ABSTRACT
Bacterial riboswitches are non-coding RNA structural elements that direct gene expression in numerous metabolic pathways. The key regulatory roles of riboswitches, and the urgent need for new classes of antibiotics to treat multi-drug resistant bacteria, has led to efforts to develop small-molecules that mimic natural riboswitch ligands to inhibit metabolic pathways and bacterial growth. Recently, we reported the results of a phenotypic screen targeting the riboflavin biosynthesis pathway in the Gram-negative bacteria Escherichia coli that led to the identification of ribocil, a small molecule inhibitor of the flavin mononucleotide (FMN) riboswitch controlling expression of this biosynthetic pathway. Although ribocil is structurally distinct from FMN, ribocil functions as a potent and highly selective synthetic mimic of the natural ligand to repress riboswitch-mediated ribB gene expression and inhibit bacterial growth both in vitro and in vivo. Herein, we expand our analysis of ribocil; including mode of binding in the FMN binding pocket of the riboswitch, mechanisms of resistance and structure-activity relationship guided efforts to generate more potent analogs.
Introduction
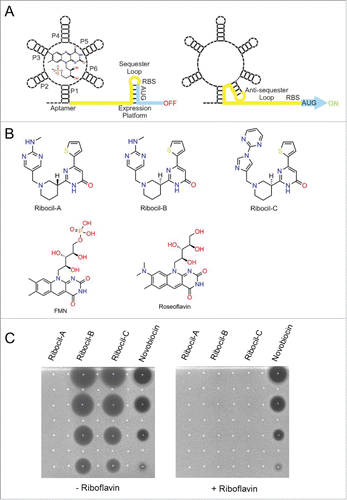
Bacterial riboswitches are a class of regulatory RNA structural elements embedded in untranslated regions of mRNAs that specifically bind natural ligands to regulate gene expression in a cis fashion.Citation1-5 Riboswitches consist of 2 functionally distinct modules an aptamer ligand binding domain and a downstream expression platform. Mechanistically, binding of the cognate ligand to the aptamer domain induces a conformational change in the expression platform that regulates gene expression typically through attenuation of transcription or inhibition of translation (). Riboswitches are thought to regulate the expression of up to 4% of all bacterial genes and more than 24 different classes of riboswitches responding to a diverse set of ligands have been identified, including; enzymatic cofactors such as thiamin pyrophosphate and flavin mononucleotide, amino acids including glycine, lysine and glutamine, the purines adenine and guanine, and inorganic ions magnesium and fluoride.Citation6-14
Figure 1. FMN riboswitch mechanism of action, ribocil chemical structures, and suppression of ribocil activity by riboflavin (A) Diagram of the FMN riboswitch including the 5′ mRNA aptamer with bound FMN and the 3′ expression platform which regulates expression of the downstream ribB gene open reading frame (blue). In the FMN ligand bound form (left panel) the FMN aptamer induces formation of the sequester loop in the expression platform that inhibits ribB expression (OFF) through early termination of transcription of the ribB ORF and sequestration of the Shine-Dalgarno ribosome binding sequence to prevent translation of fully transcribed ribB mRNAs.Citation30 Alternatively, in the absence of FMN, the FMN aptamer adopts an alternative structural conformation (ON) that induces an anti-sequester loop in the expression platform enabling uninterrupted ribB expression (right panel). (B) Chemical structures of the ribocil enantiomers ribocil-A (R isomer), ribocil-B (S isomer) and of the ribocil analog ribocil-C (S isomer). (C) Anti-bacterial activity of ribocil A, B, C spotted on top of Mueller Hinton agar plates embedded with the E. coli strain MB5746 either in the absence (left panel) or presence (right panel) of riboflavin (20 μM). Compounds were suspended in DMSO and 5 μl was spotted after 2-fold dilutions starting at 512 μg/ml for ribocil A, B and novobiocin (negative control) and at 64 μg/ml for ribocil-C.
