ABSTRACT
Survival Motor Neuron (SMN) protein localizes to both the nucleus and the cytoplasm. Cytoplasmic SMN is diffusely localized in large oligomeric complexes with core member proteins, called Gemins. Biochemical and cell biological studies have demonstrated that the SMN complex is required for the cytoplasmic assembly and nuclear transport of Sm-class ribonucleoproteins (RNPs). Nuclear SMN accumulates with spliceosomal small nuclear (sn)RNPs in Cajal bodies, sub-domains involved in multiple facets of snRNP maturation. Thus, the SMN complex forms stable associations with both nuclear and cytoplasmic snRNPs, and plays a critical role in their biogenesis. In this review, we focus on potential functions of the nuclear SMN complex, with particular emphasis on its role within the Cajal body.
Background
Cajal bodies and their components
Cajal bodies were first discovered over one hundred years ago by Santiago Ramon y Cajal.Citation1,2 He adapted Golgi's silver staining technique to identify a variety of structures within mammalian nuclei, one of which he termed the nucleolar “accessory body.” These structures were ‘rediscovered’ over the ensuing decades as they were studied in increasing detail using more advanced microscopy techniques.Citation3 However, it was not until the 1990s that a molecular characterization of Cajal bodies began.Citation4-7 Since that time, the number of known cellular components that are enriched in the Cajal body has been growing.Citation3,8-10 These components include factors important for the maturation and function of Sm-class small nuclear ribonucleoproteins (snRNPs), core particles of the spliceosome. The colocalization of Cajal bodies with relevant small nuclear RNA (snRNA) gene clusters, RNAs that guide post-transcriptional modification of snRNPs, and snRNP specific proteins provide important clues regarding the functions of Cajal bodies.Citation11-14 Additional studies in mouse embryonic fibroblasts revealed that a primary protein constituent of these structures, called coilin, was responsible for recruitment of another important component, the survival motor neuron (SMN) protein.Citation15,16
SMN and spinal muscular atrophy (SMA)
Reduced levels of SMN protein cause SMA,Citation17 whereas complete loss of Smn expression is embryonic lethal.Citation18 SMA is a common neuromuscular disorder, recognized as the most prevalent genetic cause of early childhood mortality.Citation19 Clinically, SMA patients show progressive muscle weakness of proximal muscle groups, ultimately leading to paralysis; death is typically caused by progressive and restrictive respiratory failure. Patients with the most severe (and most common) form of SMA become symptomatic in the first 6 months of life and rarely live past 2 y.Citation20-24
SMA typically results from homozygous deletion of survival motor neuron 1 (SMN1) gene; however, a small fraction of SMA patients have lost one copy of SMN1 and the remaining copy contains a point mutation.Citation25 Decreased levels of the SMN protein correlate with the phenotypic severity of SMA. Since the onset of symptoms and their severity can vary, SMA has been historically classified into 3 subtypes. More recently, clinicians have recognized that SMA is better characterized as a continuous spectrum disorder, ranging from severe (prenatal onset) to nearly asymptomatic.Citation20 While the genetic etiology of the disease is well-established, the molecular role of SMN in the disease is largely unknown and is the topic of many reviews.Citation25-34
SMN and cytoplasmic snRNP assembly
The best-characterized function for the SMN protein, which is expressed in all tissues of metazoan organisms, is in the cytoplasmic assembly of Sm-class snRNPs, core particles of the spliceosome.Citation34-37 The Sm-class snRNPs consist of uridine-rich snRNAs (e.g. U1, U2, U4, U5), several specific proteins that are unique to each snRNA, and a set of 7 common Sm proteins (B/B’, D1, D2, D3, E, F, and G). Sm-class snRNAs are transcribed by RNA polymerase II as precursors that contain additional nucleotides at the 3′ end and a monomethylated m7GpppG (m7G) cap structure at the 5′ end. The pre-snRNA transcripts are then exported from the nucleus by a distinct set of factors that includes the cap-binding complex (CBP80 and CBP20), the snRNA-specific export adaptor phosphorylated adaptor RNA export (PHAX), and arsenite resistance 2 (ARS2).Citation38 These proteins form a link between the 5′ cap and the export receptor chromosome region maintenance 1 (CRM1/Exportin1), which interacts with nuclear pore proteins to promote export.Citation39 Various reviews focus on this function of SMN.Citation34,36,37
Once the pre-snRNAs translocate to the cytoplasm, the snRNA nuclear export complex dissociates.Citation40,41 The SMN complex, which includes SMN and several tightly associated proteins, collectively called Gemins binds newly exported snRNAs.Citation42-46 The SMN protein complex regulates the entire cytoplasmic phase of the snRNP cycle including Sm core assembly, trimethylguanosine (TMG) cap formation, and snurportin1 binding to the TMG cap structure.
The SMN complex combines newly exported snRNAs with a set of Sm proteins to form a 7-membered ring around a binding site that is present within each of the Sm-class snRNAs.Citation47,48 The Sm proteins are delivered to the SMN complex via the activity of the PRMT5 complex, which methylates C-terminal arginine residues within SmB, SmD1, and SmD3Citation42,49 and then chaperones delivery of partially assembled Sm subcomplexes.Citation50,51 Gemin5, a component of the SMN complex, is thought to be the factor responsible for recognition of Sm-class snRNAs.Citation52 Thus, the SMN complex is thought to provide specificity, to avoid assembly of Sm cores onto non-target RNAs,Citation43,52 and to accelerate formation of the final product from kinetically trapped intermediates.Citation51 Assembly of the Sm core not only stabilizes the snRNA by protecting it from nucleases, but also is required for the downstream RNA-processing steps.
Following Sm-core assembly, an RNA methyltransferase called trimethylguanosine synthase (Tgs1) is recruited to the m7G cap, whereupon the RNA is hypermethylated to form a 2,2,7-trimethylguanosine (TMG) cap structure.Citation53 A properly assembled Sm core is required for cap hypermethylation and 3′-end maturation.Citation54-56 Tgs1 directly interacts with SMN both in vivo and in vitro.Citation46 Furthermore, the SMN complex does not immediately dissociate from the RNA after Sm-core assembly, suggesting that SMN may even recruit Tgs1 to the complex. Subsequently, the RNP is imported back into the nucleus by specific snRNP import factors.Citation57 Once in the nucleus the RNPs undergo additional assembly and maturation steps. provides a summary of the major events in U snRNP biogenesis.
