ABSTRACT
The expression of a gene is a tightly regulated process and is exerted by a myriad of different mechanisms. Recently, RNA modifications located in coding sequences of mRNAs, have been identified as potential regulators of gene expression. N6-methyladenosine (m6A), 5-methylcytosine (m5C), pseudouridine (Ψ) and N1-methyladenosine (m1A) have been found within open reading frames of mRNAs. The presence of these mRNA modifications has been implicated to modulate the fate of an mRNA, ranging from maturation to its translation and even degradation. However, many aspects concerning the biological functions of mRNA modifications remain elusive. Recently, systematic in vitro studies allowed a first glimpse of the direct interplay of mRNA modifications and the efficiency and fidelity of ribosomal translation. It thereby became evident that the effects of mRNA modifications were, astonishingly versatile, depending on the type, position or sequence context. The incorporation of a single modification could either prematurely terminate protein synthesis, reduce the peptide yield or alter the amino acid sequence identity. These results implicate that mRNA modifications are a powerful mechanism to post-transcriptionally regulate gene expression.
RNAs involved in the regulation of gene expression
Regulation of gene expression is a complex multistep process. The synthesis of a functional protein is subject to several layers of regulation, starting from the synthesis of various transcription factors up to the correct assembly of the nascent protein by chaperones. The most direct mechanism of regulating protein synthesis is the modulation of the amounts of messenger RNAs (mRNAs) within a cell. However, a direct correlation between the amounts of an mRNA and its corresponding protein is not always observed.Citation1,2 Hence, protein synthesis is the target of various highly sophisticated regulatory mechanisms, of which more and more have been identified in the last decade.Citation3-9
Generally, protein synthesis can be divided into 4 stages: initiation, elongation, termination, and ribosome recycling. Traditionally, the initiation step is viewed as one key feature of regulation and numerous factors and mechanisms have been described that lead either to a global or an mRNA-specific initiation control.Citation3,4,10 Not only altering amounts and activities of initiation factors, but also the presence of regulatory sequences or structural motifs in the 5′ or 3′ untranslated regions (UTRs) of mRNAs, are now well-understood factors for regulating initiation of protein synthesis.Citation3,4,11,12
Equally important for regulation of translation are RNA binding proteins (RBPs) and non-coding RNAs (ncRNAs). As soon as ncRNAs, such as miRNAs and siRNAs, had been identified, it became evident that these small ncRNA species are directly involved in modulating gene expression.Citation13,14 Although siRNA and miRNA differ in their origin and function, both guide the RNA Induced Silencing Complex (RISC) to their target mRNAs and thus induce cleavage or degradation of mRNAs, respectively. Thereby, miRNAs were reported to modulate translation initiation as well as elongation, thus rendering small ncRNAs as versatile molecules for modulating gene expression.Citation15
Whereas RBPs and ncRNAs bind to mRNAs, a recently discovered class of regulatory RNAs directly binds to the ribosome, thereby affecting protein synthesis. These ribosomal associated ncRNAs (rancRNAs) interfere with protein synthesis in a stress-dependent manner.Citation16-18 Thereby, RNA fragments derived from mRNAs or transfer RNAs (tRNAs) have been identified to bind to the ribosome and globally inhibit translation. For example, 5′-tRNA fragments, identified in Haloferax volcanii, downregulate protein synthesis globally by binding to the small ribosomal subunit, thereby competing with binding of mRNAs. Subsequently, additional tRNA fragments have been identified to interfere with translation,Citation19-21 but their biological roles and mechanisms of action are still not completely understood.Citation22
Since also full-length tRNAs interact with the ribosome and thus play a central role in translation, it seems inevitable that they would be also involved in modulating translation. Thereby, it is has been reported that the abundance, the availability of tRNAs and the codon usage within mRNAs strongly influences the speed and efficiency of translation (Citation23,24 and reviewed inCitation25,26). Recently, tRNA modifications were also revealed to be linked to translation efficiency and decoding fidelity.Citation26-28 In addition, modifications of tRNAs have been found to be involved in fine tuning of stress-related genes by driving codon biased translation.Citation7,29
In addition, mRNAs are not just mere templates for translation, but harbor essential regulatory elements. As mentioned above, specific regions within UTRs of mRNAs might be involved in regulation of translation.Citation3,4,11,12 These regulatory elements, such as structural RNA motifs or binding sites for proteins or for regulatory RNAs, are found primarily in 5′ and 3′ UTRs of mRNAs. However, also ORFs are able to influence the efficiency and speed of translation.Citation30-32 Thereby, specific codons and sequence elements cause ribosomal stalling and consequently the folding and activity of the produced proteins might be affected.Citation24,33-36 Codon-optimized sequences might result in higher product yields, but also lead to lower enzyme activities.Citation34
Recently, a mechanism in regulating ribosomal translation has been identified: thereby, modifications of RNA nucleotides within coding sequences of mRNAs were reported to directly interfere with elongation and decoding of the ribosomal translation machinery.Citation37-39 Herein, we summarize current findings of co- and post-transcriptional mRNA modifications affecting translation and anticipate what lessons these modifications might teach us (i.e. what biological roles they might exert in different organisms).