In bacteria, riboflavin (vitamin B2) concentrations are regulated by FMN riboswitches, also known as RFN elements, which control expression of genes required for biosynthesis and transport of this essential vitamin.Citation15,16 Riboflavin (RF) is the immediate precursor of the metabolites flavin mononucleotide (FMN) and, flavin adenine dinucleotide (FAD), which serve as the primary cofactors of the ubiquitous flavoenzymes that play diverse and central roles in intermediary metabolism.Citation17 FMN is the primary regulatory ligand of FMN riboswitches and although RF, and FAD, can also associate with the FMN riboswitch aptamer, their affinity is comparatively low and they do not play an important role in regulation.Citation6 Most pathogenic bacteria can synthesize RF de novo utilizing 5 enzymes encoded by the rib biosynthetic gene family and, depending on the bacterial strain FMN riboswitches control either a single rib gene or operon of several rib genes.Citation16 Many Gram-positive bacterial species and some Gram-negatives can also acquire RF by active transport from environmental sources and the expression of such RF transporter genes is similarly regulated by FMN riboswitches. Indeed, intracellular RF concentrations mediated from de novo synthesis or active transport are both regulated by FMN riboswitches, thus reflecting a potentially attractive bacterial-specific target for antibiotic development.Citation18
To date, approaches taken to identify riboswitch inhibitors have largely utilized target-based methods including high-throughput and fragment based screening and structure-guided ligand design.Citation19-22 Although such inhibitors often demonstrate in vitro activity and riboswitch selectivity, seldom is whole cell growth inhibitory activity achieved. A notable exception is roseoflavin (RoF), a natural product analog of RF originally isolated from Streptomyces davawensis.Citation23 RoF is a prodrug which is converted to roseoflavin mononucleotide (RoFMN) and roseoflavin adenine dinucleotide (RoFAD).Citation24 RF is actively transported into Gram-positive bacteria by RF transporters and RoFMN can efficiently bind FMN riboswitches, inhibit RF synthesis and transport gene expression, and suppress the growth of a number of bacterial strains.Citation20,23-26 However, the antibacterial activity of RoF is not solely the result of inhibition of FMN riboswitches as RoFMN and RoFAD have also been demonstrated to associate with numerous flavoenzymes encoded by E. coli and it is has been demonstrated that some of these enzymes are also inactivated in the RoFMN/RoFAD bound forms.Citation27 In addition, RoF-mediated E. coli antibacterial activity is not fully suppressed by even high concentrations of exogenously-added RF, supporting the hypothesis that inhibition of flavoproteins by RoF is partially responsible for antibacterial activity.Citation28 Therefore, although such a ‘multitarget’ mechanism of action for RoFMN is likely to mitigate potential issues of target-based drug resistance, the high level of structural and functional conservation between bacterial and human flavoenzymes does raise considerable safety and toxicity concerns from the perspective of developing RoF as a new antibiotic. More recently an additional riboflavin analog has been reported in an effort to identify antibacterials that inhibit the FMN riboswitch.Citation29
Validation of the riboflavin pathway and discovery of ribocil
To explore the potential suitability of RF biosynthetic pathway as a new target for antibiotic discovery, we first employed a genetic approach by constructing E. coli strains deleted for ribA, or ribB that are unable to synthesize RF de novo and must be propagated in media supplemented with RF to sustain growth. Both ΔribA and ΔribB strains when tested is a murine septicemia model were found to be dramatically attenuated in virulence, causing limited morbidity, no mortality, and yielding bacterial burdens reduced by approximately 3 x log10 as compared to the wild-type control.Citation28 Therefore, E. coli is unable to scavenge sufficient RF to establish a robust infection and consequently, inhibitors of E. coli RF biosynthesis are genetically predicted to display antibacterial efficacy. To identify cognate inhibitors of RF biosynthesis, a focused library of 57,000 synthetic small molecules with intrinsic antibacterial activity was screened for compounds whose ability to inhibit bacterial growth was specifically suppressed by RF supplementation.Citation28 Among the small molecules tested, only a single compound, later named ribocil (), was fully inhibited specifically in the presence of exogenously added riboflavin () and demonstrated to cause a dose-dependent reduction in RF levels in E. coli (IC50=0.3μM).Citation28 After 24 hours of treatment with ribocil, RF, FMN and FAD levels in E.coli were all reduced to residual levels, faithfully reflecting a phenocopy of ΔribA and ΔribB strains propagated in the absence of exogenous RF.Citation28 Interestingly, ribocil was found to be a racemic mixture of isomers which after separation led to isolation of ribocil-A (R-enantiomer) and ribocil-B (S-enantiomer) with ribocil-B found to be nearly completely responsible for inhibition of RF synthesis () and growth inhibitory activity.Citation28
