ABSTRACT
Translational regulation is a critical step in the production of proteins from genomic material in both human and viruses. However, unlike other steps of the central dogma, such as transcriptional regulation, little is known about the cis-regulatory elements involved. In a recent study we devised a high-throughput bicistronic reporter assay for the discovery and the characterization of thousands of novel Internal Ribosome Entry Sites (IRESs) in human and hundreds of viral genomes. Our results provide insights into the landscape of IRES elements in human and viral transcripts and the cis-regulatory sequences underlying their activity. Here, we discuss these results as well as emerging insights from other studies, providing new views about translational regulation in human and viruses. In addition, we highlight recent high-throughput technologies in the field and discuss how combining insights from high- and low-throughput approaches can illuminate yet uncovered mechanisms of translational regulation.
Introduction
Despite the importance of mRNA translation in regulating gene expression across all kingdoms of life, we have a relatively poor understanding of how ribosome specificity to its target mRNAs is conferred. Systematic methods to investigate mRNA translation have lagged far behind other fields of gene expression such as transcriptional control. Thus, while the major challenge in the field of transcriptional control is to decipher how the thoroughly investigated cis-regulatory elements are combined to orchestrate a transcriptional output,Citation1-4 the identity of the most basic building blocks that regulate the recognition and translation of mRNA by the ribosome remain uncharacterized.
Several mechanisms were described for translation initiation, which are commonly classified as cap-dependent or cap-independent. Cap-dependent translation involves the recognition of the m7GpppX cap structure at the 5′ end of the translated transcript and thus does not rely on sequence specificity for ribosome recruitment. The most investigated cap-independent mechanism involves the recruitment of the ribosome to a cis-regulatory element, originally characterized by a complex structure, in the translated transcript termed Internal Ribosome Entry Site (IRES). IRESs were initially discovered in picornaviruses,Citation5,6 positive-strand RNA viruses ([+]ssRNA) that do not acquire a cap structure during their life cycle in the cytoplasm. The observation that poliovirus infection is accompanied by a reduction in host genes translation with no impact on viral genesCitation7 together with lacking a 5′ cap structure to support cap-dependent translation led researches to hypothesize that polioviruses employ alternative mechanism for translation initiation and to the revolutionary discovery of IRES-dependent translation. The finding of IRES elements later in capped cellular transcripts led to the identification of additional role of IRESs in facilitating the ongoing synthesis of specific proteins under stress conditions in which cap-dependent translation is compromised. Accordingly, key genes in the response of cells to various stresses, such as ER stress, apoptosis and amino-acid starvation, harbor IRES elements in their 5′UTR.Citation8 Moreover, IRESs can also enhance the repertoire of the synthesized proteins from a single transcript. They enable the production of a protein from an independent open reading frame (ORF) in a bicistronic transcriptCitation9 and guide the ribosome to produce an N-truncated isoform from an alternative AUG located downstream to the authentic start codon.Citation10-13 Another cap-independent mechanism was described in plant viruses, many of which lack the 5′ cap structure. Here too, a cis-regulatory element termed cap-independent translation enhancer (CITE) facilitates the recruitment of the 40S ribosomal subunit to the translated transcript.Citation14,15 Most of the studied CITE elements reside within the 3′UTR region of viral transcripts and relocate the recruited ribosome to the 5′ end via long-distance, kissing-loop interactions with a hairpin loop located in the 5′UTR.Citation16 Unlike IRESs, CITE-mediated translation starts at the 5′ end of the mRNA and requires ribosome scanning for the recognition of the start codon.Citation14,17 Thus, these 2 types of elements are distinguished in their ability to facilitate translation of an ORF that has no free 5′ end such as in the case of bicistronic transcripts.
