abstract
Circular RNAs (circRNAs) are a novel class of non-coding RNA that assumes a covalently closed continuous conformation. CircRNAs were previously thought to be the byproducts of splicing errors caused by low abundance and the technological limitations. With the recent development of high-throughput sequencing technology, numerous circRNAs have been discovered in many species. Recent studies have revealed that circRNAs are stable and widely expressed, and often exhibit cell type-specific or tissue-specific expression. Most circRNAs can be generated from exons, introns, or both. Remarkably, emerging evidence indicates that some circRNAs can serve as microRNA (miRNA) sponges, regulate transcription or splicing, and can interact with RNA binding proteins (RBPs). Moreover, circRNAs have been reported to play essential roles in myriad life processes, such as aging, insulin secretion, tissue development, atherosclerotic vascular disease risk, cardiac hypertrophy and cancer. Although circRNAs are ancient molecules, they represent a newly appreciated form of noncoding RNA and as such have great potential implications in clinical and research fields. Here, we review the current understanding of circRNA classification, function and significance in physiological and pathological processes. We believe that future research will increase our understanding of the regulation and function of these novel molecules.
Introduction
Circular RNA (circRNA) is a novel type of noncoding RNA (ncRNA). Unlike traditionally linear RNA molecules, circRNA is a covalently closed circular molecule without 5’–3’ polarities or a polyadenylated tail.Citation1
CircRNA was first identified in RNA viruses via electron microscope as early as the 1970s.Citation2 Unfortunately, only a handful of such circRNAs, including hepatitis delta virus (HDV), DCC transcript, ETS-1, Sry and cytochrome P450 2C24 (CYPIIC24), have been sporadically reported over the past 30 y.Citation3-9 Hence, circRNAs were typically considered as a byproduct of errant splicing or mRNA process due to low transcript abundance.Citation6 With the development of high-throughput sequencing technology and bioinformatics, many circRNAs have been discovered and identified,Citation10-16 leading to a resurgence of interest in these molecules. Recent studies have revealed that circRNAs are stable, conserved and diverse, and often exhibit tissue- or developmental stage-specific expression.Citation10,11,17-23 To date, the known functions of circRNAs include roles as microRNA (miRNA) spongesCitation18,24-27 and splicing or transcriptional regulators.Citation19,28-30 In addition, circRNAs can interact with RNA binding proteins (RBPs).Citation28,31 Recent studies have implicated circRNAs in the physiological process of aging,Citation32,33 insulin secretionCitation34 and tissue development.Citation35-37 Moreover, circRNAs also have been reported to play crucial roles in the pathological process such as atherosclerotic vascular disease risk,Citation38 neurological disorders,Citation1,39 cardiac hypertrophyCitation27 and cancer.Citation25,26,40-44 Taken together, these finding indicate that circRNAs might play important roles in fundamental life processes and serve as novel clinical molecular markers, thereby providing new insights into the treatment of diseases.
In this review, we briefly discuss the current understanding of classification and function of circRNAs and highlight their roles in physiology and pathology.
Categories of circRNAs
To provide greater insight into the function of circRNAs, understanding their structure features is critical. Due to their short history of study relative to mRNA, the classification of the vast majority of circRNAs relies on empirical attributes that are based on those of long noncoding RNAs (lncRNAs); this classification system provides a convenient basis on which to classify the former uncharacterized RNA species.
CircRNAs can be generated from any region of the genome, and all result in a great diversity of lengths.Citation17-20,30,35,45,46 The majority of circRNAs are generated from coding exons: some are derived from the 5’ or 3’ untranslated regions (5’UTRs or 3’UTRs), whereas others originate from ncRNAs.Citation18,38 Some circRNAs are derived from introns and are usually excised from pre-mRNA; these species are termed circular intronic RNAs (ciRNAs).Citation18,19 Interestingly, ciRNAs formation depends on a key motif containing a 7-nt GU-rich element close to the 5splice site and an 11-nt C-rich element near the branch point.19 Finally, circRNAs can be composed of an exon and retained introns, a species termed an exon–intron circRNA, or EIciRNA.Citation30
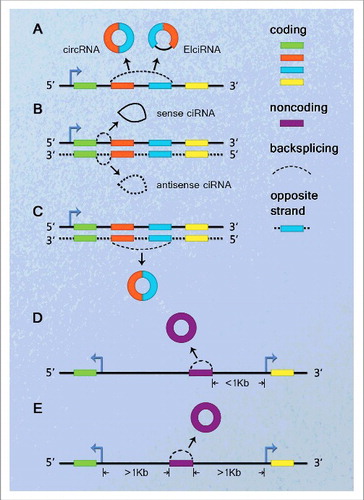
As with lncRNAs,Citation47,48 we attempt to classify circRNAs based on their genomic proximity to a neighboring gene (). A circRNA can usually be placed into one or more of 5 broad categories: (1) sense or exonic, when arising from one or more exons of the linear transcript on the same strand; (2) intronic, when it is derived wholly from an intron of the linear transcript; (3) antisense, when it overlaps one or more exons of the linear transcript on the opposite strand; (4) bidirectional or intragenic, when it is transcribed from same gene locus of the linear transcript but in close genomic proximity and not classified as ‘sense’ and ‘intronic’; and (5) intergenic, when it is located in the genomic interval between 2 genes.