Old RNA modifications - new perspectives
Modifications within the ORFs of mRNAs have already been described in the 1970s, demonstrating the occurrence of N6-methyladenosine (m6A)Citation40-44 and 5-methylcytosine (m5C).Citation45 Due to technical limitations, however, the identification of distinct nucleotide modifications was not possible. Thus, neither the detailed localization of modifications within transcripts, nor their biological role has been elucidated. Hence, these early studies were sometimes controversially interpreted in respect to a potential function of mRNA modifications, mainly due to their presumably low abundance and their identification in distinct cell types only.Citation40,44 But subsequent to uncovering specific enzymes, capable of converting adenosines (A) to inosines (I) within dsRNA (ADARs),Citation46-51 the field of mRNA modifications re-gained attention. This A-to-I editing process has been unveiled as the most prevalent form of mRNA editing, leading to a new and prospering research field.
The interest in mRNA nucleotide modifications once again increased through the invention of RNA mass spectrometry and next generation sequencing (NGS) techniques, which equipped researchers with powerful tools to identify and map modifications within transcripts. Several groups have contributed to our current understanding of when, where, and most importantly how mRNA modifications regulate gene expression. To date, the repertoire of naturally occurring eukaryal mRNA modifications (besides the 5´ cap and inosine) is comprised of m6A, pseudoruridine (Ψ), m5C and N1-methyladenosine (m1A).Citation52-59 Thereby, m6A is not only the most abundant, but also the best-characterized internal mRNA modification so far. The dynamic nature of the presence of m6A within mRNAs and its involvement in various biological functions is remarkable, ranging from splicing, regulation of translation to mRNA decay.Citation60-63 Also Ψ is dynamically deposited in mRNA transcripts, but its function within mRNAs is still elusive.Citation53-55
Even less is known about the possible role of m5C within mRNAs. m5C was reported to be present in humanCitation56 as well as in archaealCitation59 mRNAs. Due to its reported enrichment within the UTRs of human mRNAs and its location in the vicinity of Argonaute binding sites, m5C was suggested to be involved in translation regulation.Citation56 The most recent modification, which has joined the mRNA repertoire, is m1A.Citation57,58 Thereby, m1A is predominantly found in structured regions of the 5′ UTR of mRNAs and in the vicinity of canonical and alternative translation initiation sites. Most interestingly, the presence of m1A in mRNAs is connected to elevated translation rates.Citation57 Modifications of the ribose, such as 2′-O-methylations, have not yet been unambiguously identified within coding sequences of mRNAs. 2′-O-methylations are commonly found in ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs, reviewed inCitation64) or tRNAs (reviewed inCitation65) and are also speculated to be introduced into ORFs by small nucleolar RNA-guided complexes.Citation66,67
Translational regulation mediated by mRNA modifications
Until recently, mRNA modifications have only been indirectly linked to ribosomal translation.Citation61,62,68-71 An emerging body of evidence implies that the effect of modifications is not only dependent on its type but also on the translation system. Thereby, mRNAs harboring Ψs within coding sequences increased the protein yield in rabbit reticulocyte extracts but not in wheat germ extracts.Citation70 However, in bacterial translation systems mRNAs with randomly incorporated Ψs strongly inhibited protein synthesis.Citation70 Similar observations have been made with other mRNA modifications as well.Citation68,70 These findings were supported by cell culture-based approaches revealing a significant cell type dependency.Citation57,70,71
Additionally, the sequence context affects the impact of mRNA modifications. Thereby, protein expression strongly depends on the corresponding mRNA sequence, thus making it even more complicated to univocally draw conclusions about the impact of specific modifications on the translation machinery.Citation70,71
The majority of these studies employed randomly modified mRNAs.Citation68-70 Alternatively, mRNAs with a complete substitution of the unmodified nucleotide by a modified version were applied.Citation71 Thus, in order to obtain a more detailed picture of the direct effect of single mRNA modifications on protein synthesis, a refined systematic approach was applied. Employing a splinted ligation protocol, RNA modifications were site-specifically incorporated into reporter mRNAs.Citation37 Thereby, the nucleotide derivatives were positioned at the 1st, 2nd or 3rd position of the codon, respectively, and subsequently peptide products of the corresponding mRNA construct were analyzed.
