ABSTRACT
Pre-mRNAs from thousands of eukaryotic genes can be non-canonically spliced to generate circular RNAs, some of which accumulate to higher levels than their associated linear mRNA. Recent work has revealed widespread mechanisms that dictate whether the spliceosome generates a linear or circular RNA. For most genes, circular RNA biogenesis via backsplicing is far less efficient than canonical splicing, but circular RNAs can accumulate due to their long half-lives. Backsplicing is often initiated when complementary sequences from different introns base pair and bring the intervening splice sites close together. This process is further regulated by the combinatorial action of RNA binding proteins, which allow circular RNAs to be expressed in unique patterns. Some genes do not require complementary sequences to generate RNA circles and instead take advantage of exon skipping events. It is still unclear what most mature circular RNAs do, but future investigations into their functions will be facilitated by recently described methods to modulate circular RNA levels.
As originally described in Crick's central dogma of molecular biology,Citation1 the information encoded in protein-coding genes is sequentially transferred from DNA to RNA to protein, with proteins performing most of the structural and functional roles in cells. This protein-centric view has been tempered by the realization that eukaryotic genomes are extensively transcribed to yield a plethora of noncoding RNAs (For a review see refs.Citation2-5). In particular, thousands of circular RNAs with little or no protein-coding capacity are generated from genes in a variety of eukaryotes, including humans, mice, C. elegans, D. melanogaster, S. pombe, and plants (For a review see refs.Citation6-12). Some of these circular RNAs are expressed at much higher levels than their associated linear mRNAs,Citation13,14 suggesting that the main function of some protein-coding genes may be to produce circular noncoding RNAs rather than proteins.
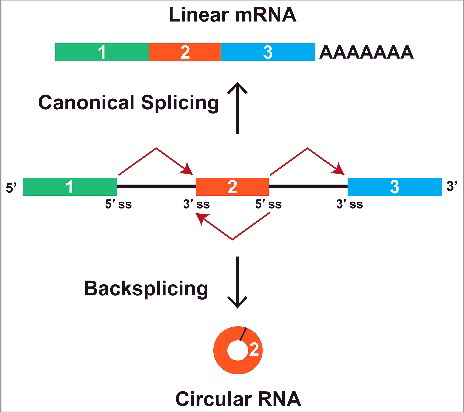
Most genes in eukaryotes are disrupted by intronic sequences, and their precursor mRNAs (pre-mRNAs) must be spliced so that introns are removed and exons joined together (For a review see Citation15). When a pre-mRNA is spliced in the canonical manner, exons are sequentially joined 5′ to 3′ (e.g. exon 1 is joined to exon 2, which is joined to exon 3, etc.) to produce a linear mRNA that can then be translated (). Nearly all genes are subjected to alternative splicing events, such as exon skipping (e.g., exon 1 is joined to exon 3), but the functional output of these events has always been thought to be a linear mRNA.Citation16,17 In stark contrast, a circular RNA with covalently linked ends is generated when a pre-mRNA undergoes “backsplicing” to join a splice donor to an upstream splice acceptor (e.g. the end of exon 2 is joined to the beginning of exon 2) ().
Figure 1. A pre-mRNA can be spliced to generate a linear or circular RNA. When the pre-mRNA splice sites (ss) are joined in the canonical order, a linear mRNA is generated that is subsequently polyadenylated and exported to the cytoplasm for translation (top). Alternatively, backsplicing can join a 5′ ss to an upstream 3′ ss to generate a circular RNA whose ends are covalently linked by a 3′-5′ phosphodiester bond (bottom). This competition between canonical splicing and backsplicing helps determine which mature RNAs are generated from a gene.
As all internal exons (excluding the first and last exon) of a gene have splicing signals at their 5′ and 3′ ends, they all theoretically can circularize. Nevertheless, only a small subset of possible backsplicing events actually occurs in cells. This is, in part, because the splicing machinery acts in a co-transcriptional manner to quickly remove most introns as soon as they have been fully transcribed by RNA polymerase II.Citation18-20 By the time a given splice donor (5′ splice site) is transcribed, all of the upstream splice acceptors (3′ splice sites) in that pre-mRNA have usually been removed, making backsplicing impossible. However, there are an increasing number of introns that are known to be slowly or post-transcriptionally spliced.Citation21-24 At these genes, there is the opportunity for direct competition between canonical splicing and backsplicing.Citation25 Depending on how a pre-mRNA is ultimately spliced, a variety of linear or circular RNAs can be produced, each potentially with a unique function. Circular RNAs are naturally resistant to degradation by exonucleases and thus accumulate as stable transcripts. In at least 2 cases (CDR1as/ciRS-7 and Sry), circular RNAs function to sponge specific microRNAs,Citation26-28 although most circular RNAs (outside of DrosophilaCitation29) contain few microRNA binding sitesCitation30,31 and likely have a different function. For example, circular RNAs may allow the formation of large RNA-protein complexes, e.g., at neuronal synapses,Citation32,33 or possibly be translated.Citation34-37
This review highlights recent advances that help explain how the choice between linear vs. circular RNA production is made. Many circular RNAs are expressed in a tissue-specific manner and at low levels,Citation29,31-33,38 likely because backsplicing is far less efficient than canonical splicing.Citation39 In most cases, production of a circular RNA is facilitated by intronic complementary sequences (such as repetitive elements), which base pair to bring the intervening splice sites into close proximity. Repetitive elements are commonplace in introns, and various linear or circular RNAs can be generated depending on which repeats base pair to one another.Citation40 RNA binding proteins further regulate circular RNA biogenesis in a combinatorial manner, allowing tight control over which mature transcripts are generated.Citation37 At certain genes, circular RNAs are produced independently of repetitive elements and the backsplicing reaction is instead coupled to an exon skipping event.Citation41 In addition, a separate class of circular intronic RNAs are made not by backsplicing but by a failure to debranch intron lariats.Citation42 It is thus becoming increasingly clear that circular RNAs can be generated via multiple, tightly regulated strategies. Characterization of these biogenesis mechanisms has allowed the recent development of methods to efficiently produce circular RNAs in vivo, which will help reveal how this large class of transcripts fits into the regulatory landscape of the cell.