Elucidating in a whole cell context the MOA of ribocil was facilitated by the isolation of multiple independent ribocil-resistant (ribocilR) E. coli mutants followed by whole genome sequencing to map the mutations to their target.Citation28 Remarkably, none of the mutations mapped to the open reading frame of any of the rib genes encoding riboflavin biosynthetic enzymes; instead all 19 mutations causal for drug resistance were located within the FMN riboswitch located immediately 5′ of ribB. Therefore, genetic data predicted ribocil inhibited RF synthesis exclusively by binding to the FMN riboswitch and potentially mimics the effects of the natural FMN ligand to inhibit ribB expression. Demonstration that ribocil inhibited FMN riboswitch controlled plasmid-based reporter gene expression (EC50 = 0.3 μM), and that ribocil tightly binds purified in vitro synthesized E. coli FMN riboswitch aptamer RNA (KD = 13 nM) further confirmed that ribocil specifically interdicts FMN riboswitch-mediated gene expression.Citation28 Pharmacological validation of both the RF pathway as a suitable target for drug development as well as the efficacy of ribocil was also obtained by administering ribocil-C (), a more active analog of ribocil (), in a murine septicemia model of E. coli infection, where bacterial growth was inhibited up to 3 x log10 versus the placebo control.Citation28
The RFN element of the Fusobacterium nucleatum impX riboflavin transporter conforms to the FMN riboswitch family at the levels of nucleotide sequence and secondary structure and is the only riboswitch for which a co-crystal structure has been solved in complex with its natural ligand.Citation25 In the F. nuclelateum FMN co-crystal structure the central FMN binding site is located in the junctional region, or butterfly fold, of the 2 peripheral domains that form as a result of tertiary interactions between the P2-P6 or P3-P5 stem loops.Citation25 Based on this co-crystal structure, and to more deeply understand the molecular basis of ribocil's growth inhibitory activity, we solved a 2.95-Å co-crystal structure of ribocil engaged with the F. nucleatum impX FMN riboswitch.Citation28 The key findings from the structure were that in complex with the F. nucleatum riboswitch ribocil adopts a constrained U-shaped conformation and forms strong and favorable pi-stacking and H-bond interactions within the FMN binding site.Citation28
Molecular interactions of ribocil with the FMN riboswitch and stereochemistry of binding
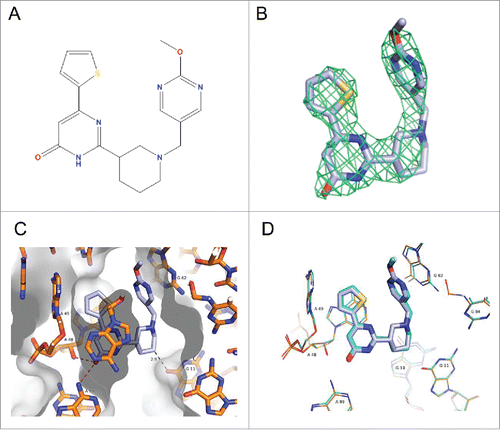
To further confirm the binding mode and biologically relevant chirality of ribocil, we have solved a 2.95-Å crystal co-structure with the related ribocil-D analog () by X-ray crystallography using the same methodologies as described previously with the racemic mixture of ribocil.Citation28 The electron density allows for the determination of the binding mode of the compound (). The same key interactions () are found with the compound environment, in particular an extended network of face to face and face to edge pi-stacking interactions and 2 key H-bonds between the carbonyl and the 2′OH of A48 and the exocyclic NH2 of A99. These key H-bonds imply ribocil-D adopts the low energy pyrimidinone form, over its several other tautomers, to bind to the FMN riboswitch. A comparison of the 2 structures () shows that both the coordinates of the 2 aptamer-ligand complex superpose very closely and are identical within the experimental errors.
Figure 2. Co-crystal structure of ribocil-D bound to the F. nucleatum FMN riboswitch. (A) Chemical structure of ribocil-D. (B) Electron density difference map of ribocil-D displayed as a grid at 3.0σ level. (C) Ribocil-D in the FMN binding site of the F. nucleatum riboswitch with RNA and ligand structures represented as sticks. Carbon atoms are colored slate blue for the ligand and orange for the RNA nucleotide bases. Solvent-accessible surface is represented as gray, with darker gradations representing surfaces facing up. Key bases in the binding site are labeled and key H-bond is indicated by the red dashed lines. The weak H-bond formed by a methyl group is indicated with a gray dashed line. (D) Overlay of the of the X-ray co-crystal structures of ribocil-D, which is represented as sticks colored slate blue, and ribocil-B.Citation28 (PBD entry 5C45), which is represented as sticks colored cyan. The nucleotide number for RNA bases interacting with ribocil-D and ribocil-B is indicated.