Strikingly, the resulting effects on translation were not only strongly dependent on the type but also on the position of the modifications. Thereby, 2′-O-methylated nucleosides at the 1st codon position only marginally affected translation, however, when placed at the 2nd position they caused an almost complete termination of protein synthesis at the modified nucleotide.Citation37 On the contrary, m6A revealed the strongest inhibition at the 1st and Ψ at the 3rd codon position.Citation37,38 In addition, also the sequence context seemed to exert a significant influence on translation. Whereas the 2′-O-methyl group at the second codon position was independent of the codon and the sequence context,Citation37 m6A exhibited a strong sequence dependence.Citation38
In these studies, not only the efficiency of translation was investigated but also a long-standing question concerning the ability of mRNA modifications to rewire the genetic code was addressed. Thereby, the best-known example of recoding the genetic information at the RNA level is A-to-I editing.Citation72,73 Within coding sequences I is read as a G by the translation machinery and therefore can lead to an amino acid change, dependent on its position within a codon.
It has been speculated that Ψ within coding sequences might also possess the potential to rewire the genetic code.Citation53,74-76 This speculation was based on observations that Ψ, located within a stop codon caused partial read-through of translation.Citation75 However, in coding sequences a recoding event, induced by Ψs could not be detected.Citation37 In contrast, m5C induced, to some extent, an amino acid substitution.Citation37 Thereby, m5C placed at the 2nd position of a CCC codon, resulted in a substitution of proline by leucine. Importantly, this partial “recoding” was also strongly dependent on the position of the modification within the codon and might also be influenced by the sequence context, similar to m6A.Citation38 Thus, additional experiments will be required to identify mechanisms how m5C interferes with ribosomal decoding. It further needs to be demonstrated, if this rather weak recoding effect is of biological relevance. Nevertheless, it is remarkable that the decoding process of the bacterial translation machinery is affected by m5C in a codon position-dependent manner.
Potential functions of internal mRNA modifications
Dependent on the modification and the position within a codon, a variety of effects on protein synthesis have been observed. Introducing a 2′-O-methyl group at the 2nd codon position prematurely terminated translation efficiently at the site of modification. This poses the question, as to the function of translation termination at the modification site and the associated peptide fragments. As a very direct consequence, modifications might merely reduce the amounts of a protein produced by the modified mRNA, thereby regulating its expression in analogy to miRNAs, for example. It is thereby interesting to note that in eukarya, mRNAs containing premature stop codons, which would result in such shortened peptides, are immediately degraded by the nonsense mediated decay machinery (NMD, reviewed inCitation77).
In bacteria, mRNAs lacking a stop codon due to shortening of the mRNA by degradation or cleavage, are subject to the tmRNA pathway (reviewed inCitation78). Thereby, the transfer-messenger RNA (tmRNA), binds to the ribosomal A site, by first structurally mimicking an alanine tRNA, and adding alanine to the peptide chain.Citation79,80 Subsequently, the tmRNA acts as an mRNA adding a specific protein sequence, encoded within the tmRNA, to the truncated peptide that is subsequently recognized and degraded by a protease.Citation81 Thus, in both eukarya and bacteria, these mechanisms prevent the generation of shortened peptides that might be harmful to cells.