Circular RNAs are generated from many eukaryotic genes
Viroids, which are plant pathogens, were the first circular RNAs to be identified in 1976.Citation43 Sänger and colleagues surprisingly found that viroid RNA was unable to be labeled by polynucleotide kinase or degraded by snake venom phosphodiesterase, suggesting that it lacked 5′ or 3′ termini and was instead a covalently closed molecule.Citation43 Subsequent work showed that hepatitis δ virus, a satellite virus of hepatitis B virus that causes severe liver disease in humans, likewise has a circular RNA genome.Citation44 It was not until the 1990s that the first circular RNAs generated from eukaryotic genes were serendipitously identified. While characterizing the gene structures of human DCC,Citation45 human ETS-1,Citation46,47 and mouse Sry,Citation48 several circular RNAs were found that contained exons joined precisely at consensus splice sites, but in an order different from that encoded in the pre-mRNA. This suggested that the pre-mRNA splicing machinery was involved in their biogenesis. Nevertheless, these circular RNAs were largely interpreted as random errors since the DCC and ETS-1 circles were expressed at much lower levels (∼0.01%) than their associated linear mRNAs.Citation45-47 The Sry circular RNA was also undetectable or expressed at low levels in most tissues, but it remarkably represented more than 90% of Sry transcripts in mouse testes. This was the first hint that the dominant output of a gene could, in some cases, be a circular RNA rather than a linear mRNA.Citation48
A handful of additional circular RNAs were identified over the ensuing yearsCitation49-56 until deep sequencing efforts combined with new computational algorithms all of a sudden revealed thousands of previously missed circular RNAs from eukaryotic cells.Citation13,14,27,29,31,33,40,57,58 Over 25,000 putative circular RNAs, derived from ∼15% of actively transcribed genes, were identified in human fibroblasts alone.Citation14 Like the DCC, ETS-1, and Sry transcripts, the vast majority of these circular RNAs are generated in a tissue-specific manner using canonical splice sites, consist almost exclusively of exonic sequences, lack poly(A) tails, and localize in the cytoplasm.Citation13,14,27,31,38 Recent work has revealed that alternative splicing events commonly occur within circular RNAs, which further increases the complexity of circular transcripts present in cells.Citation59,60 For example, a subset of circular RNAs known as EIciRNAs (exon-intron circular RNAs) retain an intron between exonic sequences, and this particular class of circular transcripts are retained in the nucleus.Citation29,61 Most circular RNAs are present at low levels,Citation31 and have been estimated to be, on average, <3% of the abundance of the canonical linear transcript from the same gene.Citation30 Nevertheless, there are hundreds of circular RNAs that are expressed at levels >10-fold higher than their corresponding linear transcript,Citation14 especially in the brain.Citation25,29,32,33,62
Circular RNAs are stable, but their biogenesis is slow
Given that some circular RNAs are the predominant outputs of their host genes, an obvious question is whether this is due to the preferential synthesis of circular RNAs and/or differences in linear vs. circular RNA half-lives. Due to their natural resistance to exonucleases, circular RNAs appear to be very stable transcripts with half-lives greater than 24–48 hr.Citation14,30 This is significantly longer than the half-life of an average mRNA (8-9 hr in human cells).Citation63,64 Nevertheless, the efficiency of backsplicing is far less than canonical splicing,Citation39 which makes many circular RNAs infrequently generated and unable to be rapidly induced, e.g. in response to stimulation by growth factors.Citation30 Backsplicing, like many alternative splicing events,Citation23 appears to largely occur post-transcriptionally after RNA polymerase II has reached its termination site.Citation39 Consistent with these observations, removal of the downstream polyadenylation signal eliminates circular RNA production from some expression plasmids.Citation65 A subset of circular RNAs are likely made co-transcriptionally as they can be found in chromatin fractions and their biogenesis is affected by transcription elongation rates.Citation25,39 The molecular details are still unclear, but it appears that the timing of backsplicing is governed by how quickly the splicing machinery can be assembled. When exons are flanked by long complementary repeats that efficiently bring the splice sites into close proximity (see below), backsplicing can occur more rapidly and pre-mRNA 3′ end processing is not required.Citation37 How then does the pre-mRNA splicing machinery “know” whether a pre-mRNA should be spliced to generate a linear mRNA or any of its possible circular RNAs?
Base pairing between intronic repeat elements facilitates circular RNA biogenesis
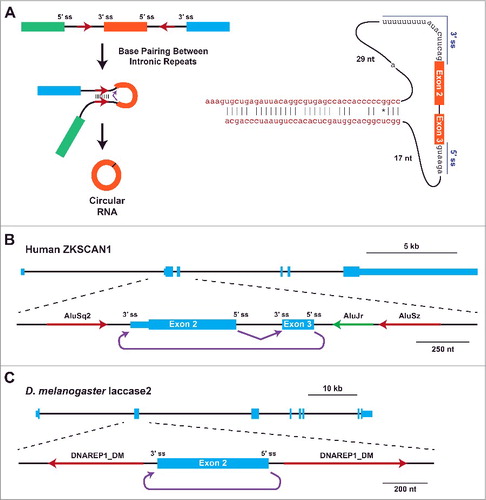
Upon searching for sequence motifs that distinguish regions that generate circular RNAs from those that do not, Jeck et al. noticed that pairs of Alu repeat elements (which are each ∼300-nt in length) are statistically enriched in the introns that flank human circular RNAs.Citation14 In particular, the pairs of Alu elements were more likely to be in an inverted, complementary orientation (). This observation was reminiscent of early studies on the mouse Sry circular RNA, which has ∼50-kb of near perfectly complementary sequences (>99.7% identity) in the flanking introns.Citation48 As similar repeat structures are not present at the human Sry locus, which does not generate a circular RNA, a connection between intronic repeats and circular RNA biogenesis was proposed.Citation48 Subsequent experiments revealed that complete removal of either repeat from mouse Sry expression plasmids eliminated circular RNA production, whereas the inclusion of ∼400-nt of the flanking repeats was sufficient.Citation66 This suggested a model in which Sry backsplicing is trigged when the flanking intronic sequences base pair and bring the intervening splice sites close together,Citation48,66 which was supported using proof-of-principle in vitro splicing substrates ().Citation67
Figure 2. Base pairing between intronic complementary sequences facilitates circular RNA biogenesis. (A) Backsplicing is commonly induced when inverted repeats (red arrows) in the flanking introns base pair to one another. This brings the intervening splice sites (ss) into close proximity, facilitating catalysis (left). Extensive mutagenesis of the human ZKSCAN1 locus revealed minimal introns that are sufficient for the generation of a circular RNA from exons 2 and 3.Citation65 Besides the splice sites, ∼40-nt complementary repeats (red) are needed for backsplicing. (B) Exon/intron structure of the human ZKSCAN1 locus, highlighting the region that contains exons 2 and 3. Complementary AluS elements (red) immediately flank these exons, and facilitate backsplicing from the end of exon 3 to the beginning of exon 2 (purple).Citation65 (C) Exon/intron structure of the D. melanogaster Laccase2 locus, highlighting a region that contains exon 2. A pair of DNAREP1_DM family transposons are close to the circularizing exon and facilitate backsplicing.Citation37
Although very few exons are flanked by repeats as long as those at the mouse Sry locus, most (>70 %)Citation39 exons that generate circular RNAs in human cells have complementary repeats, usually Alu elements, in their flanking introns.Citation14,40,68 Circular RNAs in other species, including mice, C. elegans,Citation68 and Drosophila,Citation37 are often also flanked by complementary intronic repeats (but not in all casesCitation25,29; see below). As the sequences of these repeats are generally quite different from human Alu elements, backsplicing does not appear to be dependent on the presence of particular sequence motifs (beyond the splice sites). For example, non-repetitive complementary sequences flank the exons that generate the abundant human GCN1L1Citation40 and SMARCA5 circular RNAs.