The most common technique to detect novel IRESs is a bicistronic reporter assay in which the tested sequence is inserted between 2 cistrons (encoding 2 different reporter proteins) so that the first cistron is translated via the canonical cap-dependent mechanism and the second cistron is translated if the inserted sequence is a functional IRES sequence (but not CITE). The evaluation of each candidate requires amplification of the tested sequence, cloning it to a bicistronic vector, transfection of the vector into cells, and performing a functional assay to detect the expression levels of the 2 reporter proteins (e.g. dual luciferase assay). Thus, although IRESs are essential for the synthesis of many human and viral proteins and take part in a variety of biological processes, such as viral infections,Citation5,6 the response of cells to stress,Citation8 and organismal development,Citation18 only few dozens of IRESs were identified to dateCitation19 and little is known about their position in the human and viral genomes and the mechanisms by which they recruit the ribosome. In a recent study we devised a high-throughput bicistronic assay to obtain quantitative and accurate measurements of IRES activity for 55,000 fully designed oligonucleotides representing native and synthetic sequences from the human genome as well as hundreds of viral genomes.Citation20 To this end, we employed a fluorescent-based bicistronic reporter carrying mRFP and eGFP as the first and second cistrons, respectively, and used a fluorescence-activated cell sorting together with deep sequencing (FACS-seq)Citation21 to facilitate the measurements of IRES activity in mass. Our assay uncovered thousands of novel IRESs, identified distinct regions in human and viral transcripts that are enriched in IRESs, and several regulatory elements underlying IRES activity
Here, we discuss these results that together with recent discoveries in the field of translational control suggest the existence of novel mechanisms such as the recruitments of the ribosome to the 3′UTR of human transcripts and regulated translation of individual proteins from [+]ssRNA viral genomes. In addition, we survey recent high-throughput technologies such as ribosome profiling and mRNA display and discuss how combining insights from systematic studies with low-throughput experiments can enhance our understanding of translational regulation in human and viruses.
Identification of hundreds of cellular IRESs
While there is strong consensus in the scientific community about viral IRESs, the existence of cellular IRESs is subject to a long lasting debate. The most common argument is that the experimental design of a bicistronic DNA reporter is prone to artifacts such as cryptic promoter or cryptic splice site that give raise to capped monocistronic transcripts.Citation22,23 These transcripts in turn can be efficiently translated via the canonical cap-dependent mechanism leading to false positive results.Citation24 To account for these potential artifacts we devised 2 additional high-throughput assays: (a) high-throughput assay for promoter activity in which the entire library (55,000) was cloned into a bicistronic plasmid lacking endogenous promoter; (b) high-throughput assay for splicing activity in which reduction in the intact bicistronic mRNA transcript was quantitatively measured using deep sequencing of the cDNA and gDNA. In addition to these 2 assays we also performed a set of controls including: qRT-PCR on the entire eGFP(+) population with 3 different sets of primers probing the mRFP cistron in various locations; qRT-PCR on isolated clones expressing individual IRESs with primers on the eGFP and the mRFP cistrons; and validation of selected IRESs in a bicistronic and monocistronic luciferase plasmids.
Using these controls our assay revealed 583 novel IRESs in the 5′UTRs of cellular genes. Although we cannot validate every individual IRES identified, we believe that most of these sequences are true IRES elements. Interestingly, systematic investigation of the cis-regulatory sequences (that we discuss below) uncovered some mutual elements underlying the activity of cellular and viral IRESs such as short complementary sequences to the 18S rRNA (18S rRNA).
The identification of this largest collection of cellular IRESs represents an advance in the research of IRES-dependent translation of mammalian genes. However, further investigation of these IRESs within their native mRNA context under stress conditions and in non-stressed cells is needed to determine the manner by which they regulate gene expression.
The landscape of IRES elements in human transcripts
Low throughput methods to investigate IRES activity are slow and labor-intensive and typically test only a few sequences at a time. Thus, the tested sequences are carefully pre-selected so that most of the studies focus on the 5′UTR region when searching for novel IRESs. In our study we exploited the power of high-throughput measurements to conduct an unbiased and systematic profiling of IRES elements across human transcripts by assaying thousands of sequences spanning 159 transcripts. Surprisingly, we found that in addition to an expected enrichment in the 5′UTR region, IRESs are highly enriched in the 3′UTR of human transcripts. Interestingly, although translation is typically considered to be a linear process in which the ribosome initiates translation at the 5′ end and drops from the massage at the 3′ end, mRNA molecules in cells are circularized through interaction between eIF4G and the Poly(A)-binding protein (PABP).Citation25 Thus, we hypothesize that IRES elements can recruit the ribosome to the 3′UTR to facilitate translation from the start codon by utilizing mRNA looping ().
Figure 1. Hypothetical model for translation initiation from the 3′UTR of human transcript. mRNA is circularized as a result of interactions between the 50cap binding protein eIF4E, the scaffold protein eIF4G and the poly(A)-binding protein PABP. According to the proposed model, ribosomes are attracted to an IRES elements in the 30UTR of the circularized transcript (1), shunted to the 50 end (2), and initiate translation at the AUG of the encoded open reading frame (ORF) (3).
