ABSTRACT
Background: In most animals, the mitochondrial genome is characterized by its small size, organization into a single circular molecule, and a relative conservation of the number of encoded genes. In box jellyfish (Cubozoa, Cnidaria), the mitochondrial genome is organized into 8 linear mito-chromosomes harboring between one and 4 genes each, including 2 extra protein-coding genes: mt-polB and orf314. Such an organization challenges the traditional view of mitochondrial DNA (mtDNA) expression in animals. In this study, we investigate the pattern of mitochondrial gene expression in the box jellyfish Alatina alata, as well as several key nuclear-encoded molecular pathways involved in the processing of mitochondrial gene transcription. Results: Read coverage of DNA-seq data is relatively uniform for all 8 mito-chromosomes, suggesting that each mito-chromosome is present in equimolar proportion in the mitochondrion. Comparison of DNA and RNA-seq based assemblies indicates that mito-chromosomes are transcribed into individual transcripts in which the beginning and ending are highly conserved. Expression levels for mt-polB and orf314 are similar to those of other mitochondrial-encoded genes, which provides further evidence for them having functional roles in the mitochondrion. Survey of the transcriptome suggests recognition of the mitochondrial tRNA-Met by the cytoplasmic aminoacyl-tRNA synthetase counterpart and C-to-U editing of the cytoplasmic tRNA-Trp after import into the mitochondrion. Moreover, several mitochondrial ribosomal proteins appear to be lost. Conclusions: This study represents the first survey of mitochondrial gene expression of the linear multi-chromosomal mtDNA in box jellyfish (Cubozoa). Future exploration of small RNAs and the proteome of the mitochondrion will test the hypotheses presented herein.
Background
The mitochondrion, largely responsible for cellular respiration, is thought to have originated through a single endosymbiotic event of an α–proteobacteriumCitation1 very early in the evolution of eukaryotes (though see Pittis and Gabaldòn (2016)Citation2 for an alternative hypothesis). Subsequent gene transfer to the nucleus of the host cell and loss of genes from the proto-mitochondrionCitation3 has rendered the typical mitochondrial genome a small molecule that encodes only a fraction of the genes necessary for the functioning and maintenance of the mitochondrion. In most animals (Metazoa), mitochondrial DNA (mtDNA) comprises a single, circular and compact molecule of ∼17 kbp in size, encoding 13 (+2/−2) protein-coding genes involved in oxidative phosphorylation, 2 rRNA (rRNA) genes, and up to 25 tRNA (tRNA) genes involved in the translation of mitochondrial-encoded proteins. While gene order in the mt genome is highly variable, most protein-coding and ribosomal genes are interspersed with tRNA genes. The replication of the typical circular mtDNA is performed through the formation of a D-loop, where the replication of the two strands is out of sync.Citation4 The “tRNA punctuation model” proposes that mtDNA is transcribed into one or two polycistronic precursor transcripts (pre-mRNA molecules) that are then excised at the tRNA sites,Citation5 releasing mostly single-gene (monocistronic) mRNAs, rRNA and tRNA transcripts.Citation6
In the early branching metazoan phylum Cnidaria (sea anemones, corals, hydras, hydroids, and jellyfish), the mt genome encodes for only one or 2 tRNA genes,Citation7-17 challenging the tRNA punctuation model of gene expression. Species from the cnidarian class Anthozoa (the clade containing reef-building corals, sea anemones, soft corals, and their relatives) possess relatively large, circular mitochondrial genomes in which protein-coding genes are interspaced with intergenic regions (IGRs) that might play a role in mitochondrial mRNA maturation (e.g.Citation18,19). The mt genome in the cnidarian subphylum Medusozoa (e.g. hydroids and jellyfish) is organized into mono-, bi-, or multi-linear chromosomes.Citation7-13,20 Medusozoan mtDNA is very compact with a few small IGRs.Citation12,15 In hydrozoans, Kayal et al. (2015)Citation15 suggested that both rRNA genes and stemloop-like secondary structures formed by IGRs could be recognized by the nucleolytic processing mechanism, producing mature often multi-cistronic mRNA transcripts.
Cubozoans (box jellyfish) have mtDNA made up of eight small linear chromosomes.Citation12,13,20 Mitochondrial chromosomes range from 4 to 6 kbp in length and harbor between one and four genes, flanked by long and conserved telomere-like inverted repeats (IRs) with opposite orientation thought to be involved in the maintenance of the linear mito-chromosomes.Citation13 In addition to the 13 protein-coding genes common in Metazoa, most medusozoan mtDNA (excluding hydroidolinan hydrozoans) encodes for two extra protein-coding genes (mt-polB and orf314). Mohammadou et al (2004)Citation21 have found that mt-polB has affinities with polymerases from the B family. Kayal et al. (2012)Citation12 have suggested that orf314 might be acting as a terminal protein by binding the 5′ single-stranded end of the linear chromosomes as described for the liverwort Marchantia polymorphaCitation22 and some yeast species.Citation23,24 mt-polB and orf314 are present in all medusozoan subclasses and lack internal stop codons; in cubozoans mt-polB and orf314 are found together on an independent linear mito-chromosome. The maintenance of these genes through long evolutionary times (acquired before the divergence of Medusozoa and maintained in several medusozoan clades) has been used as evidence to suggest that they have functional roles in the mitochondrion.Citation12,13 Additional evidence on the expression of these genes is needed to confirm and refine this hypothesis and to assess specific functions.
Here, we surveyed recently released RNA-seq data from the box jellyfish Alatina alata (SRR1952741) and assembled the near complete multi-chromosomal mitochondrial genome, as well as several mitochondrial-targeted genes involved in mtDNA expression, including but not limited to mitochondrial aminoacyl-tRNA synthases (mt-aaRS), polymerases, and ribosomal proteins. Our analysis offers insights into the expression of mtDNA genes and provides additional evidence about the functional roles of mt-polB and orf314.
