ABSTRACT
Circular RNA from backspliced exons (or exonic circular RNA, circRNA) is a type of covalently closed non-colinear RNA that was recently rediscovered in eukaryotes. Although circRNAs are often expressed at low levels, emerging evidence indicates that many circRNAs are linked to physiological development and various diseases. Notably, circRNAs have been shown to serve as oncogenic stimuli or tumor suppressors in cancer. circRNAs may regulate gene expression through different mechanisms. In addition, circRNAs have been shown to be useful as biomarkers of diseases due to their stability, specific expression and relation to diseases both in cells and in extracellular fluid. This review summarizes current knowledge of human circRNAs and discusses the emerging role and clinical implication of these multifarious transcripts.
Introduction
RNA circles are covalently closed and single-stranded transcripts that were first observed in the late 1970s.Citation1 There are 5 general categories of RNA circles, including genomic circular RNA (viroids and hepatitis delta virus), circular intron RNA (circular intronic RNAs and excised introns), circular processing intermediate RNA (rRNA precursors and permuted tRNAs), circular housekeeping noncoding RNA (some snoRNAs and RNase P), and circular spliced-exon RNA.Citation2 Circular RNA, derived from back-spliced exons anchored by a breakpoint with flanking GU/AG sequence, is essentially exonic circular RNA (circRNA). For nearly 40 years, a systematic and comprehensive understanding of these covalently closed non-colinear RNAs has been lacking because traditional RNA analysis could not provide reliable high-throughput detection methods. Only a small number of circRNAs in different organisms have been identified, and they were generally disregarded as the results of mis-splicing and as by-products of pre-mRNA processing.Citation3-6 However, recent advances using biochemical enrichment strategies and novel bioinformatics approaches coupled with deep sequencing have allowed comprehensive investigation of circRNAs.Citation7-10 Specifically, a large number of circRNAs have been successfully identified in various cell lines and across different species.Citation11-16 Moreover, many circRNAs have been found to be expressed in a cell type-specific or tissue-specific manner,Citation13-16 suggesting they might have biological functions.
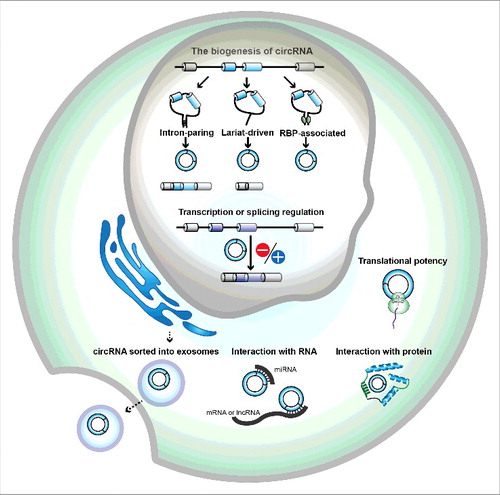
circRNAs are mainly derived from precursor mRNAs (pre-mRNAs), which are transcribed by RNA polymerase II (Pol II). Typically, a circRNA in eukaryotes, including human cells, comes from a backsplicing event of pre-mRNA with spliceosome-mediated circularization.Citation17 The backsplice joins a 5′ splice site (splice donor) of a downstream exon with a 3′ splice site (splice acceptor) of an upstream exon to yield a circular RNA with a junction between exons. A proportion of circRNAs consist of a single exon, in which it is the joining of downstream splice donor with upstream splice accepter. There are 3 different models of circRNA formation that have been described: intron-paring, lariat-driven circularization and RNA binding protein-driven circularization (). In the intron-paring model, circularization via back-splicing is associated with base-pairing across different introns especially between repetitive sequences such as ALU repeats, while the lariat-driven circularization occurs by joining the splice sites of the exons that are skipped during linear RNA formation.Citation8,18,19 circRNA biogenesis is also regulated by trans-factors, such as RNA-binding proteins (RBPs).Citation20 RBPs bind to the introns near splice sites, which may facilitate the production of circRNA. Genome-wide RNA-sequencing analyses have now identified a considerable number of circRNAs derived from backsplicing. Although they are generally expressed at low levels, increasing evidence has shown that circRNAs are linked to physiological developmentCitation15 and different diseases, such as neurological dystrophy,Citation21 cardiovascular diseases,Citation22-25 and cancer.Citation26-28 Recently, circRNAs have also been shown to be enriched and stable in plasma,Citation29 saliva,Citation30 and even in serum exosomes,Citation31 indicating the potential of circRNAs as readable biomarkers.