Figure 1. (A) Sm-class snRNAs are transcribed by RNA polymerase II as precursors that contain additional nucleotides at the 3′ end and a monomethylated m7GpppG (m7G) cap structure at the 5′ end. The pre-snRNA transcripts are then exported from the nucleus, often passing through the Cajal body. Once the pre-snRNAs translocate to the cytoplasm, the snRNA nuclear export complex dissociates. The SMN complex combines newly exported snRNAs with a set of Sm in a process known as Sm core assembly. Then, the snRNA is hypermethylated to form a 2,2,7-trimethylguanosine (TMG) cap structure in the pre-import complex. Finally, the RNP is imported back into the nucleus by specific snRNP import factors shown in B. Once in the nucleus, the partially assembled RNPs can transit through Cajal bodies where they undergo additional assembly and maturation steps. (B) The adaptor for the TMG cap is called snurportin1 (SPN1); this protein binds directly to the TMG cap and significantly improves the kinetics of snRNP import. SMN interacts with both SPN1 and Importin β. SMN functions to bridge the gap between the Sm core and the import machinery. These proteins, together with the snRNA and associated snRNP proteins, constitute the pre-import RNP complex.
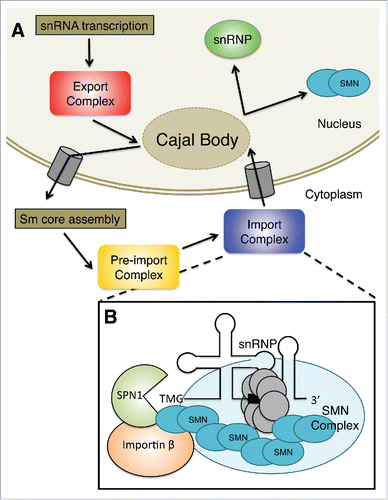
The SMN complex as a nuclear import adaptor
Nuclear transport of snRNPs involves the use of a bipartite nuclear localization signal (NLS), comprised of the TMG cap and the Sm core.Citation58-60 In vitro, these NLS pathways are independent, enlisting discrete import adaptersCitation61,62 but sharing a common import receptor, importin β.Citation63 In vivo, the situation likely involves the synergistic use of both pathways, although additional studies are required in order to clarify this issue. In any event, the adaptor for the TMG cap is called snurportin1 (SPN1); this protein binds directly to the TMG cap and significantly improves the kinetics of snRNP import.Citation61,64,65 At the time, relatively little was known about the factor that mediates the interaction between the Sm core and importin β. Previous studies had indicated that the Sm core adaptor is a cytosolic factor that binds to Sm proteins and importin β.Citation63 Subsequently, SMN was shown to directly interact with these proteins.Citation45,66-68 Furthermore, co-fractionation studies with HeLa cytosolic extracts demonstrated that SMN, SPN1 and importin β were present in a novel, pre-import RNP complex.Citation45,62 Along with several other hints in the literature,Citation32,44,69 these findings suggested that SMN functions to bridge the gap between the Sm core and the import machinery.
Functional studies of this pre-import RNP complex were carried out using digitonin-permeabilised HeLa cells. The results demonstrated that import of SMN and splicing snRNPs are coupled in somatic cells.Citation62,65 Two lines of evidence substantiated this conclusion. First, purified SMN complex was required for in vitro snRNP import and vice versa. Second, an excess of snRNPs significantly improved SMN import kinetics; the converse also held true. Notably, the SMN complex (but not SMN alone) was sufficient to restore snRNP import defects caused by depletion of SMN from the reconstituted cytosol. Furthermore, mutations in the YG Box self-oligomerization domain of SMN disrupted nuclear import,Citation62 suggesting the importance of oligomeric SMN in the active import complex. Collectively, these data indicated that additional SMN complex members (e.g., Gemins) are required for cap-independent snRNP import.Citation62
SMN import regulation
A model of the putative U snRNP import complex is shown in . The precise composition of the Gemin proteins and the overall stoichiometry of importin β within this complex is completely unknown. Given the independence of the 2 import pathways in vitro,Citation61,62,65 the question remains whether the Sm-core and TMG cap import adaptors act synergistically in vivo. Moreover, the SMN complex might only play a role in import of newly-assembled snRNPs. In such a scenario, post-mitotic snRNPs would strictly utilize the TMG cap-dependent pathway. Future studies will be needed to clarify this important issue.
Of the various factors that control SMN and U snRNP nuclear import, very little is known. However, a zinc finger protein called ZPR1 was reported to participate in this process.Citation70 ZPR1 is an essential, highly-conserved protein in eukaryotes.Citation71,72 The protein was originally identified as a factor that binds to the cytoplasmic tyrosine kinase domains of epidermal growth factor (EGF)-like receptors in the absence of mitogens.Citation73 Ligand binding and autophosphorylation of the receptor causes ZPR1 to be released from the receptor and subsequently accumulate in the nucleus.Citation74
The C-terminal tail of ZPR1 interacts indirectly with SMN, forming cytoplasmic snRNP-containing complexes.Citation70 Given that ZPR1, SMN and SPN can be detected in a pre-import RNP complex,Citation45 and that anti-ZPR1 antibodies co-deplete Sm proteins from cytoplasmic lysates,Citation62 it appears that ZPR1 is involved in snRNP biogenesis. Although ZPR1 forms complexes with U snRNPs, it is not directly required for SMN nuclear transport in vitro.Citation62 However, these complexes are disrupted when there are low levels of SMN, and both ZPR1 and SMN fail to accumulate in nuclear bodies. Moreover, depletion of ZPR1 results in disruption of Cajal bodies and defective nuclear localization of SMN.Citation75 Overexpression of ZPR1 in SMA patient fibroblasts restores the subnuclear accumulation of SMN and increases overall levels of SMN protein.Citation76 Because ZPR1 also interacts with EGF-type receptor proteins that regulate cellular proliferation, the most likely scenario places ZPR1 in a signaling cascade upstream of SMN and U snRNP nuclear import.
