ABSTRACT
Spliceosomal snRNPs are complex particles that proceed through a fascinating maturation pathway. Several steps of this pathway are closely linked to nuclear non-membrane structures called Cajal bodies. In this review, I summarize the last 20 y of research in this field. I primarily focus on snRNP biogenesis, specifically on the steps that involve Cajal bodies. I also evaluate the contribution of the Cajal body in snRNP quality control and discuss the role of snRNPs in Cajal body formation.
Biogenesis of snRNPs
Spliceosomal small nuclear ribonucleoprotein particles (snRNPs) are complex particles that consist of small (< 200 nt) nuclear RNA (snRNA), a heptameric ring of Sm or Like-Sm (LSm) proteins and 1 to 12 proteins that are specific for each snRNP. There have been 5 major and 4 minor snRNPs that have been described, each named according to the snRNA that they contain. The major snRNPs are U1, U2, U4, U5 and U6 and the minor snRNPs have been designated U11, U12, U4atac and U6atac. It is of note that the U5 snRNP is common to both major and minor splicing pathways.
All snRNPs proceed through an interesting assembly pathway that involves several cellular compartments. Spliceosomal snRNPs follow a general rule, which states that composite particle maturation occurs primarily in compartments, which differ from the final destination of the mature particle. Biogenesis of snRNPs has been covered by several comprehensive reviews.Citation1-6 In this review, I will specifically focus on the role of Cajal bodies (CBs) in snRNP maturation and the emerging function of CBs in quality control of the assembly process.
With the exception of U6 and U6atac, all snRNAs are transcribed by RNA polymerase II, however they are not polyadenylated. Their 3′ end processing is closely coupled to transcription and depends on the 3′ box located 9–19 nt downstream of the mature 3′ end.Citation7 The first processing step involves endonucleolytic cleavage upstream of the 3′ box, which leaves a tail of a couple nucleotides that requires further trimming. The Integrator complex, first isolated and described by Ramin Shiekhattar and colleagues was shown to play an essential role in initial 3′ end processing.Citation8 3′ end cleavage is coupled to proper transcription termination via the cap-binding complex and SRRT (ARS2).Citation9,10 After transcription and initial 3′ end processing, partially processed snRNAs are exported to the cytoplasm in a m7G-cap dependent manner. Export is facilitated by the exportin XPO1 (CRM1), RanGTP as well as the adaptor proteins NCBP20 (CBP20) /NCBP1 (CBP80), PHAX and SRRT.Citation9,11 In addition, NONO and SFPQ (PSF) promote the association of PHAX with snRNAs.Citation12 In the cytoplasm, a heptameric ring of 7 Sm proteins is assembled around the consensus Sm-binding sequence (AUUUUUG), resulting in the formation of the core snRNP. Assembly of the Sm ring is promoted and controlled by PRMT5 and SMN complexes (reviewed inCitation2 and ref. therein). After Sm ring formation, the m7G-cap is hypermethylated by TGS1 followed by the final 3′ end trimming.Citation13-15 However, localization of the final trimming step has not been clearly resolved. Studies preformed in human cell culture have shown that the trimming activity resides in the cytoplasm,Citation13,16 while results in Xenopus leavis oocyte placed final snRNA trimming in the nucleus after import from the cytoplasm.Citation17 The trimethylguanosine cap, together with the Sm-ring, serve as signals for nuclear import. The trimethylguanosine dependent import pathway has been well described and involves the protein Snurportin-1, which binds the trimethylguanosine cap and acts as a bridge between the trimethylguanosine cap and importin-β.Citation18-20 Although the exact molecular mechanism of the pathway dependent on Sm proteins has yet to be revealed, it has been determined that the SMN complex interacts in the cytoplasm with the core snRNP and importin-β which lead to the proposal that the SMN complex is involved in snRNP nuclear import.Citation21,22 In addition, Importin 7 was shown to participate in the snRNP import pathway in Drosophila melanogaster.Citation23 After re-import, several snRNA nucleotides are modified by scaRNA-directed methylation and pseudouridylation.Citation24
In regards to the final steps of snRNP biogenesis and the addition of snRNP-specific proteins, available information is rather sparse. However, several findings indicate that the addition of snRNP-specific proteins occurs after import of the core snRNP into the cell nucleus. Both U1 and U2 snRNP-specific proteins have been shown to be imported into the nucleus independently of snRNAs.Citation25-27 Similarly, U4 and U5 snRNAs reach the nucleus after the depletion of key snRNP-specific proteins Prpf8 and Prpf31.Citation28,29 Together, these findings imply that snRNP-specific proteins are added after the core snRNP has entered the nucleus. Injection of fluorescently labeled U4 and U5 snRNAs into the Xenopus laevis oocyte revealed the occurrence of transient nucleolar localization.Citation30 However, nucleolar localization has not been observed in human cells (our unpublished data andCitation31).