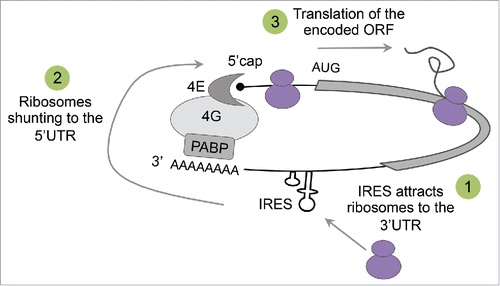
Although more experiments are needed in order to determine the existence of such a mechanism, several findings from other studies support this hypothesis. These include: (a) Emerging ribosome profiling experiments from various studies demonstrate high abundance of ribosomes at the 3′UTR region of eukaryotic transcripts.Citation26-28 (b) Recent work by Paek et al.Citation29 showing that tethering eIFG4 to the 3′UTR of a transcript via artificial MS2-binding sites enhances translation of the upstream coding sequence. Moreover, the authors obtained similar effect on translation when planting the natural EMCV IRES in the 3′UTR. Thus, this study provides direct evidence that IRESs can enhance the translation of a message when placed in the 3′UTR. (c) Finally, it is known for decades that some plant viruses utilize CITE elements in their 3′UTR to recruit translation initiation factors in a cap-independent manner.Citation23,30,31 Long distance interactions between the 3′ and the 5′ ends in these viruses result in mRNA circularization and positioning of the scanning machinery near the 5′ end. Since the mechanisms governing gene expression of plant viruses were evolved in eukaryotes within the same environment as cellular transcripts, it is possible that cellular transcripts also evolved similar translational cis-regulatory elements in their 3′UTR. Although IRESs and CITEs differ in the mechanism by which they initiate translation, with the latter requiring an additional step of scanning from a free 5′ end, they both share similar properties with respect to their ability to recruit the translation machinery. Since our assay relies on a bicistronic construct, in which the eGFP has no free 5′ end, it detects IRESs specifically, i.e. elements that can both recruit the ribosome and initiate translation locally. However, we cannot determine whether in the native context of the 3′UTR these elements lead to both recruitment and initiation or just recruitment followed by shunting of the translation machinery to the 5′UTR in a similar fashion to CITE elements. To answer this important question further investigation of the identified IRESs within their native context in required.
The landscape of IRES elements in [+]ssRNA viral genomes
In the absence of a 5′ cap to support cap-dependent translation, IRESs are essential for the life cycle of uncapped [+]ssRNA viruses. Moreover, some of these viruses, such as poliovirus, cleave the cellular scaffold protein eIF4G that facilitates the ribosome recruitment to the 5′ cap structure resulting in a global shift from cap-dependent to IRES-dependent translation in cells.Citation32 Thus, the investigation of IRESs in [+]ssRNA viral genomes is critical for understanding their regulation.
Many [+]ssRNA viruses translate their entire genome as one single polyprotein. Then, viral or cellular proteases cleave the polyprotein precursor to give rise to the mature and functional proteins. While this mechanism is highly efficient for producing equimolar amounts of the viral proteome, it can be a waste of resources if only part of the viral proteins is needed. Thus, we hypothesized that [+]ssRNA viruses, which evolved IRES elements in their 5′UTR to facilitate the translation of the full polyprotein, may also utilize similar elements to translate only part of their proteins from the same transcript. However, as in the case of human transcripts most of the studies had focused on the 5′UTR so that little is known about IRES activity in other regions.
One advantage of using synthetically designed oligos is the ability to assay in a single experiment the activity of sequences from hundreds of different organisms. As part of our study we conducted systematic and unbiased profiling of IRES activity across the genomes of 131 different [+]ssRNA viruses. Our measurements uncovered that IRESs are highly abundant across the polyprotein region of uncapped [+]ssRNA viruses, with similar levels to those obtained at the 5′UTR. Remarkably, this high abundance is specific for uncapped [+]ssRNA viruses and were not obtained in the polyprotein region of capped [+]ssRNA viruses and the coding sequence of human transcripts. These results suggest that in addition to the characterized mechanism of post-translational cleavage of the polyprotein, viruses can also employ IRESs to regulate the translation of only part of their genome (). Notably, viral proteins typically appear in a specific order in the genome such that structural proteins are encoded at the beginning of the polyprotein and non-structural proteins (e.g., viral polymerases) are encoded after the structural proteins. Thus, the presence of IRES elements along the polyprotein region can exploit this spatial organization to provide an efficient mechanism for coordinated translation of either structural or non-structural proteins.