Nuclear functions of the SMN complex
Following import and localization of newly assembled snRNPs to the Cajal body, coilin may function to disrupt the SMN-snRNP complex and facilitate higher-order snRNP formation.Citation16,45,77,78 SMN is thought to dissociate from snRNPs soon after their import, as the protein does not co-purify with mature snRNP mono-particles.Citation79,80 Once released from the SMN complex, the newly assembled snRNP is then free to diffuse throughout the interchromatin space. The fate of the SMN complex following snRNP release in the Cajal body is mostly unknown. In most cell types, the nuclear fraction of the SMN complex localizes primarily within Cajal bodies. However, a small fraction of nuclear SMN protein has been found to co-immunoprecipitate with spliceosomal subcomplexes.Citation81,82 SMN also accumulates in distinct nuclear substructures called Gemini bodies, or Gems.Citation83 Cajal bodies contain a plethora of RNAs and their associated proteins, but components of Gems have thus far been limited to constituents of the SMN complex.Citation9,83 A U1 snRNP component, U1-70K, also localizes to gems, but its interaction with SMN is thought to be snRNP-independent.Citation84 Given its recently described role in human U1 snRNP assembly, U1-70K protein might be considered as an auxiliary member of the SMN complex.Citation85 Notably, a decrease in SMN protein levels (and thus an increase in SMA severity) is correlated with the loss of SMN nuclear foci.Citation24,86 Thus, the nuclear functions of SMN may well turn out to be important for understanding disease etiology.
Inter- and intra-molecular interactions within the Cajal body
Photobleaching studies in living cells have shown that there are distinct kinetic groups of Cajal body components.Citation87,88 The proteins with the longest Cajal body residence times are coilin, SMN, Gemin3 and Tgs1. These factors typically reside in Cajal bodies on the order of minutes, whereas other groups of proteins have much shorter residence times.Citation87 However, despite the direct physical interaction of coilin and SMN, Cajal bodies and Gems are kinetically autonomous compartments.Citation87 The basis for this kinetic autonomy is likely due to the ability of SMN and coilin to maintain homo-typic interactions (coilin-coilin and SMN-SMN) within the separated nuclear bodies (). Hetero-typic interactions are therefore likely to regulate overall composition of a given nuclear body.Citation89 Indeed, fluorescence resonance energy transfer (FRET) by acceptor photobleaching experiments in living nuclei showed that coilin and SMN interact with themselves and with each other inside the Cajal body.Citation87
Figure 2. (A) In SMN(+) Cajal bodies, coilin and SMN physically interact. These heterotypic interactions are likely to regulate the overall composition of the nuclear body. Cajal bodies and Gems are kinetically autonomous compartments as illustrated by the Gem and SMN(−) Cajal body. The basis for this kinetic autonomy is likely due to the ability of SMN and coilin to maintain homotypic interactions (coilin-coilin and SMN-SMN) within the separated nuclear bodies. (B) SMN-Gem2 exists as a stable heterodimer that, for purposes of discussing higher order oligomerization, has been shown as a single structural unit (labeled “SMN”). At varying concentrations, this complex exists in an equilibrium mixture containing dimers, tetramers (dimer of dimers), and octamers of SMN-Gem2. Human SMN-Gem2 forms dimers to octamers and possibly even larger complexes. Octamers appear to form via self-association of tetramers. (C) The concentration of SMN within certain subcellular compartments (e.g. Cajal bodies, snRNA gene clusters or cytoplasmic stress granules) is significantly greater than that of the surrounding regions of the cell. High concentrations of SMN are also thought to be found in mRNP transport particles along axons as well as at neuromuscular junctions. If there are unique functions of SMN that can only be performed by higher-order oligomers it is likely that such functions are being carried out in specific cellular locales that contain high concentrations of the SMN complex, as illustrated by the bright green circles.
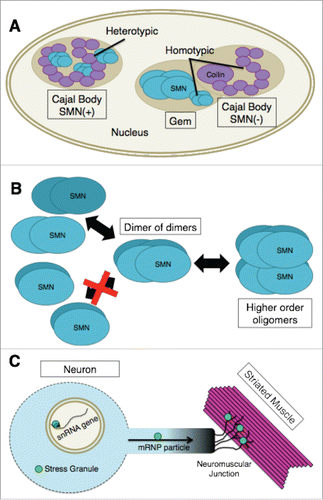
Cajal body homeostasis requires ongoing snRNP biogenesis, as perturbation of SMN or SPN1 function results in disassembly of Cajal bodies and relocalization of coilin to nucleoli.Citation90 Similarly, depletion of other factors involved in small RNP biogenesis, such as Tgs1, WDR79/WRAP53, PHAX, INTS4 and USPL1 also cause Cajal bodies to disassemble.Citation91-95 In addition to overall protein levels, post-translational modifications (PTMs) of resident proteins, including SMN and coilin, are almost certainly important for establishing and maintaining inter- and intra-molecular interactions within Cajal bodies.Citation96-100 Disruption of these PTMs can result in mislocalization of SMN and alterations in Cajal body integrity. These and other factors will need to be addressed by future studies.
Oligomeric properties of SMN complexes
X-ray crystallographic studies of the SMN C-terminal region show that the YG box forms a helical structure whose dimerization is driven by a network of hydrophobic interactions similar to those found in glycine zipper domains of certain transmembrane channel proteins.Citation101,102 The core of this helical domain contains a highly conserved YxxxYxxxY motif.Citation101 Notably, phylogenetic analysis reveals that this motif can be extended on both ends to form a longer LxxxLxxxYxxxYxxxYxxxL helix (A.G.M., unpublished observations). Structural analysis of the SMN Tudor domain has also been carried out,Citation35,103 revealing the existence of an “aromatic cage” of β-sheets that mediates recognition of dimethylarginine residues present on Sm proteins. Future studies will be required to determine how these 2 regions, as well as the α-helical Gemin2 (Gem2) binding domain at the N-terminusCitation50,104 all fit together in space to carry out SMN's various functions.Citation105
The functional importance of the SMN C-terminal domain has been well-documented. More than half of the known SMA patient missense mutations cluster within the YG box.Citation25,106,107 Furthermore, mutations that completely disrupt SMN's ability to self-oligomerize display severe phenotypes in human SMA patients as well as in animal models.Citation86,108-113 Despite this strong correlation, the composition and stoichiometry of the various complexes formed by SMN are not well understood. In vitro, SMN-Gem2 exists as a stable heterodimer that, for purposes of discussing higher order oligomerization, can be considered as a single structural unit.Citation66,114 As the concentration increases, this unit exists in an equilibrium mixture containing dimers, tetramers and octamers of SMN-Gem2.Citation102 Importantly, the SMN-Gem2 complex does not multimerize by forming symmetric bundles.Citation102 Instead, SMN tetramers are formed by a dimer of dimers (). In the fission yeast system, dimers and tetramers are the only species observed.Citation102 Human SMN-Gem2 forms dimers to octamers and possibly even larger complexes (). Octamers appear to form via self-association of tetramers, although the existence of a hexameric SMN complex cannot be ruled out.Citation102 In vivo, human SMN-Gem2 cosediments with Gemins3–8,Citation115 however, the relative stoichiometries of these proteins are completely unknown.