Material and methods
We obtained a recently published transcriptome assembly for Alatina alataCitation25 as well as 2 publicly available short read archive (SRA) datasets from the National Center for Biotechnology Information (NCBI; ncbi.nlm.nih.gov/sra/): SRR217435 containing SOLID DNA-seq reads and SRR1952741 containing ILLUMINA HiSeq 2500 RNA-seq reads. The RNA-seq library for the data set used in this study was prepared using the TruSeq mRNA enrichment protocol, where oligo-dT beads capture RNA molecules with poly(A) tail, before random reverse transcription is performed.Citation25 Tissue on which the transcriptome assembly was based did not include any gut tissue, lessening concerns about contamination from other organisms. We mapped the SOLID DNA-seq data to a published mt genome of Alatina alata (JN642330-JN642344) using Bowtie2Citation26 with –end-to-end and –very-sensitive options to estimate individual chromosome and gene coverage.
Fig. S1 details the bioinformatics workflow we used in this study. In short, we captured and assembled mitochondrial contigs from the assembled transcriptome using the BLAST plugin in Geneious v.9.Citation27 We filtered raw reads from the ILLUMINA HiSeq RNA-seq data with Trimmomatic v.0.32Citation28 and mapped filtered reads to the assembled mitochondrial chromosomes using Bowtie2, with default settings. We used Geneious to visualize and manually check the assemblies, and to build consensus sequences. We annotated individual chromosomes following the reference mitogenome and predicted the origin and end of transcription. Finally, we mapped trimmed RNA-seq reads back to the mitochondrial consensus sequences to estimate individual chromosome and gene coverage with Bowtie2 using –end-to-end and –very-sensitive options (Fig. S1).
We also investigated several genes involved in the expression and function of the mitochondrion from the RNA-seq data: the glutamyl-tRNA amidotransferase subunits (GatA-C) that are absent in other cnidarian classes,Citation29 the mitochondrial ribonucleases complex P (MRPP1-3) and Z (RNase Z) responsible for the cleavage of the 5′ and 3′ ends of the tRNA, the mitochondrial poly(A) RNA polymerase (MTPAP) that creates the 3′ poly(A) tails of mitochondrial transcripts, the full set of aminoacyl-tRNA synthetases (aaRSs), the mitochondrial DNA-directed RNA polymerase (POLRMT), and mitochondrial ribosomal proteins involved in transcription. First, we identified homologs of the genes of interest in several species from the Uniprot database (http://www.uniprot.org/; last accessed 3/5/2016; the list of query sequences is found in Table S1). We then used the BLAST plugin in Geneious to search the transcriptome assembly for A. alata for putative homologs. All BLAST hits with a max e-value of 1e-1 were retained. Retained hits were were aligned and assembled when possible into consensus contigs. The resulting contigs were individually checked against the NCBI protein database (https://blast.ncbi.nlm.nih.gov/Blast.cgi) using blastx to remove potential bacterial contaminants (the top 10 or more hits of bacterial origin) and genes not involved in mitochondrial expression (eukaryote hits with the majority reporting other functions). When the identity of the gene was dubious (not a majority reporting a single function), the translated coding region was blasted against the Uniprot database. Incomplete transcripts were extended, when possible, to include parts of the 5′ and 3′ untranslated regions (UTRs) using a combination of the mirabait script from MIRA v.4,Citation30 MITObim v.1.8,Citation31 and the overlap-layout-consensus assembler implemented in Geneious. The presence of putative mitochondrial targeting-signal peptides was assessed using the online versions of the programs MitoProt II,Citation32 TargetP v1.1,Citation33 PredotarCitation34 and Mitofates.Citation35 Given that the bioinformatics tools for signal peptide prediction are trained on only a handful of model organisms, we considered a signal peptide present when probabilities were >0.5 for at least 2 of the 4 prediction programs. All identified mitochondrion-targeted genes were deposited in the FigShare digital repository and can be accessed at https://dx.doi.org/10.6084/m9.figshare.3380848.
Since two out of 3 components of the mitochondrial ribonucleases complex P (MRPP1-2) are part of large gene families, we used a phylogenetic approach to confirm the identifications of all 3 mitochondrial ribonucleases complexes (MRPP1-3). This approach also allowed us to test the accuracy of our BLAST-based method described above. We first downloaded the sequences that matched “trna methyltransferase 10” and “reviewed:yes” for MRPP1 (240 sequences), “3 hydroxyacyl coa dehydrogenase” and “reviewed:yes” for MRPP2 (282 sequences), and “mitochondrial ribonuclease protein p” for MRPP3 (136 sequences) from the Uniprot database (last accessed 4/5/2016). We then aligned each set with the corresponding sequence identified from Alatina alata using the online program MAFFT v.7Citation36 with the L-INSi algorithm. Gblocks v0.91bCitation37 was used on mrrp2 with gap allowed at all positions; the MRRP1 and MRRP3 alignments were not filtered since Gblocks would remove most (MRPP1) or all (MRPP3) the data given the high level of sequence variability within the alignments. Phylogenetic analyses were performed under a Maximum Likelihood framework using RAxML v.8.2.7Citation38 with the PROTGAMMAWAG model of sequence evolution (100 independent searches for the best tree; 1000 bootstraps). All alignments and trees are deposited in the FigShare digital repository and can be accessed at https://dx.doi.org/10.6084/m9.figshare.3380848.
We obtained the full set of sequences belonging to the polymerase α superfamily (DNA_pol_A PF00476) from the Pfam databaseCitation39 and created a local HMM database using HMMER v3.1b1.Citation40 We searched the transcriptome assembly using HMMER and isolated contigs matching the gamma polymerase gene (polG). We used Geneious to assemble the positive matches into a single contig and checked the assembly by mapping the filtered reads against the polG contig with Bowtie2 (see above).