Figure 2. Hypothetical model for specific proteins translation from [+]ssRNA genomes. Uncapped [+]ssRNA genomes harbor IRES elements at the 50UTR and along the polyprotein coding region. In the characterized mechanism (left) ribosomes initiate translation from the 50UTR IRES to produce the polyprotein precursor. Cleaving the polyprotein by proteases result in equimolar amounts of viral proteins. In the hypothetical model (right) ribosomes initiate translation from an IRES element along the polyprotein region resulting in the production of specific subset of the viral proteome.
![Figure 2. Hypothetical model for specific proteins translation from [+]ssRNA genomes. Uncapped [+]ssRNA genomes harbor IRES elements at the 50UTR and along the polyprotein coding region. In the characterized mechanism (left) ribosomes initiate translation from the 50UTR IRES to produce the polyprotein precursor. Cleaving the polyprotein by proteases result in equimolar amounts of viral proteins. In the hypothetical model (right) ribosomes initiate translation from an IRES element along the polyprotein region resulting in the production of specific subset of the viral proteome.](/cms/asset/36690573-7f62-4d08-94c0-295bdb373c19/krnb_a_1212802_f0002_oc.gif)
Interestingly, in a recent study Pirakitikulr et al.Citation33 found regulatory RNA structures along the polyprotein region of Hepatitis C Virus (HCV) genome. These structures are conserved across multiple genotypes and genetic manipulation in their sequence led to reduction in virus replication and infection. Since many IRESs act through structural RNA elements and in light of our findings of IRESs along the coding sequences of [+]ssRNA viruses, it will be interesting to investigate whether the structural elements described in Pirakitikulr et al. are indeed functional IRES elements, which proteins are translated from these IRESs, and how their translation affect virus replication and infection.
The cis-regulatory sequences underlying IRES activity
IRESs can act by various mechanisms to recruit the ribosome to the mRNA including the formation of secondary structures, attracting IRES Trans-Acting Factors (ITAFs) to specific RNA sequence elements and complementarity to the 18S rRNA.Citation34-36 However, many of the sequence elements underlying IRES activity are uncharacterized and thus, the relationship between RNA sequence and IRES activity remains mostly unknown and current bioinformatics predictions are not available. The reason for this relatively poor characterization is the low number of IRESs identified to date and lacking large-scale permutation and mutagenesis studies in which the native sequence of a specific IRES is systematically mutated and the resulting effect on expression is measured. In that sense, the state of the field of translational control is very different from other fields of gene expression such as transcriptional control in which many high-throughput studies investigating native and synthetic regulatory sequences were performed so that the basic “building blocks,” i.e., transcription factors binding-sites,Citation37,38 core-promoter elementsCitation39 and nucleosome disfavoring sequences,Citation40 are thoroughly characterized.
We set out to increase our understanding of the cis-regulatory elements facilitating IRES-dependent translation by: (a) Elaborating the collection of known IRESs by thousands of novel sequences, providing 50-fold increase over current data.Citation19 This largest collection of IRESs facilitates for the first time in-depth computational analysis as was previously done to decipher the sequence features predictive of promoter and enhancer activities.Citation41,42 (b) Conducting careful and systematic interference in the native sequences of ∼100 known and novel IRESs by mutating one region at a time and measuring its effect on expression. By doing so we were able to detect the position and sequence of the functional cis-regulatory elements governing the activity of each of the investigated IRESs. Analysis of this data revealed 2 classes of IRESs: structural and non-structural, which act through local cis-regulatory elements typically located in the vicinity of the start codon (up to ∼60nt) and enriched in poly(U) sequences. These short regulatory elements can represent either ITAF binding-sitesCitation43 or complementary sequences to the 18S rRNA that attract the ribosome via Watson-Crick base pairing in a similar fashion to Shaine-Delgarno sequences in bacteria.Citation36,44-48 Since rRNA-IRES interactions rely on the primary sequence of the message one can speculate that extracting all possible short k-mers with complementary sequence to the 18S rRNA can enhance our ability to predict IRES activity. However, with no prior knowledge of the regions in the 18S rRNA that can facilitate rRNA-IRES interactions we cannot tell which of the complementary k-mers can act as functional IRES element. To address this important question we performed systematic mapping of the functional regions of the 18S rRNA and uncovered one distinct active region and hundreds of k-mers with complementary sequence to this region that can act as short IRES elements. Indeed, mutating these novel k-mers in native IRESs led to reduction in their activity.
Despite this advance toward deciphering the sequences that underlie IRES activity, additional experimental and computational studies are needed to achieve a comprehensive understanding of this complex mode of regulation.