Studies in fission yeast suggest that the SMN dimer [i.e. (SMN-Gem2)2] is the basal functional unit. Using in vivo estimates of SMN protein concentration,Citation116 Van Duyne and colleagues calculated that the concentration of the dimer (∼15 nM) lies far, far below the dissociation constant of SMN tetramers (∼1 uM).Citation102 Immunofluorescence studies showed that yeast SMN is diffusely distributed throughout the nucleus and cytoplasm, with no discernible foci.Citation117 Thus, the SMN dimer is the most abundant species in vivo, and is presumed to be functionally active in snRNP biogenesis.Citation102
Although changes in the composition and organization of the SMN complex are likely to be regulated by various PTMs, the studies of yeast SMN have important consequences for metazoans, particularly when it comes to the oligomerization status of the protein. For example, the concentration of SMN within nuclear Cajal bodies or cytoplasmic stress granules is significantly greater than that of the surrounding nucleoplasm or cytosol, respectively. Hence, if there are unique functions of SMN that can only be performed by higher-order oligomers, then it is therefore likely that such functions are being carried out in specific cellular locales that contain high concentrations of the SMN complex ().
SMN and Cajal Body structure
As mentioned above, loss of SMN is detrimental to Cajal body integrity.Citation90,91,95,100 Cajal bodies consistently associate with specific loci on multiple chromosomes, many of which include snRNA gene arrays, snoRNAs, as well as other small U RNA and histone gene clusters.Citation11,12,118-120 Following siRNA knockdown of essential Cajal body components, WDR79/WRAP53 or USPL1, these chromosomal regions were no longer clustered, and the expression of many of the associated small U RNA loci were significantly reduced. Thus, disruption of Cajal bodies leads to a loss of genome conformation and this may lead to inefficient expression of snRNAs. On the other hand, factors like USPL1 have been shown to participate directly in processing of pre-snRNA transcripts,Citation94 so it is difficult to distinguish between the function of the protein and that of the nuclear body.
While there is not a direct connection between SMN and genome conformation disruption, SMN is important for maintaining the structure of the Cajal body. Previously, low levels of SMN have been linked to widespread changes in expression patterns,Citation121-128 but it is currently an open question as to how these changes might be linked to Cajal bodies. In addition, the mechanism by which these conformational changes in the nucleus affect the rest of the cell and which cellular components and processes are most severely affected has yet to be studied.
SMN and transcriptional regulation
While SMN is definitively important for maintaining the structure of the Cajal body, there is emerging evidence suggesting the involvement of SMN in other processes while in this nuclear body. One potential role for SMN is in RNA Polymerase II (RNAPII) termination. Zhao et al.Citation129 recently showed that SMN binds to the symmetric dimethylation of arginine-1810 (R1810me2s) of the RNAPII C-terminal domain (CTD). Asymmetric dimethylation of this residue leads to reduced expression of snRNAs and snoRNAs when recognized by TDRD3.Citation130
In addition to the interaction of SMN with R1810me2s, SMN interacts with senataxin (SETX) (). Binding of SMN at the R1810me2s stabilizes interaction of SETX with the CTD. SETX is a RNA/DNA helicase that is responsible for unwinding R-loops around transcription termination sites.Citation131 This unwinding allows the 5′-to-3′ exonuclease XRN2 to be recruited, thus terminating transcription.Citation132 Mutation of arginine to alanine at this residue and knockdown of SMN cause accumulation of RNAPII at termination regions of active genes across the genome (). A CRISPR knockout of SMN led to increased R-loops in termination regions and γ-H2AX, a marker of DNA damage, accumulates at these sites.
Figure 3. (A) The binding of SMN to R1810me2s on RNAPII CTD is shown to stabilize the binding of an RNA/DNA helicase, SETX, to the CTD. SETX prevents R-loops at termination sites and allows the exonuclease XRN2 to terminate the transcript, and RNAPII is released. The prevention of R-loops by SETX also allows the spliceosome to access the RNA and modify the transcript through splicing. (B) When SMN is reduced, or the R1810 residue is lost, this results in reduced recruitment of SETX and an increase in R-loops and DNA damage. RNAPII also accumulates at the transcription termination site.
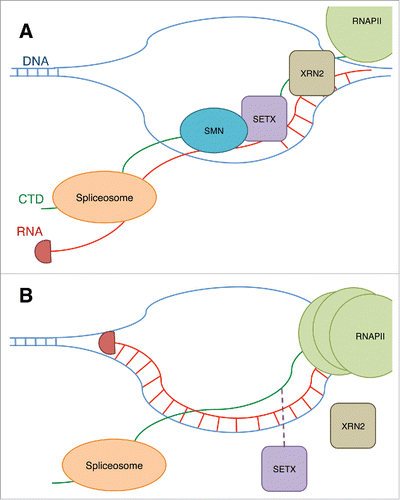
R-loops have been shown to both stabilize and destabilize the genome depending on the context.Citation133,134 The characteristics and mechanisms that determine whether an R-loop is beneficial or detrimental to the genome are unknown. SMN may contribute to this determination through its interaction with SETX.
If SMN does indeed affect RNAPII termination through its interaction with senataxin, it's possible that its structural and transcriptional functions coincide adjacent to the Cajal body. It has not been shown that this active role of SMN is associated with the Cajal body, but this would provide a parsimonious model for efficient transcript processing of chromosomal regions associated with the Cajal body, including snRNAs gene arrays. This provides a potential mechanism for how these genes are expressed and regulated in a finely tuned manner. It has been shown that snRNAs with extended 3′ regions associate with Cajal bodiesCitation135 and that snRNAs accumulate in the Cajal body before they are exported to the cytoplasm.Citation136 Conveniently, snRNA export proteins PHAX and CRM1 are also concentrated at Cajal bodies.Citation13,137 Inhibition of PHAX leads to accumulation of snRNAs in the Cajal body.Citation136 The return of snRNPs to their initial site of biogenesis is also thought to be a mechanism for feedback regulation of snRNA transcription and gene dosage compensation.Citation138 It is clear that the Cajal body is used as a hub for snRNAs as they transition from transcription to export, but whether this nuclear body is also directly associated with the active transcriptional regulation of snRNAs remains to be studied.