Figure 1. Schematic presentation of circRNA classification. (A) Sense or exonic, when overlapping one or more exons of the linear transcript on the same strand. (B) Intronic, when arising from an intron of the linear transcript in either sense or antisense orientation. (C) Antisense, when overlapping one or more exons of the linear transcript, as they transcribe from the opposite strand. (D) Bidirectional or intragenic, when transcribing from same gene locus of the linear transcript, but in close genomic proximity within 1 kb and not classified into ‘sense’ and ‘intronic’. (E) Intergenic, when it locates outside at least 1kb away from known gene locus.
Biological functions of circRNAs
Although the abundance of circRNAs is now recognized, the potential function of these molecules remains the issue of debate. Recent studies have revealed 3 possible biological functions of circRNAs. Specifically, they can function as miRNA sponges, regulate transcription or splicing, and interact with RBPs ().Citation1,49-52
Figure 2. Biological functions of circular RNAs. CircRNAs contain miRNA binding sites to act as competitive endogenous RNA. CircRNAs sequester miRNAs from binding mRNA targets. EIciRNAs can enhance gene transcription via interacting with U1 snRNP and RNA Polymerase II in the promoter region of the host gene. GU-rich sequences near the 5’ splice site (red box) and C-rich sequences near branch point (purple box) are minimally sufficient for ciRNA formation. The stable ciRNA binds to elongating RNA Pol II and promotes transcription. CircRNA biogenesis competes with linear splicing. Circularization and splicing compete against each other to keep the transcripts dynamic balance. CircRNA formation act as ‘mRNA trap’ to make linear transcripts untranslated by sequestering the translation start site or break the integrity of mature linear RNA. CircRNAs can also function as RNA binding protein (RBP) sponge to interact with RBPs, such as MBL, p21 and CDK2.
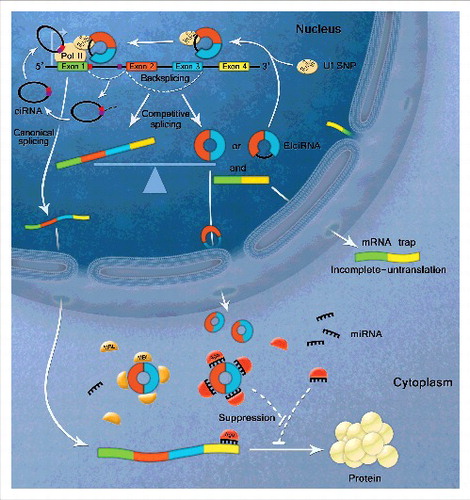
Considering the stability of their circular structure and their conserved sequences, circRNAs are ideal to act as miRNA sponges or competitive endogenous RNAs (ceRNAs). Seven circRNAs - ciRS-7/CDR1as (for circular RNA sponge for miR-7 or CDR1 antisense),Citation18,24 Sry circRNA (circ-Sry),Citation24 circ-ITCH,Citation25,43 circ-Foxo3,Citation26 circRNA HRCR,Citation27 circHIPK3Citation53 and hsa_circ_001569Citation54 - have been verified to function as miRNA sponges. The ciRS-7/CDR1as molecule contains more than 70 miR-7 binding sites and can adsorb miR-7 to reduce its miRNA activity.Citation24 circ-Sry is highly expressed in adult mouse testis;Citation7 it harbors 16 miR-138 binding sites and interacts with miR-138.Citation24 Although these 2 examples are striking, the 2 circRNAs have a larger number of putative miRNA binding site. It is currently debated whether the function of circRNA as an miRNA sponge is a general (but not exclusive) feature.Citation49 Two studies indicated that most circRNAs do not function as miRNA sponges as a large majority of circRNAs does not possess more miRNA binding sites than do co-linear mRNAs.Citation20,29 However, with the advance of circRNA research, increasing evidence indicate that circRNA can act as miRNA sponge, which may not depend on the numbers of miRNA binding site. For example, circ-ITCH was also found to function as a sponge of miR-7, miR-17 and miR-124 via luciferase reporter assays.Citation25 Yang and coworkers recently discovered that circ-Foxo3 could serve as an effective miRNA sponge through its interaction with miR-22, miR-136*, miR-138, miR-149*, miR-433, miR-762, miR-3614–5p and miR-3622b–5p.Citation26 Moreover, 3 recent studies just found that circRNA HRCR,Citation27 circHIPK3Citation53 and hsa_circ_001569Citation54 could act as miRNA sponge, such as miR-223, miR-124 and miR-145. These findings support the idea that circRNA acting as miRNA sponges is a widespread phenomenon in many eukaryotes.