Results and discussion
Expression of mt genes in Alatina alata
We mapped SOLID DNA-seq data from the SRR archive (SRR217435) to published mitochondrial chromosomes obtained from these data (JN642330-JN642344Citation13) and found that all eight mito-chromosomes have similar but unequal (variance 18%) read coverage. That is, no chromosome is found to have considerably more coverage than the others (; Table S2). Interestingly, we found the genes cox1, cox2, mt-polB, nad4 and orf314 having the highest coverage in the DNA-seq SOLID data (). The variation in the coverage between various mito-chromosomes in the DNA-seq SOLID data does not appear to correspond to the functional requirements of mitochondrial genes. For instance, despite the key role played by mt-rRNAs, rnl and rns display no noticeable deviation from the average read coverage, in stark contrast with RNA-seq coverage (, ). It is very likely that all eight mito-chromosomes are in equimolar proportion in the mitochondrion of A. alata.
Figure 1. Mitochondrial read coverage values in the mtDNA of Alatina alata. (A) Nucleotide coverage of the unique regions of individual mitochondrial genes from the DNA-seq and RNA-seq data. (B) Nucleotide coverage of the unique regions of mitochondrial chromosomes from the DNA-seq and RNA-seq data. The Y axis represents mean coverage in kbp.
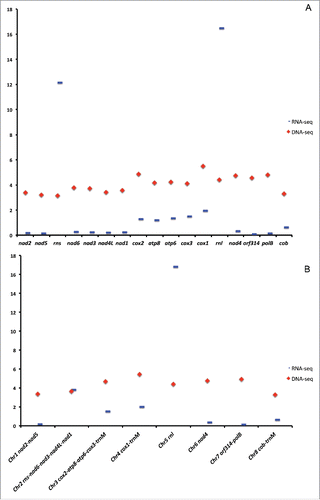
Table 1. Read coverage of the uniq regions of the mitochondrial genes in the DNA-seq and RNA-seq datasets.
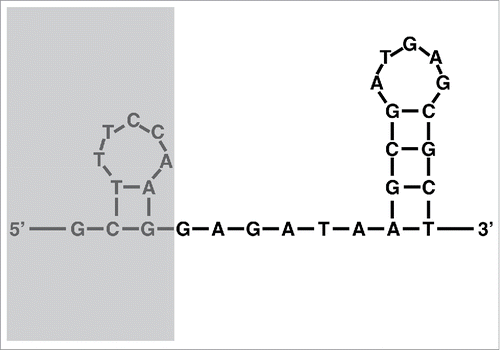
We recovered nearly complete mitochondrial chromosomes from the RNA-seq reads, including part of the terminal inverted repeats (IRs). Compared to the reference mito-genomes, 10 bp are missing in the RNA-based assemblies, which we define as the origin of transcription (OT). The OT is part of a 30 bp region present on all mito-chromosomes with putative stem-loop structures predicted by the DNA folding program from the mfold Web Server with default parametersCitation41 (). The complete conservation of the OT and its putative secondary structure support its role as the origin of transcription for the mito-chromosomes. Smith et al (2012)Citation13 described a gene-conversion model for the presence of partial duplication at the subtelomeric regions of all mito-chromosomes. The level of conservation of subtelomeric regions varies among mito-chromosomes: the coding regions of seven out of the eight mito-chromosomes start with the 5′end of rnl, with the highest conservation found in nad2 (Fig. S2). A small (∼5 bp) sequence conservation was found between the 5′end of rnl and rns.Citation13
Figure 2. Secondary structure of the putative origin of translation (OT) found on all 8 chromosomes of the mt genome of Alatina alata using the DNA folding program from the mfold Web Server with default parameters. The greyed area is present at the DNA level but absent from the RNA-seq data.
The pattern of gene expression corresponds to the organization of the genes on each chromosome, with no break within mitochondrial contigs, as described in other medusozoans,Citation15 where ≥10 bp stem-loop structures might be responsible in mitochondrial pre-mRNA processing. Such a pattern suggests that mito-chromosomes are expressed into single per-chromosome transcripts () starting at the OT region a few base pairs before the IR upstream of the coding region, and ending 161 bp within the IR downstream of the coding region. Average coverage ranged from ∼130 to ∼16,800 nucleotides per base of the unique regions of the genes (the large regions of genes not duplicated on other chromosomes), and the highest values were observed for the mito-chromosomes harboring the rRNA genes (; ). High-level representation of mt-rRNA in the RNA-seq data is expected given that ribosomes are essential in the expression of the mitochondrial genes. Interestingly, the 2 chromosomes containing cox1-3 and atp6 and 8 genes are also highly expressed in the RNA-seq data (; Table S2).
Figure 3. Gene expression in the mitochondrion of Alatina alata. (A) Pattern of gene expression and their coverage in the DNA-seq and RNA-seq data sets for a model mito-chromosome. Graphs of read coverage do not correspond to the true values, but rather approximate graphical representations for visualization purposes. CR = putative control region; IGR = inter-genic region; EF? = putative elongation factor involved in the expansion of the rRNA; RNase? = putative enzyme responsible for recognizing and excising stemloop structures found at the 3′end of rRNA and 5′ of tRNA genes. Colored arrows correspond to protein coding genes (green), ribosomal gene (red), tRNA gene (purple), and inverted repeats or IR (cyan). (B) Nuclear genes involved in the maturation of the mitochondrial tRNA-Met: RNase P complex made of three subunits MRPP1-3 responsible for excising extra nucleotides at the 5′end of the tRNA; RNase Z responsible for excising extra nucleotides at the 3′end of the tRNA; CCAse responsible for adding CCA to the 3′end of the tRNA. (C) Import and editing of cytoplasmic tRNA-Trp, where the CCA anticodon is edited via an as yet unknown C-to-U editing enzyme (?). (D) Composition of the mitochondrial ribosome large (39S) and small (28S) subunits. Secondary structures of the large (rnl) and small (rns) rRNA subunits are in red; numbers correspond to individual mitochondrial ribosomal proteins (MRPs) with identified (in black) or lacking (in red) mitochondrial-targeting signal peptides. Number in bold is the newly identified mitochondrial ribosome small subunit S39.