Figure 1. The biogenesis and potential mechanisms of human circular RNAs. circRNAs in human cells mianly come from backsplicing events of pre-mRNAs with spliceosome-mediated circularization. There are 3 different models of circularization: intron-paring, lariat-driven circularization and RNA binding protein-driven circularization. Intron-paring circularization is the base-pairing across different introns to form a circular RNA. Lariat-driven circularization is the joining of splice sites of the exons that are skipped during linear RNA formation. RBP (RNA binding protein) – associated circularization is the binding of RBP to the introns near splice sites to form a circular RNA. The potential mechanisms of circRNAs include RNA interaction, protein interaction, transcription or splicing regulation, and translation. circRNAs can be also packaged in exosomes, which are cell-derived vesicles, and be secreted into extracellular space.
In this review, we summarize the current knowledge on human circRNAs, and present an overview of the emerging roles of human circRNAs and their potential clinical implications.
Identification and properties of human circRNAs
Detecting human circRNAs
Pervasive detection of circRNAs with high-throughput RNA sequencing (RNA-seq) in human cells has been achieved in various types of cells and tissues. Most circRNAs are derived from pre-mRNA and are expressed at low levels, but some circRNAs have a higher abundance than their linear counterparts. Notably, there are at least 10 thousand unique human circRNA candidates, which indicates that circRNAs may comprise one of the largest RNA families in human RNA transcriptome. Several databases and web tools have been developed to explore circRNAs and their potential regulatory networks (). Cirbase is a comprehensive database to explore public circRNA datasets.Citation11 Circ2Traits is a database for circRNA potentially associated with disease and traits.Citation32 Circnet provides circRNA expression profiles and circRNA-miRNA-gene regulatory networks.Citation33 CircInteractome is a web-tool for exploring circular RNAs and their interacting proteins and miRNAs.Citation34 The design of divergent primers and specific siRNA for circRNA was also provided by circInteractome. CIRCpedia is an integrative database, aiming to annotating alternative back-splicing and alternative splicing in circRNAs across different cell lines.Citation35
Table 1. Online circRNA databases and webtools.
The landscape of human circRNAs has been explored in different cells and tissues. However, it should be noted that there are always false positives regardless of the algorithm and materials used. The observable non-colinear junction sites may also arise from trans-splicing or genetic rearrangements or may simply be due to sequencing and alignment errors.Citation36 These weaknesses in circRNA analyses necessitate unbiased identification of circRNAs from different bioinformatics methods and pipelines and further removal of false positives arising from other non-colinear transcript, sequencing and alignment errors.Citation37 Several pipelines have been developed to specifically identify these non-colinear reads. A statistical and comprehensive analysis led by Hansen et al. evaluated the output from 5 different algorithms (circRNA_finder,Citation38 find_circ,Citation9 CIRCexplorer,Citation18 CIRI,Citation39 and MapSpliceCitation40) using common data sets generated from deep-sequencing of eukaryotic rRNA-depleted RNACitation37. In their overview of examined algorithms, CIRCexplorer and Mapsplice showed the best accuracy and good sensitivity with respect to circRNA detection. However, these 2 algorithms required gene annotation, and the processing speed is low. The other algorithms had a fast processing speed with low sensitivity and poor accuracy. For the maximum degree of unbiased analyses, it is crucial to take additional measures, such as overlapping the results of several algorithms.
Properties of human circRNAs
circRNAs have been identified that have several important biological properties associated strongly with their functions and implications. For instance, circRNAs comprise over 14% of the transcribed genes in fibroblasts,Citation8 and an estimate of more than 100,000 different circRNAs have been identified in human cells,Citation11 suggesting the abundance of circRNAs. They are predominant in the cytoplasm, but some were found to be enriched in the nucleus. Among the abundant circular transcripts, analysis revealed that there is always a predominant circular isoform from the transcription event in a given gene locus.Citation16 The length of most human exonic circRNAs was less than 1,500 nucleotides (nt), and the median length was 500 nt. Interestingly, circRNAs with high abundance were markedly longer than other circRNAs in terms of their flanking introns, suggesting long flanking introns might facilitate the formation of circRNAs.