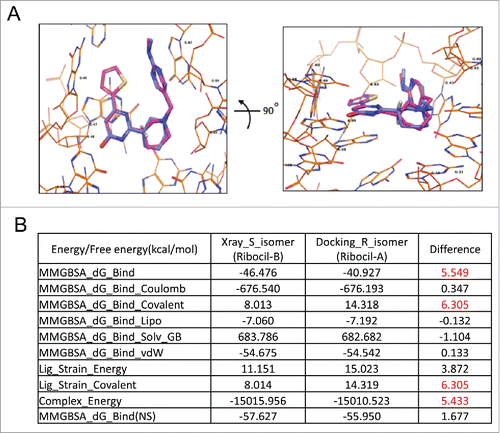
The R stereoisomer of ribocil (ribocil-A) was modeled in the ligand binding site of E. coli FMN riboswitch, and superimposed with the S stereoisomer (ribocil-B). The binding modes of 2 isomers are very similar (), raising the question as to why ribocil-B (S-isomer) is a tight binder to the FMN riboswitch and ribocil-A (R-isomer) does not similarly interactCitation28 and display antibiotic activity (see also ). To answer this question, we conducted an energetic analysis using the MMGBSACitation31 method with the binding structures of the 2 isomers. The various energy and free energy terms are reported (). Interestingly the difference in binding free energy between 2 isomers is 5.549 kcal/mol. The R-isomer (ribocil-A) possess a less favorable binding free energy compared to the S-isomer (ribocil-B) that is mainly due to a higher ligand strain energy as described by MMGBSA_dG_Bind_Covalent and Lig_Strain_Covalent terms with a difference in values for both terms of 6.305 kcal/mol. When considering the energy terms alone, the binding energy difference is 5.433 kcal/mol, thus the binding free energy difference observed is largely from enthalpy and very little from entropy. Furthermore, these modeled energetic impacts of the R and S stereoisomer conformations corresponds well with our measurements of ribocil-riboswitch binding energetics, as shown in . A marked, > 4.3 kcal/mol, difference was observed in aptamer-binding free energy between ribocil-A and ribocil-B (KD > 10,000 and 6.6 nM, respectively), a value entirely consistent with the modeled binding energy difference.
Figure 3. Analyzing binding differences of ribocil-A (non-binder) and ribocil-B (tight binder). (A) Structure models of ribocil-A and ribocil-B in E. coli FMN aptamer in 2 orientations, ribocil-B carbon atoms colored in light blue, ribocil-A in magenta, hydrogen atom at the chiral center of 2 compounds in white. (B) Energetic analysis of ribocil-A and ribocil-B binding toward E. coli riboswitch. Notable differences in binding energy are highlighted in red.
Consequences of ribocil resistance riboswitch mutations on riboswitch binding to FMN and ribocil
To ascertain the effects of riboswitch mutations on the interactions of the riboswitch with FMN and ribocil we characterized the effects of riboswitch point mutants C111U, C100U, G93U, G37U, and C33U and Δ94–102 on riboswitch secondary structure, binding to FMN and the ability of ribocil to compete with FMN for the riboswitch binding site (, ). Despite the potential for these mutations to disrupt key structural contacts within the riboswitch they do not appear to significantly alter the overall secondary structure of the riboswitch. As shown in , similar thermal denaturation profiles are observed for each of the wild-type and mutant riboswitches. These data indicate that many of the stabilizing intramolecular secondary structural interactions, such as base stacking and base pairing, are maintained globally in the mutant riboswitches.
Figure 4. Binding of FMN and ribocil-B, and ribocil-C to wild-type and ribocilR mutant RNA apatmers. (A) Temperature dependent changes in absorbance for the wild-type riboswitch and mutants. (B) Fluorescence based analysis of binding of wild-type and mutant riboswitches to FMN. Solid lines represent the nonlinear least square analysis for direct FMN binding to riboswitches according to a quadratic equation. (C) Analysis of FMN competition binding by ribocil for wild-type and mutant riboswitches. For all panels in A, B and C, wild-type and mutant riboswitches are denoted with wild-type riboswitch (black), while mutants are shown in the following colors: C100U (red), C111U (green), C33U (blue), G37U (orange), G93U (purple), and Δ94–102 (cyan). (D) FMN-competitive binding of ribocil (black circles), ribocil A (blue diamonds), ribocil B (red squares), and ribocil C (green triangles) to the FMN riboswitch. Solid lines in C and D represent the nonlinear least squares analyses for the competition binding of each compound to the riboswitch aptamer against FMN according to a cubic equation fully describing the competition equilibria.
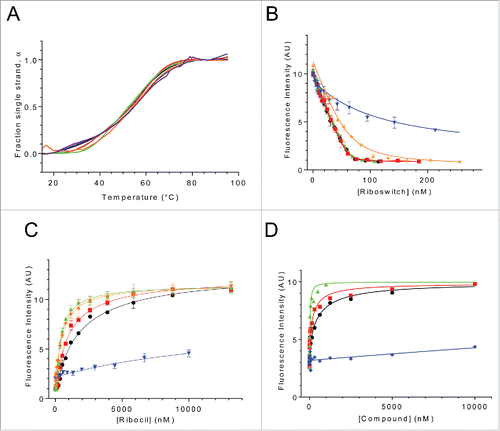
Table 1. Binding parameters underlying the interactions of FMN and ribocil with the E. coli FMN riboswitch aptamer and ribocilR mutant aptamers.