To directly test whether intronic repeats regulate circular RNA biogenesis, we and others generated and mutated plasmids that express various circular RNAsCitation26,37,40,65,69 or removed repeats from endogenous gene loci using CRISPR-Cas9 genome editing.Citation39,70 In particular, our group has extensively characterized the human ZKSCAN1 locus, which produces a 668-nt circular RNA comprised of exons 2 and 3 (), and the Drosophila Laccase2 locus, which produces a 490-nt circular RNA comprised of exon 2 ().Citation37,65 Like most loci that generate circular RNAs, the ZKSCAN1 and Laccase2 circularizing exons are flanked by longer than average introns.Citation13,14,29,40,62 Nevertheless, long flanking introns are not required for ZKSCAN1 or Laccase2 backsplicing as miniature introns containing only the splice sites and flanking inverted repeats are sufficient for robust circular RNA expression from plasmids ().Citation37,65 Mutating the splice sites completely eliminates circular RNA production,Citation25,35,65,69 as does treatment with an inhibitor of spliceosome assembly.Citation69
Flanking intronic repeats as short as 30–40 nt were surprisingly found to be sufficient for exon circularization ().Citation65 Disrupting the base pairing between the repeats eliminates ZKSCAN1 and Laccase2 backsplicing, while the introduction of compensatory mutations in the repeats is sufficient to rescue circular RNA production.Citation37,65 It is important to note that not all short repeat sequences support circularization, likely due to thermodynamics and/or recognition by RNA binding proteins.Citation65 Nevertheless, (CA)n simple repeats that are complementary over a <30-nt region appear to be sufficient to drive Drosophila Semaphorin-2b circular RNA biogenesis.Citation37 These data, as well as genome-wide analysesCitation14,40,62,68 and detailed studies on the human EPHB4,Citation65 GCN1L1,Citation40,65 HIPK3,Citation65 LPAR1,Citation69 and POLR2ACitation40 circular RNAs, confirm the original Sry circular RNA biogenesis modelCitation48 () and suggest that it is applicable to thousands of eukaryotic genes. Indeed, one can accurately predict many circular RNAs, especially from highly transcribed genes, by searching for pairs of complementary sequences in the flanking introns.Citation68
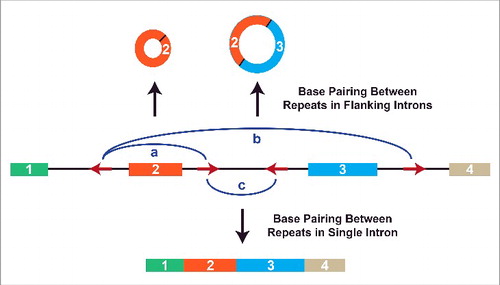
However, the presence of inverted intronic repeats is not sufficient for exon circularization. This is, in part, because of the co-transcriptional nature of splicing, but also because introns commonly contain many repetitive elements that compete for base pairing () (For a review see ref.Citation8). Approximately 17% and 10% of the human genome is derived from LINE-1 (L1) retrotransposons and Alu elements, respectively,Citation71,72 and repetitive sequences can affect a transcript's fate in multiple ways (For a review see ref.Citation73,74). Within a given pre-mRNA, the number of repeats, the distance between them, and their degree of complementarity all affect which repeats base pair to one another, which in turn modulates pre-mRNA splicing patterns.Citation40 If base pairing occurs between repeats in separate introns, backsplicing is induced (as discussed above). Different circular RNAs can even be produced from a single gene depending on which repeats in separate introns base pair to one another (). In contrast, canonical splicing to yield a linear mRNA occurs if repeats within a single intron base pair to one another ().Citation40 The efficiency of backsplicing is further affected by exon length, with longer exons (>300-nt) able to form circular RNAs better than short exons.Citation37,41,65
Figure 3. Alternative circularization is driven by competition for base pairing between intronic repeats. Multiple complementary repeat elements (red arrows) are often present in a pre-mRNA. Depending on which repeats base pair to one another (denoted by blue arcs), different pre-mRNA splicing patterns are triggered.Citation40 (a) If the repeats flanking exon 2 base pair to one another, backsplicing is induced to generate a circular RNA comprised of exon 2. (b) A larger circular RNA can be generated if the repeats flanking exons 2 and 3 base pair. This allows backsplicing from the end of exon 3 to the beginning of exon 2. Canonical splicing removes the intron between exons 2 and 3 to yield the mature circular RNA. (c) Alternatively, base pairing between repeats in a single intron leads to canonical splicing and the generation of a linear mRNA.
The genomic repeat landscape varies significantly across species,Citation75 which causes different populations of circular RNAs to be expressed. As pointed out above, the Sry circular RNA is expressed in mouse but not humans.Citation48 Likewise, the Laccase2 circular RNA is expressed in D. melanogaster but not other Drosophilids, such as D. yakuba, as the flanking repeats are not evolutionarily conserved.Citation37 Interestingly, humans use the equivalent splice sites to express homologs of ∼20% of mouse circular RNAs,Citation14,31,33 and the expression of some mammalian circular RNAs is conserved back to flies.Citation25,33 This suggests the existence of conserved routes of circular RNA production (e.g., using repeat sequences that were present in the last common ancestor), but the mechanistic details remain to be fleshed out. It is also possible that convergent evolution has selected for the production of these particular circular RNAs. In total, current data indicate that the RNA output of a protein-coding gene can be very different depending on the organism, even if the open reading frame is conserved. There is nevertheless a population of circular RNAs that are evolutionarily conserved and likely to be functional.