Because of the correlation between circRNAs and their parent gene, the formed are thought to positively regulate parent gene expression in cis- or trans-acting manner. ciRNA was found to localize to the nucleus and to interact with the Pol II machinery to activate its parent gene transcription.Citation19 Similarly, EIciRNAs was shown to localize predominantly to the nucleus and to interact with U1 snRNP and Pol II through complementary sequence pairing between EIciRNA and U1 snRNA (snRNPs), thereby enhancing the transcription of its parental gene.Citation30 Later, circ-ITCH was reported to act as a sponge of miR-7, miR-17 and miR-138 to increase the level of ITCH.Citation25 Notably, The pocessing of circRNAs also affects splicing. There is a negative correlation between circRNA formation and alternative splicing, potentially leading to altered gene expression. circMbl formation competes with mbl pre-mRNA splicing. circMbl and circMbl flanking introns harbor conserved MBL protein binding sites that bind strongly to MBL. The alteration of MBL level significantly affects circMbl formation, an effect dependent on MBL binding sites.Citation28 Moreover, backsplicing may produce circRNA and a corresponding linear RNA.Citation28 The circRNA may contain a translation start site, leaving the corresponding linear RNA untranslated.Citation17,29,55 Alternatively, circRNA formation may disrupt the integrity of mature linear RNA, also leaving it untranslated.Citation56 This phenomenon is known as an mRNA trap.Citation55
Like lncRNA, circRNAs can interact with RBPs or function as RBP sponges.Citation28,31,57 As described above, circMbl contains conserved MBL protein binding sites. MBL can interact with circMbl to balance the efficiency between circMbl and mbl mRNA production.Citation28 Moreover, Du and others found that circ-Foxo3 could interact with both CDK2 and p21 forming a ternary complex, blocking cell cycle progression,Citation31 and that circ-Foxo3 could promote cardiac senescence by modulating multiple factors associated with stress and senescence responses, such as ID-1, E2F1, FAK, and HIF1α.Citation57
CircRNAs in the life processes
CircRNAs are stable, conserved noncoding RNAs with cell-, tissue-, and developmental stage-specific patterns of expression. Because circRNAs have the above functions, they are the subject of much recent attention. Recent studies have suggested that circRNAs play important roles in biological processes and have the potential to become clinical molecular biomarkers ().
Table 1. circRNAs in the life processes.
CircRNAs in the physiological processes
Recent studies have indicated that circRNAs may be involved in essential physiological processes. For example, CDR1as expression was up-regulated in islet cells by the PMA and long-term forskolin stimulation but not high glucose levels. Strikingly, CDR1as overexpression significantly promotes insulin biosynthesis and secretion through the CDR1as/miR-7 pathway, which provides a potential target for improving β cell function in diabetic patients.Citation34 Boeckel et al. found that circular RNAs are expressed in endothelial cells and are regulated by hypoxia. The circular RNA cZNF292 controls the proliferation and angiogenic sprouting of endothelial cells. These findings indicate that endothelial circRNAs are regulated by hypoxia and have a biological function.Citation58 Moreover, a large number of circRNAs were shown to be dysregulated in the absence of CD28 in CD8(+) T cells during aging using a circRNA microarray approach. The identification of a circRNA-miRNA-gene network suggested that circRNA100783 might regulate phosphoprotein-related signal transduction in the context of CD28-dependent CD8(+) T cell aging.Citation33
Interestingly, circRNAs play crucial roles in physiological processes in non-human mammals. CDR1as harbors 74 miR-7 seed matches that are often highly conserved. Moreover, CDR1as is expressed at higher levels in nervous tissue.Citation18,59 The overexpression of CDR1as in zebrafish embryos substantially reduces the size of the midbrain, phenocopying the morphological defects in the midbrain the accompany miR-7 loss-of-function.Citation18 Some studies have shown that circRNAs are highly abundant in the mammalian brainCitation35,37,60 and are especially enriched in synaptoneurosomes.Citation36 Many circRNAs alter their abundance during synaptogenesis and neuronal differentiation.Citation35,36 Moreover, Westholm et al. reported that circRNA levels increased substantially in the central nervous system during aging compared with the levels of the corresponding co-linear isoforms and might serve as aging biomarkers in Drosophila.Citation32 These data indicated that circRNAs could be involved in neuronal differentiation or development and might act to regulate synaptic function. Remarkably, Abdelmohsen and others have identified circRNAs that are abundantly expressed in the skeletal muscle tissues of Rhesus monkey. Among these, they found that subsets of circRNAs exhibited age-dependent expression patterns.Citation61 Taken together, these findings suggest roles for circRNAs in the physiological processes.
CircRNAs in the pathological processes
A growing body of research has demonstrated that circRNAs play crucial roles in the initiation and development of diseases and might function as novel biomarkers for disease diagnosis and therapy.
CircRNAs and neurological diseases
For example, Satoh et al. reported that the stable overexpression of prion protein (PrPC) in HEK293 cells induced the expression of CDR1as but not CDR1.Citation62 Hence, PrPC might be involved in the regulation of CDR1as expression. It would be worth elucidating the function of CDR1as in prion diseases. Besides, Kuliw detected that the level of ciRS-7 is significantly reduced in sporadic Alzheimer disease (AD) compared to its expression in age-matched control hippocampal CA1.Citation39 Intriguingly, previous studies found that miR-7 can act as a direct regulator of ubiquitin protein ligase A (UBE2A), which is essential for the clearance of amyloid peptides in AD,Citation39 and miR-7 can down-regulate α-synuclein expression, which is implicated in Parkinson disease (PD).Citation63 Considering ciRS-7 can function as miR-7 sponge, these findings suggests that ciRS-7 may play a role in AD and PD.