Several chaperones have been shown to assist in the addition of snRNP-specific proteins to the core snRNP. In yeast, Aar2p interacts with Prp8p before the addition of other U5-specific proteins promoting U5 snRNP formation.Citation32 Structural studies have revealed a mutually exclusive binding of Prp8p protein with Aar2p and Brr2p.Citation33,34 The Prp8p-Aar2p interaction is disrupted upon Aar2p phosphorylation which represents the molecular switch between Aar2p and Brr2p binding to Prp8p.Citation33 In Drosophila melanogaster, the ECD protein interacts with U5 snRNP proteins and likely participates in U5 snRNP assembly.Citation35 Finally, NUFIP and the HSP90/R2TP chaperone system were found to interact with U4-specific Prpf31 and assist with U4 snRNP biogenesis.Citation28 Less is known about chaperons involved in U1 and U2 snRNP assembly. U1-specific SNRNP70 (U1-70K) has been recently found to interact with the SMN complex and promote Sm ring assembly specifically on the U1 snRNA,Citation36,37 which suggests that SNRNP70 interacts with U1 snRNA in the cytoplasm. Both U1 snRNA and SNRNP70 have been detected in gems, nuclear structures that are rich in SMN complexes.Citation36 SNRNP70 interacts with the SMN complex also in the cell nucleus suggesting that nuclear SMN is involved in U1 snRNP maturation and/or recycling.Citation36,38 Alternatively, nuclear SNRNP70-SMN complexes could represent post-imported U1 particles. U2 snRNP biogenesis proceed through 3 distinct complexes: the 12S complex, the 15S complex and 17S complex. These complexes reflect addition of SNRNPA1 (U2A) and SNRPB2 (U2B”) proteins, the SF3b complex, and the SF3a trimeric complex, respectivelly.Citation39 17S U2 snRNP associates with additional proteins including Tudor domain containing SMNDC1 (SPF30), DEAH-box helicases DHX15 (hPrp43) and DDX46 (hPrp5) and the U2AF dimer.Citation40
The U6 snRNP follows a different biogenesis pathway. U6 snRNA is synthesized by RNA polymerase III and remains in the cell nucleus. New U6 transcripts contain an extended tail of uridines, which are added by the TUTase TUT1.Citation41,42 Like other RNA polymerase III transcripts, the U-stretch at the 3′ end of the U6 snRNA is bound by the La protein that protects RNAs from degradation and facilitates their further maturation.Citation43 The mature U6 snRNA associates with a ring of 7 LSm 2–8 proteins. The LSm2-8 ring is pre-assembled without RNA in the cytoplasm and is localized to the nucleus by the LSm8 protein.Citation44,45 The 5′ end of U6 contains a γ-monomethyl cap while the 3′ end ribose is protected by cyclic 2′-3′ phosphate which is formed by USB1, a protein that is mutated in patients with the rare hereditary disease, Clericuzio-type poikiloderma with neutropenia.Citation46 The formation of the cyclic phosphate at the 3′ end destabilizes La binding and facilitates LSm2-8 association with the U6 snRNA.Citation47 U6 snRNA is also modified by methylation as well as the isomerization of uridines to pseudouridines, but these modifications are guided by snoRNAs and occur in the nucleolus.Citation48,49 The key role of the nucleolus in U6 biogenesis has been further documented by following the path of microinjected U6 snRNA through the nucleolus and nucleolar localization of TUT1.Citation42,50 U6 snRNA further interacts with SART3, which targets U6 snRNA to CBs and promotes U4/U6 annealing.Citation51-53 A new U6 snRNA modifying TUTase USIP-1 has been recently identified in Caenorhabditis elegans. This enzyme interacts with an LSm-free U6 snRNP containing SART3, which lead to the speculation that USIP-1 specifically associates and modifies the U6 snRNA released after splicing.Citation54
The completion of snRNP biogenesis is finalized with the formation of the composite particles, U4/U6 and U4/U6•U5. First, U4 and U6 snRNAs are brought together by the combined action of SART3 and LSm 2–8 proteins.Citation53,55 After U4/U6 duplex formation, U4/U6 specific proteins are added creating the U4/U6 particle.Citation56 The U4/U6 snRNP further associates with the U5 snRNP to form the mature U4/U6•U5 tri-snRNP. The interaction between U4-specific Prpf31 and U5-specific Prpf6 is essential for tri-snRNP assembly,Citation57,58 however, other snRNP proteins, namely Prpf3 and EFTUD2 (hSnu114), participate and secure tri-snRNP stability as well.Citation59,60 Structural studies of yeast and human tri-snRNPs revealed a beautiful arch that consists of EFTUD2 TPR repeats and connects U5 and U4 proteins.Citation61,62 The structure of the human tri-snRNP also points to the important role of USP39 (Sad1, U4/U6•U5-65kD subunit) in sealing the tri-snRNP structure.Citation62 Finally, the splicing competent U4/U6•U5 snRNP is assembled and ready for splicing.
There is less known about the biogenesis of minor snRNPs. U4atac/U6atac snRNP contains the same set of proteins as the U4/U6 snRNP and likely follows the same maturation pathway as its major counterparts. U11 and U12 snRNPs enter the splicing reaction as a pre-assembled complex with the assistance of 2 important proteins that contribute to U11/U12 snRNP formation, RNPC3 (U11/U12-65K) and PDCD7 (U11-59K).Citation63
Cajal bodies and snRNPs - a close relationship
CB is closely linked to spliceosomal snRNAs and snRNPs. Shortly after the discovery of coilin, a marker and scaffolding protein of CBs (reviewed inCitation64), snRNAs were detected in these nuclear compartments.Citation65,66 Since this important finding, the debate regarding the function of CBs in snRNA/snRNP metabolism has been ongoing. Soon afterwards, the hypothesis that splicing takes place in CBs was neglected. It has since become clear that CBs are involved in many aspects of short non-coding RNA metabolism including snRNP biogenesis, but we still lack a comprehensive and detailed understanding of the exact role of CBs in this process. The importance of coilin has been shown in several studies including the reduction of litter size in coilin knockout mice suggesting problems with embryogenesis and/or fertility.Citation67,68 Similarly, coilin depletion in fish embryos has been shown to be lethal.Citation69 Flies without coilin are fully viable and fertileCitation70 but have problems with egg-laying regulation (Joe Gall, personal communication). Similarly, plants are also viable without coilin, however coilin mutations, which disrupted CBs, showed changes in the expression pattern of a subset of genes.Citation71,72 It should be noted that changes in mRNA expression were also observed after disruption of CBs in human cell culture.Citation73 Despite significant progress, we are still far from complete understanding of CB molecular function. In the text that follows, readers can find an up-to-date description of our current knowledge of the role of CB in snRNA and snRNP.