Conclusions and perspectives
Following its discovery as the product of the SMA-determining gene,Citation17 the SMN protein was quickly shown to accumulate in nuclear Cajal bodies.Citation67,139,140 Understanding the function of the SMN complex within the Cajal body has been a much slower process. As discussed in the preceding paragraphs, the nuclear fraction of SMN may well participate in both early and late events in the biogenesis of spliceosomal snRNPs. Particularly appealing is the idea that SMN might also chaperone assembly of other types of RNPs, wherever they may happen to congregate inside the cell. What sets the Cajal body bound fraction of SMN apart from the nucleoplasmic SMN complex? The simple answer is that the apparent protein concentration difference between the 2 subcellular compartments determines the oligomerization potential of nucleoplasmic SMN. Is there a specific molecular function that is performed only by the larger SMN multimers? If so, is a similar function carried out by SMN in stress granules, where the cytoplasmic fraction of SMN is most concentrated?Citation141-143 Additional studies will be needed to answer these questions.
One possibility is that the larger SMN oligomers might serve as intracellular signaling hubs. Interestingly, SMN depletion is associated with innate immune and stress signaling.Citation144-146 Moreover, hypomorphic SMN mutations that cause milder forms of SMA in humans and significant viability defects in SMA model flies are neither associated with defects in pre-mRNA splicing nor spliceosomal snRNP biogenesis.Citation147 Although potential defects in snRNP biogenesis at specific developmental time points are possible, these findings indicate that snRNP-dependent RNA processing changes are unlikely to be primary drivers of SMA pathology. Rather, SMN-dependent activation of innate immune and other types of stress signaling pathways may be more important to neuromuscular pathophysiology than previously envisioned.Citation147,148,149
As the storehouse of nuclear SMN protein, the Cajal body likely plays an important role in its nuclear activities. Learning more about mechanisms of SMN nuclear-cytoplasmic trafficking and its various functions within the Cajal body should help elucidate SMN's role in organismal development and SMA etiology. The mysteries surrounding the connection between SMN and Cajal bodies, and the implications of this relationship for human disease are a perfect example of an observation that Cajal himself made in Advice for a Young Investigator, saying that, “each problem solved stimulates an infinite number of new questions, and that today's discovery contains the seed of tomorrow's [idea].”
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Cajal SR. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol 1903; 2:129-221
- Cajal SR. El núcleo de las células piramidales del cerebro humano y de algunos mamíferos. Trab Lab Invest Biol 1910; 8:27-62
- Gall JG. C AJAL B ODIES: The First 100 Years. Annu Rev Cell Dev Biol 2000; 16:273-300; PMID:11031238
- Raska I, Ochs RL, Andrade LEC, Chan EKL, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Struct Biol 1990; 104:120-7; PMID:2088441; https://doi.org/10.1016/1047-8477(90)90066-L
- Andrade LE, Tan EM, Chan EK. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci U S A 1993; 90:1947-51; PMID:8446613; https://doi.org/10.1073/pnas.90.5.1947
- Tuma RS, Stolk JA, Roth MB. Identification and characterization of a sphere organelle protein. J Cell Biol 1993; 122:767-73; PMID:8349728; https://doi.org/10.1083/jcb.122.4.767
- Chan EKL, Takano S, Andrade LEC, Hamel JC, Matera AG. Structure, expression and chromosomal localization of human p80-coilin gene. Nucleic Acids Res 1994; 22:4462-9; PMID:7971277; https://doi.org/10.1093/nar/22.21.4462
- Matera AG, Shpargel KB. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr Opin Cell Biol 2006; 18:317-24; PMID:16632338; https://doi.org/10.1016/j.ceb.2006.03.005
- Staněk D, Neugebauer KM. The Cajal body: A meeting place for spliceosomal snRNPs in the nuclear maze. Chromosoma 2006; 115:343-54; PMID:16575476; https://doi.org/10.1007/s00412-006-0056-6
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 2009; 17:639-47; PMID:19922869; https://doi.org/10.1016/j.devcel.2009.10.017
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A 1995; 92:5915-9; PMID:7597053; https://doi.org/10.1073/pnas.92.13.5915
- Frey MR, Bailey AD, Weiner AM, Matera AG. Association of snRNA genes with coiled bodies is mediated by nascent snRNA transcripts. Curr Biol 1999; 9:126-35; PMID:10021385; https://doi.org/10.1016/S0960-9822(99)80066-9
- Frey MR, Matera AG. RNA-mediated interaction of Cajal bodies and U2 snRNA genes. J Cell Biol 2001; 154:499-509; PMID:11489914; https://doi.org/10.1083/jcb.200105084
- Jády BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 2003; 22:1878-88; https://doi.org/10.1093/emboj/cdg187
- Tucker KE. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 2001; 154:293-308; PMID:11470819; https://doi.org/10.1083/jcb.200104083
- Hebert MD, Szymczyk PW, Shpargel KB, Gregory Matera A. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev 2001; 15:2720-9; PMID:11641277; https://doi.org/10.1101/gad.908401
- Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80:155-65; PMID:7813012; https://doi.org/10.1016/0092-8674(95)90460-3
- Schrank B, Götz R, Gunnersen JM, Ure JM, Toyka KV, Smith AG, Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc Natl Acad Sci U S A 1997; 94:9920-5; PMID:9275227; https://doi.org/10.1073/pnas.94.18.9920
- Pearn J. Classification of spinal muscular atrophies. Lancet 1980; 315:919-22; https://doi.org/10.1016/S0140-6736(80)90847-8
- Tiziano FD, Melki J, Simard LR. Solving the puzzle of spinal muscular atrophy: What are the missing pieces? Am J Med Genet A 2013; 161A(11):2836-45; PMID:24124019