CircRNAs and cardiovascular diseases
cANRIL (circular antisense non-coding RNA in the INK4 locus, cANRIL) is an antisense transcript synthesized from the INK4/ARF locus. Burd and others have found that cANRIL isoform expression is associated with INK4/ARF transcription and atherosclerotic vascular disease (ASVD) risk. Interestingly, 9p21 polymorphisms within the ASVD risk interval may regulate ANRIL splicing and cANRIL production, as determined using next-generation DNA sequencing and splice prediction algorithms.Citation38 It would be interesting to unveil the function of cANRIL in ASVD.
Wang et al. found that circRNA HRCR inhibit the development of cardiac hypertrophy and heart failure induced in the mice by isoproterenol and transverse aortic construction via miR-223.Citation27 Moreover, ectopic expressed circ-Foxo3 could bind to senescence-related proteins ID1 and E2F1, and stress related proteins HIF1α and FAK in cytoplasm, promote cardiac senescence.Citation57 These results provide new insights for understanding the pathogenesis of heart diseases.
CircRNAs and degenerative diseases
Both circMbl and its flanking intron sequences can be bound by MBL, and the modulation of MBL level strongly affects circMbl formation. circRNA biogenesis competes with canonical mbl pre-mRNA splicing.Citation28 MBL has been shown to regulate the mbl pre-mRNA splicing efficiency of circMbl and mbl mRNA. Moreover, circMbl can serve as a sponge for MBL, thereby regulating the production of MBL protein. However, MBL functional deficiency is known to be involved in a severe degenerative disease called myotonic dystrophy.Citation1 We therefore speculate that circMbl could be associated with the initiation and development of myotonic dystrophy. In addition, Liu and others demonstrated that some circRNAs were differently expressed in osteoarthritis, and that circRNA-CER could regulate MMP13 expression by competitively binding miR-136 in human cartilage degradation. Citation64
CircRNAs are dysregulated in cancer
In cancer biology, it is very important to identify gene expression differences between tumor and normal samples, as knowledge of circRNA expression signatures in tumor and normal samples is necessary. Moreover, recent research has demonstrated that circRNAs are globally down-regulated in colorectal cancer (CRC) tissues compared to matched normal tissues and are even more reduced in CRC cell lines. Interestingly, a negative correlation between global circular RNA abundance and proliferation has been reported.Citation41 In addition, recent studies also found that circRNA expression patterns were dysregulated in pancreatic cancer,Citation40 basal cell carcinoma,Citation65 cutaneous squamous cell carcinoma,Citation66 laryngeal squamous cell carcinoma,Citation67 hepatocellular carcinomaCitation68 and gliomaCitation69 via microarray or sequencing technology. These findings indicate that dysregulated circRNAs may be involved in the development of some cancer. However, whether circRNAs are dysregulated in other tumor remains to be clarified.
Remarkably, Guarnerio and othersCitation70 found a novel type of circRNA derived from cancer-associated chromosomal translocations: aberrant fusion-circRNAs (f-circRNA). F-circRNAs are oncogenic and contribute to cellular transformation. Especially, F-circM9 contributes to leukemia progression and confers resistance to treatment in leukemic cells.
CircRNA–miRNA axes are involved in cancers
CircRNAs, which have emerged as new transcriptome regulators, constitute an exciting new field of biological inquiry. Recent studies have demonstrated that circRNAs are associated with disease-related miRNAs. It is clear that miRNAs are involved in nearly all aspects of cellular functions and play important roles in disease initiation and progression.Citation71,72 Because many circRNAs contain several conserved binding sites for miRNAs,Citation32,73 circRNAs could interact with miRNAs to control their biological function. This crosstalk between circRNAs and miRNAs is the key to understanding the role of circRNAs in carcinogenesis and other diseases.Citation74,75 For example, CDR1as has been confirmed to bind specifically to miR-7 and to inhibit its biological function.Citation24 Furthermore, miR-7 has been shown to play oncogenic or tumor-suppressive roles in cancers.Citation76-79 Hence, the CDR1as/miR-7 axis may perform an essential regulatory role in cancer-associated pathways. In parallel, circ-Sry controls the biological functions of miR-138 by binding to its 16 binding sites.Citation24 miR-138 has also been shown to play important roles in cancer.Citation80 It would therefore be interesting to explore the functions of the circ-Sry/miR-138 axis in cancer.