Pre-export events: snRNA transcription and 3′ end processing
Both major and minor U snRNA genes are often found in the vicinity of nuclear CBs.Citation73-76 Several studies have provided evidence that snRNA transcription and nascent snRNAs are necessary for the association of CBs with U snRNA gene.Citation77-80 It was recently suggested that CBs are involved in the organization of snRNA genes within the nuclear space.Citation73 The tight connection between CBs and snRNA genes was further demonstrated by Karla Neugebauer and colleagues. They used unbiased chromatin immunoprecipitation followed by DNA-seq to show the association between coilin and snRNA genes.Citation81 CBs are thus connected to snRNAs from their birth and knockdown of proteins that are essential for CB integrity (WRAP53 and USPL1), result in lower expression of all snRNAs.Citation73 How exactly CBs enhance snRNA expression is currently unclear. However, CBs could directly regulate snRNA transcription via a regulatory feedback mechanism where they “sense” the amount of total snRNAs and regulate snRNA transcription of nearby snRNA genes accordingly.Citation82
The U2 pre-snRNA is highly concentrated in CBs, which points to the role of CBs in snRNA 3′ end processing.Citation83 In addition, Int4 and Int11 subunits of the Integrator complex are important for CB integrity, which further supports a connection between Integrator and CBs.Citation84 The Int4 subunit was localized to CBs,Citation84 however other Integrator subunits do not specifically accumulate in CBs.Citation85 Considering that coilin directly associates with snRNAs and might even have a direct role in their processing,Citation81,86 it is feasible to conclude that CBs are involved in 3′ end processing of pre-snRNAs. However, the exact role and the importance of CBs in this process have yet to be clarified.
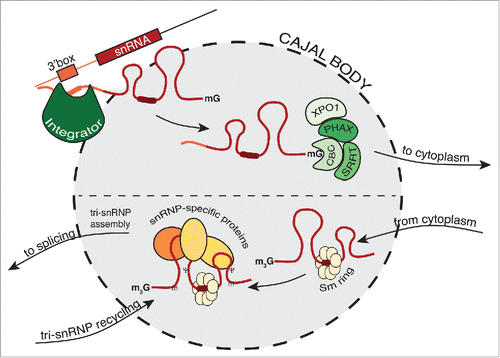
After 3′ end cleavage, snRNAs are exported to the cytoplasm by a complex, which contains, among other proteins, XPO1 and PHAX, which are both located in CBs.Citation78,87,88 In addition, inhibition of PHAX results in the accumulation of U1 snRNA in Xenopus laevis oocyte CBs.Citation89 These findings together suggest that newly transcribed snRNAs pass through the CBs on their way out of the nucleus supporting the idea that the export complex is assembled in CBs ().
Figure 1. Cajal body is involved in different steps of snRNP biogenesis. First, the Integrator cleaves the 3′ end of snRNA. Then, the export complex forms at the 5′ end and snRNA is exported to the cytoplasm. After Sm ring assembly and cap hypermethylation, the core snRNP returns to the Cajal body where snRNP-specific proteins are added and U4/U6•U5 tri-snRNP forms.
Post-import events: snRNA modification, snRNP assembly and recycling
After completing the cytoplasmic phase, the core snRNPs (snRNAs+Sm proteins) return to the nucleus and first accumulate in CBs.Citation90 However, the precise molecular signals that navigate core snRNPs to CBs are unknown. Coilin interacts directly with snRNAs and the Sm proteins, thus both possibly serving as a CB targeting signals.Citation81,91 It was recently shown that SART3 is important for proper CB localization of U4 and U5 snRNAs but the molecular mechanism has been elusive.Citation29 Also the SMN complex, which interacts with the Sm ring in the cytoplasm, was suggested to target new snRNPs to CBs through the interaction of the SMN Tudor domain with the coilin RG-box.Citation21,22,88,92 However, the key experiments that would identify the CB targeting signal have yet to be performed.
After import to the CB, new snRNA is modified by 2′-O-ribose methylation and by isomerization of uridines to pseudouridines. Modified nucleotides are selected by short non-coding RNAs - scaRNAs which are specifically localized in CBs.Citation93 In Drosophila melanogaster, scaRNAs are also localized in CBs but a coilin null mutant lacking CBs exhibited normal levels of snRNA methylation and pseudouridylation.Citation94 These results indicate that post-transcriptional modification of snRNAs primarily occurs in CBs but these structures are not essential, at least in flies.
Core snRNPs must further associate with snRNP-specific proteins. When this process is inhibited, snRNAs accumulate in CBs, which strongly suggests that core snRNPs meet their specific protein partners primarily here.Citation28,29 The final step in snRNP maturation is the formation of the splicing competent U2 and U4/U6•U5 snRNPs. U2 snRNP maturation, specifically the formation of the 17S complex, has been shown to occur in CBs.Citation27,95 Also U4/U6 and U4/U6•U5 snRNP assembly has been placed in CBs.Citation51,52,57
During splicing, the tri-snRNP undergoes a remarkable rearrangement, which results in the release of individual snRNPs from the post-splicing spliceosome. However, the exact molecular composition of snRNPs after splicing is not well defined. The U6 protein, SART3, is released well ahead of splicing during tri-snRNP assembly.Citation53 U6-specific LSm proteins are destabilized during splicing,Citation96 which suggests that U6 snRNA is released as naked snRNA! U4/U6 snRNP-specific proteins leave the spliceosome at the same time as U4 snRNA although it is not known whether these proteins remain bound to the U4 snRNA. However, it is reasonable to speculate that SNU13, which directly binds the U4 kink-turn motif,Citation97 stays stably associated with the released U4 snRNA. Also Prpf31, which is bound to the SNU13-U4 snRNA complex, remains likely linked to the released U4 snRNP.Citation98 The only molecularly characterized post-splicing snRNP is the 35S U5 snRNP, which contains 4 of the 6 U5-specific proteins, proteins from the NTC complex and step 2 factors.Citation99 Little is known about the factors that recycle the post-splicing snRNP and reassemble functional snRNPs. SART3, a protein that is highly accumulated in CBs,Citation52 was shown to act in U4/U6 reannealing.Citation53 Because snRNPs repeatedly cycle through CBs and U4/U6 snRNPs accumulate in CBs after inhibition of spliceosome recycling it is reasonable to speculate that CB are involved in snRNP re-assembly after splicing.Citation100
There is virtually no information available that addresses the stability of snRNPs in vivo or the requirements to repair the Sm-ring and/or snRNP-specific proteins. The SMN complex has been also found in the nucleus (gems and CBs) and has been suggested to be important for snRNP recycling.Citation101 Thus, it is plausible that the nuclear SMN complex is involved in the repair of damaged Sm rings, which might take place in CBs. An alternative hypothesis suggests that the SMN complex disassembles the Sm ring during snRNP degradation.