- Monani UR, De Vivo DC. Neurodegeneration in spinal muscular atrophy: from disease phenotype and animal models to therapeutic strategies and beyond. Future Neurol 2014; 9:49-65; PMID:24648831; https://doi.org/10.2217/fnl.13.58
- Lewelt A, Newcomb TM, Swoboda KJ. New therapeutic approaches to spinal muscular atrophy. Curr Neurol Neurosci Rep 2012; 12:42-53; PMID:22134788; https://doi.org/10.1007/s11910-011-0240-9
- Prior TW, Russman B. Spinal muscular atrophy. Society 2008; 5:499-506
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AHM. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet 1997; 6:1205-14; PMID:9259265; https://doi.org/10.1093/hmg/6.8.1205
- Burghes AHM, Beattie CE. Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 2009; 10:597-609; PMID:19584893; https://doi.org/10.1038/nrn2670
- Kolb SJ, Kissel JT. Spinal muscular atrophy: A timely review. Arch Neurol 2011; 68:979-84; PMID:21482919; https://doi.org/10.1001/archneurol.2011.74
- Alatorre-Jiménez M, Sánchez-Luna J, González-Renovato E, Sánchez-López A, Sánchez-Luna S, Hernández-Navarro V, Pacheco-Moises F, Ortiz G. Spinal muscular atrophy: review of a child onset disease. Br J Med Med Res 2015; 6:647-60; https://doi.org/10.9734/BJMMR/2015/14251
- Tisdale S, Pellizzoni L. Disease mechanisms and therapeutic approaches in spinal muscular atrophy. J Neurosci 2015; 35:8691-700; PMID:26063904; https://doi.org/10.1523/JNEUROSCI.0417-15.2015
- Ahmad S, Bhatia K, Kannan A, Gangwani L. Molecular mechanisms of neurodegeneration in spinal muscular atrophy. J Exp Neurosci 2016; 10:39-49; PMID:27042141
- Awano T, Kim J, Monani UR. Spinal muscular atrophy: Journeying from bench to bedside. Neurotherapeutics 2014; 11(4):786-95; PMID:24990202
- Fallini C, Bassell GJ, Rossoll W. Spinal muscular atrophy: The role of SMN in axonal mRNA regulation. Brain Res 2012; 1462:81-92; PMID:22330725; https://doi.org/10.1016/j.brainres.2012.01.044
- Sleeman J. Small nuclear RNAs and mRNAs: linking RNA processing and transport to spinal muscular atrophy. Biochem Soc Trans 2013; 41:871-5; PMID:23863147; https://doi.org/10.1042/BST20120016
- Chari A, Paknia E, Fischer U. The role of RNP biogenesis in spinal muscular atrophy. Curr Opin Cell Biol 2009; 21:387-93; PMID:19286363; https://doi.org/10.1016/j.ceb.2009.02.004
- Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 2014; 15:108-21; PMID:24452469; https://doi.org/10.1038/nrm3742
- Tripsianes K, Madl T, Machyna M, Fessas D, Englbrecht C, Fischer U, Neugebauer KM, Sattler M. Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. Nat Struct Mol Biol 2011; 18:1414-20; PMID:22101937; https://doi.org/10.1038/nsmb.2185
- Kolb SJ, Battle DJ, Dreyfuss G. Molecular functions of the SMN complex. J Child Neurol 2007; 22:990-4; PMID:17761654; https://doi.org/10.1177/0883073807305666
- Li DK, Tisdale S, Lotti F, Pellizzoni L. SMN control of RNP assembly: From post-transcriptional gene regulation to motor neuron disease. Semin Cell Dev Biol 2014; 32:22-9; PMID:24769255; https://doi.org/10.1016/j.semcdb.2014.04.026
- Hallais M, Pontvianne F, Andersen PR, Clerici M, Lener D, Benbahouche NEH, Gostan T, Vandermoere F, Robert MC, Cusack S, et al. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat Struct Mol Biol 2013; 20:1358-66; PMID:24270878; https://doi.org/10.1038/nsmb.2720
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997; 90:1051-60; PMID:9323133; https://doi.org/10.1016/S0092-8674(00)80371-2
- Kitao S, Segref A, Kast J, Wilm M, Mattaj IW, Ohno M. A compartmentalized phosphorylation/dephosphorylation system that regulates U snRNA export from the nucleus. Mol Cell Biol 2008; 28:487-97; PMID:17967890; https://doi.org/10.1128/MCB.01189-07
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 2000; 101:187-98; PMID:10786834; https://doi.org/10.1016/S0092-8674(00)80829-6
- Meister G, Bühler D, Pillai R, Lottspeich F, Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat Cell Biol 2001; 3:945-9; PMID:11715014; https://doi.org/10.1038/ncb1101-945
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science 2002; 298:1775-9; PMID:12459587; https://doi.org/10.1126/science.1074962
- Massenet S, Pellizzoni L, Paushkin S, Mattaj IW, Dreyfuss G. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol Cell Biol 2002; 22:6533-41; PMID:12192051; https://doi.org/10.1128/MCB.22.18.6533-6541.2002
- Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin β. Hum Mol Genet 2002; 11:1785-95; PMID:12095920; https://doi.org/10.1093/hmg/11.15.1785
- Mouaikel J, Narayanan U, Verheggen C, Matera AG, Bertrand E, Tazi J, Bordonné R. Interaction between the small‐nuclear‐RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep 2003; 4:616-22; PMID:12776181; https://doi.org/10.1038/sj.embor.embor863
- Kambach C, Walke S, Nagai K. Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr Opin Struct Biol 1999; 9:222-30; PMID:10322216; https://doi.org/10.1016/S0959-440X(99)80032-3
- Will CL, Lührmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol 2001; 13:290-301; PMID:11343899; https://doi.org/10.1016/S0955-0674(00)00211-8
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell 2001; 7:1111-7; PMID:11389857; https://doi.org/10.1016/S1097-2765(01)00244-1
- Grimm C, Chari A, Pelz JP, Kuper J, Kisker C, Diedrichs K, Stark H, Schindelin H, Fischer U. Structural basis of assembly chaperone- mediated snRNP Formation. Mol Cell 2013; 49:692-703; PMID:23333303; https://doi.org/10.1016/j.molcel.2012.12.009
- Chari A, Golas MM, Klingenhäger M, Neuenkirchen N, Sander B, Englbrecht C, Sickmann A, Stark H, Fischer U. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell 2008; 135:497-509; PMID:18984161; https://doi.org/10.1016/j.cell.2008.09.020
- Yong J, Kasim M, Bachorik JL, Wan L, Dreyfuss G. Gemin5 delivers snRNA precursors to the SMN complex for snRNP Biogenesis. Mol Cell 2010; 38:551-62; PMID:20513430; https://doi.org/10.1016/j.molcel.2010.03.014
- Mouaikel J, Verheggen C, Bertrand E, Tazi J, Bordonné R. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol Cell 2002; 9:891-901; PMID:11983179; https://doi.org/10.1016/S1097-2765(02)00484-7