Moreover, discoveries of circRNAs in different species have shown they are evolutionarily conserved, indicating selection for persistence of theses RNAs. circRNAs also present stability in both cells and serum exosomes in comparison to mRNA and resistance to RNase R activity, an exonuclease that degrades linear molecules. As circRNAs are prevalent in many human cells and tissues, many of them showed cell- or tissue-type specificity. Together, the abundance, conservation and tissue/cell type specificity indicate that a proportion of circRNAs might have biological functions or clinical implications.
The emerging roles of human circRNAs
Given the widespread and dynamic expression patterns of circRNAs, an increasing number of studies have focused on the potential role and function of circRNAs in human development and diseases.
Brain development and related disease
Several studies have demonstrated that circRNAs are strikingly enriched in the mammalian brain.Citation9,13,15 circRNAs are upregulated overall during brain differentiation,Citation13 are highly enriched in synapses,Citation15 and are often differentially expressed compared to their mRNA isoforms, indicating that some of these circRNAs may have functions in the brain. CDR1as circRNA, one of the best known circRNAs, functions as an miR-7 sponge in neuronal tissues.Citation9,41 The researchers assayed CDR1as and miR-7 in mouse brains with in situ hybridization, and their high co-expression pattern was revealed specifically in areas of the mesencephalon, also known as the developing midbrain. They further examined the biological effects of CDR1as by interaction with miR-7 in a Danio rerio (zebrafish) model. Of note, a loss-of-function phenotype with clear reduction in the zebrafish midbrain size can be induced by knockdown of mature miR-7 expression or injection of CDR1as transcripts, suggesting CDR1as can suppress normal miR-7 functions. Additionally, CDR1as is significantly reduced in Alzheimer patients, indicating that the CDR1as–miR-7 axis is deregulated and may have a function in Alzheimer disease.Citation21
Cardiovascular disease
Some studies have focused on the role of circRNAs in cardiovascular diseases. A circRNA from long non-coding RNA ANRIL (antisense non-coding RNA in the INK4 locus) was positively correlated with INK4/ARF expression and atherosclerosis risk.Citation22 Although this study suggested that circular ANRIL expression and structure may accidentally arise from by-products of mRNA splicing instead of a heavily regulated event related to very low copy numbers, this causal variant can still modulate ANRIL or even INK4/ARF expression, which correlates with atherosclerotic vascular disease risk. The previously mentioned CDR1as, which mediates the regulation of miR-7 on its target gene expression, has also shown evidence of promoting myocardial infarctions (MIs). A study using MI mice to examine CDR1as and miR-7 demonstrated that CDR1as overexpression in myocardial muscle cells promoted cell apoptosis and that this effect can be reversed by miR-7 overexpressionCitation24. However, further mechanism to illustrate the CDR1as/miR-7 pathway during MI-induced apoptotic process need to be performed.
Another study also investigated the promotion of cardiac cell death by circRNAs. Du et al. discovered that the role of Foxo3 circular RNA (circ-Foxo3) in cellular senescence was to retain the anti-senescent protein ID-1, the transcription factor E2F1, and the anti-stress proteins FAK and HIF1a in the cytoplasm, thus inhibiting their functions in anti-stress and anti-senescent processes.Citation25 Strikingly, in their study, in vivo delivery of circ-Foxo3 targeted siRNA into mouse embryonic fibroblasts and primary cardiac myocytes resulted in lower levels of circ-Foxo3 expression and β-gal staining, which is a standard marker for senescence, suggesting a potential therapeutic approach for myocardial protection. While circ-Foxo3 promotes cardiac cell death, a circular RNA (HRCR) was found to protect the heart from pathological hypertrophy and heart failure by sequestering and inhibiting miR-223 activity as an endogenous miRNA sponge.Citation23 In general, circRNAs in the cardiovascular system seem promising as therapeutic targets.