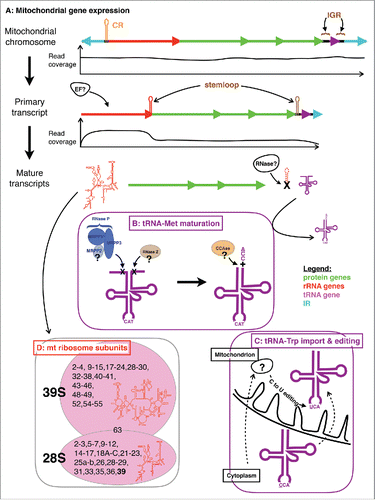
Conservation of subtelomeric regions suggests that transcription can start on any mito-chromosome and an as yet unknown elongation factor is required to recognize the correct gene (). Mitochondrial ribosomal proteins or rRNA genes might act as elongation factors in the transcription. Mito-chromosome 2 harbors rns along with four protein-coding genes (nad1, 3, 4L and 6), and displays the highest coverage of RNA reads (mean coverage ∼4 kbp; ). Read coverage is not evenly distributed along this mito-chromosome: the protein-coding genes have read coverage ∼55 times lower than rns (). The dramatic drop in read coverage is observed at the 3′end of rns, which can fold into a stem-loop structure (Fig. S2). We propose that the secondary structure at the 3′end of rns is recognized by some factor(s) and helps regulate transcription by detaching the RNA POLRMT from its template.
We found no read spanning beyond one-fourth (161 out of 702 bp) of the IR downstream of the coding region of the chromosomes. Studies have shown polyadenylation of both rRNA and mRNA in mammalian mitochondria.Citation42 In the transcriptome of Alatina alata, we identified and annotated several complete sequences (including both the 5′ and 3′ UTR regions) homologous to the poly(A) polymerase (PAP) gene, one of which (MTPAP PIP5K1A) likely contains an N-terminal mitochondrial-targeting signal peptide (). Assuming similar polyadenylation is taking place in the mitochondria of all animal species, we posit that the recognition site for MTATP in Alatina is about 1/4th of the way into the IR of the mitochondrial chromosomes.
Table 2. Read coverage of some mitochondrial-targeted genes in the RNA-seq data set.
A functional mitochondrial-encoded type B DNA polymerase
We recovered the complete sequence of the polG gene whose product is responsible for mtDNA replication, including the mitochondrial-targeting signal peptide (). The product of this gene is similar in size to its counterparts in other animals. Medusozoan mito-genomes (excluding hydroidolinan hydrozoans) harbor two additional protein-coding genes orf314 and mt-polB. Our previous studiesCitation12,13 have suggested that orf314 and mt-polB play roles in the maintenance and replication of the mitochondrial telomeres: the product of mt-polB has affinities with members of the DNA-directed DNA polymerase type B found in plant and fungal mitochondria, and viruses; orf314 was hypothesized to encode for a DNA-binding protein that could sit at the 5′ single-stranded end of mitochondrial telomeres to prevent their degradation by exonucleases and may be involved in the maintenance of the integrity of the mito-chromosomes during replication.Citation7,12 It has been posited that the linearization of medusozoan mtDNA originated in association with the incorporation of a linear plasmid containing both mt-polB and orf314.Citation12 The presence of these genes on a separate mito-chromosome in A. alata was already suggested as evidence of positive selection for their maintenance.Citation13 We recovered the chromosome containing these genes from the RNA-seq data with per-gene coverage values similar but below that of the other protein coding genes ( and S2), indicating that these genes are expressed and polyadenylated (i.e., present in the mRNA pool). These data provide further evidence that both orf314 and mt-polB are functional in the mitochondrion.
Near-parallel loss of mt-tRNA and mt-aaRS in Cubozoa suggests tRNA editing
The only complete mitochondrial genome for Cubozoa published to date, that of Alatina alata (JN642330-JN642344), encodes one single mitochondrial tRNA for methionine (mt-tRNAMet or trnM),Citation13 suggesting that the other tRNAs are imported from the cytoplasm. We recovered the complete set of cytoplasmic aminoacyl-tRNA synthetases (nc-aaRS) but only one mitochondrial aminoacyl-tRNA synthetase (mt-PheRS) from the RNA-seq data (). This pattern is in accord with the evolutionary scenario of parallel loss of mt-tRNAs and mt-aaRS, excluding mt-PheRS, which is made of a single protein as opposed to nc-PheRS composed of 2 subunits encoded by 2 separate genes.Citation29,43 Three full transcripts (Glu/ProRS, IleRS and ValRS) did not harbor a recognizable 5′ mitochondrial-targeting signal peptide (), while the sequence for HisRS was missing the 5′ end where the mt-targeting signal peptide is expected to be found. The absence of mt-targeting signal peptides in Glu/ProRS, IleRS and ValRS suggests some post-transcriptional modification that might be responsible for the transport of these nc-aaRSs into the mitochondrion, though the possibility of a deviant targeting signal not identified by the prediction programs cannot be ruled out.
Table 3. List of aminoacyl-tRNA synthetases (aaRS) found in the RNA-seq data of Alatina alata.