- Mattaj IW. Cap trimethylation of U snRNA is cytoplasmic and dependent on U snRNP protein binding. Cell 1986; 46:905-11; PMID:2944599; https://doi.org/10.1016/0092-8674(86)90072-3
- Neuman de Vegvar HE, Dahlberg JE. Nucleocytoplasmic transport and processing of small nuclear RNA precursors. Mol Cell Biol 1990; 10:3365-75; PMID:2355910; https://doi.org/10.1128/MCB.10.7.3365
- Plessel G, Fischer U, Lührmann R. m3G cap hypermethylation of U1 small nuclear ribonucleoprotein (snRNP) in vitro: evidence that the U1 small nuclear RNA-(guanosine-N2)-methyltransferase is a non-snRNP cytoplasmic protein that requires a binding site on the Sm core domain. Mol Cell Biol 1994; 14:4160-72; PMID:8196654; https://doi.org/10.1128/MCB.14.6.4160
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 1999; 15:607-60; PMID:10611974; https://doi.org/10.1146/annurev.cellbio.15.1.607
- Fischer U, Lührmann R. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science 1990; 249:786-90; PMID:2143847; https://doi.org/10.1126/science.2143847
- Fischer U, Sumpter V, Sekine M, Satoh T, Luhrmann R. Nucleo-cytoplasmic transport of U snRNPs: definition of a nuclear location signal in the Sm core domain that binds a transport receptor independently of the m3G cap. 1993; 12:573-83; PMID:7679989
- Hamm J, Darzynkiewicz E, Tahara SM, Mattaj IW. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell 1990; 62:569-77; PMID:2143105; https://doi.org/10.1016/0092-8674(90)90021-6
- Huber J, Dickmanns A, Lührmann R. The importin-β binding domain of snurportin1 is responsible for the Ran- and energy-independent nuclear import of spliceosomal U snRNPs in vitro. J Cell Biol 2002; 156:467-79; PMID:11815630; https://doi.org/10.1083/jcb.200108114
- Narayanan U, Achsel T, Lührmann R, Matera AG. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol Cell 2004; 16:223-34; PMID:15494309; https://doi.org/10.1016/j.molcel.2004.09.024
- Palacios I, Hetzer M, Adam SA, Mattaj IW. Nuclear import of U snRNPs requires importin β. EMBO J 1997; 16:6783-92; PMID:9362492; https://doi.org/10.1093/emboj/16.22.6783
- Huber J, Cronshagen U, Kadokura M, Wada T, Sekine M, Lu R. Snurportin1, an m 3 G-cap-specific nuclear import receptor with a novel domain structure. EMBO J 1998; 17:4114-26; PMID:9670026; https://doi.org/10.1093/emboj/17.14.4114
- Ospina JK, Gonsalvez GB, Janna B, Darzynkiewicz E, Gerace L, Matera AG. Cross-Talk between Snurportin1 subdomains. Mol Biol Cell 2005; 16:4660-71; PMID:16030253; https://doi.org/10.1091/mbc.E05-04-0316
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP Biogenesis. Cell 1997; 90:1023-9; PMID:9323130; https://doi.org/10.1016/S0092-8674(00)80368-2
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J 1996; 15:3555-65; PMID:8670859
- Liu Q, Fischer U, Wang F, Dreyfuss G. The spinal muscular atrophy disease gene product, SMN, and its associated protein SIP1 are in a complex with spliceosomal snRNP proteins. Cell 1997; 90:1013-21; PMID:9323129; https://doi.org/10.1016/S0092-8674(00)80367-0
- Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci 2001; 114:4407-19; PMID:11792806
- Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat Cell Biol 2001; 3:376-83; PMID:11283611; https://doi.org/10.1038/35070059
- Galcheva-Gargova Z, Gangwani L, Konstantinov KN, Mikrut M, Theroux SJ, Enoch T, Davis RJ. The cytoplasmic zinc finger protein ZPR1 accumulates in the nucleolus of proliferating cells. Mol Biol Cell 1998; 9:2963-71; PMID:9763455; https://doi.org/10.1091/mbc.9.10.2963
- Gangwani L, Mikrut M, Galcheva-Gargova Z, Davis RJ. Interaction of ZPR1 with translation elongation factor-1alpha in proliferating cells. J Cell Biol 1998; 143:1471-84; PMID:9852145; https://doi.org/10.1083/jcb.143.6.1471
- Galcheva-gargova AZ, Konstantinov KN, Wu I, George F, Barrett T, Davis RJ, Galcheva-gargova Z, Konstantinov KN, Klier FG, Barrett T, et al. Binding of zinc finger protein ZPR1 to the epidermal growth factor receptor. Science 1996; 272:1797-802; PMID:8650580; https://doi.org/10.1126/science.272.5269.1797
- Matera AG, Hebert MD. The survival motor neurons protein uses its ZPR for nuclear localization. Nat Cell Biol 2001; 3:E93-4; PMID:11283627; https://doi.org/10.1038/35070157
- Gangwani L, Flavell RA, Davis RJ. ZPR1 is essential for survival and is required for localization of the survival motor neurons (SMN) protein to Cajal bodies. Mol Cell Biol 2005; 25:2744-56; PMID:15767679; https://doi.org/10.1128/MCB.25.7.2744-2756.2005
- Ahmad S, Wang Y, Shaik GM, Burghes AH, Gangwani L. The zinc finger protein ZPR1 is a potential modifier of spinal muscular atrophy. Hum Mol Genet 2012; 21:2745-58; PMID:22422766; https://doi.org/10.1093/hmg/dds102
- Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schmperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 2005; 114:155-66; PMID:16003501; https://doi.org/10.1007/s00412-005-0003-y
- Strzelecka M, Trowitzsch S, Weber G, Lührmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. TL - 17. Nat Struct Mol Biol 2010; 17:403-9; PMID:20357773; https://doi.org/10.1038/nsmb.1783
- Trinkle-Mulcahy L, Boulon S, Lam YW, Urcia R, Boisvert FM, Vandermoere F, Morrice NA, Swift S, Rothbauer U, Leonhardt H, et al. Identifying specific protein interaction partners using quantitative mass spectrometry and bead proteomes. J Cell Biol 2008; 183:223-39; PMID:18936248; https://doi.org/10.1083/jcb.200805092
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Lührmann R, Lu R. Conservation of the protein composition and electron microscopy structure of drosophila melanogaster and human spliceosomal complexes conservation of the protein composition and electron microscopy structure of Drosophila melanogaster and human spliceosom. Mol Cell Biol 2009; 29:281-301; PMID:18981222; https://doi.org/10.1128/MCB.01415-08
- Neubauer G, Gottschalk A, Fabrizio P, Séraphin B, Lührmann R, Mann M. Identification of the proteins of the yeast U1 small nuclear ribonucleoprotein complex by mass spectrometry. Proc Natl Acad Sci U S A 1997; 94:385-90; PMID:9012791; https://doi.org/10.1073/pnas.94.2.385