Quality control in Cajal bodies
The quality of snRNP formation is apparently controlled at several checkpoints along the assembly pathway (). First, it has been suggested that the exosome, TRAMP and the NEXT complex are involved in recognition and degradation of aberrant snRNAs in the nucleus but a molecular mechanism that describes how misfolded snRNAs are recognized is unknown.Citation102,103 Furthermore, it has been shown that inhibition of the snRNP export complex formation leads to the accumulation of snRNAs in CBs of the Xenopus laevis oocyte, which suggests the existence of a quality control checkpoint, which monitors snRNA before their export from the nucleus.Citation89 In the cytoplasm, U1 snRNAs, which lack the functional Sm-binding sequence are degraded by Rrp6- or by Dcp2-dependent pathways.Citation104 As I have previously mentioned, the formation of the Sm-ring and the trimethylguanosine cap both serve as import signals and thus represent an important quality control point that ensures that only properly assembled and modified core snRNPs return to the nucleus.Citation2 It is worth mentioning that while the Sm-ring is indispensable for the import, the importance of the trimethylguanosine cap varies among individual snRNPs and cell types.Citation105-107
Figure 2. SnRNP biogenesis is closely controlled along the maturation pathway. Misfolded snRNAs are degraded by the exosome/TRAMP complex dependent pathway. Next, formation of the snRNP export complex is monitored and only the properly assembled export complex is transported to the cytoplasm. In the cytoplasm, snRNAs with non-functional Sm-binding site are degraded. Sm-ring assembly and cap trimethylation serve as an additional checkpoint and only properly formed snRNPs are imported back to the nucleus. The core snRNP is targeted to the Cajal body where snRNP-specific proteins are added and di- and tri-snRNPs are formed. If either of these last steps fails, immature snRNPs are retained in the Cajal body.
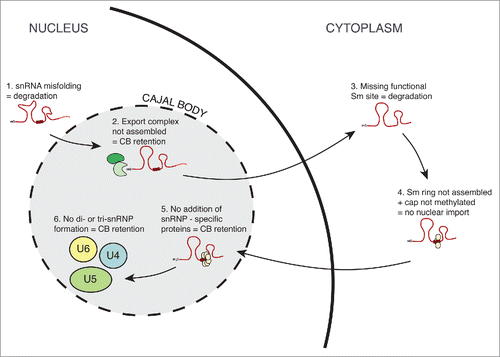
After reaching the nucleus, core snRNPs first accumulate in CBs. When snRNP-specific proteins are depleted or the formation of di- or tri-snRNP is blocked, U4, U5 and U6 snRNAs highly accumulate in CBs.Citation28,29,31,57 This demonstrates the presence of an additional checkpoint that controls the final steps of snRNP maturation, sequesters immature snRNPs and allows only mature particles to enter the splicing reaction. SnRNP sequestration in CBs depends on SART3, which interacts with coilin and thus provides a molecular bridge between snRNPs and coilin.Citation29 However, the exact molecular mechanism of how SART3 recognizes immature snRNPs is unknown.
Cajal bodies need snRNPs
CBs in cell culture are sensitive to inhibition of transcription, splicing and snRNP biogenesis.Citation108 First, snRNPs disappear from CBs after transcription and splicing inhibition, followed by CB disassembly shortly after. This shows that constant production and recycling of snRNPs is essential for CB integrity. Recently, our laboratory showed that CBs can be induced in primary fibroblasts that normally lack them, by inhibition of tri-snRNP assembly.Citation29 In addition, overexpression of coilin does not induce CB formation but overexpression of snRNP components does.Citation109 Finally, snRNP-specific proteins are potent inducers of CBs when artificially immobilized to chromatin.Citation110 These results demonstrate that CB formation requires at least 2 components: coilin and snRNPs. Findings that coilin directly interacts with snRNAs in vivo and that snRNAs induce coilin aggregation in vitro supports this 2-composite model of CB formation.Citation81,111 In the future, we have to uncover rules that govern and control protein-protein and RNA-protein interaction in the CB microenvironment to reveal the basic principles of how snRNPs drive CB formation.
Final remarks
Many steps in snRNP biogenesis are closely associated with CBs but these microscopically visible CBs are not present in all cell types. This signifies that CBs are not absolutely essential and that snRNPs are able to mature and recycle without them. However, the presence of CBs in embryonic, highly dividing and metabolically active cells shows that CBs are important when metabolism is high. Recent evidence that displacement of histone mRNA processing factors from nuclear bodies reduces the efficiency of histone mRNA 3′ end processing shows the importance of nuclear structures and the concentration of factors within them.Citation112 I conclude that the CB and snRNP are necessary for one another. The concentration of snRNPs in the CB enhances snRNP maturation by providing the appropriate chemical environment as well as the required snRNP biogenesis factors.Citation31,113,114 In turn, snRNPs promote coilin aggregation and CB formation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
I would like to thank Anna Malinova for critical comments on the manuscript and Jasper Manning for English correction.
Funding
This work was supported by grants from the Czech Academy of Sciences (RVO68378050) and the Czech Science Foundation (15-00790S).