Cancer
Bachmayr-Heyda and colleagues were the first to report a global reduction of circRNA abundance in colorectal cancer cell lines and cancer tissues compared to normal tissues, and they discovered a negative correlation between global circular RNA abundance and proliferation.Citation42 Further study by RNA-seq analyses of normal and cancerous tissues identified a large number of circRNA candidates.Citation16 Among these candidates, many were differently expressed between normal and cancerous tissues, suggesting a potential role of circRNA in cancer. Importantly, a study carried out by Guarnerio et al. demonstrated the existence of circRNAs derived from transcribed exons of distinct genes affected by the translocations; namely, fusion circRNAs (f-circRNA).Citation43 More than one f-circRNA was discovered in cancer cells, including bone marrow-derived leukemic cells of acute promyelocytic leukemia (APL) patients, SK-NEP-1 sarcoma cell lines, and H3122 lung cancer cell lines. The researchers further explored the contributions of 2 f-circRNAs, f-circPR and f-circM9, to cellular transformation and tumor progression. The transduction of the f-circRNA vectors into mouse embryonic fibroblasts (MEFs) can increase the cell proliferation rate and the ability to form loci, whereas silencing of the fusion circRNAs can reverse the increased proliferation rate. In the presence of the fusion protein derived from the same genes, transducing f-circM9 into leukemic cells increased proliferation and colony formation compared to f-circM9 negative cells, suggesting that fusion circRNAs could contribute to leukemogenic processes when coupled with other oncogenic stimuli. Moreover, f-circRNA also contributed to resistance to chemotherapy in leukemic cells. On the other hand, the knockdown of f-circPR in NB4 cells triggered apoptosis, indicating a critical role for fusion circRNAs in maintaining the viability of leukemic cells. The role of these f-circRNAs in tumorigenesis and cancer progression suggests circRNA could be a new type of tumor oncogene (onco-circRNA). Although the mechanism of these f-circRNAs remains unknown, this work sheds new light on the role of circRNAs in cancer onset and progression, with potential diagnostic and therapeutic implications.
Additionally, circRNA has also been shown to be an inhibitory factor in tumor growth. It was shown that cir-ITCH expression is typically downregulated in esophageal squamous cell carcinoma.Citation44 This circular RNA could increase the level of ITCH by acting as miRNA sponge. In another study, cir-ITCH was shown to play an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway.Citation28 While circ-Foxo3 exhibits a senescence function in normal cells, it has been shown that the ectopic expression of circ-Foxo3 could suppress tumor growth and cancer cell proliferation, as it has a sponge effect on multiple miRNAs.Citation26 Similarly, the researchers delivered siRNA that specifically silenced circ-Foxo3, and it also decreased cell apoptosis, adding evidence to its potential implication as a therapeutic target.
Other studies
In addition, several studies focused on circRNAs in clinically important areas not discussed above. For example, circRNAs have been observed in human epithelial-mesenchymal transition (EMT), and the production of approximately one-third of these circRNAs was dynamically regulated by Quaking, an RNA binding protein.Citation20 Further experiments on Quaking binding motifs with circRNAs demonstrated the purposeful synthesis of circRNAs during EMT, suggesting that these circRNAs may affect EMT-related roles such as cell migration, invasion and metastasis. In addition, comprehensive circRNA profiling revealed that circular RNA100783 was involved in chronic CD28-associated CD8(+) T cell aging.Citation45 Another study discovered a circRNA that functioned as a miR-136 sponge related to human cartilage degradation.Citation46
The mechanism of circRNAs
Though the general mechanisms of circRNAs remain elusive, recent findings have indicated that circRNAs may interact with RNA or proteins and may serve as transcription or splicing regulators.