In this context, site-specific incorporation of certain modifications within mRNAs might have a function, in addition to merely reducing protein levels: in particular, they might enable the generation of shortened peptides with novel functions, beneficial for eukarya or bacteria, respectively. Dependent on the level of modification within an mRNA and its position, various amounts of these peptides can be generated and peptides of various sizes, exhibiting different functions, might be produced. The benefit of such a mechanism is that, despite the presence of the modification and its impact on translation, in addition the full-length protein could still be synthesized in sufficient levels, thereby increasing protein diversity.
In fact, there are examples which support such a model: apoliprotein B (apoB) is synthesized in the liver as apoB100, whereas in the small intestine the apoB48 variant is present.Citation82,83 Both proteins are produced by the same gene, but through C-to-U editing in the small intestine a UAA codon is generated, resulting in the truncated form of the protein with distinct functions compared with full-length apoB100.Citation84
In addition, regulatory mechanisms exist that induce these mRNA modifications only in response to certain environmental cues. Such a dependency has been demonstrated for Ψ, m6A and very recently for m1A.Citation52,53,57 These mechanisms thus might reflect an epigenetic regulation of gene expression on the level of RNA, where signals from the environment of a cell are transferred to RNAs. However, if mRNA modifications in fact represent such a so far unidentified mechanism to generate smaller, but functional, peptides of various length, still the question has to be answered, how the ribosome deals with such a premature termination, in particular as release factors are required to free the premature terminated peptide from the ribosome.
In summary, the site-specific incorporation of modified RNA nucleotides into coding regions of mRNAs revealed astonishingly versatile effects on protein synthesis depending not only on the type of the RNA modification but also on the codon position (). In addition, various organisms and cell types potentially cope differently with the presence of modifications within mRNAs. Their biological function can range from fine-tuning translational rates to premature termination of protein synthesis. Post-transcriptional mRNA modifications might even possess the potential to expand the diversity of proteins through recoding. Therefore, it is of utmost importance to elucidate all mechanisms behind.
Figure 1. The potential cellular functions mediated by mRNA modifications are versatile. Modifications can be co- or post-transcriptionally incorporated into mRNAs (green) in response to environmental stimuli, such as changes in growth conditions or stress (e.g. starvation, radiation, temperature shifts, nutrition deprivation etc.). Single modifications or combinations thereof within an mRNA might regulate the rate of translation, the actual peptide length, or the identity of a protein. Thus, mRNA modifications represent a highly sophisticated biological mechanism to regulate gene expression.
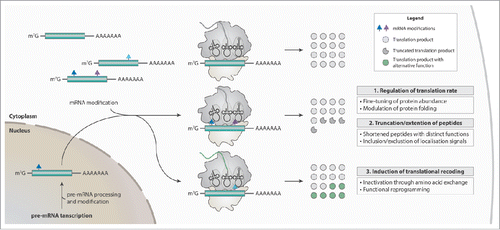
mRNA modifications not only affect translation, but can also act as markers to provide landing platforms for proteinsCitation61,62,85,86 or stimulate other regulatory processes like mRNA degradationCitation60 or localization.Citation87 Their role as markers is reminiscent of the regulation of gene expression through epigenetic DNA and histone modifications. In line with that, not single modifications but a combination thereof might collectively mediate biological functions. Such modification patterns could serve as landmarks to stimulate or trigger down-stream effects. However, this is purely speculative and many aspects of mRNA modifications are still far from being completely understood. Elucidating the regulation of mRNA modifications and their cellular functions will open up a completely new way in understanding gene regulation on the level of RNA.
Disclosure of potential confllicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We apologize for the omission of many important references due to space limitations. We would like to thank Nina Clementi for critically reading the manuscript and helpful discussions.
Funding
This work was funded by the Austrian Science Fund (FWF) (P 22658-B12 and P 28494-BBL to M.E; SFB F4411 to A.H) and the European Commission (GA N 602133 ncRNAPain to A.H).
