. J Biol Chem 2005; 280:16368-76; PMID:15713661; http://dx.doi.org/10.1074/jbc.M500912200
- Nguyen VT, Kiss T, Michels AA, Bensaude O. 7SK small nuclear RNA binds to and inhibits the activity of CDK9/cyclin T complexes. Nature 2001; 414:322-5; PMID:11713533; http://dx.doi.org/10.1038/35104581
- Wassarman DA, Steitz JA. Structural analyses of the 7SK ribonucleoprotein (RNP), the most abundant human small RNP of unknown function. Mol Cell Biol 1991; 11:3432-45; PMID:1646389; http://dx.doi.org/10.1128/MCB.11.7.3432
- Egloff S, Van Herreweghe E, Kiss T. Regulation of polymerase II transcription by 7SK snRNA: two distinct RNA elements direct P-TEFb and HEXIM1 binding. Mol Cell Biol 2006; 26:630-42; PMID:16382153; http://dx.doi.org/10.1128/MCB.26.2.630-642.2006
- Muniz L, Egloff S, Kiss T. RNA elements directing in vivo assembly of the 7SK/MePCE/Larp7 transcriptional regulatory snRNP. Nucleic Acids Res 2013; 41:4686-98; PMID:23471002; http://dx.doi.org/10.1093/nar/gkt159
- Muniz L, Egloff S, Ughy B, Jady BE, Kiss T. Controlling cellular P-TEFb activity by the HIV-1 transcriptional transactivator Tat. PLoS Pathog 2010; 6:e1001152; PMID:20976203; http://dx.doi.org/10.1371/journal.ppat.1001152
- Xue Y, Yang Z, Chen R, Zhou Q. A capping-independent function of MePCE in stabilizing 7SK snRNA and facilitating the assembly of 7SK snRNP. Nucleic Acids Res 2010; 38:360-9; PMID:19906723; http://dx.doi.org/10.1093/nar/gkp977
- Barboric M, Lenasi T, Chen H, Johansen EB, Guo S, Peterlin BM. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc Natl Acad Sci U S A 2009; 106:7798-803; PMID:19416841; http://dx.doi.org/10.1073/pnas.0903188106
- Blazek D, Barboric M, Kohoutek J, Oven I, Peterlin BM. Oligomerization of HEXIM1 via 7SK snRNA and coiled-coil region directs the inhibition of P-TEFb. Nucleic Acids Res 2005; 33:7000-10; PMID:16377779; http://dx.doi.org/10.1093/nar/gki997
- Markert A, Grimm M, Martinez J, Wiesner J, Meyerhans A, Meyuhas O, Sickmann A, Fischer U. The La-related protein LARP7 is a component of the 7SK ribonucleoprotein and affects transcription of cellular and viral polymerase II genes. EMBO Rep 2008; 9:569-75; PMID:18483487; http://dx.doi.org/10.1038/embor.2008.72
- Chen R, Yang Z, Zhou Q. Phosphorylated positive transcription elongation factor b (P-TEFb) is tagged for inhibition through association with 7SK snRNA. J Biol Chem 2004; 279:4153-60; PMID:14627702; http://dx.doi.org/10.1074/jbc.M310044200
- Fisher RP. The CDK network: linking cycles of cell division and gene expression. Genes Cancer 2012; 3:731-8; PMID:23634260; http://dx.doi.org/10.1177/1947601912473308
- Garber ME, Mayall TP, Suess EM, Meisenhelder J, Thompson NE, Jones KA. CDK9 autophosphorylation regulates high-affinity binding of the human immunodeficiency virus type 1 tat-P-TEFb complex to TAR RNA. Mol Cell Biol 2000; 20:6958-69; PMID:10958691; http://dx.doi.org/10.1128/MCB.20.18.6958-6969.2000
- McNamara RP, McCann JL, Gudipaty SA, D'Orso I. Transcription factors mediate the enzymatic disassembly of promoter-bound 7SK snRNP to locally recruit P-TEFb for transcription elongation. Cell Rep 2013; 5:1256-68; PMID:24316072; http://dx.doi.org/10.1016/j.celrep.2013.11.003