Translocations shuffle intronic repeats to cause aberrant circular RNA expression in cancer cells
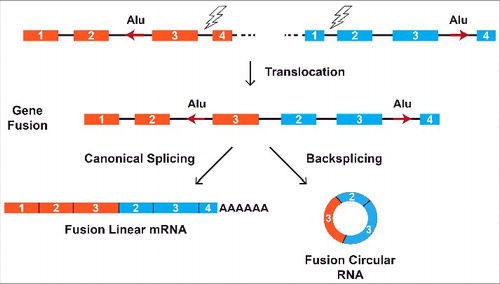
Chromosomal translocations are commonly observed in multiple cancers, especially hematological malignancies as well as a growing number of solid tumors.Citation76,77 These translocations rearrange the genome and join regions that are normally separated, resulting in fusion genes that can deregulate cell growth. When these fusion genes are transcribed, intronic sequences that flank the translocation breakpoint are juxtaposed, which can lead to the formation of aberrant circular RNAs if complementary repeat sequences base pair to one another (). Recent work by Guarnerio and colleagues revealed that the PML/RARα translocation in acute promyelocytic leukemia (APL), MLL/AF9 translocation in acute myeloid leukemia (AML), EWSR1/FL1 translocation in Ewing sarcoma, and EML4/ALK1 translocation in lung cancer all generate aberrant circular RNAs via this mechanism.Citation78 These fusion circular RNAs, which are not expressed in normal cells, promote cancer development in part by contributing to cellular transformation and protecting cancer cells from drug-induced apoptosis. Circular RNAs may thus represent promising new therapeutic targets. Chromosomal translocations can also theoretically cause the loss of a normal circular RNA if the rearrangement causes a pair of flanking intronic repeats to become uncoupled.
Figure 4. Chromosomal translocations can lead to aberrant circular RNA expression. Rearrangements between nonhomologous regions can result in the joining of 2 previously separate genes (colored orange and blue). Regulatory sequences, such as intronic Alu repeats (red arrows), that flank the translocation breakpoint become juxtaposed when the fusion gene is transcribed. This can lead to the formation of aberrant circular RNAs when complementary repeat sequences base pair to one another. Recent work has demonstrated that these fusion circular RNAs then act with the fusion mRNA/protein to drive cancer development.Citation78.
Combinatorial control of circular RNA expression by RNA binding proteins
Although intronic repeat sequences are critical regulators of circular RNA biogenesis, it is clear that more than these cis-acting sequences are at play. Many circular RNAs are expressed in tissue-specific patterns or are induced/repressed as cells change their state.Citation29,32,33,38,62,70,79-84 For example, hundreds of circular RNAs are regulated during epithelial-mesenchymal transition (EMT), with the majority increasing in abundance.Citation82 The expression of the corresponding linear mRNA for many of these circular RNAs does not change, suggesting that the differences in RNA circles are not due to changes in overall gene transcription levels. Instead, the RNA binding protein Quaking (QKI) is induced during EMT and acts to promote circular RNA biogenesis from many pre-mRNAs.Citation82 Because QKI can form dimers, it was proposed that QKI binds to flanking introns and brings the intervening splice sites close together (analogous to how inverted repeats in flanking introns promote backsplicing) (). Consistent with this model, insertion of QKI binding sites into a pair of flanking introns was sufficient to induce circular RNA production from an exon that does not normally circularize.Citation82 This indicates that QKI binding alone can be sufficient to promote circular RNA biogenesis. It should, however, be noted that many QKI-regulated circular RNAs (e.g. SMARCA5) are also flanked by complementary intronic sequences, suggesting the regulation is more complicated than currently appreciated.
The Drosophila Muscleblind (Mbl) RNA binding protein appears to regulate the production of circular RNAs from its own pre-mRNA in a manner similar to QKI.Citation25 When Mbl protein levels are in excess, it binds to both introns flanking exon 2 of the Mbl pre-mRNA, triggering the production of an exon 2 circular RNA. Mbl protein is critical for Mbl backsplicing as the circular RNA is not observed when Mbl protein is depleted from cells using RNAi.Citation25,37 Auto-regulation of many splicing factor genes has been previously observed, although the generation of a circular RNA is a twist on the commonly used mechanism: most splicing factors bind their own pre-mRNAs to cause an alternative splicing event that leads to nonsense-mediated decay (NMD).Citation85 The Mbl locus is thus regulated by a bit different strategy, but the outcome is the same as it ensures Mbl protein expression is maintained within a tight range. Interestingly, the Mbl circular RNA is also able to bind Mbl protein, which likely further limits the ability of excess Mbl protein to regulate other RNAs in trans.Citation25
ADAR (adenosine deaminase acting on RNA) enzymes, which can unwind double-stranded regions by converting adenosines to inosines,Citation73 inhibit the expression of over 80 circular RNAs in human HEK293 cells.Citation33,68 This is likely because ADAR activity disrupts base pairing between the intronic repeats, thereby directly preventing circular RNA production.
The focus so far has been on how individual RNA binding proteins can regulate circular RNA levels, but there is emerging evidence that backsplicing is regulated in a combinatorial manner by multiple factors acting at once.Citation37 The Drosophila Laccase2 and PlexA circular RNAs are flanked by inverted repeats, but their expression levels are also controlled by multiple hnRNP (heterogeneous nuclear ribonucleoprotein) and SR (serine-arginine) proteins. Depletion of any of these well-characterized splicing factors by RNAi is sufficient to alter endogenous circular RNA levels, whereas simultaneous depletion of factors results in additive effects on expression.Citation37 This indicates that each factor plays a non-redundant role and that backsplicing at the Laccase2 and PlexA loci is regulated by the combined activities of intronic repeats and splicing regulatory proteins. A model has been proposed in which base pairing between the intronic repeats promotes circularization, but protein binding helps ensure that the appropriate amounts of linear and circular RNAs are produced.Citation37 As exons (along with their flanking introns) contain unique sets of hnRNP and SR protein binding sites, each of which aids or blocks spliceosome assembly, distinct expression patterns of circular RNAs are observed. Future studies will hopefully reveal details of how this “backsplicing code” fits in with the canonical splicing codeCitation86 to dictate splicing outcomes.
Circular RNAs generated via exon skipping
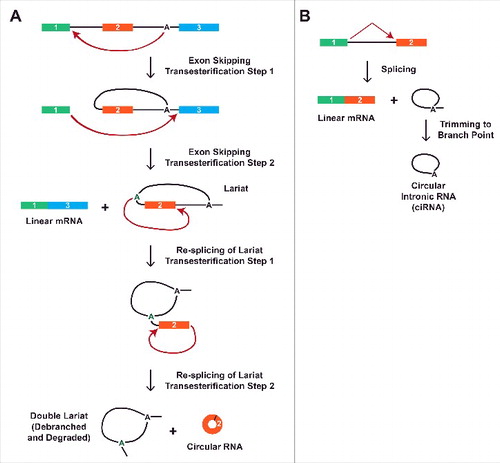
Repetitive elements are widespread in mammalian genomes, but are much less common in simple eukaryotes, such as Schizosaccharomyces pombe, that generate circular RNAs.Citation87 It has additionally been proposed that many Drosophila circular RNAs are generated independently of repeats.Citation29 This suggests the existence of alternative mechanisms for circular RNA biogenesis. Indeed, the S. pombe mrps16 pre-mRNA lacks repetitive elements, and instead takes advantage of exon skipping to generate a circular RNA.Citation41 By splicing exon 1 to exon 3, an intron lariat containing exon 2 is released, which is subsequently re-spliced to covalently join the beginning and end of exon 2 together (). The mature circular RNA consisting of mrps16 exon 2 then accumulates in cells, while the double lariat from the re-splicing event and the skipped linear mRNA are both rapidly degraded.Citation41,88
Figure 5. Generation of circular RNAs via exon skipping or a failure to debranch introns. (A) Circular RNA biogenesis can proceed through an exon-containing lariat. Via an alternative splicing event, exon 2 can first be skipped to generate a linear mRNA consisting of exons 1 and 3 as well as an intron lariat intermediate. The lariat can then be re-spliced to generate a circular RNA comprised of exon 2 along with a double lariat, which is subsequently debranched and degraded. (B) Although most intron lariats are rapidly debranched, some are only trimmed to their branch point. This generates a circular intronic RNA that is covalently joined by a 2′-5′ phosphodiester bond between the 5′ end of the intron and the branch point adenosine.