References
- Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett 2009; 583:3966-73; PMID:19850042; http://dx.doi.org/10.1016/j.febslet.2009.10.036
- Taniguchi Y, Choi PJ, Li GW, Chen H, Babu M, Hearn J, Emili A, Xie XS. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 2010; 329:533-8; PMID:20671182; http://dx.doi.org/10.1126/science.1188308
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/10.1016/j.cell.2009.01.042
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 2004; 5:827-35; PMID:15459663; http://dx.doi.org/10.1038/nrm1488
- Wilson DN, Nierhaus KH. The weird and wonderful world of bacterial ribosome regulation. Crit Rev Biochem Mol Biol 2007; 42:187-219; PMID:17562451; http://dx.doi.org/10.1080/10409230701360843
- Wilson DN, Arenz S, Beckmann R. Translation regulation via nascent polypeptide-mediated ribosome stalling. Curr Opin Struct Biol 2016; 37:123-33; PMID:26859868; http://dx.doi.org/10.1016/j.sbi.2016.01.008
- Endres L, Dedon PC, Begley TJ. Codon-biased translation can be regulated by wobble-base tRNA modification systems during cellular stress responses. RNA Biol 2015; 12:603-14; PMID:25892531; http://dx.doi.org/10.1080/15476286.2015.1031947
- Meister G. miRNAs get an early start on translational silencing. Cell 2007; 131:25-8; PMID:17923084; http://dx.doi.org/10.1016/j.cell.2007.09.021
- Sofos N, Xu K, Dedic E, Brodersen DE. Cut to the chase–Regulating translation through RNA cleavage. Biochimie 2015; 114:10-7; PMID:25633441; http://dx.doi.org/10.1016/j.biochi.2015.01.009
- Duval M, Simonetti A, Caldelari I, Marzi S. Multiple ways to regulate translation initiation in bacteria: Mechanisms, regulatory circuits, dynamics. Biochimie 2015; 114:18-29; PMID:25792421; http://dx.doi.org/10.1016/j.biochi.2015.03.007
- Xue S, Barna M Specialized ribosomes: a new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol 2012; 13:355-69; PMID:22617470; http://dx.doi.org/10.1038/nrm3359
- Xue S, Barna M. Cis-regulatory RNA elements that regulate specialized ribosome activity. RNA Biol 2015; 12:1083-7; PMID:26327194; http://dx.doi.org/10.1080/15476286.2015.1085149
- Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009; 136:642-55; PMID:19239886; http://dx.doi.org/10.1016/j.cell.2009.01.035
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993; 75:843-54; PMID:8252621; http://dx.doi.org/10.1016/0092-8674(93)90529-Y
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010; 79:351-79; PMID:20533884; http://dx.doi.org/10.1146/annurev-biochem-060308-103103
- Pircher A, Bakowska-Zywicka K, Schneider L, Zywicki M, Polacek N. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol Cell 2014; 54:147-55; PMID:24685157; http://dx.doi.org/10.1016/j.molcel.2014.02.024
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 2013; 10:1798-806; PMID:24351723; http://dx.doi.org/10.4161/rna.27177
- Gebetsberger J, Zywicki M, Künzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012; 2012:260909; PMID:23326205; http://dx.doi.org/10.1155/2012/260909
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011; 43:613-23; PMID:21855800; http://dx.doi.org/10.1016/j.molcel.2011.06.022
- Jöchl C, Rederstorff M, Hertel J, Stadler PF, Hofacker IL, Schrettl M, Haas H, Hüttenhofer A. Small ncRNA transcriptome analysis from Aspergillus fumigatus suggests a novel mechanism for regulation of protein synthesis. Nucleic Acids Res 2008; 36:2677-89; PMID:18346967; http://dx.doi.org/10.1093/nar/gkn123
- Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 2009; 185:35-42; PMID:19332886; http://dx.doi.org/10.1083/jcb.200811106
- Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol 2013; 10:553-63; PMID:23563448; http://dx.doi.org/10.4161/rna.24285
- Pedersen S. Escherichia coli ribosomes translate in vivo with variable rate. EMBO J 1984; 3:2895-8; PMID:6396082
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol 2009; 16:274-80; PMID:19198590; http://dx.doi.org/10.1038/nsmb.1554
- Fredrick K, Ibba M. How the sequence of a gene can tune its translation. Cell 2010; 141:227-9; PMID:20403320; http://dx.doi.org/10.1016/j.cell.2010.03.033
- Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nat Rev Genet 2015; 16:98-112; PMID:25534324; http://dx.doi.org/10.1038/nrg3861
- Manickam N, Joshi K, Bhatt MJ, Farabaugh PJ. Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res 2016; 44:1871-81; PMID:26704976; http://dx.doi.org/10.1093/nar/gkv1506
- Deng W, Babu IR, Su D, Yin S, Begley TJ, Dedon PC. Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet 2015; 13:6557-67
- Chan CT, Pang YL, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 2012; 3:937; PMID:22760636; http://dx.doi.org/10.1038/ncomms1938
- Chaney JL, Clark PL. Roles for Synonymous Codon Usage in Protein Biogenesis. Annu Rev Biophys 2015; 44:143-66; PMID:25747594; http://dx.doi.org/10.1146/annurev-biophys-060414-034333
- Chevance FF, Le Guyon S, Hughes KT. The effects of codon context on in vivo translation speed. PLoS Genet 2014; 10:e1004392; PMID:24901308; http://dx.doi.org/10.1371/journal.pgen.1004392
- Wen JD, Lancaster L, Hodges C, Zeri AC, Yoshimura SH, Noller HF, Bustamante C, Tinoco I. Following translation by single ribosomes one codon at a time. Nature 2008; 452:598-603; PMID:18327250; http://dx.doi.org/10.1038/nature06716
- Peil L, Starosta AL, Lassak J, Atkinson GC, Virumäe K, Spitzer M, Tenson T, Jung K, Remme J, Wilson DN. Distinct XPPX sequence motifs induce ribosome stalling, which is rescued by the translation elongation factor EF-P. Proc Natl Acad Sci U S A 2013; 110:15265-70; PMID:24003132; http://dx.doi.org/10.1073/pnas.1310642110
- Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett 1999; 462:387-91; PMID:10622731; http://dx.doi.org/10.1016/S0014-5793(99)01566-5
- Cortazzo P, Cerveñansky C, Marín M, Reiss C, Ehrlich R, Deana A. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem Biophys Res Commun 2002; 293:537-41; PMID:12054634; http://dx.doi.org/10.1016/S0006-291X(02)00226-7
- Starosta AL, Lassak J, Peil L, Atkinson GC, Virumäe K, Tenson T, Remme J, Jung K, Wilson DN. Translational stalling at polyproline stretches is modulated by the sequence context upstream of the stall site. Nucleic Acids Res 2014; 42:10711-9; PMID:25143529; http://dx.doi.org/10.1093/nar/gku768
- Hoernes TP, Clementi N, Faserl K, Glasner H, Breuker K, Lindner H, Hüttenhofer A, Erlacher MD. Nucleotide modifications within bacterial messenger RNAs regulate their translation and are able to rewire the genetic code. Nucleic Acids Res 2016; 44:852-62; PMID:26578598; http://dx.doi.org/10.1093/nar/gkv1182
- Choi J, Ieong KW, Demirci H, Chen J, Petrov A, Prabhakar A, O'Leary SE, Dominissini D, Rechavi G, Soltis SM, et al. N(6)-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat Struct Mol Biol 2016; 23:110-5; PMID:26751643; http://dx.doi.org/10.1038/nsmb.3148
- Hoernes TP, Erlacher MD. Translating the epitranscriptome. Wiley Interdiscip Rev RNA 2016 June 27; http://dx.doi.org/10.1002/wrna.1375.