Previous studies have suggested that the loss of mt-tRNAs preceded that of the corresponding mt-aaRSs in CnidariaCitation43; cases where an mt-tRNA is present in the absence of the corresponding mt-aaRS can be explained by recruitment of the mt-tRNA for other functions, such as a structural component of the mitochondrial ribosome.Citation29 In mammals, where mt-tRNAVal has been found as a structural part of the mitochondrial ribosome, the tRNA gene (trnV) is encoded between the small and large ribosomal subunits (rns and rnl respectively) in the mt genome, leading to a polycistronic transcript containing all three RNAs.Citation44,45 In demosponge mtDNA, trnG (mt-tRNAGly) and trnV are the most common tRNA genes found between rns and rnl, but never trnR (mt-tRNAArg),Citation46 yet mt-ArgRS is one of the five mt-aaRSs (the others being mt-CysRS, mt-LysRS, mt-ThrRS and mt-ValRS) that seem to have been lost in this clade.Citation29 This pattern cannot explain the role of these tRNAs as structural parts of the ribosome as shown in mammals.Citation44 The presence of mt-tRNAMet on three out of the eight mitochromosomes of A. alata concurrent with the absence of mt-MetRS in the nucleus is an interesting pattern that cannot be explained by the role of this tRNA as a structural component of the mitochondrial ribosome.
We investigated several key genes responsible for mitochondrial tRNA processing, namely RNase P (for 5′-end maturation), RNase Z (for 3′-end maturation), and CCase (responsible for adding CCA to the 3′-end of the tRNA)(). In humans, RNase P is a protein complex composed of three subunits: MRPP1, one out of at least three homologs of a tRNA methyltransferase 10 (C homolog); MRPP2, a 3-hydroxyacyl-CoA dehydrogenase; MRPP3, the mitochondrial ribonuclease P protein 3.Citation47,48 We identified homologs to both MRPP2 and MRPP3 (the latter with a mitochondrial targeting signal peptide) but not to MRPP1 (). Instead, our phylogenetic analysis identified a tRNA methyltransferase 10 A homolog (Fig. S3) with a mitochondrial targeting signal peptide, while human homologs do not harbor such a signal peptide (data not shown). We therefore decided to label the transcript tRNA methyltransferase 10 homolog A as MRPP1 (). Phylogenetic analyses of gene trees for transcripts annotated MRPP1-3 confirm they being homologous to MRPP1-3 genes derived from other organisms (Figs. S3-5). Lopez Sanchez et al. (2011) recently showed that MRPP1 and MRPP3 are sufficient for processing the 5′end of mitochondrial tRNAs in humans.Citation49
The same authors and others identified the zinc phosphodiesterase ELAC protein 2 as RNase Z responsible for processing the 3′end of mitochondrial tRNAs.Citation49,50 We recovered two transcripts that encode for proteins similar to ELAC2: the shorter transcript (ELAC2a) displayed more similarity (in the form of higher matching base and alignment length BLAST results in the Uniprot database) with homologs from other taxa but was incomplete, missing the 5′ terminal; the longer transcript (ELAC2b) lacked a mitochondrial targeting signal peptide (). Finally, we identified one CCAse homolog that lacked an acceptable mitochondrial targeting signal peptide ().
The presence of mitochondrial targeting signals on the transcripts of several mitochondrial tRNA-processing genes combined with the high expression of mt-tRNAMet (Table S3) suggests that trnM requires a functional tRNA structure in the mitochondrion of this species (). Given the very distinct bacterial-like translation initiation system in the mitochondrion compared to the eukaryote-like one for the cytosol, the loss of trnM is thought to be nigh impossible.Citation51 Yet, the loss of mt-MetRS requires nc-MetRS, whose transcript harbors an mt-targeting signal peptide () to recognize and aminoacetylate mt-tRNAMet in the mitochondrion. Alternatively, trnM could play a role as initiation of translation in the mitochondrion.
Both Octocorallia and Cubozoa have lost both the mitochondrial tRNA and the mitochondrial aminoacyl-tRNA synthetase for trpyptophan (mt-tRNATrp and mt-TrpRS, respectively).Citation29 In the absence of both mt-tRNATrp and mt-TrpRS, tryptophan residues are processed by a combination of cytoplasmic nc-tRNATrp and nc-TrpRS. In the ctenophore Pleurobrachia bachei, the loss of mt-tRNATrp has resulted in the reassignment of the TGA codon to serine.Citation29,52,53 In cubozoan mtDNA, tryptophan residues are encoded by two codons TGG and TGA at a relatively similar proportion (Table S3). In octocorals, TGG is the main codon used for tryptophan, the exceptions seemingly resulting either from sequencing errors or present in the extra protein coding gene homolog to bacterial mutSCitation54(Table S3). In the cytoplasm, nc-tRNATrp only recognizes TGG, while TGA is a stop codon. In the absence of mRNA editing in Alatina (proportion of TGA was similar in RNA and DNA-seq data; Table S3), we suggest that the anticodon of nc-tRNATrp(CCA) is edited in the mitochondrion to recognize and aminoacylate TGA codons (anticodon UCA) through a C → U editing mechanism (). Anticodon C to U editing has been reported in the mitochondrion of Trypanosoma allowing the recognition of TGA as tryptophan in the absence of an mt-tRNATrp(UCA) gene,Citation55 and in marsupials where some of the mt-tRNAAsp(GCC) are edited to mt-tRNAAsp(GUC).Citation56 Alternatively, addition factors (RNA and/or proteins) would be required to guide TGA recognition by a native nc-tRNATrp. Future studies focusing on short RNAs should allow testing our hypotheses.
The mitochondrial ribosomal proteins complex
Ribosomes, essential pieces of the translation machinery, are complex molecules made of two subunits (large and small), each composed of RNA (of different origin for the cytoplasm and the mitochondrion) and many proteins. In animals, all mitochondrial proteins are encoded by the nucleus, while two RNA subunits (12S and 16S for small and large RNA subunits, respectively) are encoded by the mitochondrial DNA (rns and rnl). We used the dataset of a recent study on HolozoaCitation57 to investigate ribosomal protein coding genes in the transcriptome of A. alata. We recovered the same cnidarian set of large (40 gene transcripts) and small (29 gene transcripts) subunits of the mitochondrial ribosome, with a few exceptions, plus the unassigned MRP63 (; ).