Interestingly, there are other examples of exon skipping events in S. pombe,Citation89 yet none of these genes appear to generate circular RNAs.Citation41 This argues that the production of an exon-containing lariat is not sufficient for circular RNA biogenesis, and the mechanisms by which cells determine when to re-splice to yield a circular RNA remain unclear. The length of the skipped exon appears to be one key determinant,Citation41 but other factors such as RNA secondary structures and the speed of lariat debranching are also likely to be involved. Interestingly, a correlation between exon skipping and circular RNA biogenesis has been noted at other genesCitation49,50,52,55 and in a global transcriptome analysis of human endothelial cells.Citation90 Detailed analyses are now required to determine if re-splicing truly occurs at these loci. If it does, this could represent an important way that a single pre-mRNA is able to generate both a linear mRNA and a circular RNA.
Circular RNAs in the nucleus: Exon-intron circular RNAs and circular intronic RNAs
All of the circular RNAs discussed so far have been derived from exons and localized to the cytoplasm, but 2 classes of circular transcripts are known to localize to the nucleus and regulate the expression of their parental genes. The first class are exon-intron circular RNAs (EIciRNAs) that are circularized via backsplicing, but are not fully spliced and retain an internal intron in the mature transcript.Citation61 EIciRNAs co-localize with the promoter of their parental gene and use the 5′ splice site from their retained intron to directly bind U1 snRNP (small nuclear ribonucleoprotein) to promote transcription in cis.
A second class of nuclear circular RNAs are generated from intron lariats. The vast majority of intron lariats are rapidly debranched and degraded, but some are stable in Xenopus oocytesCitation91 and at least 100 introns fail to be debranched in humans.Citation42 These accumulate as circular intronic RNAs (ciRNAs) due to the 2′-5′ phosphodiester bond at the branch point, and can reach levels similar to that of their parental linear mRNA ().Citation42 It is still unclear why these introns escape debranching, but mutational analysis revealed a 7-nt GU-rich element near the 5′ splice site and an 11-nt C-rich element close to the branchpoint that are both critical for ciRNA biogenesis. Once generated, ciRNAs accumulate in the nucleus near their sites of transcription, where they interact with the elongating RNA polymerase II complex to promote the expression of their parental gene. Both EIciRNAs and ciRNAs additionally localize to other sites in the nucleus, suggesting they may regulate other loci in trans.Citation42
Methods for ectopic expression of circular RNAs in cells
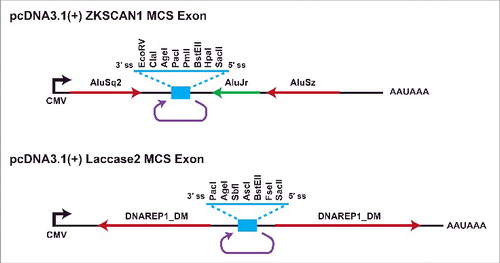
Multiple groups have now translated knowledge about circular RNA biogenesis mechanisms into methods for expressing circular RNAs from intronsCitation42 or exons in cells.Citation25,26,37,40,41,61,65,69,82,92 In most cases, exonic circular RNA expression plasmids consist of a promoter driving only the exon(s) that circularize along with their immediate flanking sequences (). This limits the types of undesired linear RNAs that can be produced. Nevertheless, we have found that most published expression plasmids backsplice at a low efficiency and generate many undesired transcripts, including linear RNAs and concatemers, that limit the utility of these plasmids for defining circular RNA functions. In an effort to improve circular RNA expression methods, we recently optimized the Drosophila Laccase2 and human ZKSCAN1 flanking intronic sequences so that they efficiently express “designer” exonic circular RNAs in human and fly cells ().Citation37 Easy-to-use restriction sites are present and various circular RNAs (ranging in size from 300 to 1500-nt in length) can be generated, including ones that are translated when an internal ribosome entry site (IRES) is present. It should be noted that these plasmids do not efficiently circularize small exons (≤300 -nt), but an alternative strategy that involves processing from tRNA introns can be used to generate small circles.Citation93,94 Methods, such as splint ligation or the use of self-cleaving ribozymes, can also be used to generate circular RNAs in vitro (For a review see ref.Citation95). Beyond allowing ectopic expression of circular RNAs to define their functions, these approaches can be used to design RNA circles that sequester microRNAs or proteins as well as identify novel IRES sequences.
Figure 6. Plasmids for ectopic expression of circular RNAs in cells. To facilitate the expression of nearly any circular RNA in mammalian cells, we optimized the ZKSCAN1 and Laccase2 flanking introns and replaced the endogenous exon with a multicloning site exon (blue).Citation37 These plasmids have been successfully used to express circular RNAs of various sizes, including ones with an IRES that enables translation.
Circular RNA degradation
Once generated, exonic circular RNAs accumulate in the cytoplasm, perhaps using the exon-junction complex to aid in their export from the nucleus (For a review see ref.Citation96). Circular RNAs have long half-lives and progressively accumulate as flies age,Citation29 likely because they are naturally resistant to degradation by exonucleases. Nevertheless, it is highly unlikely that cells allow circular RNA levels to go unchecked, and endonucleases are probably able to facilitate circular RNA decay by providing access points for exonucleases (For a review see ref.Citation97). Major RNA endonucleases in eukaryotic cells include Ago-2 (which functions in RNA silencing), angiogenin (which cleaves tRNAs during stress), CPSF73 (which functions in mRNA 3′ end formation), IRE1 (which functions in ER stress), RNase L (which is involved in innate immunity), SMG6 (which is important for nonsense mediated decay), among others. The CDR1as/ciRS-7 circular RNA contains a near perfect miR-671 target site that can be cleaved by Ago-2 to trigger transcript degradation.Citation56 This, however, appears to be an isolated mechanism, as no other circular RNA is known to contain similar sequences that induce Ago-2 slicing. Roles for other RNA endonucleases in circular RNA decay have not yet been explored.