Despite the similar structural stabilities of the riboswitch mutants only 2 of the mutants, C111U and C100U retain full FMN-binding capacity, KD-FMN 1 nM. (). G37U retains binding affinity for FMN with slight reduction relative to wild type (∼8-fold). C33U exhibits significant reduction in FMN binding, ∼80-fold less than the wild type. While G93U and Δ94–102 mutants show no binding to FMN under the conditions studied (not shown). The impact of these mutations on FMN binding correlates with their effects on ribocil binding (). As was observed for FMN binding, C111U and C100U that showed wild-type levels of FMN binding also show wild-type like levels of ribocil binding (KD-ribocil ≤13.4, ≤7, and ≤10.3, for wild type, C111U and C100U, respectively). G37U again shows only slightly weaker binding toward ribocil (KD-ribocil 25 nM, less than 2-fold). C33U which showed much weaker FMN binding than wild-type also shows dramatically reduced ribocil binding (KD-ribocil 2500 nM, ∼200-fold). Since ribocil binding was initially characterized through its ability to displace bound FMN,Citation28 ribocil binding is unable to be characterized for G93U and Δ94–102 that do not bind FMN. However, the assumption that G93U and Δ94–102 do not bind ribocil efficiently is quite strong considering ribocil is a synthetic mimic of FMN utilizing the same key RNA nucleotides as FMN to interact with the riboswitch aptamer.
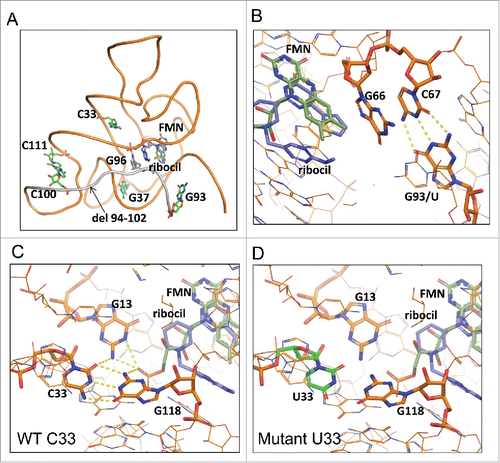
A homology model approach was used to better understand FMN and ribocil binding to the E. coli FMN riboswitch and to further explore how the ribocilR mutations affect binding potency. The modeled E. coli riboswitch aptamer exhibits a similar structural fold as the F. nucelatum template and predicts that ribocil binding is facilitated by similar aromatic stacking and H-bond interactions in the highly homologous E. coli FMN binding site.Citation28,32 In this model, ribocilR mutants are spread across the aptamer, with 3 of them, C33U, G93U, and Δ94–102, located close to the ligand binding site. Interestingly only these 3 mutants show dramatic reduction in FMN and ribocil binding. Δ94–102 affects FMN and ribocil binding directly as a result of the deletion of G96, which stacks with the methylamino-pyrimidinyl group of ribocil. As illustrated in , C33 forms a Watson-Crick base pair with G118, and together with G13, establishes an extensive hydrogen bond network, which connect the 3 bases tightly. Although C33 does not interact directly with either FMN or ribocil its 2 neighbors G118 and G13 play key roles in the interaction. The FMN tail phosphate (PO3) group makes hydrogen bonds with G118 and G13, and the ribocil methylamino-pyrimidinyl group packs with G118 sugar ring. In addition, the G13 base is in close contact with the ribocil central piperidine ring and the FMN hydroxyl tail. For the C33U ribocilR mutant () 2 of the 3 hydrogen bonds presented in C33-G118 pairing are abolished, and the mismatched oxygen atoms and nitrogen atoms should inflict columbic repulsions, forcing conformation changes at U33, G118 and G13. The altered structure at G118 and G13 leads to unfavorable interactions with FMN or ribocil and thus weakens the ligands binding significantly. Like C33U, G93U is not in direct contact with the ligand binding site either, but through C67 and G66, shown in , it can exert its effect to ligand binding: G93 base pairs with C67, which stacks against G66, and G66 base packs with FMN tricyclic and ribocil thiophene. The mutation from G93 to U93 would remove its hydrogen bond network with C67, which in turn could cause structural changes at G66, and these changes could alter the interactions with ligands resulting in reduced binding of FMN and ribocil.