Systematic investigation of translational regulation using high-throughput methods
Several technologies were recently developed for taking the field of translational regulation into the high-throughput era. The most prominent example is ribosome profiling, which allows monitoring of protein translation by deep sequencing of ribosome-protected mRNA fragments.Citation49 Since its initial development in yeast, it was rapidly applied to various organisms such as mammalian cellsCitation50 and virusesCitation51,52 and became a routine experiment in many of the studies in the field. However, although ribosome profiling uncovers the translated regions in the genome and translation efficiency, it does not provide information on the functional cis-regulatory elements that guide the ribosome to facilitate translation initiation. In that sense, ribosome profiling resembles RNA-seq but it does not provide insights on the sequences through which ribosomes are recruited to the mRNA.
To expose the cis-regulatory elements providing specificity in ribosome recruitment a functional assay aimed at measuring the effect of sequence on translation initiation should be performed. In our study we employed a massively parallel reporter assay using fluorescent-based bicistronic plasmid to facilitate quantitative measurements of IRES activity for thousands of fully designed sequences. Using a different approach, Wellensiek et al.Citation53 devised a method to assayed thousands of native genomic fragments for cap-independent translation in-vitro by combining mRNA display and deep sequencing. In addition to the technological differences, these 2 methods also differ in a couple of aspects: (a) The experimental environment (in-vitro vs. in-vivo): The mRNA display experiment, which is conducted in-vitro, is not sensitive to potential artifacts of cryptic promoter and splice site and therefore does not require additional experiments to rule out sequences with false positive IRES activity as performed in our study. In addition, an in-vitro system enables changing the trans environment, such as the concentration of specific ITAF, and to measure its effect on activity. However, while in-vitro systems work well for some IRESs, some reports suggest that other IRESs require a ‘nuclear experience’ in order to be functional.Citation24,54,55 In addition, positive activity obtained in-vitro can stem from secondary structure or interaction with the translation machinery that do not occur in the cellular environment. Thus, in-vitro systems are exposed to false negative and false positive results reducing their sensitivity and specificity in the detection of novel IRESs. (b) The input sequences measured (native genomic fragments vs. synthetically designed oligonucleotides): Native genomic fragments are not limited in length, which is a clear disadvantage of our method that uses synthetic oligos ∼200 nt in length as the input sequences. However, methods that use native sequences are limited to organisms that can be cultured in lab conditions and can usually assay a single genome at a time. In addition, native genomic sequences do not allow the detailed characterization of the cis-regulatory elements, which require systematic changes in the native sequence using synthetic manipulations. As DNA synthesis technologies constantly improve it is possible that soon designed sequences with similar lengths to those achieved when using native genomic fragments will become available.
These recent advances in technology together with high-throughput structural measurementsCitation56 and RNA-binding proteins immunoprecipitations techniquesCitation57 provide efficient tools for scientist to substantially increase the number of sequences assayed and to gain genome-wide insights on translational regulation.Citation58 However, increasing the throughput of measurements cannot entirely substitute low-throughput experiments. For example, in the functional high-throughput assays mentioned here the evaluated sequences are tested out of their native context. Thus, these assays cannot draw definitive conclusions and should be followed by detailed experiments at the gene level such as disrupting the regulatory elements at their native mRNA location (e.g. by CRISPR). Another limitation is that high-throughput assays do not facilitate high-level phenotypic measurements. Translational regulation has a critical role in organismal development and thus investigating the effect of cis-regulatory elements within the context of the entire organism is essential to uncover their phenotypic effect during embryogenesis.Citation18 Finally, although high-throughput methods provide information on thousands of sequences and the statistical power to investigate general principles, they cannot replace low-throughput experiments with respect to the certainty and accuracy at the single sequence level. Rather, combining insights from various high-throughput assays and low-throughput studies are both powerful and essential for deciphering translational control and shedding light on these yet uncovered regulatory mechanisms.
Disclosure of potential confllicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the NIH and the European Research Council (ERC) to E.S. and student research grants from the Kahn Center for Systems Biology and the Azrieli Center for Systems Biology to S.WG. S.WG. is the recipient of the Clore Ph.D. fellowship.