References
- Li DK, Tisdale S, Lotti F, Pellizzoni L. SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin Cell Dev Biol 2014; 32:22-9; PMID:24769255; https://doi.org/10.1016/j.semcdb.2014.04.026
- Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 2014; 15:108-21; PMID:24452469; https://doi.org/10.1038/nrm3742
- Patel SB, Bellini M. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res 2008; 36:6482-93; PMID:18854356; https://doi.org/10.1093/nar/gkn658
- Fischer U, Englbrecht C, Chari A. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip Rev RNA 2011; 2:718-31; PMID:21823231; https://doi.org/10.1002/wrna.87
- Sleeman J. Small nuclear RNAs and mRNAs: linking RNA processing and transport to spinal muscular atrophy. Biochem Soc Trans 2013; 41:871-5; PMID:23863147; https://doi.org/10.1042/BST20120016
- Mroczek S, Dziembowski A. U6 RNA biogenesis and disease association. Wiley Interdiscip Rev RNA 2013; 4:581-92; PMID:23776162; https://doi.org/10.1002/wrna.1181
- Hernandez N. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J 1985; 4:1827-37; PMID:2411548
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 2005; 123:265-76; PMID:16239144; https://doi.org/10.1016/j.cell.2005.08.019
- Hallais M, Pontvianne F, Andersen PR, Clerici M, Lener D, Benbahouche Nel H, Gostan T, Vandermoere F, Robert MC, Cusack S, et al. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat Struct Mol Biol 2013; 20:1358-66; PMID:24270878; https://doi.org/10.1038/nsmb.2720
- Andersen PR, Domanski M, Kristiansen MS, Storvall H, Ntini E, Verheggen C, Schein A, Bunkenborg J, Poser I, Hallais M, et al. The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat Struct Mol Biol 2013; 20:1367-76; PMID:24270879; https://doi.org/10.1038/nsmb.2703
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 2000; 101:187-98; PMID:10786834; https://doi.org/10.1016/S0092-8674(00)80829-6
- Izumi H, McCloskey A, Shinmyozu K, Ohno M. p54nrb/NonO and PSF promote U snRNA nuclear export by accelerating its export complex assembly. Nucleic Acids Res 2014; 42:3998-4007; PMID:24413662; https://doi.org/10.1093/nar/gkt1365
- Madore SJ, Wieben ED, Pederson T. Intracellular site of U1 small nuclear RNA processing and ribonucleoprotein assembly. J Cell Biol 1984; 98:188-92; PMID:6200485; https://doi.org/10.1083/jcb.98.1.188
- Madore SJ, Wieben ED, Kunkel GR, Pederson T. Precursors of U4 small nuclear RNA. J Cell Biol 1984; 99:1140-4; PMID:6206077; https://doi.org/10.1083/jcb.99.3.1140
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonne R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 2002; 9:891-901; PMID:11983179; https://doi.org/10.1016/S1097-2765(02)00484-7
- Huang Q, Jacobson MR, Pederson T. 3′ processing of human pre-U2 small nuclear RNA: a base-pairing interaction between the 3′ extension of the precursor and an internal region. Mol Cell Biol 1997; 17:7178-85; PMID:9372950; https://doi.org/10.1128/MCB.17.12.7178
- Yang H, Moss ML, Lund E, Dahlberg JE. Nuclear processing of the 3′-terminal nucleotides of pre-U1 RNA in Xenopus laevis oocytes. Mol Cell Biol 1992; 12:1553-60; PMID:1549111; https://doi.org/10.1128/MCB.12.4.1553
- Huber J, Cronshagen U, Kadokura M, Marshallsay C, Wada T, Sekine M, Lührmann R. Snurportin1, an m3G-cap-specific nuclear import receptor with a novel domain structure. EMBO J 1998; 17:4114-26; PMID:9670026; https://doi.org/10.1093/emboj/17.14.4114
- Huber J, Dickmanns A, Luhrmann R. The importin-beta binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol 2002; 156:467-79; PMID:11815630; https://doi.org/10.1083/jcb.200108114
- Ospina JK, Gonsalvez GB, Bednenko J, Darzynkiewicz E, Gerace L, Matera AG. Cross-talk between snurportin1 subdomains. Mol Biol Cell 2005; 16:4660-71; PMID:16030253; https://doi.org/10.1091/mbc.E05-04-0316
- Narayanan U, Achsel T, Luhrmann R, Matera AG. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol Cell 2004; 16:223-34; PMID:15494309; https://doi.org/10.1016/j.molcel.2004.09.024
- Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin beta. Hum Mol Genet 2002; 11:1785-95; PMID:12095920; https://doi.org/10.1093/hmg/11.15.1785
- Natalizio AH, Matera AG. Identification and characterization of Drosophila Snurportin reveals a role for the import receptor Moleskin/Importin7 in snRNP biogenesis. Mol Biol Cell 2013; PMID:23885126; https://doi.org/10.1091/mbc.E13-03-0118
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O- methylation and pseudouridylation guide RNAs. Embo J 2002; 21:2746-56; PMID:12032087; https://doi.org/10.1093/emboj/21.11.2746
- Romac JM, Graff DH, Keene JD. The U1 small nuclear ribonucleoprotein (snRNP) 70K protein is transported independently of U1 snRNP particles via a nuclear localization signal in the RNA-binding domain. Mol Cell Biol 1994; 14:4662-70; PMID:7516470; https://doi.org/10.1128/MCB.14.7.4662
- Kambach C, Mattaj IW. Nuclear transport of the U2 snRNP-specific U2B″ protein is mediated by both direct and indirect signalling mechanisms. J Cell Sci 1994; 107(Pt 7):1807-16; PMID:7983149
- Nesic D, Tanackovic G, Kramer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci 2004; 117:4423-33; PMID:15316075; https://doi.org/10.1242/jcs.01308
- Bizarro J, Dodre M, Huttin A, Charpentier B, Schlotter F, Branlant C, Verheggen C, Massenet S, Bertrand E. NUFIP and the HSP90/R2TP chaperone bind the SMN complex and facilitate assembly of U4-specific proteins. Nucleic Acids Res 2015; 43:8973-89; PMID:26275778; https://doi.org/10.1093/nar/gkv809
- Novotny I, Malinova A, Stejskalova E, Mateju D, Klimesova K, Roithova A, Švéda M, Knejzlík Z, Staněk D. SART3-dependent accumulation of incomplete spliceosomal snRNPs in cajal bodies. Cell Rep 2015; 10:429-40; PMID:25600876; https://doi.org/10.1016/j.celrep.2014.12.030