Table 4. List of mitochondrial ribosomal proteins (MRPs) found in the RNA-seq data of Alatina alata.
From the large ribosomal subunit, L1, L16, L27, L50, L53 and L56 were not recovered. From the small ribosomal subunit, only S30 was not recovered. We also found that the subunits L11, L13, L15, L19, L30, L33, L40, L43 and L52 from the large subunit, and S6, S17, S28 and S35 from the small subunit had low probability (≤0 .5 for at least three out of the four prediction software used) for possessing mitochondrial-targeting signal peptides (). Sheel et al. (2014) also found L50 and L56 to be absent from the hydrozoan Hydra magnapapillata.Citation57 Assuming the lack of these genes reflects their loss in the cubozoan A. alata, the loss of L56 could be a synapomorphy of Medusozoa (Hydra and Alatina), while the loss of L1, L16, L27 and S30 appear to be autapomorphies to Alatina.
Interestingly, we found two versions of subunits L37 and S25, suggesting gene duplication, though an assembly artifact is conceivable. Both candidates for subunit L37 had recognizable mitochondrial-targeting signal peptides but the longer transcript (L37b) displayed higher probability of export to the mitochondrion, while the shorter transcript (L37a) showed more similarity (in the form of higher matching base and alignment length BLAST results in the Uniprot database) to homologs from other taxa (). Similarly, while we identified a mitochondrial-targeting signal peptide on the two candidate transcripts for subunit S25, the longer transcript (S25b) showed more similarity to homologs from other taxa (). We also found the transcript for subunit S31 to be far larger in size than homologs from other metazoans, with an addition of ∼840 nucleotides (∼280 amino acids) on the 5′-end of the coding DNA sequence (CDS). The function of the extra amino acids in S31 was not determined.
Lastly, we identified another mitochondrial ribosomal protein subunit (S39) with similarities to pentatricopeptide repeat-containing protein 3 previously not found by Sheel and Hausdorf (2014).Citation57 Using the BLAST search tool on the Uniprot database, we identified homologs from Nematostella vectensis (Cnidaria: Anthozoa; predicted protein v1g212337), Hydra vulgaris (Cnidaria: Hydrozoa; pentatricopeptide repeat-containing protein 3, mitochondrial T2M872) and Amphimedon queenslandica (Porifera: Demospongiae; uncharacterized protein LOC100633076) with mitochondrial targeting signal peptides (data not shown).
Conclusion
Cubozoans have one of the most unusual mtDNA organizations in Metazoa, where genes are distributed on eight linear chromosomes with long terminal inverted repeats. We explored the recently published RNA-seq data from the box jellyfish Alatina alata to better understand the mechanisms involved in the expression of linear mitochondrial chromosomes. We propose a pattern of mitochondrial gene expression where each mito-chromosome is transcribed into a single transcript starting at a well conserved origin of transcription (OT; 30 bp) and ending with the polyadenylation site about 1/4th of the way into the inverted repeat (IR) downstream of the coding regions of the mito-chromosomes. We captured the two extra protein-coding genes mt-polB and orf314 in the RNA-seq data with coverage levels similar to other mitochondrial protein-coding genes, providing further evidence for the functionality of these genes. We also found that the evolution of mitochondrial translational machinery in A. alata displays similarities to what has been documented in other non-bilaterians.Citation29 Finally, we suggest that the mt-tRNAMet is likely a functional tRNA that could be recognized and aminoacetylated by the nc-MetRS, and that an imported nc-tRNATrp is edited in order to recognize TGA codons as tryptophan. Future studies exploring the repertoire of small RNAs and the proteome of the mitochondrion will be able to test these hypotheses.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplementary_Data.zip
Download Zip (1.8 MB)Acknowledgments
Some of this work was performed using resources of the Laboratories of Analytical Biology at the Smithsonian National Museum of Natural History. EK and BB were supported by a Smithsonian Institution Peter Buck postdoctoral fellowship. We also want to thank two anonymous reviewers for providing constructive criticisms to earlier versions of this manuscript.
References
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund A, Eriksson A-SS, Winkler HH, Kurland CG, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 1998; 396:133-40; PMID:9823893; http://dx.doi.org/10.1038/24094
- Pittis AA, Gabaldón T. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature 2016; 531:101-4; PMID:26840490; http://dx.doi.org/10.1038/nature16941
- Andersson SG, Karlberg O, Canbäck B, Kurland CG. On the origin of mitochondria: a genomics perspective. Philos Trans R Soc Lond B Biol Sci 2003; 358:165-77; PMID:12594925; http://dx.doi.org/10.1098/rstb.2002.1193
- Holt IJ, Reyes A. Human mitochondrial DNA Replication. Cold Spring Harb Perspect Biol 2012; 4:a012971; http://dx.doi.org/10.1101/cshperspect.a012971
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981; 290:470-4; PMID:7219536; http://dx.doi.org/10.1038/290470a0
- Brugler MR. Molecular evolution in black corals (Cnidana Anthozoa Hexacorallia) implications for antipatharian systematics. 2011.