Figure 5. Molecular modeling of ribocilR mutants in the E. coli FMN riboswitch aptamer, ligand FMN carbon atoms in light green, ribocil carbon atoms in light blue. (A) The carbon atoms of all single nucleic acid mutants are colored in green, the deletion mutant del 94–102 colored in gray, (B) G93/U mutant. G93, its counterpart C67 and ligand binding neighbor G66 are displayed in stick, hydrogen bonds between G93 and C67 are highlighted in yellow dash line; (C) and (D) C33/U mutant. (C) Wild-type C33 and the hydrogen bond network with its 2 ligand binding neighbors G118 and G13, labeled with yellow dash lines; (D) Mutant U33-G118 mismatched base pair, U33 colored in green.
Other mutations G37U, C100U and C111U are all distantly removed from the ligand binding site, and apparently the local structural changes from the mismatched base pairs due to mutations do not cause significant effects on ligand binding with at most an 8-fold reduction with FMN in G37U.
The phenotypes of the C100U and C111U resistant mutations, both in the P5 stem, cannot be explained based on ribocil binding directly as RNA aptamers containing either C100U and C111U associate with ribocil with wild-type affinity in the in vitro binding assay (). However, both C100U and C111U mutations do affect riboswitch regulated gene expression albeit with a much more severe effect elicited by C111UCitation28, which completely inhibits FMN regulated gene expression (EC50>200uM). Treatment of E. coli harboring the C100U mutation instead results in a flattening of ribocil dose response curve, with the outcome that gene expression is inhibited incompletely in C100U containing cells as compared to bacteria with wild-type FMN riboswitch.Citation28 Overall, in the context of riboflavin synthesis, E. coli containing either C100U or C111U produce flavin levels sufficient to support bacterial growth to wild-type levels.Citation28 Since these mutations do not impact ribocil binding their inability to inhibit ribB gene expression could most easily be explained if compound binding failed to trigger formation of the sequester loop but instead leaves the riboswitch in a conformation in which the anti-sequester loop was stabilized. In this case, ribocil binding observed in the in vitro binding experiment () must then be directed exclusively or preferentially to the ON state of these 2 mutant riboswitches, compared to the OFF state binding by ribocil to the wild-type and other mutant riboswitches.
C100U and C111U may be in the same class as the C219U and U218C ribocilR mutations, which are located in the proposed stem of the sequester loop and also have no effect on ribocil binding, but nevertheless enable ribB expression and riboflavin synthesis despite ribocil binding to the aptamer, most likely through inhibiting formation of the sequester loop.Citation28
SAR efforts to improve ribocil drug characteristics
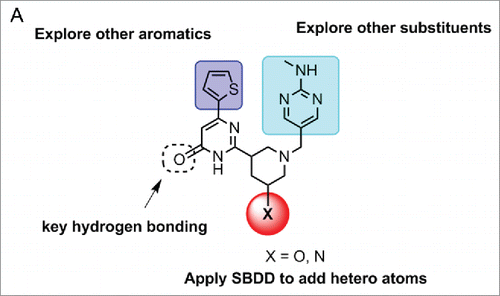
SAR efforts using principles of structure based drug design (SBDD) were initiated to improve the inhibitory activity of the ribocil lead compound (). Consistent with the FMN-riboswitch crystal structure with ribocil-B, the central hydroxy-pyrimidine piperidine core is essential for compound binding, as many analogs with different core scaffolds all lost inhibitory activity. The only modification allowed was addition of a hydroxyl group at the 3-position, potentially establishing H-bonding interaction with G11 in FMN-riboswitch. In addition, attempts to truncate either the thiophene or methylamino-pyrimidine ring led to total loss of compound activity. Thus, SAR efforts were focused on replacing the thiophene and methylamino-pyrimidine ring in the library fashion. While the SAR on the thiophene ring is quite tight, a variety of substituents can be tolerated on the methylamino-pyrimidine side. One notable compound which stood out from medicinal chemistry efforts was one in which the methylamino-pyrimidine group was replaced with a N-(2-pyrimidine) imidazole group. Further resolution of the enantiomers led to ribocil-C which displays approximately 8-fold greater potency against E. coli.
Conclusions
The discovery and mechanistic characterization of ribocil highlights the identification of the first entirely synthetic, selective, and structurally distinct small molecule mimic of a natural ligand to a bacterial riboswitch. Here we report a new co-crystal structure of the F. nucleatum FMN aptamer in complex with ribocil-D, a close analog of ribocil, which reaffirms the previously described binding mode of ribocil to the FMN aptamer. Interestingly, modeling and energetic analyses performed here provide a mechanistic basis as to why the S-isomer of ribocil specifically binds the FMN binding site of the riboswitch aptamer. Such findings underscore the actual level of difficulty (and requisite good fortune!) in discovering an unambiguous target-specific inhibitor; in this case the entire Merck corporate library was ultimately screened to identify one cognate inhibitor of the target and which only one of 2 isomers in a racemic mixture was active and conferred an antibacterial effect. Finally, extensive additional characterization of drug resistance mechanisms demonstrates that ribocilR mutations largely affect ribocil binding directly. However, less common ribocilR mutations can potentially disrupt riboswitch conformational changes so as to indirectly decouple regulation of the expression platform from aptamer ligand binding.