- Gerbi SA, Borovjagin AV, Odreman FE, Lange TS. U4 snRNA nucleolar localization requires the NHPX/15.5-kD protein binding site but not Sm protein or U6 snRNA association. J Cell Biol 2003; 162:821-32; PMID:12939253; https://doi.org/10.1083/jcb.200301071
- Klingauf M, Stanek D, Neugebauer KM. Enhancement of U4/U6 small nuclear ribonucleoprotein particle association in Cajal bodies predicted by mathematical modeling. Mol Biol Cell 2006; 17:4972-81; PMID:16987958; https://doi.org/10.1091/mbc.E06-06-0513
- Boon KL, Grainger RJ, Ehsani P, Barrass JD, Auchynnikava T, Inglehearn CF, Beggs JD. prp8 mutations that cause human retinitis pigmentosa lead to a U5 snRNP maturation defect in yeast. Nat Struct Mol Biol 2007; 14:1077-83; PMID:17934474; https://doi.org/10.1038/nsmb1303
- Weber G, Cristao VF, de LAF, Santos KF, Holton N, Rappsilber J, Beggs JD, Wahl MC. Mechanism for Aar2p function as a U5 snRNP assembly factor. Genes Dev 2011; 25:1601-12; PMID:21764848; https://doi.org/10.1101/gad.635911
- Nguyen TH, Li J, Galej WP, Oshikane H, Newman AJ, Nagai K. Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure 2013; 21:910-19; PMID:23727230; https://doi.org/10.1016/j.str.2013.04.017
- Claudius AK, Romani P, Lamkemeyer T, Jindra M, Uhlirova M. Unexpected role of the steroid-deficiency protein ecdysoneless in pre-mRNA splicing. PLoS Genet 2014; 10:e1004287; PMID:24722212; https://doi.org/10.1371/journal.pgen.1004287
- Stejskalova E, Stanek D. The splicing factor U1-70K interacts with the SMN complex and is required for nuclear gem integrity. J Cell Sci 2014; 127:3909-15; PMID:25052091; https://doi.org/10.1242/jcs.155838
- So BR, Wan L, Zhang Z, Li P, Babiash E, Duan J, Younis I, Dreyfuss G. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat Struct Mol Biol 2016; 23:225-30; PMID:26828962; https://doi.org/10.1038/nsmb.3167
- Yu Y, Chi B, Xia W, Gangopadhyay J, Yamazaki T, Winkelbauer-Hurt ME, Yin S, Eliasse Y, Adams E, Shaw CE, et al. U1 snRNP is mislocalized in ALS patient fibroblasts bearing NLS mutations in FUS and is required for motor neuron outgrowth in zebrafish. Nucleic Acids Res 2015; 43:3208-18; PMID:25735748; https://doi.org/10.1093/nar/gkv157
- Kramer A, Gruter P, Groning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol 1999; 145:1355-68; PMID:10385517; https://doi.org/10.1083/jcb.145.7.1355
- Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Luhrmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. Embo J 2002; 21:4978-88; PMID:12234937; https://doi.org/10.1093/emboj/cdf480
- Lund E, Dahlberg JE. Cyclic 2′,3′-phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science 1992; 255:327-30; PMID:1549778; https://doi.org/10.1126/science.1549778
- Trippe R, Guschina E, Hossbach M, Urlaub H, Luhrmann R, Benecke BJ. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA 2006; 12:1494-504; PMID:16790842; https://doi.org/10.1261/rna.87706
- Rinke J, Steitz JA. Association of the lupus antigen La with a subset of U6 snRNA molecules. Nucleic Acids Res 1985; 13:2617-29; PMID:2582364; https://doi.org/10.1093/nar/13.7.2617
- Zaric B, Chami M, Remigy H, Engel A, Ballmer-Hofer K, Winkler FK, Kambach C. Reconstitution of two recombinant LSm protein complexes reveals aspects of their architecture, assembly, and function. J Biol Chem 2005; 280:16066-75; PMID:15711010; https://doi.org/10.1074/jbc.M414481200
- Novotny I, Podolska K, Blazikova M, Valasek LS, Svoboda P, Stanek D. Nuclear LSm8 affects number of cytoplasmic processing bodies via controlling cellular distribution of Like-Sm proteins. Mol Biol Cell 2012; 23:3776-85; PMID:22875987; https://doi.org/10.1091/mbc.E12-02-0085
- Mroczek S, Krwawicz J, Kutner J, Lazniewski M, Kucinski I, Ginalski K, Dziembowski A. C16orf57, a gene mutated in poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible for the U6 snRNA 3′ end modification. Genes Dev 2012; 26:1911-25; PMID:22899009; https://doi.org/10.1101/gad.193169.112
- Licht K, Medenbach J, Luhrmann R, Kambach C, Bindereif A. 3′-cyclic phosphorylation of U6 snRNA leads to recruitment of recycling factor p110 through LSm proteins. RNA 2008; 14:1532-8; PMID:18567812; https://doi.org/10.1261/rna.1129608
- Ganot P, Jady BE, Bortolin ML, Darzacq X, Kiss T. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol Cell Biol 1999; 19:6906-17; PMID:10490628; https://doi.org/10.1128/MCB.19.10.6906
- Tycowski KT, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell 1998; 2:629-38; PMID:9844635; https://doi.org/10.1016/S1097-2765(00)80161-6
- Gerbi SA, Lange TS. All Small Nuclear RNAs (snRNAs) of the [U4/U6.U5] Tri-snRNP Localize to Nucleoli; Identification of the Nucleolar Localization Element of U6 snRNA. Mol Biol Cell 2002; 13:3123-37; PMID:12221120; https://doi.org/10.1091/mbc.01-12-0596
- Stanek D, Neugebauer KM. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J Cell Biol 2004; 166:1015-25; PMID:15452143; https://doi.org/10.1083/jcb.200405160
- Stanek D, Rader SD, Klingauf M, Neugebauer KM. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol 2003; 160:505-16; PMID:12578909; https://doi.org/10.1083/jcb.200210087
- Bell M, Schreiner S, Damianov A, Reddy R, Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. Embo J 2002; 21:2724-35; PMID:12032085; https://doi.org/10.1093/emboj/21.11.2724
- Ruegger S, Miki TS, Hess D, Grosshans H. The ribonucleotidyl transferase USIP-1 acts with SART3 to promote U6 snRNA recycling. Nucleic Acids Res 2015; 43:3344-57; PMID:25753661; https://doi.org/10.1093/nar/gkv196
- Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Luhrmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′- end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. Embo J 1999; 18:5789-802; PMID:10523320; https://doi.org/10.1093/emboj/18.20.5789
- Nottrott S, Urlaub H, Luhrmann R. Hierarchical, clustered protein interactions with U4/U6 snRNA: a biochemical role for U4/U6 proteins. Embo J 2002; 21:5527-38; PMID:12374753; https://doi.org/10.1093/emboj/cdf544
- Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. Embo J 2004; 23:3000-9; PMID:15257298; https://doi.org/10.1038/sj.emboj.7600296
- Makarova OV, Makarov EM, Liu S, Vornlocher HP, Luhrmann R. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6center dotU5 tri-snRNP formation and pre-mRNA splicing. Embo J 2002; 21:1148-57; PMID:11867543; https://doi.org/10.1093/emboj/21.5.1148
- Liu S, Mozaffari-Jovin S, Wollenhaupt J, Santos KF, Theuser M, Dunin-Horkawicz S, Fabrizio P, Bujnicki JM, Lührmann R, Wahl MC. A composite double-/single-stranded RNA-binding region in protein Prp3 supports tri-snRNP stability and splicing. eLife 2015; 4:e07320; PMID:26161500; https://doi.org/10.7554/eLife.07320
- Liu S, Rauhut R, Vornlocher HP, Luhrmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. RNA 2006; 12:1418-30; PMID:16723661; https://doi.org/10.1261/rna.55406
- Nguyen TH, Galej WP, Bai XC, Savva CG, Newman AJ, Scheres SH, Nagai K. The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature 2015; 523:47-52; PMID:26106855; https://doi.org/10.1038/nature14548
- Agafonov DE, Kastner B, Dybkov O, Hofele RV, Liu WT, Urlaub H, Lührmann R, Stark H. Molecular architecture of the human U4/U6.U5 tri-snRNP. Science 2016; 351:1416-20; PMID:26912367; https://doi.org/10.1126/science.aad2085
- Benecke H, Luhrmann R, Will CL. The U11/U12 snRNP 65K protein acts as a molecular bridge, binding the U12 snRNA and U11-59K protein. EMBO J 2005; 24:3057-69; PMID:16096647; https://doi.org/10.1038/sj.emboj.7600765
- Machyna M, Neugebauer KM, Stanek D. Coilin: The first 25 years. RNA Biol 2015; 12:590-6; PMID:25970135; https://doi.org/10.1080/15476286.2015.1034923
- Carmo-Fonseca M, Pepperkok R, Sproat BS, Ansorge W, Swanson MS, Lamond AI. In vivo detection of snRNP-rich organelles in the nuclei of mammalian cells. Embo J 1991; 10:1863-73; PMID:1710980
- Matera AG, Ward DC. Nucleoplasmic organization of small nuclear ribonucleoproteins in cultured human cells. J Cell Biol 1993; 121:715-27; PMID:8491767; https://doi.org/10.1083/jcb.121.4.715
- Walker MP, Tian L, Matera AG. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One 2009; 4:e6171; PMID:19587784; https://doi.org/10.1371/journal.pone.0006171
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 2001; 154:293-307; PMID:11470819; https://doi.org/10.1083/jcb.200104083
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 2010; 17:403-9; PMID:20357773; https://doi.org/10.1038/nsmb.1783
- Deryusheva S, Gall JG. Dynamics of coilin in Cajal bodies of the Xenopus germinal vesicle. Proc Natl Acad Sci U S A 2004; 101:4810-4; PMID:15044688; https://doi.org/10.1073/pnas.0401106101
- Collier S, Pendle A, Boudonck K, van Rij T, Dolan L, Shaw P. A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol Biol Cell 2006; 17:2942-51; PMID:16624863; https://doi.org/10.1091/mbc.E05-12-1157
- Kanno T, Lin WD, Fu JL, Wu MT, Yang HW, Lin SS, Matzke AJ, Matzke M. Identification of Coilin Mutants in a Screen for Enhanced Expression of an Alternatively Spliced GFP Reporter Gene in Arabidopsis thaliana. Genetics 2016; PMID:27317682; https://doi.org/10.1534/genetics.116.190751
- Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G, Hakim O, Baek S, Hager GL, Dundr M. Cajal bodies are linked to genome conformation. Nat Commun 2016; 7:10966; PMID:26997247; https://doi.org/10.1038/ncomms10966
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A 1995; 92:5915-9; PMID:7597053; https://doi.org/10.1073/pnas.92.13.5915
- Smith KP, Carter KC, Johnson CV, Lawrence JB. U2 and U1 snRNA gene loci associate with coiled bodies. J Cell Biochem 1995; 59:473-85; PMID:8749717; https://doi.org/10.1002/jcb.240590408
- Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol Biol Cell 1999; 10:1653-63; PMID:10233169; https://doi.org/10.1091/mbc.10.5.1653
- Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol 1999; 9:126-35; PMID:10021385; https://doi.org/10.1016/S0960-9822(99)80066-9
- Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 2001; 154:499-509; PMID:11489914; https://doi.org/10.1083/jcb.200105084
- Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol 2007; 179:1095-103; PMID:18070915; https://doi.org/10.1083/jcb.200710058
- Broome HJ, Hebert MD. Coilin displays differential affinity for specific RNAs in vivo and is linked to telomerase RNA biogenesis. J Mol Biol 2013; 425:713-24; PMID:23274112; https://doi.org/10.1016/j.jmb.2012.12.014
- Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler P, Neugebauer KM. Global identification of coilin binding partners reveals hundreds of small non-coding RNAs that traffic through Cajal bodies. Mol Cell 2014; 56:389-99; PMID:25514182; https://doi.org/10.1016/j.molcel.2014.10.004
- Matera AG. Of coiled bodies, gems, and salmon. J Cell Biochem 1998; 70:181-92; PMID:9671224; https://doi.org/10.1002/(SICI)1097-4644(19980801)70:2%3c181::AID-JCB4%3e3.0.CO;2-K
- Smith KP, Lawrence JB. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol Biol Cell 2000; 11:2987-98; PMID:10982395; https://doi.org/10.1091/mbc.11.9.2987
- Takata H, Nishijima H, Maeshima K, Shibahara K. The integrator complex is required for integrity of Cajal bodies. J Cell Sci 2012; 125:166-75; PMID:22250197; https://doi.org/10.1242/jcs.090837