- Shao Z, Graf S, Chaga OY, Lavrov DV. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): A linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene 2006; 381:92-101; PMID:16945488; http://dx.doi.org/10.1016/j.gene.2006.06.021
- Voigt O, Erpenbeck D, Wörheide G. A fragmented metazoan organellar genome: the two mitochondrial chromosomes of Hydra magnipapillata. BMC Genomics 2008; 9:350; PMID:18655725; http://dx.doi.org/10.1186/1471-2164-9-350
- Kayal E, Lavrov DV. The mitochondrial genome of Hydra oligactis (Cnidaria, Hydrozoa) sheds new light on animal mtDNA evolution and cnidarian phylogeny. Gene 2008; 410:177-86; PMID:18222615; http://dx.doi.org/10.1016/j.gene.2007.12.002
- Park E, Hwang D-S, Lee J-S, Song J-I, Seo T-K, Won Y-J. Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol Phylogenet Evol 2012; 62:329-45; PMID:22040765; http://dx.doi.org/10.1016/j.ympev.2011.10.008
- Zou H, Zhang J, Li W, Wu S, Wang G. Mitochondrial Genome of the Freshwater Jellyfish Craspedacusta sowerbyi and Phylogenetics of Medusozoa. PLoS One 2012; 7:e51465; PMID:23240028; http://dx.doi.org/10.1371/journal.pone.0051465
- Kayal E, Bentlage B, Collins AG, Kayal M, Pirro S, Lavrov DV. Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol Evol 2012; 4:1-12; PMID:22113796; http://dx.doi.org/10.1093/gbe/evr123
- Smith DR, Kayal E, Yanagihara AA, Collins AG, Pirro S, Keeling PJ. First complete mitochondrial genome sequence from a box jellyfish reveals a highly fragmented, linear architecture and insights into telomere evolution. Genome Biol Evol 2012; 4:52-8; PMID:22117085; http://dx.doi.org/10.1093/gbe/evr127
- Luz BLP, Capel KCC, Stampar SN, Kitahara MV. Description of the mitochondrial genome of the tree coral Dendrophyllia arbuscula (Anthozoa, Scleractinia). Mitochondrial DNA 2015; 27(4):1-2;PMID:26119126; http://dx.doi.org/10.3109/19401736.2015.1060435
- Kayal E, Bentlage B, Cartwright P, Yanagihara AA, Lindsay DJ, Hopcroft RR, Collins AG. Phylogenetic analysis of higher-level relationships within Hydroidolina (Cnidaria: Hydrozoa) using mitochondrial genome data and insight into their mitochondrial transcription. PeerJ 2015; 3:e1403; PMID:26618080; http://dx.doi.org/10.7717/peerj.1403
- Arrigoni R, Vacherie B, Benzoni F, Barbe V. The complete mitochondrial genome of Acanthastrea maxima (Cnidaria, Scleractinia, Lobophylliidae). Mitochondrial DNA 2016; 27(2):927-8; PMID:24938099; http://dx.doi.org/10.3109/19401736.2014.926489
- Wu J-S, Ju Y-M, Hsiao S-T, Hsu C-H. Complete mitochondrial genome of Junceella fragilis (Gorgonacea, Ellisellidae). Mitochondrial DNA 2016; 27(2):1229-30; PMID:25090396; http://dx.doi.org/10.3109/19401736.2014.945531
- Figueroa DF, Baco AR. Complete mitochondrial genomes elucidate phylogenetic relationships of the deep-sea octocoral families Coralliidae and Paragorgiidae. Deep Res Part II Top Stud Oceanogr 2014; 99:83-91; http://dx.doi.org/10.1016/j.dsr2.2013.06.001
- Foox J, Brugler MR, Siddall ME, Rodríguez E. Multiplexed pyrosequencing of nine sea anemone (Cnidaria: Anthozoa: Hexacorallia: Actiniaria) mitochondrial genomes. Mitochondrial DNA 2015; 27(4):1-7; PMID:26104159; http://dx.doi.org/10.3109/19401736.2015.1053114
- Ender A, Schierwater B. Placozoa are not derived cnidarians: evidence from molecular morphology. Mol Biol Evol 2003; 20:130-4; PMID:12519915; http://dx.doi.org/10.1093/molbev/msg018
- Mouhamadou B, Barroso G, Labarère J. Molecular evolution of a mitochondrial polB gene, encoding a family B DNA polymerase, towards the elimination from Agrocybe mitochondrial genomes. Mol Genet Genomics 2004; 272:257-63; PMID:15365817; http://dx.doi.org/10.1007/s00438-004-1050-4
- Oldenburg DJ, Bendich AJ. Mitochondrial DNA from the liverwort Marchantia polymorpha: circularly permuted linear molecules, head-to-tail concatemers, and a 5′ protein. J Mol Biol 2001; 310:549-62; PMID:11439023; http://dx.doi.org/10.1006/jmbi.2001.4783
- Tomáška L, Nosek J, Fukuhara H. Identification of a putative mitochondrial telomere-binding protein of the yeast Candida parapsilosis. J Biol Chem 1997; 272:3049-56; PMID:9006955; http://dx.doi.org/10.1074/jbc.272.5.3049
- Fricova D, Valach M, Farkas Z, Pfeiffer I, Kucsera J, Tomáška L, Nosek J. The mitochondrial genome of the pathogenic yeast Candida subhashii: GC-rich linear DNA with a protein covalently attached to the 5′ termini. Microbiology 2010; 156:2153-63; PMID:20395267; http://dx.doi.org/10.1099/mic.0.038646-0
- Zapata F, Goetz FE, Smith SA, Howison M, Siebert S, Church SH, Sanders SM, Ames CL, McFadden CS, France SC, et al. Phylogenomic analyses support traditional relationships within Cnidaria. PLoS One 2015; 10:e0139068; PMID:26465609; http://dx.doi.org/10.1371/journal.pone.0139068
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods 2012; 9:357-9; PMID:22388286; http://dx.doi.org/10.1038/nmeth.1923
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28:1647-9; PMID:22543367; http://dx.doi.org/10.1093/bioinformatics/bts199
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014; 30:2114-20; PMID:24695404; http://dx.doi.org/10.1093/bioinformatics/btu170
- Pett W, Lavrov DV. Cytonuclear interactions in the evolution of animal mitochondrial tRNA metabolism. Genome Biol Evol 2015; 7:2089-2101; PMID:26116918; http://dx.doi.org/10.1093/gbe/evv124