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci U S A 1974; 71:3971-5; PMID:4372599; http://dx.doi.org/10.1073/pnas.71.10.3971
- Lavi S, Shatkin AJ. Methylated simian virus 40-specific RNA from nuclei and cytoplasm of infected BSC-1 cells. Proc Natl Acad Sci U S A 1975; 72:2012-6; PMID:166375; http://dx.doi.org/10.1073/pnas.72.6.2012
- Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 1975; 4:379-86; PMID:164293; http://dx.doi.org/10.1016/0092-8674(75)90158-0
- Salditt-Georgieff M, Jelinek W, Darnell JE, Furuichi Y, Morgan M, Shatkin A. Methyl labeling of HeLa cell hnRNA: a comparison with mRNA. Cell 1976; 7:227-37; PMID:954080; http://dx.doi.org/10.1016/0092-8674(76)90022-2
- Adams JM, Cory S. Modified nucleosides and bizarre 5′-termini in mouse myeloma mRNA. Nature 1975; 255:28-33; PMID:1128665; http://dx.doi.org/10.1038/255028a0
- Dubin DT, Stollar V. Methylation of Sindbis virus “26S” messenger RNA. Biochem Biophys Res Commun 1975; 66:1373-9; PMID:1191298; http://dx.doi.org/10.1016/0006-291X(75)90511-2
- Rebagliati MR, Melton DA. Antisense RNA injections in fertilized frog eggs reveal an RNA duplex unwinding activity. Cell 1987; 48:599-605; PMID:2434240; http://dx.doi.org/10.1016/0092-8674(87)90238-8
- Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci U S A 1994; 91:11457-61; PMID:7972084; http://dx.doi.org/10.1073/pnas.91.24.11457
- Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature 1996; 379:460-4; PMID:8559253; http://dx.doi.org/10.1038/379460a0
- Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. RNA 2000; 6:755-67; PMID:10836796; http://dx.doi.org/10.1017/S1355838200000170
- Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 1988; 55:1089-98; PMID:3203381; http://dx.doi.org/10.1016/0092-8674(88)90253-X
- Hough RF, Bass BL. Purification of the Xenopus laevis double-stranded RNA adenosine deaminase. J Biol Chem 1994; 269:9933-9; PMID:8144588
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012; 485:201-6; PMID:22575960; http://dx.doi.org/10.1038/nature11112
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014; 515:143-6; PMID:25192136; http://dx.doi.org/10.1038/nature13802
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014; 159:148-62; PMID:25219674; http://dx.doi.org/10.1016/j.cell.2014.08.028
- Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 2014; 9:e110799; PMID:25353621; http://dx.doi.org/10.1371/journal.pone.0110799
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 2012; 40:5023-33; PMID:22344696; http://dx.doi.org/10.1093/nar/gks144
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016; 530:441-6; PMID:26863196; http://dx.doi.org/10.1038/nature16998
- Li X, Xiong X, Wang K, Wang L, Shu X, Ma S, Yi C. Transcriptome-wide mapping reveals reversible and dynamic N(1)-methyladenosine methylome. Nat Chem Biol 2016; 12:311-6; PMID:26863410; http://dx.doi.org/10.1038/nchembio.2040
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 2013; 9:e1003602; PMID:23825970; http://dx.doi.org/10.1371/journal.pgen.1003602
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014; 505:117-20; PMID:24284625; http://dx.doi.org/10.1038/nature12730
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, Qian SB. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 2015; 526:591-4; PMID:26458103; http://dx.doi.org/10.1038/nature15377
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015; 161:1388-99; PMID:26046440; http://dx.doi.org/10.1016/j.cell.2015.05.014
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res 2014; 24:1403-19; PMID:25412662; http://dx.doi.org/10.1038/cr.2014.151
- Bachellerie JP, Cavaillé J, Hüttenhofer A. The expanding snoRNA world. Biochimie 2002; 84:775-90; PMID:12457565; http://dx.doi.org/10.1016/S0300-9084(02)01402-5
- Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA 2011; 2:611-31; PMID:21823225; http://dx.doi.org/10.1002/wrna.79
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science 2006; 311:230-2; PMID:16357227; http://dx.doi.org/10.1126/science.1118265