Remarkably, Li et al. found that circ-ITCH expression is commonly downregulated in esophageal squamous cell carcinoma compared to paired adjacent tissues. circ-ITCH can increase the level of ITCH by acting as a sponge for miR-7, miR-17 and miR-124, which promote the ubiquitination and degradation of phosphorylated Dvl2 to inhibit the expression of the oncogene c-myc, thereby suppressing the canonical Wnt signaling pathway.Citation25 Subsequent parallel study has shown that circ-ITCH expression is also usually down-regulated in CRC, and that circ-ITCH can also increase the level of ITCH by interacting with miR-7, miR-20a and miR-214, thereby suppressing the Wnt/β-catenin pathway.Citation43 Moreover, hsa_circ_001569 is upregulated in CRC, and promotes the proliferation and invasion of CRC cells by regulating miR-145 and its targets.Citation54 Specifically, 2 recent studies respectively have demonstrated that the overexpression of circ-Foxo3 in MDA-MB-231 cells suppresses tumor growth in vivo as well as cancer cell proliferation and survival in vitro,Citation26 and that silencing of circHIPK3 RNA inhibits human Huh7, HCT-116 and HeLa cell proliferation by sponging with miR-124.Citation53
CircRNAs as a source of diagnostic markers
CircRNAs are more stable than linear RNAs due to their higher nuclease stability and exhibit long half lives in cells.Citation17,19,81 These features are an enormous clinical advantage for use as diagnostic and therapeutic biomarkers for cancers. For instance, hsa_circ_002059 was found to be commonly downregulated in gastric cancer, which was associated with TNM stage and distal metastasis. Hence, hsa_circ_002059 was regarded as a potential novel biomarker for the diagnosis of gastric cancer.Citation42 Similarly, hsa_circ_0001649 expression was significantly down-regulated in hepatocellular carcinoma (HCC), which was correlated with tumor size and the occurrence of tumor embolus. Thus, hsa_circ_0001649 is a novel putative biomarker for HCC.Citation44
Intriguingly, circular RNA has been shown to be enriched and stable in exosomes (small membrane vesicles containing endocytic origins secreted by cells). Moreover, the abundance of tumor-derived exosomal circRNAs in the serum of xenografted mice was correlated with tumor mass, which may serve as a promising biomarker for cancer detection.Citation46 Remarkably, Memczak and colleagues observed that circRNAs are reproducibly and easily detected in clinical standard blood samples. Hundreds of circRNAs are expressed at much higher levels than their corresponding linear mRNAs in human blood, suggesting that they may represent a new class of biomarkers for human disease.Citation82 Likewise, Bahn and others predicted 422 putative circRNAs in human cell-free saliva using high-throughput RNA sequencing. Some of these predicted circRNAs were validated by RT-PCR using divergent primers. All PCR products were purified and subjected to Sanger sequencing.Citation83 Considering the urgent clinical need for non-invasive biomarker detection for many disease states, circRNA biomarkers in human blood and saliva or other bodily fluids appear very promising.
Conclusion and perspective
In the past, circRNAs were considered a byproduct of aberrant splicing events or intermediates that had escaped from intron lariat debranching.Citation6,84,85 Thus, they were thought to be unlikely to play crucial roles in biological processes. Nevertheless, with advancements in high-throughput sequencing technologies and bioinformatics, circRNAs were found to be broadly expressed and to perform co- or post-transcriptional regulation in the cells and tissues of various species. Moreover, multiple circular RNAs have been identified from a single precursor RNA in a phenomenon referred to as alternative circularization.Citation21 These findings greatly expanded our knowledge of the complexity of gene regulation and have moved circRNA to the forefront of biological research. Along with in-depth studies of circRNAs, several circRNA databases have been constructed, such as circBase (http://www.circbase.org/),Citation86 circNet (http://circnet.mbc.nctu.edu.tw/),Citation87 Circ2Traits (http://gyanxet-β.com/circdb/)Citation74 and CircInteractome (http://circinteractome.nia.nih.gov).Citation88 These databases provided excellent platforms to facilitate further functional research on circRNAs.
To date, studies have revealed that circRNAs are evolutionarily conserved and are expressed in a cell-, tissue- and developmental stage-specific manner. Although the functions of circRNAs are largely unknown, accumulating evidence has shown that circRNAs function in multiple biological processes, such as miRNA binding, protein binding and regulation of transcription or splicing.Citation1,52 Remarkably, endogenous circRNAs have not been found to associate with ribosomes for translation, suggesting that they have a tendency to serve as a new class of ncRNAs. Nevertheless, exogenous circRNAs engineered to contain an internal ribosome entry site (IRES) or via a rolling circle amplification mechanism can be translated in vivo or in vitro,Citation89,90 so we cannot exclude the possibility that some circRNAs are translatable. As studies of circRNAs increase in number, it is likely that additional functions will be revealed.
Strikingly, recent studies have also reported that circRNAs play essential roles in the physiological processes of aging, insulin secretion and tissue development. Moreover, circRNAs are reported to be involved in the pathological processes, especially cancer. Hence, these findings indicate that circRNAs might play important roles in biological processes, serve as novel clinical molecular biomarkers and provide insight into the treatment of diseases. However, current studies of the relationship between circRNAs and disease physiology and pathology are limited. In the near future, if properly modified and delivered, circRNAs have the potential roles to act as molecular tools or therapies to regulate cellular homeostasis by interacting with miRNAs and other RNAs or RBPs.