More broadly, our studies firmly support an emerging view that RNA structural elements are druggable by small molecules.Citation33-35 As RNA structural elements are increasingly being recognized to play important roles in gene expression affecting both microbial and human cellular physiology, our work emphasizes a new and largely unexploited class of drug targets potentially broadly relevant to disease intervention.Citation36-38 In summary, data described from the original characterization of ribocil,Citation28 together with our continued studies here including conclusions drawn from SAR-guided medicinal chemistry underscore that small molecule discovery and development of RNA target inhibitors can be successfully achieved using analogous strategies to the traditional protein target-based drug development paradigm.
Disclosure of potential conflicts of interest
All authors are employees for Merck & Co. and may own stock in the company.
References
- Serganov A, Nudler A. A decade of riboswitches. Cell 2013; 152:17-24; PMID:23332744; http://dx.doi.org/10.1016/j.cell.2012.12.024
- Mellin JR, Cossart P. Unexpected versatility in bacterial riboswitches. Trends Genet 2015; 31:150-56; PMID:25708284; http://dx.doi.org/10.1016/j.tig.2015.01.005
- Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 2005; 59:487-517; PMID:16153177; http://dx.doi.org/10.1146/annurev.micro.59.030804.121336
- Breaker RR. 2011. Prospects for riboswitch discovery and analysis. Mol Cell 2011; 43:867-79; PMID:21925376; http://dx.doi.org/10.1016/j.molcel.2011.08.024
- Lünse CE, Schüller A, Mayer G. The promise of riboswitches as potential antibacterial drug targets. Int J Med Microbiol 2014; 304:79-92; PMID:24140145; http://dx.doi.org/10.1016/j.ijmm.2013.09.002
- Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci USA 2002; 99:15908-13; PMID:12456892; http://dx.doi.org/10.1073/pnas.212628899
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 2002; 111:747-56; PMID:12464185; http://dx.doi.org/10.1016/S0092-8674(02)01134-0
- Winkler WC, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 2002; 419:952-56; PMID:12410317; http://dx.doi.org/10.1038/nature01145
- Mandal M, Breaker RR. (2004). Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 2004; 11:29-35; PMID:14718920; http://dx.doi.org/10.1038/nsmb710
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 2004; 306:275-79; PMID:15472076; http://dx.doi.org/10.1126/science.1100829
- Grundy FJ, Lehman SC, Henkin TM. The L box regulon: lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc Natl Acad Sci USA 2003; 100:12057-62; PMID:14523230; http://dx.doi.org/10.1073/pnas.2133705100
- Ames TD, Breaker RR. Bacterial aptamers that selectively bind glutamine. RNA Biol 2011; 8:82-9; PMID:21282981; http://dx.doi.org/10.4161/rna.8.1.13864
- Dann CE 3rd, Wakeman CA, Sieling CL, Baker SC, Irnov I, Winkler WC. Structure and mechanism of a metal-sensing regulatory RNA. Cell 2007; 130:878-92; PMID:17803910; http://dx.doi.org/10.1016/j.cell.2007.06.051
- Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Widespread genetic switches and toxicity resistance proteins for fluoride. Science 2012; 335:233-35; PMID:22194412; http://dx.doi.org/10.1126/science.1215063
- Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DAA. Conserved RNA structure element involved in the regulation of bacterial RF synthesis genes. Trends Genet 1999; 15:439-42; PMID:10529804; http://dx.doi.org/10.1016/S0168-9525(99)01856-9
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Regulation of RF biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res 2002; 30:3141-51; PMID:12136096; http://dx.doi.org/10.1093/nar/gkf433
- Massey V. The chemical and biological versatility of riboflavin. Biochem Soc Trans 2000; 28:283-96; PMID:10961912; http://dx.doi.org/10.1042/bst0280283
- Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat Biotech 2006; 24:1558-64; PMID:17160062; http://dx.doi.org/10.1038/nbt1268