- Larochelle S, Amat R, Glover-Cutter K, Sanso M, Zhang C, Allen JJ, Shokat KM, Bentley DL, Fisher RP. Cyclin-dependent kinase control of the initiation-to-elongation switch of RNA polymerase II. Nat Struct Mol Biol 2012; 19:1108-15; PMID:23064645; http://dx.doi.org/10.1038/nsmb.2399
- Sedore SC, Byers SA, Biglione S, Price JP, Maury WJ, Price DH. Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR. Nucleic Acids Res 2007; 35:4347-58; PMID:17576689; http://dx.doi.org/10.1093/nar/gkm443
- Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol 2000; 1:137-43; PMID:11253366; http://dx.doi.org/10.1038/35040083
- Hager GL, McNally JG, Misteli T. Transcription dynamics. Mol Cell 2009; 35:741-53; PMID:19782025; http://dx.doi.org/10.1016/j.molcel.2009.09.005
- D'Orso I, Frankel AD. RNA-mediated displacement of an inhibitory snRNP complex activates transcription elongation. Nat Struct Mol Biol 2010; 17:815-21; PMID:20562857; http://dx.doi.org/10.1038/nsmb.1827
- Barboric M, Lenasi T. Kick-sTARting HIV-1 transcription elongation by 7SK snRNP deporTATion. Nat Struct Mol Biol 2010; 17:928-30; PMID:20683478; http://dx.doi.org/10.1038/nsmb0810-928
- Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 2013; 153:855-68; PMID:23663783; http://dx.doi.org/10.1016/j.cell.2013.04.028
- Cherrier T, Le Douce V, Eilebrecht S, Riclet R, Marban C, Dequiedt F, Goumon Y, Paillart JC, Mericskay M, Parlakian A, et al. CTIP2 is a negative regulator of P-TEFb. Proc Natl Acad Sci U S A 2013; 110:12655-60; PMID:23852730; http://dx.doi.org/10.1073/pnas.1220136110
- Gudipaty SA, McNamara RP, Morton EL, D'Orso I. PPM1G Binds 7SK RNA and Hexim1 To Block P-TEFb Assembly into the 7SK snRNP and Sustain Transcription Elongation. Mol Cell Biol 2015; 35:3810-28; PMID:26324325; http://dx.doi.org/10.1128/MCB.00226-15
- Iyengar S, Farnham PJ. KAP1 protein: an enigmatic master regulator of the genome. J Biol Chem 2011; 286:26267-76; PMID:21652716; http://dx.doi.org/10.1074/jbc.R111.252569
- Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell 2001; 8:327-37; PMID:11545735; http://dx.doi.org/10.1016/S1097-2765(01)00314-8
- Mizutani T, Ishizaka A, Suzuki Y, Iba H. 7SK small nuclear ribonucleoprotein complex is recruited to the HIV-1 promoter via short viral transcripts. FEBS Lett 2014; 588:1630-6; PMID:24607481; http://dx.doi.org/10.1016/j.febslet.2014.01.067
- Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, et al. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 2013; 153:1327-39; PMID:23746844; http://dx.doi.org/10.1016/j.cell.2013.04.048
- Dolken L, Ruzsics Z, Radle B, Friedel CC, Zimmer R, Mages J, Hoffmann R, Dickinson P, Forster T, Ghazal P, et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA 2008; 14:1959-72; PMID:18658122; http://dx.doi.org/10.1261/rna.1136108
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 2008; 322:1845-8; PMID:19056941; http://dx.doi.org/10.1126/science.1162228
- Bai X, Kim J, Yang Z, Jurynec MJ, Akie TE, Lee J, LeBlanc J, Sessa A, Jiang H, DiBiase A, et al. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 2010; 142:133-43; PMID:20603019; http://dx.doi.org/10.1016/j.cell.2010.05.028
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 2010; 463:237-40; PMID:20075919; http://dx.doi.org/10.1038/nature08674