The study of circRNAs has gradually become one of the hottest topics in the field of genetics. However, the field of circular RNA presents a wealth of unanswered questions. Currently, several bioinformatic algorithms such as find-circ,Citation18 MapSplice,Citation91 CIRCexplorer,Citation21 circRNAfinderCitation32 and CIRICitation92 are available to predict circRNAs. However, the consistency of these computational tools remains relatively low, and no systematic approach is available for identifying circRNAs in the human transcriptome. There remains a need for a robust algorithm to achieve more reliable predictions with a better balance between precision and sensitivity.Citation51,93 Given the huge number of circRNAs and the empirical classification of circRNAs based on lncRNA categories, it is important to develop a standard nomenclature or a comprehensive classification system to enable future circRNAs research. Moreover, current studies of circRNAs have mainly focused on circRNA formation, whereas little is known about the degradation of most circRNAs. A possible mechanism for circRNA clearance is to eliminate them via releasing vesicles such as microvesicles and exosomes.Citation94 The expression profiles of circRNAs maybe depend on a balance between biogenesis and degradation. Moreover, given that cellular localization is often associated with physiological function and mechanism, it is a topic worth exploring.
Taken together, although circRNAs are currently in the spotlight, the field is still in its infancy. Further intensive studies are needed to explore the molecular and biological functions of circRNAs. The world of the transcriptome may be more complicated than previously thought. circRNA, as the new star of noncoding RNA, would be widely involved in the regulation of physiological and pathophysiological processes, serve as stable clinical biomarkers of disease, and also provide new potential therapeutic targets.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
Thanks for anonymous reviewers’ constructive comments and suggestions. This work was supported by grant from the National Natural Science Foundation of China (No. 81672339), and Health public welfare industry special scientific research projects of China (No.201202007).
References
- Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett 2015; 365:141–8; PMID:26052092; https://doi.org/10.1016/j.canlet.2015.06.003
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Pro Natl Acad Sci U S A 1976; 73:3852–6; PMID:1069269; https://doi.org/10.1073/pnas.73.11.3852
- Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986; 323:558–60; PMID:2429192; https://doi.org/10.1038/323558a0
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell 1991; 64:607–13; PMID:1991322; https://doi.org/10.1016/0092-8674(91)90244-S
- Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J 1992; 11:1095–8; PMID:1339341
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155–60; PMID:7678559
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993; 73:1019–30; PMID:7684656; https://doi.org/10.1016/0092-8674(93)90279-Y
- Zaphiropoulos PG. Differential expression of cytochrome P450 2C24 transcripts in rat kidney and prostate: evidence indicative of alternative and possibly trans splicing events. Biochem Biophys Res Commun 1993; 192:778–86; PMID:7916606; https://doi.org/10.1006/bbrc.1993.1482
- Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Pro Natl Acad Sci U S A 1996; 93:6536–41; PMID:8692851; https://doi.org/10.1073/pnas.93.13.6536
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7:e30733; PMID:22319583; https://doi.org/10.1371/journal.pone.0030733
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9:e1003777; PMID:24039610; https://doi.org/10.1371/journal.pgen.1003777
- Fan X, Zhang X, Wu X, Guo H, Hu Y, Tang F, Huang Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol 2015; 16:148; PMID:26201400; https://doi.org/10.1186/s13059-015-0706-1
- Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol 2011; 12:R16; PMID:21324177; https://doi.org/10.1186/gb-2011-12-2-r16
- Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015; 21:2076–87; PMID:26464523; https://doi.org/10.1261/rna.052282.115
- Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res 2012; 40:3131–42; PMID:22140119; https://doi.org/10.1093/nar/gkr1009
- Broadbent KM, Broadbent JC, Ribacke U, Wirth D, Rinn JL, Sabeti PC. Strand-specific RNA sequencing in Plasmodium falciparum malaria identifies developmentally regulated long non-coding RNA and circular RNA. BMC Genomics 2015; 16:454; PMID:26070627; https://doi.org/10.1186/s12864-015-1603-4
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141–57; PMID:23249747; https://doi.org/10.1261/rna.035667.112
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333–8; PMID:23446348; https://doi.org/10.1038/nature11928
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792–806; PMID:24035497; https://doi.org/10.1016/j.molcel.2013.08.017
- Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15:409; PMID:25070500; https://doi.org/10.1186/s13059-014-0409-z
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell 2014; 159:134–47; PMID:25242744; https://doi.org/10.1016/j.cell.2014.09.001
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015; 10:170–7; PMID:25558066; https://doi.org/10.1016/j.celrep.2014.12.019
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015; 160:1125–34; PMID:25768908; https://doi.org/10.1016/j.cell.2015.02.014
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384–8; PMID:23446346; https://doi.org/10.1038/nature11993
- Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015; 6:6001–13; PMID:25749389; https://doi.org/10.18632/oncotarget.3469
- Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 2016; 35:3919-31; PMID:26657152; https://doi.org/10.1038/onc.2015.460
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016; pii: ehv713; PMID: 26802132; https://doi.org/10.1093/eurheartj/ehv713
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56:55–66; PMID:25242144; https://doi.org/10.1016/j.molcel.2014.08.019
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453–61; PMID:24811520; https://doi.org/10.1038/nbt.2890
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22:256–64; PMID:25664725; https://doi.org/10.1038/nsmb.2959
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016; 44:2846–58; PMID:26861625; https://doi.org/10.1093/nar/gkw027
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014; 9:1966–80; PMID:25544350; https://doi.org/10.1016/j.celrep.2014.10.062
- Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing 2015; 12:17; PMID:26451160; https://doi.org/10.1186/s12979-015-0042-z
- Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 2015; 5:12453; PMID:26211738; https://doi.org/10.1038/srep12453
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015; 58:870–85; PMID:25921068; https://doi.org/10.1016/j.molcel.2015.03.027
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 2015; 18:603–10; PMID:25714049; https://doi.org/10.1038/nn.3975
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 2015; 16:126; PMID:26076956; https://doi.org/10.1186/s13059-015-0690-5
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010; 6:e1001233; PMID:21151960; https://doi.org/10.1371/journal.pgen.1001233
- Lukiw WJ. Circular RNA (circRNA) in Alzheimer disease (AD). Front Genet 2013; 4:307; PMID:24427167; https://doi.org/10.3389/fgene.2013.00307
- Qu S, Song W, Yang X, Wang J, Zhang R, Zhang Z, Zhang H, Li H. Microarray expression profile of circular RNAs in human pancreatic ductal adenocarcinoma. Genom Data 2015; 5:385–7; PMID:26484292; https://doi.org/10.1016/j.gdata.2015.07.017
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 2015; 5:8057; PMID:25624062; https://doi.org/10.1038/srep08057
- Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015; 444:132–6; PMID:25689795; https://doi.org/10.1016/j.cca.2015.02.018
- Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PLoS One 2015; 10:e0131225; PMID:26110611; https://doi.org/10.1371/journal.pone.0131225
- Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark 2016; 16:161–9; PMID:26600397; https://doi.org/10.3233/CBM-150552
- Shen T, Han M, Wei G, Ni T. An intriguing RNA species-perspectives of circularized RNA. Protein Cell 2015; 6:871–80; PMID:26349458; https://doi.org/10.1007/s13238-015-0202-0
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015; 25:981–4; PMID:26138677; https://doi.org/10.1038/cr.2015.82
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136:629–41; PMID:19239885; https://doi.org/10.1016/j.cell.2009.02.006
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10:155–9; PMID:19188922; https://doi.org/10.1038/nrg2521
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 2016; 1859:163–8; PMID:26171810; https://doi.org/10.1016/j.bbagrm.2015.07.007
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014; 20:1829–42; PMID:25404635; https://doi.org/10.1261/rna.047126.114
- Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 2015; 6:563–79; PMID:26230526; https://doi.org/10.1002/wrna.1294
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol cell Biol 2016; 17:205–11; PMID:26908011; https://doi.org/10.1038/nrm.2015.32
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016; 7:11215; PMID:27050392; https://doi.org/10.1038/ncomms11215
- Xie H, Ren X, Xin S, Lan X, Lu G, Y L, Ss Y, Zc Z, Wt L, Ding YQ, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016; 7:26680-91; PMID:27058418; https://doi.org/10.18632/oncotarget.8589
- Chao CW, Chan DC, Kuo A, Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med 1998; 4:614–28; PMID:9848078
- Gualandi F, Trabanelli C, Rimessi P, Calzolari E, Toffolatti L, Patarnello T, Kunz G, Muntoni F, Ferlini A. Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J Med Genet 2003; 40:e100; PMID:12920092; https://doi.org/10.1136/jmg.40.8.e100
- Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 2016; PMID:26873092; https://doi.org/10.1093/eurheartj/ehw001
- Boeckel JN, Jae N, Heumuller AW, Chen W, Boon RA, Stellos K, Zeiher AM, John D, Uchida S, Dimmeler S. Identification and Characterization of Hypoxia-Regulated Endothelial Circular RNA. Circ Res 2015; 117:884–90; PMID:26377962; https://doi.org/10.1161/CIRCRESAHA.115.306319
- Dropcho EJ, Chen YT, Posner JB, Old LJ. Cloning of a brain protein identified by autoantibodies from a patient with paraneoplastic cerebellar degeneration. Pro Natl Acad Sci U S A 1987; 84:4552–6; PMID:3474619; https://doi.org/10.1073/pnas.84.13.4552
- Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 2015; 16:245; PMID:26541409; https://doi.org/10.1186/s13059-015-0801-3
- Abdelmohsen K, Panda AC, De S, Grammatikakis I, Kim J, Ding J, Noh JH, Kim KM, Mattison JA, de Cabo R, et al. Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany NY) 2015; 7:903–10; PMID:26546448; https://doi.org/10.18632/aging.100834
- Satoh J, Yamamura T. Gene expression profile following stable expression of the cellular prion protein. Cell Mol Neurobiol 2004; 24:793–814; PMID:15672681; https://doi.org/10.1007/s10571-004-6920-0