References
- Mogno I, Kwasnieski JC, Cohen BA. Massively parallel synthetic promoter assays reveal the in vivo effects of binding site variants. Genome Res 2013; 23:1908-15; PMID:23921661; http://dx.doi.org/10.1101/gr.157891.113
- Weingarten-Gabbay S, Segal E. The grammar of transcriptional regulation. Hum Genet 2014; 133(6):701-11; PMID:24390306; http://dx.doi.org/10.1007/s00439-013-1413-1
- Smith RP, Taher L, Patwardhan RP, Kim MJ, Inoue F, Shendure J, Ovcharenko I, Ahituv N. Massively parallel decoding of mammalian regulatory sequences supports a flexible organizational model. Nat Genet 2013; 45:1021-8; PMID:23892608; http://dx.doi.org/10.1038/ng.2713
- Yanez-Cuna JO, Kvon EZ, Stark A. Deciphering the transcriptional cis-regulatory code. Trends Genet 2013; 29:11-22; PMID:23102583; http://dx.doi.org/10.1016/j.tig.2012.09.007
- Jang SK, Kräusslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol 1988; 62:2636-43; PMID:2839690
- Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 1988; 334:320-5; PMID:2839775; http://dx.doi.org/10.1038/334320a0
- Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem 1982; 257:14806-10; PMID:6294080
- Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol 2005; 6:318-27; PMID:15803138; http://dx.doi.org/10.1038/nrm1618
- Du X, Wang J, Zhu H, Rinaldo L, Lamar KM, Palmenberg AC, Hansel C, Gomez CM. Second cistron in CACNA1A gene encodes a transcription factor mediating cerebellar development and SCA6. Cell 2013; 154:118-33; PMID:23827678; http://dx.doi.org/10.1016/j.cell.2013.05.059
- Cornelis S, Bruynooghe Y, Denecker G, Van Huffel S, Tinton S, Beyaert R. Identification and characterization of a novel cell cycle-regulated internal ribosome entry site. Mol Cell 2000; 5:597-605; PMID:10882096; http://dx.doi.org/10.1016/S1097-2765(00)80239-7
- Candeias MM, Powell DJ, Roubalova E, Apcher S, Bourougaa K, Vojtesek B, Bruzzoni-Giovanelli H, Fåhraeus R. Expression of p53 and p53/47 are controlled by alternative mechanisms of messenger RNA translation initiation. Oncogene 2006; 25:6936-47; PMID:16983332; http://dx.doi.org/10.1038/sj.onc.1209996
- Herbreteau CH, Weill L, Décimo D, Prévôt D, Darlix JL, Sargueil B, Ohlmann T. HIV-2 genomic RNA contains a novel type of IRES located downstream of its initiation codon. Nat Struct Mol Biol 2005; 12:1001-7; PMID:16244661; http://dx.doi.org/10.1038/nsmb1011
- Vagner S, Touriol C, Galy B, Audigier S, Gensac MC, Amalric F, Bayard F, Prats H, Prats AC. Translation of CUG- but not AUG-initiated forms of human fibroblast growth factor 2 is activated in transformed and stressed cells. J Cell Biol 1996; 135:1391-402; PMID:8947560; http://dx.doi.org/10.1083/jcb.135.5.1391
- Shatsky IN, Dmitriev SE, Andreev DE, Terenin IM. Transcriptome-wide studies uncover the diversity of modes of mRNA recruitment to eukaryotic ribosomes. Crit Rev Biochem Mol Biol 2014; 49:164-77; PMID:24520918; http://dx.doi.org/10.3109/10409238.2014.887051
- Simon AE, Miller WA. 3′ cap-independent translation enhancers of plant viruses. Annu Rev Microbiol 2013; 67:21-42; PMID:23682606; http://dx.doi.org/10.1146/annurev-micro-092412-155609
- Gao F, Kasprzak WK, Szarko C, Shapiro BA, Simon AE. The 3′ untranslated region of Pea Enation Mosaic Virus contains two T-shaped, ribosome-binding, cap-independent translation enhancers. J Virol 2014; 88:11696-712; PMID:25100834; http://dx.doi.org/10.1128/JVI.01433-14
- Sharma SD, Kraft JJ, Miller WA, Goss DJ. Recruitment of the 40S ribosome subunit to the 3′-untranslated region (UTR) of a viral mRNA, via the eIF4 complex, facilitates cap-independent translation. J Biol Chem 2015; 290:11268-81; PMID:25792742; http://dx.doi.org/10.1074/jbc.M115.645002
- Xue S, Tian S, Fujii K, Kladwang W, Das R, Barna M. RNA regulons in Hox 5′ UTRs confer ribosome specificity to gene regulation. Nature 2015; 517:33-8; PMID:25409156; http://dx.doi.org/10.1038/nature14010