Recent work suggests that circular RNAs may also be eliminated from cells by packaging them into extracellular vesicles, such as exosomes and microvesicles.Citation98,99 More than 1,000 circular RNAs were identified in human serum exosomesCitation99 and blood,Citation100 and circular RNAs appear to be preferentially packaged into extracellular vesicles over their linear counterparts.Citation98 Besides allowing cells to possibly eliminate excess circular RNAs, the packaging of circular RNAs may contribute to cell-to-cell communication.
Conclusions and perspectives
Although most long transcripts made by RNA polymerase II have a 5′ cap and a poly(A) tail, recent efforts have identified a number of RNAs, including circular RNAs, that defy this dogma (For a review see refs.Citation101-103). At this point, the expression of most circular RNAs has only been supported by a handful of deep sequencing reads, and it remains possible that many lowly expressed circular RNAs are sequencing artifacts or splicing noise that is non-functional.Citation104 Nevertheless, a growing number of circular RNAs have been validated using orthogonal techniques (e.g., Northern blots) and shown to be the predominant outputs of their host genes. This strongly suggests circular RNA functionality, which will hopefully be confirmed with over-expression studies and the characterization of circular RNA knockouts generated via CRISPR-Cas9 genome editing. Deletion of one or both of the flanking intronic repeats should prevent the production of the nearby circular RNA, while having minimal or no effect on linear mRNA processing.
Base pairing between intronic repeat elements is a critical step in the biogenesis of many RNA circles, but it is still unknown precisely how base pairing directs spliceosome assembly toward backsplicing. The assembly process is likely regulated by multiple cis sequences and trans-acting factors, including well-characterized splicing factors like hnRNPs and SR proteins, that ultimately allow circular RNAs to be expressed in tissue-specific patterns. RNAi or CRISPR-based genetic screens coupled to detailed biochemical studies are needed to reveal further details of the backsplicing mechanism. Interestingly, a number of genes generate circular RNAs in the absence of repetitive elements,Citation29,41,105 but almost nothing is known about these biogenesis mechanisms. In some cases, circular RNA biogenesis may be linked to exon skipping events, but it is equally likely that RNA binding proteins may trigger backsplicing via mechanisms similar to those proposed for MblCitation25 and QKI.Citation82 It is also possible that complementary sequences within an exon could help bring the splice sites at the ends of that exon closer together, facilitating backsplicing.
In general, a clearer understanding of the interplay between co-transcriptional splicing, well-studied alternative splicing events (exon skipping, alternative 5′ or 3′ splice sites, etc.), and backsplicing is needed. This will help reveal how the cell ensures that certain introns are slowly spliced and how, in some cases, this facilitates the production of circular RNAs. Once an RNA circle has been generated, a number of important questions about its function and post-transcriptional fate remain to be addressed. Analogous to how bacterial operons function, a circular RNA might act in the same pathway as the protein produced from its parental gene. For example, aberrant fusion circular RNAs expressed from chromosomal translocations have been demonstrated to work with the linear fusion mRNA to drive tumorigenesis.Citation78 Alternatively, circular RNAs may allow the formation of large RNA-protein complexes or regulate gene expression patterns. In addition to detailed characterization of circular RNA functions, further studies are needed to address: (i) how are circular RNAs exported to the cytoplasm?; (ii) do endogenous circular RNAs ever associate with ribosomes to be translated?; and (iii) how are circular RNAs degraded? Answers to each of these questions will help reveal new insights into how circular RNAs are regulated similarly to as well as differently from linear mRNAs that are capped and polyadenylated. Considering that circular RNAs were largely considered a rare oddity just a few years ago, it seems certain that future research will continue its torrid pace and reveal many more unexpected insights into these widespread outputs of eukaryotic genes.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
I thank Jeff Wilusz and members of my laboratory for suggestions and discussions. Supported by NIH R00-GM104166 and R35-GM119735. J.E.W. is a Rita Allen Foundation Scholar.
References
- Crick FH. On protein synthesis. Symp Soc Exp Biol 1958; 12:138-63; PMID:13580867
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009; 23:1494-504; PMID:19571179; https://doi.org/10.1101/gad.1800909
- Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell 2014; 157:77-94; PMID:24679528; https://doi.org/10.1016/j.cell.2014.03.008
- Yang L, Froberg JE, Lee JT. Long noncoding RNAs: fresh perspectives into the RNA world. Trends Biochem Sci 2014; 39:35-43; PMID:24290031; https://doi.org/10.1016/j.tibs.2013.10.002
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet 2007; 8:413-23; PMID:17486121; https://doi.org/10.1038/nrg2083
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nature Biotechnol 2014; 32:453-61; PMID:24811520; https://doi.org/10.1038/nbt.2890
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016; 17:205-11; PMID:26908011; https://doi.org/10.1038/nrm.2015.32
- Wilusz JE. Repetitive elements regulate circular RNA biogenesis. Mob Genet Elements 2015; 5:1-7; PMID:26442181; https://doi.org/10.1080/2159256X.2015.1045682
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development 2016; 143:1838-47; PMID:27246710; https://doi.org/10.1242/dev.128074
- Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: Identification, biogenesis and function. Biochim Biophys Acta 2016; 1859:163-8; PMID:26171810; https://doi.org/10.1016/j.bbagrm.2015.07.007
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014; 20:1829-42; PMID:25404635; https://doi.org/10.1261/rna.047126.114
- Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip Rev RNA 2015; 6:563-79; PMID:26230526; https://doi.org/10.1002/wrna.1294
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012; 7:e30733; PMID:22319583; https://doi.org/10.1371/journal.pone.0030733
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141-57; PMID:23249747; https://doi.org/10.1261/rna.035667.112
- Fu XD, Ares M, Jr. Context-dependent control of alternative splicing by RNA-binding proteins. Nat Rev Genet 2014; 15:689-701; PMID:25112293; https://doi.org/10.1038/nrg3778
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature 2008; 456:470-6; PMID:18978772; https://doi.org/10.1038/nature07509