RNA interaction
CircRNAs have been found to interact with miRNA and function as miRNA sponges. The best example is CDR1as circRNA, which harbors more than 60 conserved binding sites for miR-7.Citation9,41 CDR1as is highly expressed, localized in the cytoplasm and has the capacity to bind to up to 20,000 miR-7 molcules per cell in brain tissue. Inhibition of CDR1as expression resulted in the reduced expression of miR-7 targeting mRNAs, suggesting that CDR1as competes with mRNAs for miR-7 binding. Other than CDR1as, only a few circRNAs in mammals are known to function as potential miRNA sponges. The testis-specific circRNA, sex-determining region Y (circSRY) contains 16 target sites for miR-138 in mice.Citation41 Notably, most recent publications have suggested that circRNAs do not necessarily function as miRNA sponges in human and mouse cells.Citation12,13,15,20 However, there might be a portion of circRNAs that are highly abundant and can bind to miRNAs. Moreover, one circRNA may be associated with a variety of miRNAs, as shown with circHIPK3 and circ-Foxo3, which can bind to multiple miRNAs.Citation16,26 Such an interaction would be more common for circRNAs that are associated with miRNAs. Thus, circRNA may, but not generally, function as sponge for miRNA. In addition to miRNA, circRNA can probably bind to mRNA or lncRNA to regulate their activities, although evidence for this has not been discovered yet.
Protein interaction
It has been shown that RNA-binding proteins (RBPs), such as Argonaute,Citation9,16,41 RNA polymerase II,Citation47 and MBL,Citation48 can bind to circRNAs. Certain circRNAs can probably regulate the function of RBPs by acting as competing elements, similar to the way they modulate miRNA activity.
An interesting example of the capacity of circRNA to interact with proteins is circ-Foxo3, which is generated from Foxo3, a member of the fork-head family of transcription factors. circ-Foxo3 was found to interact with the anti-senescence proteins ID1 and E2F1 and the anti-stress proteins FAK and HIF1α, causing them to be retained in the cytoplasm.Citation25 The effect of circ-Foxo3 on the subcellular translocation of these proteins blocked their anti-senescent function. Another study by the same research group showed that circ-Foxo3 could also bind to the cell cycle proteins cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21), resulting in the formation of a ternary complex.Citation49 The formation of this circ-Foxo3-p21-CDK2 ternary complex arrested the function of CDK2 and blocked cell cycle progression, adding support to circ-Foxo3 acting as a protein's scaffold to modulate certain cellular activities. In summary, circ-Foxo3 seems to be able to bind to many different proteins. However, how circ-Foxo3 directly interacts with these proteins without an RNA binding domain is not yet understood.
Transcription or splicing regulation
Although circRNAs are predominately located in the cytoplasm, a group of circRNAs with retained introns, named exon-intron circRNAs (EIciRNAs), have been shown to be localized in the nucleus and have been implicated in transcription regulation of their parental genes.Citation47 The EIciRNAs are enriched at the site of transcription and have been associated with RNA polymerase II through an interaction with U1 small nuclear ribonucleoprotein (snRNP), which can promote transcription of their host genes. These observations suggested that circRNA may function as a transcriptional regulator through a scaffold of protein in the nucleus. Another potential function of circRNAs could be to interrupt the splicing of an mRNA. Because circRNA often consists of exons that are also included in mRNA, both of which are derived from a pre-mRNA, the production of circRNA is expected to interrupt the splicing of mRNA. The canonical pre-mRNA splicing can compete with circularization of exons.Citation48 Mechanisms of this competition are tissue specific and conserved from flies to humans. In addition, the host gene can also regulate its circRNA biogenesis. The second exon of the splicing factor muscleblind (MBL/MBNL1) is circularized in flies and humans.Citation48 This circRNA (circMbl) and its flanking introns contain conserved muscleblind binding sites, which are strongly and specifically bound by MBL. Modulation of MBL levels strongly affects circMbl biosynthesis, and this effect is dependent on the MBL binding sites, indicating that muscleblind acts as a factor involved in circMbl biogenesis.Citation48
Translation
circRNAs are generally believed to function as noncoding RNAs because they have not been found to associate with ribosomes for translation.Citation8,12 However, circRNAs engineered with an internal ribosome entry site (IRES) can be efficiently translated in vitro and in vivo,Citation50,51 indicating that circRNAs have the potential to participate in translation. The possibility that some circRNAs are translatable is further exemplified by a recent systematic discovery that thousands of novel cap-independent translation sequences are found in the human genome.Citation52 Moreover, circular RNA has been reported to be translated in living human cells without any translational elements by a rolling circle amplification (RCA) mechanism.Citation53 Nevertheless, direct evidence of the translation of endogenous circRNAs has not been found yet.