- Vitali P, Basyuk E, Le Meur E, Bertrand E, Muscatelli F, Cavaillé J, Huttenhofer A. ADAR2-mediated editing of RNA substrates in the nucleolus is inhibited by C/D small nucleolar RNAs. J Cell Biol 2005; 169:745-53; PMID:15939761; http://dx.doi.org/10.1083/jcb.200411129
- Heilman KL, Leach RA, Tuck MT. Internal 6-methyladenine residues increase the in vitro translation efficiency of dihydrofolate reductase messenger RNA. Int J Biochem Cell Biol 1996; 28:823-9; PMID:8925412; http://dx.doi.org/10.1016/1357-2725(96)00014-3
- Karikó K, Muramatsu H, Keller JM, Weissman D. Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther 2012; 20:948-53; http://dx.doi.org/10.1038/mt.2012.7
- Karikó K, Muramatsu H, Welsh FA, Ludwig J, Kato H, Akira S, Weissman D. Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 2008; 16:1833-40; http://dx.doi.org/10.1038/mt.2008.200
- Li B, Luo X, Dong Y. Effects of Chemically Modified Messenger RNA on Protein Expression. Bioconjug Chem 2016; 27:849-53; PMID:26906521; http://dx.doi.org/10.1021/acs.bioconjchem.6b00090
- Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol 2016; 17:83-96; PMID:26648264; http://dx.doi.org/10.1038/nrm.2015.4
- Mallela A, Nishikura K. A-to-I editing of protein coding and noncoding RNAs. Crit Rev Biochem Mol Biol 2012; 47:493-501; PMID:22988838; http://dx.doi.org/10.3109/10409238.2012.714350
- Fernández IS, Ng CL, Kelley AC, Wu G, Yu YT, Ramakrishnan V. Unusual base pairing during the decoding of a stop codon by the ribosome. Nature 2013; 500:107-10; http://dx.doi.org/10.1038/nature12302
- Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 2011; 474:395-8; PMID:21677757; http://dx.doi.org/10.1038/nature10165
- Parisien M, Yi C, Pan T. Rationalization and prediction of selective decoding of pseudouridine-modified nonsense and sense codons. RNA 2012; 18:355-67; PMID:22282339; http://dx.doi.org/10.1261/rna.031351.111
- Lykke-Andersen S, Jensen TH. Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 2015; 16:665-77; PMID:26397022; http://dx.doi.org/10.1038/nrm4063
- Keiler KC. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol 2015; 13:285-97; PMID:25874843; http://dx.doi.org/10.1038/nrmicro3438
- Bessho Y, Shibata R, Sekine S, Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, Yokoyama S. Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc Natl Acad Sci U S A 2007; 104:8293-8; PMID:17488812; http://dx.doi.org/10.1073/pnas.0700402104
- Komine Y, Kitabatake M, Yokogawa T, Nishikawa K, Inokuchi H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli. Proc Natl Acad Sci U S A 1994; 91:9223-7; PMID:7524073; http://dx.doi.org/10.1073/pnas.91.20.9223
- Keiler KC, Waller PR, Sauer RT. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 1996; 271:990-3; PMID:8584937; http://dx.doi.org/10.1126/science.271.5251.990
- Chen SH, Habib G, Yang CY, Gu ZW, Lee BR, Weng SA, Silberman SR, Cai SJ, Deslypere JP, Rosseneu M, et al. Apolipoprotein B-48 is the product of a messenger RNA with an organ-specific in-frame stop codon. Science 1987; 238:363-6; PMID:3659919; http://dx.doi.org/10.1126/science.3659919
- Powell LM, Wallis SC, Pease RJ, Edwards YH, Knott TJ, Scott J. A novel form of tissue-specific RNA processing produces apolipoprotein-B48 in intestine. Cell 1987; 50:831-40; PMID:3621347; http://dx.doi.org/10.1016/0092-8674(87)90510-1
- Olofsson SO, Bòren J. Apolipoprotein B: a clinically important apolipoprotein which assembles atherogenic lipoproteins and promotes the development of atherosclerosis. J Intern Med 2005; 258:395-410; PMID:16238675; http://dx.doi.org/10.1111/j.1365-2796.2005.01556.x
- Xu C, Wang X, Liu K, Roundtree IA, Tempel W, Li Y, Lu Z, He C, Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol 2014; 10:927-9; PMID:25242552; http://dx.doi.org/10.1038/nchembio.1654
- Liu N, Dai Q, Zheng G, He C, Parisien M, Pan T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015; 518:560-4; PMID:25719671; http://dx.doi.org/10.1038/nature14234
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013; 155:793-806; PMID:24209618; http://dx.doi.org/10.1016/j.cell.2013.10.026