- Junn E, Lee KW, Jeong BS, Chan TW, Im JY, Mouradian MM. Repression of α-synuclein expression and toxicity by microRNA-7. Pro Natl Aca Sci U S A 2009; 106:13052–7; PMID:19628698; https://doi.org/10.1073/pnas.0906277106
- Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, Ao Y. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 'Sponge' in Human Cartilage Degradation. Sci Rep 2016; 6:22572; PMID:26931159; https://doi.org/10.1038/srep22572
- Sand M, Bechara FG, Sand D, Gambichler T, Hahn SA, Bromba M, Stockfleth E, Hessam S. Circular RNA expression in basal cell carcinoma. Epigenomics 2016; 8:619–32; PMID:27097056; https://doi.org/10.2217/epi-2015-0019
- Sand M, Bechara FG, Gambichler T, Sand D, Bromba M, Hahn SA, Stockfleth E, Hessam S. Circular RNA expression in cutaneous squamous cell carcinoma. J Dermatol Sci 2016; 83:210-8; PMID:27298156; http://dx/doi.org/10.1016/j.jdermsci.2016.05.012
- Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T, Wang X, Li Q, Tian L, Liu M, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res 2016; 8:932–9; PMID:27158380
- Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive Circular RNA Profiling Reveals That hsa_circ_0005075, a New Circular RNA Biomarker, Is Involved in Hepatocellular Crcinoma Development. Medicine 2016; 95:e3811; PMID:27258521; https://doi.org/10.1097/MD.0000000000003811
- Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 2016; 44:e87; PMID:26873924; https://doi.org/10.1093/nar/gkw075
- Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016; 165:289–302; PMID:27040497; https://doi.org/10.1016/j.cell.2016.03.020
- Ruan K, Fang X, Ouyang G. MicroRNAs: novel regulators in the hallmarks of human cancer. Cancer Lett 2009; 285:116–26; PMID:19464788; https://doi.org/10.1016/j.canlet.2009.04.031
- Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med 2009; 60:167–79; PMID:19630570; https://doi.org/10.1146/annurev.med.59.053006.104707
- Thomas LF, Saetrom P. Circular RNAs are depleted of polymorphisms at microRNA binding sites. Bioinformatics 2014; 30:2243–6; PMID:24764460; https://doi.org/10.1093/bioinformatics/btu257
- Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 2013; 4:283; PMID:24339831; https://doi.org/10.3389/fgene.2013.00283
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013; 73:5609–12; PMID:24014594; https://doi.org/10.1158/0008-5472.CAN-13-1568
- Meza-Sosa KF, Perez-Garcia EI, Camacho-Concha N, Lopez-Gutierrez O, Pedraza-Alva G, Perez-Martinez L. MiR-7 promotes epithelial cell transformation by targeting the tumor suppressor KLF4. PLoS One 2014; 9:e103987; PMID:25181544; https://doi.org/10.1371/journal.pone.0103987
- Suto T, Yokobori T, Yajima R, Morita H, Fujii T, Yamaguchi S, Altan B, Tsutsumi S, Asao T, Kuwano H. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis 2015; 36:338–45; PMID:25503932; https://doi.org/10.1093/carcin/bgu242
- Hao Z, Yang J, Wang C, Li Y, Zhang Y, Dong X, Zhou L, Liu J, Qian J. MicroRNA-7 inhibits metastasis and invasion through targeting focal adhesion kinase in cervical cancer. Int J Clin Exp Med 2015; 8:480–7; PMID:25785020
- Kalinowski FC, Brown RA, Ganda C, Giles KM, Epis MR, Horsham J, Leedman PJ. microRNA-7: a tumor suppressor miRNA with therapeutic potential. Int J Biochem Cell Biol 2014; 54:312–7; PMID:24907395; https://doi.org/10.1016/j.biocel.2014.05.040
- Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol 2015; 9:1–8; PMID:26649774; https://doi.org/10.1080/15476286.2015.1122162
- Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 2014; 9:e90859; PMID:24609083; https://doi.org/10.1371/journal.pone.0090859
- Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PLoS One 2015; 10:e0141214; PMID:26485708; https://doi.org/10.1371/journal.pone.0141214
- Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 2015; 61:221–30; PMID:25376581; https://doi.org/10.1373/clinchem.2014.230433
- Kopczynski CC, Muskavitch MA. Introns excised from the Delta primary transcript are localized near sites of Delta transcription. J Cell Biol 1992; 119:503–12; PMID:1383233; https://doi.org/10.1083/jcb.119.3.503
- Qian L, Vu MN, Carter M, Wilkinson MF. A spliced intron accumulates as a lariat in the nucleus of T cells. Nucleic Acids Res 1992; 20:5345–50; PMID:1437551; https://doi.org/10.1093/nar/20.20.5345
- Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014; 20:1666–70; PMID:25234927; https://doi.org/10.1261/rna.043687.113
- Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 2016; 44:D209–15; PMID:26450965; https://doi.org/10.1093/nar/gkv940
- Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA biology 2016; 13:34–42; PMID:26669964; https://doi.org/10.1080/15476286.2015.1128065
- Perriman R, Ares M, Jr. Circular mRNA can direct translation of extremely long repeating-sequence proteins in vivo. RNA 1998; 4:1047–54; PMID:9740124; https://doi.org/10.1017/S135583829898061X
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015; 21:172–9; PMID:25449546; https://doi.org/10.1261/rna.048272.114
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 2010; 38:e178; PMID:20802226; https://doi.org/10.1093/nar/gkq622
- Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015; 16:4; PMID:25583365; https://doi.org/10.1186/s13059-014-0571-3
- Hansen TB, Veno MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res 2016; 44:e58; PMID:26657634; https://doi.org/10.1093/nar/gkv1458
- Lasda E, Parker R. Circular RNAs Co-Precipitate with Extracellular Vesicles: A Possible Mechanism for circRNA Clearance. PLoS One 2016; 11:e0148407; PMID:26848835; https://doi.org/10.1371/journal.pone.0148407