Clinical implication of human circRNAs
A promising area of research focuses on using circRNA molecules as biomarkers due to their stability and specific expression both in cells and in extracellular fluid. circRNAs are more stable in cells than mRNA because of their non-polyadenylated structure and the absence of 5′ to 3′ polarity. Recent studies also showed that circRNAs are abundant in saliva, blood and even in exosomes.Citation16,29,30 Moreover, some potentially unique circRNAs (such as f-circRNAs) were found to be expressed only in pathological conditions.Citation27 For example, a circRNA named circ_101222 was found to be more significantly enriched in the blood corpuscles of patients with pre-eclampsia than in those of corresponding healthy controls.Citation54 Using the plasma protein factor endoglin (ENG) in combination with circ_101222 resulted in a sensitivity of 0.7073, a specificity of 0.8049, and overall area under the curve of 0.876 (95% confidence interval 0.816–0.922) for the prediction of pre-eclampsia.Citation54 Although the results need to be validated in a larger population, they have been used to create an early prediction model with low-cost strategies, such as microarrays and enzyme linked immunosorbent assays (ELISAs). A recent work demonstrated that serum exosomal circRNAs (exo-circRNAs) were able to distinguish patients with colorectal cancer (CRC) from healthy controls, and the abundance of tumor-derived exo-circRNA in the serum of xenograft mice was correlated with tumor mass.Citation31 Another study had also successfully explored the potential of circRNA in the predictions of gastric cancer.Citation55 In addition, circRNAs showed their suitability as biomarkers by having a more different expression pattern than mRNAs across patients with Epithelial Ovarian Carcinoma (EOC).Citation56 Several circRNA biomarkers have also been proposed, as hsa_circRNA_100855 and hsa_circRNA_104912 for laryngeal cancer (LC),Citation57 as hsa_circ_0001649 for hepatocellular carcinoma (HCC),Citation58 and as circRNA_001569 and hsa_circ_001988 for CRC.Citation59,60 Together, circRNAs in general seem to be potential biomarkers for diseases.
Beyond these features, circRNAs may be exploited for therapeutic applications. Because the expression of f-circRNAs in cancer cells is essential for their maintenance and confers resistance to chemotherapy, it has been suggested that pharmaceutical interventions that are aimed at blocking f-circRNAs or their down-stream effectors could prove to be beneficial as therapeutic targets to eradicate diseases.Citation27 Another study that employed in vivo delivery of siRNA-targeting endogenous circ-Foxo3 showed that it abrogated the effect of doxorubicin-induced cardiomyopathy, suggesting a potential therapeutic approach for myocardial protection.Citation25
Future considerations
Surveillance of the human transcriptome by RNA-seq and bioinformatic approaches has identified more than 10 thousand different circRNAs in human cells. Although the functions of the majority of circRNAs remain unknown, studies are beginning to elucidate the functional roles of a handful of circRNAs. The potential biogenesis and mechanisms have been suggested (). However, there are still many unanswered questions regarding their biogenesis, nuclear export and decay. The particular characteristics of circRNAs as long circularized RNA molecules that are typically expressed at low levels and largely overlap with linear mRNAs pose technical and conceptual challenges to the study of circRNA regulation and function. The mechanisms of circRNA are still far from fully understood. Unlike miRNA, circRNAs may have more than one mechanism of action due to their diverse lengths and structures. It is likely that circRNA may regulate gene expression or function at multiple levels, similar to lncRNAs, which have diverse mechanisms of action. In addition, it opens up a new range of possibilities for diagnostics and therapies for diseases involving circRNA. However, the field is still in its infancy, and we are still far being able to incorporate circRNA into clinical practice. It is will be essential to obtain a more thorough understanding of circRNA functions and mechanisms.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported in part by grants from the National Natural Science Foundation of China (81472617, 81672779) and Shanghai Pujiang Program (14PJ1401900).