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. TRIM24 links a non-canonical histone signature to breast cancer. Nature 2010; 468:927-32; PMID:21164480; http://dx.doi.org/10.1038/nature09542
- Bunch H, Zheng X, Burkholder A, Dillon ST, Motola S, Birrane G, Ebmeier CC, Levine S, Fargo D, Hu G, et al. TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat Struct Mol Biol 2014; 21:876-83; PMID:25173174; http://dx.doi.org/10.1038/nsmb.2878
- Hu G, Kim J, Xu Q, Leng Y, Orkin SH, Elledge SJ. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev 2009; 23:837-48; PMID:19339689; http://dx.doi.org/10.1101/gad.1769609
- Najafabadi HS, Mnaimneh S, Schmitges FW, Garton M, Lam KN, Yang A, Albu M, Weirauch MT, Radovani E, Kim PM, et al. C2H2 zinc finger proteins greatly expand the human regulatory lexicon. Nat Biotechnol 2015; 33:555-62; PMID:25690854; http://dx.doi.org/10.1038/nbt.3128
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol 2006; 8:870-6; PMID:16862143; http://dx.doi.org/10.1038/ncb1446
- Sanso M, Levin RS, Lipp JJ, Wang VY, Greifenberg AK, Quezada EM, Ali A, Ghosh A, Larochelle S, Rana TM, et al. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev 2016; 30:117-31; PMID:26728557; http://dx.doi.org/10.1101/gad.269589.115
- Prasanth KV, Camiolo M, Chan G, Tripathi V, Denis L, Nakamura T, Hübner MR, Spector DL. Nuclear organization and dynamics of 7SK RNA in regulating gene expression. Mol Biol Cell 2010; 21:4184-96; PMID:20881057; http://dx.doi.org/10.1091/mbc.E10-02-0105
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 2010; 37:429-37; PMID:20159561; http://dx.doi.org/10.1016/j.molcel.2010.01.026
- Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, Marshall S, Florens L, Washburn MP, Shilatifard A. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol Cell Biol 2012; 32:2608-17; PMID:22547686; http://dx.doi.org/10.1128/MCB.00182-12
- Luo Z, Lin C, Shilatifard A. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 2012; 13:543-7; PMID:22895430; http://dx.doi.org/10.1038/nrm3417
- Flynn RA, Almada AE, Zamudio JR, Sharp PA. Antisense RNA polymerase II divergent transcripts are P-TEFb dependent and substrates for the RNA exosome. Proc Natl Acad Sci U S A 2011; 108:10460-5; PMID:21670248; http://dx.doi.org/10.1073/pnas.1106630108
- Liu W, Ma Q, Wong K, Li W, Ohgi K, Zhang J, Aggarwal AK, Rosenfeld MG. Brd4 and JMJD6-associated anti-pause enhancers in regulation of transcriptional pause release. Cell 2013; 155:1581-95; PMID:24360279; http://dx.doi.org/10.1016/j.cell.2013.10.056
- Kim TK, Shiekhattar R. Architectural and functional commonalities between enhancers and promoters. Cell 2015; 162:948-59; PMID:26317464; http://dx.doi.org/10.1016/j.cell.2015.08.008
- Chen R, Liu M, Li H, Xue Y, Ramey WN, He N, Ai N, Luo H, Zhu Y, Zhou N, et al. PP2B and PP1alpha cooperatively disrupt 7SK snRNP to release P-TEFb for transcription in response to Ca2+ signaling. Genes Dev 2008; 22:1356-68; PMID:18483222; http://dx.doi.org/10.1101/gad.1636008
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 2005; 19:535-45; PMID:16109377; http://dx.doi.org/10.1016/j.molcel.2005.06.029
- Patel MC, Debrosse M, Smith M, Dey A, Huynh W, Sarai N, Heightman TD, Tamura T, Ozato K. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol Cell Biol 2013; 33:2497-507; PMID:23589332; http://dx.doi.org/10.1128/MCB.01180-12