- Smith CW, Valcarcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 2000; 25:381-8; PMID:10916158; https://doi.org/10.1016/S0968-0004(00)01604-2
- Carrillo Oesterreich F, Herzel L, Straube K, Hujer K, Howard J, Neugebauer KM. Splicing of Nascent RNA Coincides with Intron Exit from RNA Polymerase II. Cell 2016; 165:372-81; PMID:27020755; https://doi.org/10.1016/j.cell.2016.02.045
- Bentley DL. Coupling mRNA processing with transcription in time and space. Nat Rev Genet 2014; 15:163-75; PMID:24514444; https://doi.org/10.1038/nrg3662
- Pandya-Jones A, Black DL. Co-transcriptional splicing of constitutive and alternative exons. RNA 2009; 15:1896-908; PMID:19656867; https://doi.org/10.1261/rna.1714509
- Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev 2015; 29:63-80; PMID:25561496; https://doi.org/10.1101/gad.247361.114
- Vargas DY, Shah K, Batish M, Levandoski M, Sinha S, Marras SA, Schedl P, Tyagi S. Single-molecule imaging of transcriptionally coupled and uncoupled splicing. Cell 2011; 147:1054-65; PMID:22118462; https://doi.org/10.1016/j.cell.2011.10.024
- Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell 2013; 152:1252-69; PMID:23498935; https://doi.org/10.1016/j.cell.2013.02.034
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 2014; 24:1774-86; PMID:25258385; https://doi.org/10.1101/gr.177790.114
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56:55-66; PMID:25242144; https://doi.org/10.1016/j.molcel.2014.08.019
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; https://doi.org/10.1038/nature11993
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; https://doi.org/10.1038/nature11928
- Wilusz JE, Sharp PA. Molecular biology. A circuitous route to noncoding RNA. Science 2013; 340:440-1; PMID:23620042; https://doi.org/10.1126/science.1238522
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014; 9:1966-80; PMID:25544350; https://doi.org/10.1016/j.celrep.2014.10.062
- Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res 2016; 44:1370-83; PMID:26657629; https://doi.org/10.1093/nar/gkv1367
- Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15:409; PMID:25070500; https://doi.org/10.1186/s13059-014-0409-z
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nature Neurosci 2015; 18:603-10; PMID:25714049; https://doi.org/10.1038/nn.3975
- Rybak-Wolf A, Stottmeister C, Glazar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2015; 58:870-85; PMID:25921068; https://doi.org/10.1016/j.molcel.2015.03.027
- Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995; 268:415-7; PMID:7536344; https://doi.org/10.1126/science.7536344
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015; 21:172-9; PMID:25449546; https://doi.org/10.1261/rna.048272.114
- Abe N, Matsumoto K, Nishihara M, Nakano Y, Shibata A, Maruyama H, Shuto S, Matsuda A, Yoshida M, Ito Y, et al. Rolling Circle Translation of Circular RNA in Living Human Cells. Sci Rep 2015; 5:16435; PMID:26553571; https://doi.org/10.1038/srep16435
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 2015; 29:2168-82; PMID:26450910; https://doi.org/10.1101/gad.270421.115
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9:e1003777; PMID:24039610; https://doi.org/10.1371/journal.pgen.1003777
- Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, Yang L, Chen LL. The biogenesis of nascent circular RNAs. Cell Rep 2016; 15:611-24; PMID:27068474; https://doi.org/10.1016/j.celrep.2016.03.058
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell 2014; 159:134-47; PMID:25242744; https://doi.org/10.1016/j.cell.2014.09.001
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 2015; 4:e07540; PMID:26057830; https://doi.org/10.7554/eLife.07540
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792-806; PMID:24035497; https://doi.org/10.1016/j.molcel.2013.08.017
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73:3852-6; PMID:1069269; https://doi.org/10.1073/pnas.73.11.3852
- Kos A, Dijkema R, Arnberg AC, van der Meide PH, Schellekens H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986; 323:558-60; PMID:2429192; https://doi.org/10.1038/323558a0
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell 1991; 64:607-13; PMID:1991322; https://doi.org/10.1016/0092-8674(91)90244-S
- Cocquerelle C, Daubersies P, Majerus MA, Kerckaert JP, Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J 1992; 11:1095-8; PMID:1339341
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155-60; PMID:7678559
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993; 73:1019-30; PMID:7684656; https://doi.org/10.1016/0092-8674(93)90279-Y
- Zaphiropoulos PG. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc Natl Acad Sci U S A 1996; 93:6536-41; PMID:8692851; https://doi.org/10.1073/pnas.93.13.6536
- Zaphiropoulos PG. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol 1997; 17:2985-93; PMID:9154796; https://doi.org/10.1128/MCB.17.6.2985
- Chao CW, Chan DC, Kuo A, Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol Med 1998; 4:614-28; PMID:9848078
- Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum Mol Genet 1999; 8:493-500; PMID:9949208; https://doi.org/10.1093/hmg/8.3.493
- Li XF, Lytton J. A circularized sodium-calcium exchanger exon 2 transcript. J Biol Chem 1999; 274:8153-60; PMID:10075718; https://doi.org/10.1074/jbc.274.12.8153
- Houseley JM, Garcia-Casado Z, Pascual M, Paricio N, O'Dell KM, Monckton DG, Artero RD. Noncanonical RNAs from transcripts of the Drosophila muscleblind gene. J Hered 2006; 97:253-60; PMID:16714427; https://doi.org/10.1093/jhered/esj037
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010; 6:e1001233
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011; 30:4414-22; PMID:21964070; https://doi.org/10.1038/emboj.2011.359
- Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014; 20:1666-70; PMID:25234927; https://doi.org/10.1261/rna.043687.113
- Hansen TB, Veno MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res 2016; 44:e58; PMID:26657634; https://doi.org/10.1093/nar/gkv1458
- Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, Chen LL, Yang L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 2016; 26:1277-87; PMID: 27365365; https://doi.org/10.1101/gr.202895.115
- Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nature Commun 2016; 7:12060; PMID: 27350239; https://doi.org/10.1038/ncomms12060
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22:256-64; PMID:25664725; https://doi.org/10.1038/nsmb.2959
- Veno MT, Hansen TB, Veno ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 2015; 16:245; PMID:26541409; https://doi.org/10.1186/s13059-015-0801-3
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 2007; 8:113-26; PMID:17245413; https://doi.org/10.1038/nrm2104
- Neff AT, Lee JY, Wilusz J, Tian B, Wilusz CJ. Global analysis reveals multiple pathways for unique regulation of mRNA decay in induced pluripotent stem cells. Genome Res 2012; 22:1457-67; PMID:22534399; https://doi.org/10.1101/gr.134312.111
- Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28:2233-47; PMID:25281217; https://doi.org/10.1101/gad.251926.114
- Dubin RA, Kazmi MA, Ostrer H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 1995; 167:245-8; PMID:8566785; https://doi.org/10.1016/0378-1119(95)00639-7
- Pasman Z, Been MD, Garcia-Blanco MA. Exon circularization in mammalian nuclear extracts. RNA 1996; 2:603-10; PMID:8718689
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015; 10:170-7; PMID:25558066; https://doi.org/10.1016/j.celrep.2014.12.019
- Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep 2015; 10:103-11; PMID:25543144; https://doi.org/10.1016/j.celrep.2014.12.002
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nature Commun 2016; 7:11215; PMID:27050392; https://doi.org/10.1038/ncomms11215
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature 2001; 409:860-921; PMID:11237011; https://doi.org/10.1038/35057062
- de Koning AP, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet 2011; 7:e1002384; PMID:22144907; https://doi.org/10.1371/journal.pgen.1002384
- Daniel C, Behm M, Ohman M. The role of Alu elements in the cis-regulation of RNA processing. Cell Mol Life Sci 2015; 72:4063-76; PMID:26223268; https://doi.org/10.1007/s00018-015-1990-3
- Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science 2016; 351:aac7247; PMID:26912865; https://doi.org/10.1126/science.aac7247
- Shedlock AM, Okada N. SINE insertions: powerful tools for molecular systematics. BioEssays 2000; 22:148-60; PMID:10655034; https://doi.org/10.1002/(SICI)1521-1878(200002)22:2%3c148::AID-BIES6%3e3.0.CO;2-Z
- Bunting SF, Nussenzweig A. End-joining, translocations and cancer. Nat Rev Cancer 2013; 13:443-54; PMID:23760025; https://doi.org/10.1038/nrc3537
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer 2007; 7:233-45; PMID:17361217; https://doi.org/10.1038/nrc2091
- Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi PP. Oncogenic Role of Fusion-circRNAs Derived from Cancer-Associated Chromosomal Translocations. Cell 2016; 165:289-302; PMID:27040497; https://doi.org/10.1016/j.cell.2016.03.020
- Song X, Zhang N, Han P, Moon BS, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res 2016; 44:e87; PMID:26873924; https://doi.org/10.1093/nar/gkw075
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation - exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci Rep 2015; 5:8057; PMID:25624062; https://doi.org/10.1038/srep08057
- Ahmed I, Karedath T, Andrews SS, Al-Azwani IK, Ali Mohamoud Y, Querleu D, Rafii A, Malek JA. Altered expression pattern of circular RNAs in primary and metastatic sites of epithelial ovarian carcinoma. Oncotarget 2016; PMID:27119352; https://doi.org/10.18632/oncotarget.8917
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015; 160:1125-34; PMID:25768908; https://doi.org/10.1016/j.cell.2015.02.014
- Dang Y, Yan L, Hu B, Fan X, Ren Y, Li R, Lian Y, Yan J, Li Q, Zhang Y, et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol 2016; 17:130; PMID:27315811; https://doi.org/10.1186/s13059-016-0991-3
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 2015; 16:126; PMID:26076956; https://doi.org/10.1186/s13059-015-0690-5
- Lareau LF, Brooks AN, Soergel DA, Meng Q, Brenner SE. The coupling of alternative splicing and nonsense-mediated mRNA decay. Adv Exp Med Biol 2007; 623:190-211; PMID:18380348; https://doi.org/10.1007/978-0-387-77374-2_12
- Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature 2010; 465:53-9; PMID:20445623; https://doi.org/10.1038/nature09000
- Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 2014; 9:e90859; PMID:24609083; https://doi.org/10.1371/journal.pone.0090859
- Bitton DA, Atkinson SR, Rallis C, Smith GC, Ellis DA, Chen YY, Malecki M, Codlin S, Lemay JF, Cotobal C, et al. Widespread exon skipping triggers degradation by nuclear RNA surveillance in fission yeast. Genome Res 2015; 25:884-96; PMID:25883323; https://doi.org/10.1101/gr.185371.114
- Awan AR, Manfredo A, Pleiss JA. Lariat sequencing in a unicellular yeast identifies regulated alternative splicing of exons that are evolutionarily conserved with humans. Proc Natl Acad Sci U S A 2013; 110:12762-7; PMID:23861491; https://doi.org/10.1073/pnas.1218353110
- Kelly S, Greenman C, Cook PR, Papantonis A. Exon skipping is correlated with exon circularization. J Mol Biol 2015; 427:2414-7; PMID:25728652; https://doi.org/10.1016/j.jmb.2015.02.018
- Gardner EJ, Nizami ZF, Talbot CC, Jr., Gall JG. Stable intronic sequence RNA (sisRNA), a new class of noncoding RNA from the oocyte nucleus of Xenopus tropicalis. Genes Dev 2012; 26:2550-9; PMID:23154985; https://doi.org/10.1101/gad.202184.112
- Ford E, Ares M, Jr. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc Natl Acad Sci U S A 1994; 91:3117-21; PMID:7512723; https://doi.org/10.1073/pnas.91.8.3117
- Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 2015; 21:1554-65; PMID:26194134; https://doi.org/10.1261/rna.052944.115
- Schmidt CA, Noto JJ, Filonov GS, Matera AG. A Method for Expressing and Imaging Abundant, Stable, Circular RNAs In Vivo Using tRNA Splicing. Methods Enzymol 2016; 572:215-36; PMID:27241756; https://doi.org/10.1016/bs.mie.2016.02.018
- Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res 2015; 43:2454-65; PMID:25662225; https://doi.org/10.1093/nar/gkv045
- Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci 2009; 122:1933-7; PMID:19494120; https://doi.org/10.1242/jcs.041236
- Schoenberg DR. Mechanisms of endonuclease-mediated mRNA decay. Wiley Interdiscip Rev RNA 2011; 2:582-600; PMID:21957046; https://doi.org/10.1002/wrna.78
- Lasda E, Parker R. Circular RNAs Co-precipitate with extracellular vesicles: A possible mechanism for circRNA clearance. PLoS One 2016; 11:e0148407; PMID:26848835; https://doi.org/10.1371/journal.pone.0148407
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015; 25:981-4; PMID:26138677; https://doi.org/10.1038/cr.2015.82
- Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 2015; 10:e0141214; PMID:26485708; https://doi.org/10.1371/journal.pone.0141214
- Wilusz JE. Long noncoding RNAs: Re-writing dogmas of RNA processing and stability. Biochim Biophys Acta 2016; 1859:128-38; PMID:26073320; https://doi.org/10.1016/j.bbagrm.2015.06.003
- Zhang Y, Yang L, Chen LL. Life without A tail: new formats of long noncoding RNAs. Int J Biochem Cell Biol 2014; 54:338-49; PMID:24513732; https://doi.org/10.1016/j.biocel.2013.10.009
- Peart N, Sataluri A, Baillat D, Wagner EJ. Non-mRNA 3′ end formation: how the other half lives. Wiley Interdiscip Rev RNA 2013; 4:491-506; PMID:23754627; https://doi.org/10.1002/wrna.1174
- Pickrell JK, Pai AA, Gilad Y, Pritchard JK. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet 2010; 6:e1001236; PMID:21151575; https://doi.org/10.1371/journal.pgen.1001236
- Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015; 21:2076-87; PMID:26464523; https://doi.org/10.1261/rna.052282.115