References
- Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 1979; 280:339-40; PMID:460409; https://doi.org/10.1038/280339a0
- Lasda E, Parker R. Circular RNAs: diversity of form and function. Rna 2014; 20:1829-42; PMID:25404635; https://doi.org/10.1261/rna.047126.114
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993; 73:1019-30; PMID:7684656; https://doi.org/10.1016/0092-8674(93)90279-Y
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155-60; PMID:7678559; http://www.fasebj.org
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell 1991; 64:607-13; PMID:1991322; https://doi.org/10.1016/0092-8674(91)90244-S
- Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 1996; 93:6536-41; PMID:8692851; https://doi.org/10.1073/pnas.93.13.6536
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453-61; PMID:24811520; https://doi.org/10.1038/nbt.2890
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna 2013; 19:141-57; PMID:23249747; https://doi.org/10.1261/rna.035667.112
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; https://doi.org/10.1038/nature11928
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One 2012; 7:e30733; PMID:22319583; https://doi.org/10.1371/journal.pone.0030733
- Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. Rna 2014; 20:1666-70; PMID:25234927; https://doi.org/10.1261/rna.043687.113
- Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15:409; PMID:25070500; https://doi.org/10.1186/s13059-014-0409-z
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol Cell 2015; 58:870-85; PMID:25921068; https://doi.org/10.1016/j.molcel.2015.03.027
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9:e1003777; PMID:24039610; https://doi.org/10.1371/journal.pgen.1003777
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 2015; 18:603-10; PMID:25714049; https://doi.org/10.1038/nn.3975
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016; 7:11215; PMID:27050392; https://doi.org/10.1038/ncomms11215
- Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep 2015; 10:103-11; PMID:25543144; https://doi.org/10.1016/j.celrep.2014.12.002
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell 2014; 159:134-47; PMID:25242744; https://doi.org/10.1016/j.cell.2014.09.001
- Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28:2233-47; PMID:25281217; https://doi.org/10.1101/gad.251926.114
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015; 160:1125-34; PMID:25768908; https://doi.org/10.1016/j.cell.2015.02.014
- Lukiw WJ. Circular RNA (circRNA) in Alzheimer disease (AD). Front Genet 2013; 4:307; PMID:24427167; https://doi.org/10.3389/fgene.2013.00307
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010; 6:e1001233; PMID:21151960; https://doi.org/10.1371/journal.pgen.1001233
- Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016; 37:2602-2611; PMID:26802132; https://doi.org/10.1093/eurheartj/ehv713
- Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP. The Circular RNA Cdr1as Promotes Myocardial Infarction by Mediating the Regulation of miR-7a on Its Target Genes Expression. PloS One 2016; 11:e0151753; PMID:26998750; https://doi.org/10.1371/journal.pone.0151753
- Du WW, Yang W, Chen Y, Wu ZK, Foster FS, Yang Z, Li X, Yang BB. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 2016; 2:ehw001; PMID:26873092; https://doi.org/10.1093/eurheartj/ehw001
- Yang W, Du WW, Li X, Yee AJ, Yang BB. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 2015; 35:3919-31; PMID: 26657152; https://doi.org/10.1038/onc.2015.460
- Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016; 165:289-302; PMID:27040497; https://doi.org/10.1016/j.cell.2016.03.020
- Huang G, Zhu H, Shi Y, Wu W, Cai H, Chen X. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/β-catenin pathway. PloS One 2015; 10:e0131225; PMID:26110611; https://doi.org/10.1371/journal.pone.0131225
- Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and Characterization of Circular RNAs As a New Class of Putative Biomarkers in Human Blood. PloS One 2015; 10:e0141214; PMID:26485708; https://doi.org/10.1371/journal.pone.0141214
- Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DT, Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 2015; 61:221-30; PMID:25376581; https://doi.org/10.1373/clinchem.2014.230433
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015; 25:981-4; PMID:26138677; https://doi.org/10.1038/cr.2015.82