- Chevreux B, Wetter T, Suhai S. Genome sequence assembly using trace signals and additional sequence information. Comput Sci Biol Proc Ger Conf Bioinforma 1999; 99:45-56
- Hahn C, Bachmann L, Chevreux B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads–a baiting and iterative mapping approach. Nucleic Acids Res 2013; 41:e129; PMID:23661685; http://dx.doi.org/10.1093/nar/gkt371
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 1996; 241:779-86; PMID:8944766; http://dx.doi.org/10.1111/j.1432-1033.1996.00779.x
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2007; 2:953-71; PMID:17446895; http://dx.doi.org/10.1038/nprot.2007.131
- Small I, Peeters N, Legeai F, Lurin C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 2004; 4:1581-90; PMID:15174128; http://dx.doi.org/10.1002/pmic.200300776
- Fukasawa Y, Tsuji J, Fu S-C, Tomii K, Horton P, Imai K. MitoFates: Improved Prediction of Mitochondrial Targeting Sequences and Their Cleavage Sites. Mol Cell Proteomics 2015; 14:1113-26; PMID:25670805; http://dx.doi.org/10.1074/mcp.M114.043083
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013; 30:772-80; PMID:23329690; http://dx.doi.org/10.1093/molbev/mst010
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 2000; 17:540-52; PMID:10742046; http://dx.doi.org/10.1093/oxfordjournals.molbev.a026334
- Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014; 30:1312-3; PMID:24451623; http://dx.doi.org/10.1093/bioinformatics/btu033
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 2015; 44:D279-85; PMID:26673716; http://dx.doi.org/10.1093/nar/gkv1344
- Eddy SR. Profile hidden Markov models. Bioinformatics 1998; 14:755-63; PMID:9918945; http://dx.doi.org/10.1093/bioinformatics/14.9.755
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 2003; 31:3406-15; PMID:12824337; http://dx.doi.org/10.1093/nar/gkg595
- Fernández-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Exp Physiol 2003; 88:41-56; PMID:12525854; http://dx.doi.org/10.1113/eph8802514
- Haen KM, Pett W, Lavrov DV. Parallel loss of nuclear-encoded mitochondrial aminoacyl-tRNA synthetases and mtDNA-encoded tRNAs in Cnidaria. Mol Biol Evol 2010; 27:2216-9; PMID:20439315; http://dx.doi.org/10.1093/molbev/msq112
- Brown A, Amunts A, Bai X, Sugimoto Y, Edwards PC, Murshudov G, Scheres SH, Ramakrishnan V. Structure of the large ribosomal subunit from human mitochondria. Science 2014; 346:718-22; PMID:25278503; http://dx.doi.org/10.1126/science.1258026
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature 2014; 505:515-9; PMID:24362565; http://dx.doi.org/10.1038/nature12890
- Wang X, Lavrov DV. Seventeen new complete mtDNA sequences reveal extensive mitochondrial genome evolution within the demospongiae. PLoS One 2008; 3:e2723; PMID:18628961; http://dx.doi.org/10.1371/journal.pone.0002723
- Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 2008; 135:462-74; PMID:18984158; http://dx.doi.org/10.1016/j.cell.2008.09.013
- Rossmanith W, Holzmann J. Processing mitochondrial (t)RNAs: New enzyme, old job. Cell Cycle 2009; 8:1650-3; PMID:19411849; http://dx.doi.org/10.4161/cc.8.11.8502
- Lopez Sanchez MI, Mercer TR, Davies SM, Shearwood A-MJ, Nygård KK, Richman TR, Mattick JS, Rackham O, Filipovska A. RNA processing in human mitochondria. Cell Cycle 2011; 10:2904-16; PMID:21857155; http://dx.doi.org/10.4161/cc.10.17.17060
- Brzezniak LK, Bijata M, Szczesny RJ, Stepien PP. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol 2011; 8:616-26; PMID:21593607; http://dx.doi.org/10.4161/rna.8.4.15393
- Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem 2010; 80:1-21PMID: 21417719; http://dx.doi.org/10.1146/annurev-biochem-060109-092838;
- Kohn AB, Citarella MR, Kocot KM, Bobkova YV, Halanych KM, Moroz LL. Rapid evolution of the compact and unusual mitochondrial genome in the ctenophore, Pleurobrachia bachei. Mol Phylogenet Evol 2011; 63:203-7; PMID:22201557; http://dx.doi.org/10.1016/j.ympev.2011.12.009
- Pett W, Ryan JF, Pang K, Mullikin JC, Martindale MQ, Baxevanis AD, Lavrov DV. Extreme mitochondrial evolution in the ctenophore Mnemiopsis leidyi: Insight from mtDNA and the nuclear genome. Mitochondrial DNA 2011; 22(4):1-13; PMID:21985407; http://dx.doi.org/10.3109/19401736.2011.624611
- Brugler MR, France SC. The mitochondrial genome of a deep-sea bamboo coral (Cnidaria, Anthozoa, Octocorallia, Isididae): genome structure and putative origins of replication are not conserved among octocorals. J Mol Evol 2008; 67:125-36; PMID:18506498; http://dx.doi.org/10.1007/s00239-008-9116-2
- Alfonzo JD, Blanc V, Estévez AM, Rubio MA, Simpson L. C to U editing of the anticodon of imported mitochondrial tRNA(Trp) allows decoding of the UGA stop codon in Leishmania tarentolae. EMBO J 1999; 18:7056-62; PMID:10601027; http://dx.doi.org/10.1093/emboj/18.24.7056
- Janke A, Pääbo S. Editing of a tRNA anticodon in marsupial mitochondria changes its codon recognition. Nucleic Acids Res 1993; 21:1523-5; PMID:8479901; http://dx.doi.org/10.1093/nar/21.7.1523
- Scheel BM, Hausdorf B. Dynamic evolution of mitochondrial ribosomal proteins in Holozoa. Mol Phylogenet Evol 2014; 76:67-74; PMID:24631858; http://dx.doi.org/10.1016/j.ympev.2014.03.005