- Jodoin JN, Sitaram P, Albrecht TR, May SB, Shboul M, Lee E, Reversade B, Wagner EJ, Lee LA. Nuclear-localized Asunder regulates cytoplasmic dynein localization via its role in the integrator complex. Mol Biol Cell 2013; 24:2954-65; PMID:23904267; https://doi.org/10.1091/mbc.E13-05-0254
- Broome HJ, Hebert MD. In vitro RNase and nucleic acid binding activities implicate coilin in U snRNA processing. PLoS One 2012; 7:e36300; PMID:22558428; https://doi.org/10.1371/journal.pone.0036300
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonné R, et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 2004; 16:777-87; PMID:15574332; https://doi.org/10.1016/j.molcel.2004.11.013
- Massenet S, Pellizzoni L, Paushkin S, Mattaj IW, Dreyfuss G. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol Cell Biol 2002; 22:6533-41; PMID:12192051; https://doi.org/10.1128/MCB.22.18.6533-6541.2002
- Suzuki T, Izumi H, Ohno M. Cajal body surveillance of U snRNA export complex assembly. J Cell Biol 2010; 190:603-12; PMID:20733056; https://doi.org/10.1083/jcb.201004109
- Sleeman JE, Lamond AI. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol 1999; 9:1065-74; PMID:10531003; https://doi.org/10.1016/S0960-9822(99)80475-8
- Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schümperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 2005; 114:155-66; PMID:16003501; https://doi.org/10.1007/s00412-005-0003-y
- Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev 2001; 15:2720-9; PMID:11641277; https://doi.org/10.1101/gad.908401
- Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. Embo J 2003; 22:1878-88; PMID:12682020; https://doi.org/10.1093/emboj/cdg187
- Deryusheva S, Gall JG. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol Biol Cell 2009; 20:5250-9; PMID:19846657; https://doi.org/10.1091/mbc.E09-09-0777
- Tanackovic G, Kramer A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol Biol Cell 2005; 16:1366-77; PMID:15647371; https://doi.org/10.1091/mbc.E04-11-1034
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 2009; 36:593-608; PMID:19941820; https://doi.org/10.1016/j.molcel.2009.09.040
- Schultz A, Nottrott S, Hartmuth K, Luhrmann R. RNA structural requirements for the association of the spliceosomal hPrp31 protein with the U4 and U4atac small nuclear ribonucleoproteins. J Biol Chem 2006; 281:28278-86; PMID:16857676; https://doi.org/10.1074/jbc.M603350200
- Liu S, Li P, Dybkov O, Nottrott S, Hartmuth K, Luhrmann R, Carlomagno T, Wahl MC. Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP. Science 2007; 316:115-20; PMID:17412961; https://doi.org/10.1126/science.1137924
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 2002; 298:2205-8; PMID:12411573; https://doi.org/10.1126/science.1077783
- Stanek D, Pridalova-Hnilicova J, Novotny I, Huranova M, Blazikova M, Wen X, Sapra AK, Neugebauer KM. Spliceosomal small nuclear ribonucleoprotein particles repeatedly cycle through Cajal bodies. Mol Biol Cell 2008; 19:2534-43; PMID:18367544; https://doi.org/10.1091/mbc.E07-12-1259
- Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 1998; 95:615-24; PMID:9845364; https://doi.org/10.1016/S0092-8674(00)81632-3
- Vanacova S, Stefl R. The exosome and RNA quality control in the nucleus. EMBO Rep 2007; 8:651-7; PMID:17603538; https://doi.org/10.1038/sj.embor.7401005
- Hrossova D, Sikorsky T, Potesil D, Bartosovic M, Pasulka J, Zdrahal Z, Stefl R, Vanacova S. RBM7 subunit of the NEXT complex binds U-rich sequences and targets 3′-end extended forms of snRNAs. Nucleic Acids Res 2015; 43:4236-48; PMID:25852104; https://doi.org/10.1093/nar/gkv240
- Shukla S, Parker R. Quality control of assembly-defective U1 snRNAs by decapping and 5′-to-3′ exonucleolytic digestion. Proc Natl Acad Sci U S A 2014; 111:E3277-86; PMID:25071210; https://doi.org/10.1073/pnas.1412614111
- Wersig C, Guddat U, Pieler T, Bindereif A. Assembly and nuclear transport of the U4 and U4/U6 snRNPs. Exp Cell Res 1992; 199:373-7; PMID:1371962; https://doi.org/10.1016/0014-4827(92)90447-G
- Fischer U, Sumpter V, Sekine M, Satoh T, Luhrmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. EMBO J 1993; 12:573-83; PMID:7679989
- Fischer U, Heinrich J, van Zee K, Fanning E, Luhrmann R. Nuclear transport of U1 snRNP in somatic cells: differences in signal requirement compared with Xenopus laevis oocytes. J Cell Biol 1994; 125:971-80; PMID:8195300; https://doi.org/10.1083/jcb.125.5.971
- Stanek D, Neugebauer KM. The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma 2006; 115:343-54; PMID:16575476; https://doi.org/10.1007/s00412-006-0056-6
- Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 2001; 114:4407-19; PMID:11792806
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science 2008; 322:1713-7; PMID:18948503; https://doi.org/10.1126/science.1165216
- Makarov V, Rakitina D, Protopopova A, Yaminsky I, Arutiunian A, Love AJ, Taliansky M, Kalinina N. Plant coilin: structural characteristics and RNA-binding properties. PLoS One 2013; 8:e53571; PMID:23320094; https://doi.org/10.1371/journal.pone.0053571
- Tatomer DC, Terzo E, Curry KP, Salzler H, Sabath I, Zapotoczny G, McKay DJ, Dominski Z, Marzluff WF, Duronio RJ. Concentrating pre-mRNA processing factors in the histone locus body facilitates efficient histone mRNA biogenesis. J Cell Biol 2016; 213:557-70; PMID:27241916; https://doi.org/10.1083/jcb.201504043
- Novotny I, Blazikova M, Stanek D, Herman P, Malinsky J. In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol Biol Cell 2011; 22:513-23; PMID:21177826; https://doi.org/10.1091/mbc.E10-07-0560
- Sawyer IA, Dundr M. Nuclear bodies: Built to boost. J Cell Biol 2016; 213:509-11; PMID:27241912; https://doi.org/10.1083/jcb.201605049