- Ghosal S, Das S, Sen R, Basak P, Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front Genet 2013; 4:283; PMID:24339831; https://doi.org/10.3389/fgene.2013.00283
- Liu YC, Li JR, Sun CH, Andrews E, Chao RF, Lin FM, Weng SL, Hsu SD, Huang CC, Cheng C, et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res 2016; 44:D209-15; PMID:26450965; https://doi.org/10.1093/nar/gkv940
- Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol 2016; 13:34-42; PMID:26669964; https://doi.org/10.1080/15476286.2015.1128065
- Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 2016; 26:1277-1287; PMID: 27365365; https://doi.org/10.1101/gr.202895.115
- Yu CY, Liu HJ, Hung LY, Kuo HC, Chuang TJ. Is an observed non-co-linear RNA product spliced in trans, in cis or just in vitro? Nucleic Acids Res 2014; 42:9410-23; PMID:25053845; https://doi.org/10.1093/nar/gku643
- Hansen TB, Veno MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res 2016; 44:e58; PMID:26657634; https://doi.org/10.1093/nar/gkv1458
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014; 9:1966-80; PMID:25544350; https://doi.org/10.1016/j.celrep.2014.10.062
- Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015; 16:4; PMID:25583365; https://doi.org/10.1186/s13059-014-0571-3
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 2010; 38:e178; PMID:20802226; https://doi.org/10.1093/nar/gkq622
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; https://doi.org/10.1038/nature11993
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Scientific Rep 2015; 5:8057; PMID:25624062; https://doi.org/10.1038/srep08057
- Oncogenic Circular RNAs Arise from Chromosomal Translocations. Cancer Discovery 2016; 6:OF20; PMID: 27080338; https://doi.org/10.1158/2159-8290.CD-RW2016-068
- Li F, Zhang L, Li W, Deng J, Zheng J, An M, Lu J, Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015; 6:6001-13; PMID:25749389; https://doi.org/10.18632/oncotarget.3469
- Wang YH, Yu XH, Luo SS, Han H. Comprehensive circular RNA profiling reveals that circular RNA100783 is involved in chronic CD28-associated CD8(+)T cell ageing. Immun Ageing 2015; 12:17; PMID:26451160; https://doi.org/10.1186/s12979-015-0042-z
- Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, Ao Y. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 ‘Sponge’ in Human Cartilage Degradation. Scientific Rep 2016; 6:22572; PMID:26931159; https://doi.org/10.1038/srep22572
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Bio 2015; 22:256-64; PMID:25664725; https://doi.org/10.1038/nsmb.2959
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Molecular Cell 2014; 56:55-66; PMID:25242144; https://doi.org/10.1016/j.molcel.2014.08.019
- Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 2016; 44:2846-58; PMID:26861625; https://doi.org/10.1093/nar/gkw027
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. Rna 2015; 21:172-9; PMID:25449546; https://doi.org/10.1261/rna.048272.114
- Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995; 268:415-7; PMID:7536344; https://doi.org/10.1126/science.7536344
- Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science 2016; 351:aad4939; PMID:26816383; https://doi.org/10.1126/science.aad4939
- Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Scientific Rep 2015; 5:16435; PMID:26553571; https://doi.org/10.1038/srep16435
- Zhang YG, Yang HL, Long Y, Li WL. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG 2016; PMID:26846540; https://doi.org/10.1111/1471-0528.13897
- Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015; 444:132-6; PMID:25689795; https://doi.org/10.1016/j.cca.2015.02.018
- Ahmed I, Karedath T, Andrews SS, Al-Azwani IK, Ali Mohamoud Y, Querleu D, Rafii A, Malek JA. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget 2016; 7:36366-36381; PMID: 27119352; https://doi.org/10.18632/oncotarget.8917
- Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T, Wang X, Li Q, Tian L, Liu M, et al. Circular RNA: a novel biomarker for progressive laryngeal cancer. Am J Transl Res 2016; 8:932-9; PMID:27158380; http://www.ajtr.org/contents.html
- Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark 2016; 16:161-9; PMID:26600397; https://doi.org/10.3233/CBM-150552
- Wang X, Zhang Y, Huang L, Zhang J, Pan F, Li B, Yan Y, Jia B, Liu H, Li S, et al. Decreased expression of hsa_circ_001988 in colorectal cancer and its clinical significances. Int J Clin Exp Pathol 2015; 8:16020-5; PMID:26884878
- Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget 2016; 7:26680-91; PMID: 27058418; https://doi.org/10.18632/oncotarget.8589