- Mokrejs M, Masek T, Vopálensky V, Hlubucek P, Delbos P, Pospísek M. IRESite–a tool for the examination of viral and cellular internal ribosome entry sites. Nucleic Acids Res 2010; 38:D131-6; PMID:19917642; http://dx.doi.org/10.1093/nar/gkp981
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 2016; 351:pii: aad4939; PMID:26816383; http://dx.doi.org/10.1126/science.aad4939
- Sharon E, Kalma Y, Sharp A, Raveh-Sadka T, Levo M, Zeevi D, Keren L, Yakhini Z, Weinberger A, Segal E. Inferring gene regulatory logic from high-throughput measurements of thousands of systematically designed promoters. Nat Biotechnol 2012; 30:521-30; PMID:22609971; http://dx.doi.org/10.1038/nbt.2205
- Gilbert WV. Alternative ways to think about cellular internal ribosome entry. J Biol Chem 2010; 285:29033-8; PMID:20576611; http://dx.doi.org/10.1074/jbc.R110.150532
- Shatsky IN, Dmitriev SE, Terenin IM, Andreev DE. Cap- and IRES-independent scanning mechanism of translation initiation as an alternative to the concept of cellular IRESs. Mol Cells 2010; 30:285-93; PMID:21052925; http://dx.doi.org/10.1007/s10059-010-0149-1
- Thompson SR. So you want to know if your message has an IRES?. Wiley Interdiscip Rev RNA 2012; 3:697-705; PMID:22733589; http://dx.doi.org/10.1002/wrna.1129
- Wells SE, Hillner PE, Vale RD, Sachs AB. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 1998; 2:135-40; PMID:9702200; http://dx.doi.org/10.1016/S1097-2765(00)80122-7
- Miettinen TP, Bjorklund M. Modified ribosome profiling reveals high abundance of ribosome protected mRNA fragments derived from 3′ untranslated regions. Nucleic Acids Res 2015; 43:1019-34; PMID:25550424; http://dx.doi.org/10.1093/nar/gku1310
- Guydosh NR, Green R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell 2014; 156:950-62; PMID:24581494; http://dx.doi.org/10.1016/j.cell.2014.02.006
- Young DJ, Guydosh NR, Zhang F, Hinnebusch AG, Green R. Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3′UTRs In Vivo. Cell 2015; 162:872-84; PMID:26276635; http://dx.doi.org/10.1016/j.cell.2015.07.041
- Paek KY, Hong KY, Ryu I, Park SM, Keum SJ, Kwon OS, Jang SK. Translation initiation mediated by RNA looping. Proc Natl Acad Sci U S A 2015; 112:1041-6; PMID:25583496; http://dx.doi.org/10.1073/pnas.1416883112
- Treder K, Kneller EL, Allen EM, Wang Z, Browning KS, Miller WA. The 3′ cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA 2008; 14:134-47; PMID:18025255; http://dx.doi.org/10.1261/rna.777308
- Gazo BM, Murphy P, Gatchel JR, Browning KS. A novel interaction of Cap-binding protein complexes eukaryotic initiation factor (eIF) 4F and eIF(iso)4F with a region in the 3′-untranslated region of satellite tobacco necrosis virus. J Biol Chem 2004; 279:13584-92; PMID:14729906; http://dx.doi.org/10.1074/jbc.M311361200
- Lamphear BJ, Kirchweger R, Skern T, Rhoads RE. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J Biol Chem 1995; 270:21975-83; PMID:7665619; http://dx.doi.org/10.1074/jbc.270.37.21975
- Pirakitikulr N, Kohlway A, Lindenbach BD, Pyle AM. The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures. Mol Cell 2016; 62:111-20; PMID:26924328; http://dx.doi.org/10.1016/j.molcel.2016.01.024
- Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell 1997; 89:831-8; PMID:9200601; http://dx.doi.org/10.1016/S0092-8674(00)80268-8
- Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. Structural and functional diversity of viral IRESes. Biochim Biophys Acta 2009; 1789:542-57; PMID:19632368; http://dx.doi.org/10.1016/j.bbagrm.2009.07.005
- Dresios J, Chappell SA, Zhou W, Mauro VP. An mRNA-rRNA base-pairing mechanism for translation initiation in eukaryotes. Nat Struct Mol Biol 2006; 13:30-4; PMID:16341227; http://dx.doi.org/10.1038/nsmb1031
- Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, Enge M, Taipale M, Wei G, et al. DNA-binding specificities of human transcription factors. Cell 2013; 152:327-39; PMID:23332764; http://dx.doi.org/10.1016/j.cell.2012.12.009
- Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature 2012; 489:57-74; PMID:22955616; http://dx.doi.org/10.1038/nature11247
- Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev Biol 2010; 339:225-9; PMID:19682982; http://dx.doi.org/10.1016/j.ydbio.2009.08.009
- Segal E, Widom J. What controls nucleosome positions?. Trends Genet 2009; 25:335-43; PMID:19596482; http://dx.doi.org/10.1016/j.tig.2009.06.002
- Lubliner S, Keren L, Segal E. Sequence features of yeast and human core promoters that are predictive of maximal promoter activity. Nucleic Acids Res 2013; 41:5569-81; PMID:23599004; http://dx.doi.org/10.1093/nar/gkt256
- Kleftogiannis D, Kalnis P, Bajic VB. DEEP: a general computational framework for predicting enhancers. Nucleic Acids Res 2015; 43:e6; PMID:25378307; http://dx.doi.org/10.1093/nar/gku1058
- King HA, Cobbold LC, Willis AE. The role of IRES trans-acting factors in regulating translation initiation. Biochem Soc Trans 2010; 38:1581-6; PMID:21118130; http://dx.doi.org/10.1042/BST0381581
- Zeenko V, Gallie DR. Cap-independent translation of tobacco etch virus is conferred by an RNA pseudoknot in the 5′-leader. J Biol Chem 2005; 280:26813-24; PMID:15911616; http://dx.doi.org/10.1074/jbc.M503576200
- Owens GC, Chappell SA, Mauro VP, Edelman GM. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc Natl Acad Sci U S A 2001; 98:1471-6; PMID:11171975; http://dx.doi.org/10.1073/pnas.98.4.1471
- Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc Natl Acad Sci U S A 2000; 97:1536-41; PMID:10677496; http://dx.doi.org/10.1073/pnas.97.4.1536
- Nicholson R, Pelletier J, Le SY, Sonenberg N. Structural and functional analysis of the ribosome landing pad of poliovirus type 2: in vivo translation studies. J Virol 1991; 65:5886-94; PMID:1656077
- Chappell SA, Mauro VP. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J Biol Chem 2003; 278:33793-800; PMID:12824175; http://dx.doi.org/10.1074/jbc.M303495200
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 2009; 324:218-23; PMID:19213877; http://dx.doi.org/10.1126/science.1168978
- Ingolia NT, Lareau LF, Weissman JS. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011; 147:789-802; PMID:22056041; http://dx.doi.org/10.1016/j.cell.2011.10.002
- Stern-Ginossar N, Weisburd B, Michalski A, Le VT, Hein MY, Huang SX, Ma M, Shen B, Qian SB, Hengel H, et al. Decoding human cytomegalovirus. Science 2012; 338:1088-93; PMID:23180859; http://dx.doi.org/10.1126/science.1227919
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, Ganem D. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog 2014; 10:e1003847; PMID:24453964; http://dx.doi.org/10.1371/journal.ppat.1003847
- Wellensiek BP, Larsen AC, Stephens B, Kukurba K, Waern K, Briones N, Liu L, Snyder M, Jacobs BL, Kumar S, et al. Genome-wide profiling of human cap-independent translation-enhancing elements. Nat Methods 2013; 10:747-50; PMID:23770754; http://dx.doi.org/10.1038/nmeth.2522
- Stoneley M, Subkhankulova T, Le Quesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res 2000; 28:687-94; PMID:10637319; http://dx.doi.org/10.1093/nar/28.3.687
- Semler BL, Waterman ML. IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol 2008; 16:1-5; PMID:18083033; http://dx.doi.org/10.1016/j.tim.2007.11.001
- Flynn RA, Zhang QC, Spitale RC, Lee B, Mumbach MR, Chang HY. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat Protoc 2016; 11:273-90; PMID:26766114; http://dx.doi.org/10.1038/nprot.2016.011
- Darnell JC, Richter JD. Cytoplasmic RNA-binding proteins and the control of complex brain function. Cold Spring Harb Perspect Biol 2012; 4:a012344; PMID:22723494; http://dx.doi.org/10.1101/cshperspect.a012344
- Larsson , OTian B, Sonenberg N. Toward a genome-wide landscape of translational control. Cold Spring Harb Perspect Biol 2013; 5:a012302; PMID:23209130; http://dx.doi.org/10.1101/cshperspect.a012302