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol 2005; 12:1325-35; PMID:16356850; http://dx.doi.org/10.1016/j.chembiol.2005.10.007
- Lee ER, Blount KF, Breaker RR. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol 2009; 6:187-94; PMID:19246992; http://dx.doi.org/10.4161/rna.6.2.7727
- Mulhbacher J, Brouillette E, Allard M, Fortier LC, Malouin F, Lafontaine DA. Novel riboswitch ligand analogs as selective inhibitors of guanine-related metabolic pathways. PLoS Pathog 2010; 6(4):e1000865; PMID:20421948; http://dx.doi.org/10.1371/journal.ppat.1000865
- Matzner D, Mayer G. (Dis)similar analogues of riboswitch metabolites as antibacterial lead compounds. J Med Chem 2015; 58:3275-86; PMID:25603286; http://dx.doi.org/10.1021/jm500868e
- Otani S, Takatsu M, Nakano M, Kasai S, Miura R. Roseoflavin, a new antimicrobial pigment from Streptomyces. J. Antibiot. (Tokyo) 1974; 27:86-87; PMID:4843053; http://dx.doi.org/10.7164/antibiotics.27.88
- Ott E, Stolz J, Lehmann M, Mack, M. The RFN riboswitch of Bacillus subtilis is a target for the antibiotic roseoflavin produced by Streptomyces davawensis. RNA Biol 2009; 6:276-280; PMID:19333008; http://dx.doi.org/10.4161/rna.6.3.8342
- Serganov A, Huang L, Patel DJ. Coenzyme recognition and gene regulation by a flavin mononucleotide riboswitch. Nature 2009; 458:233-237; PMID:19169240; http://dx.doi.org/10.1038/nature07642
- Pedrolli DB, Jankowitsch F, Schwarz J, Langer S, Nakanishi S, Frei E, Mack M. RF analogs as antiinfectives: occurrence, mode of action, metabolism and resistance. Curr Pharm Des 2013; 19:2552-2560; PMID:23116394; http://dx.doi.org/10.2174/1381612811319140006
- Langer, S, Hashimoto, M, Hobl, B, Mathes, T, Mack, M. Flavoproteins are potential targets for the antibiotic roseoflavin in Escherichia coli. J Bacteriol 2013; 195:4037-45; PMID:23836860; http://dx.doi.org/10.1128/JB.00646-13
- Howe JA, Wang H, Fischmann TO, Balibar CJ, Xiao L, Galgoci AM, Malinverni JC, Mayhood T, Villafania A, Nahvi A, et al. Selective small-molecule inhibition of an RNA structural element. Nature 2015; 526:672-7; PMID:26416753; http://dx.doi.org/10.1038/nature15542
- Blount KF, Megyola C, Plummer M, Osterman D, O'Connell T, Aristoff P, Quinn C, Chrusciel RA, Poel TJ, Schostarez HJ, et al. Novel Riboswitch-binding flavin analog that protects mice against clostridium difficile infection without inhibiting cecal flora. Antimicrob Agents Chemother 2015; 59:5736-46; PMID:26169403; http://dx.doi.org/10.1128/AAC.01282-15
- Pedrolli D, Langer S, Hobl B, Schwarz J, Hashimoto M, Mack M. The ribB FMN riboswitch from Escherichia coli operates at the transcriptional and translational level and regulates riboflavin biosynthesis. FEBS J. 2015; 282:3230-42; PMID:25661987; http://dx.doi.org/10.1111/febs.13226
- Schrödinger Release 2016-2: Prime, version 4.4. Schrödinger, LLC: New York, NY, 2016
- Quentin V, Estefanía M, Batey RT. Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nuc Acids Res 2011; 39:8586-98; PMID:21745821; http://dx.doi.org/10.1093/nar/gkr565
- Guan L, Disney MD. Recent advances in developing small molecules targeting RNA. ACS Chem Biol 2012; 7:73-86; PMID:22185671; http://dx.doi.org/10.1021/cb200447r
- Herman T. Small molecules targeting viral RNA. Wiley Interdiscip Rev RNA 2016 (Epub ahead of print)
- Velagapudi SP, Cameron MD, Haga CL, Rosenberg LH, Lafitte M, Duckett DR, Phinney DG, Disney MD. Design of a small molecule against an oncogenic noncoding RNA. Proc Nat Acad Sci 2016; 113:5898-903; PMID:27170187; http://dx.doi.org/10.1073/pnas.1523975113
- Carnici P, et al. The transcriptional landscape of the mammalian genome. Science 2005; 309:1559-63; PMID:16141072; http://dx.doi.org/10.1126/science.1112014
- Djebali S, et al. Landscape of transcription in human cells. Nature 2012; 489:101-8; PMID:22955620; http://dx.doi.org/10.1038/nature11233
- Morris KV, Mattick JS. The rise of regulatory RNA. Nat Rev Genet 2014; 15:423-37; PMID:24776770; http://dx.doi.org/10.1038/nrg3722