- Makarov EM, Owen N, Bottrill A, Makarova OV. Functional mammalian spliceosomal complex e contains SMN complex proteins in addition to U1 and U2 snRNPs. Nucleic Acids Res 2012; 40:2639-52; PMID:22110043; https://doi.org/10.1093/nar/gkr1056
- Matera AG. Drosophila Cajal bodies: accessories not included. J Cell Biol 2006; 172:791-3; PMID:16533940; https://doi.org/10.1083/jcb.200602002
- Stejskalová E, Staněk D. Splicing factor U1-70K interacts with the SMN complex and is required for nuclear Gem integrity. J Cell Sci 2014; 127:3909-15; https://doi.org/10.1242/jcs.155838
- So BR, Wan L, Zhang Z, Li P, Babiash E, Duan J, Younis I, Dreyfuss G. A U1 snRNP–specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat Publ Gr 2016; 23:1-7
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 1997; 16:265-9; PMID:9207792; https://doi.org/10.1038/ng0797-265
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol 2004; 164:831-42; PMID:15024031; https://doi.org/10.1083/jcb.200311121
- Sleeman JE, Trinkle-Mulcahy L, Prescott AR, Ogg SC, Lamond AI. Cajal body proteins SMN and Coilin show differential dynamic behaviour in vivo. J Cell Sci 2003; 116:2039-50; PMID:12679382; https://doi.org/10.1242/jcs.00400
- Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell 2002; 3:329-37; PMID:12361597; https://doi.org/10.1016/S1534-5807(02)00222-8
- Shpargel KB, Matera a G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc Natl Acad Sci U S A 2005; 102:17372-7; PMID:16301532; https://doi.org/10.1073/pnas.0508947102
- Girard C, Neel H, Bertrand E, Bordonné R. Depletion of SMN by RNA interference in HeLa cells induces defects in Cajal body formation. Nucleic Acids Res 2006; 34:2925-32; PMID:16738131; https://doi.org/10.1093/nar/gkl374
- Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Söderberg O, Strömblad S, Wiman KG, Farnebo M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol 2010; 8:e1000521; PMID:21072240; https://doi.org/10.1371/journal.pbio.1000521
- Takata H, Nishijima H, Maeshima K, Shibahara KI. The integrator complex is required for integrity of Cajal bodies. J Cell Sci 2012; 125:166-75; PMID:22250197; https://doi.org/10.1242/jcs.090837
- Hutten S, Chachami G, Winter U, Melchior F, Lamond AI. A role for the Cajal-body-associated SUMO isopeptidase USPL1 in snRNA transcription mediated by RNA polymerase II. J Cell Sci 2014; 127:1065-78; PMID:24413172; https://doi.org/10.1242/jcs.141788
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonné R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 2006; 17:3221-31; PMID:16687569; https://doi.org/10.1091/mbc.E06-03-0247
- Navascues J, Bengoechea R, Tapia O, Casafont I, Berciano MT, Lafarga M. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J Struct Biol 2008; 163:137-46; PMID:18571432; https://doi.org/10.1016/j.jsb.2008.04.013
- Renvoisé B, Quérol G, Verrier ER, Burlet P, Lefebvre S. A role for protein phosphatase PP1γ in SMN complex formation and subnuclear localization to Cajal bodies. J Cell Sci 2012; 125:2862-74; https://doi.org/10.1242/jcs.096255
- Husedzinovic A, Oppermann F, Draeger-Meurer S, Chari A, Fischer U, Daub H, Gruss OJ. Phosphoregulation of the human SMN complex. Eur J Cell Biol 2014; 93:106-17; PMID:24602413; https://doi.org/10.1016/j.ejcb.2014.01.006
- Tapia O, Lafarga V, Bengoechea R, Palanca A, Lafarga M, Berciano MT. The SMN Tudor SIM-like domain is key to SmD1 and coilin interactions and to Cajal body biogenesis. J Cell Sci 2014; 127:939-46; PMID:24413165; https://doi.org/10.1242/jcs.138537
- Han KJ, Foster D, Harhaj EW, Dzieciatkowska M, Hansen K, Liu CW. Monoubiquitination of survival motor neuron regulates its cellular localization and Cajal body integrity. Hum Mol Genet 2016; 25:1392-405; PMID:26908624; https://doi.org/10.1093/hmg/ddw021
- Martin R, Gupta K, Ninan NS, Perry K, Van Duyne GD. The survival motor neuron protein forms soluble glycine zipper oligomers. Structure 2012; 20:1929-39; PMID:23022347; https://doi.org/10.1016/j.str.2012.08.024
- Gupta K, Martin R, Sharp R, Sarachan KL, Ninan NS, Van Duyne GD. Oligomeric properties of survival motor neuron·Gemin2 complexes. J Biol Chem 2015; 290(33):20185-99
- Sprangers R, Selenko P, Sattler M, Sinning I, Groves MR. Definition of domain boundaries and crystallization of the SMN Tudor domain. Acta Crystallogr - Sect D Biol Crystallogr 2003; 59:366-8; https://doi.org/10.1107/S0907444902021406
- Zhang R, So BR, Li P, Yong J, Gilsovic T, Wan L, Dreyfuss G. Structure of a key intermediate of the SMN complex reveals Gemin2s Crucial function in snRNP Assembly. Cell 2011; 146:384-95; PMID:21816274; https://doi.org/10.1016/j.cell.2011.06.043
- Weiss MS, Diederichs K, Read RJ, Panjikar S, Van Duyne GD, Matera AG, Fischer U, Grimm C. A critical examination of the recently reported crystal structures of the human SMN protein. Hum Mol Genet 2016
- Li DK, Tisdale S, Lotti F, Pellizzoni L. SMN control of RNP assembly: from post-transcriptional gene regulation to motor neuron disease. Semin Cell Dev Biol 2014; 32:22-9
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat 2000; 15:228-37; PMID:10679938; https://doi.org/10.1002/(SICI)1098-1004(200003)15:3%3c228::AID-HUMU3%3e3.0.CO;2-9
- Lorson CL, Strasswimmer J, Yao JM, Baleja JD, Hahnen E, Wirth B, Le T, Burghes AHM, Androphy EJ. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet 1998; 19:63-6; PMID:9590291; https://doi.org/10.1038/ng0598-63
- Praveen K, Wen Y, Gray KM, Noto JJ, Patlolla AR, Van Duyne GD, Matera AG. SMA-Causing Missense Mutations in survival motor neuron (Smn) display a wide range of phenotypes when modeled in drosophila. PLoS Genet 2014; 10:e1004489; PMID:25144193; https://doi.org/10.1371/journal.pgen.1004489
- Clermont O, Burlet P, Benit P, Chanterau D, Saugier-Veber P, Munnich A, Cusin V. Molecular analysis of SMA patients without homozygous SMN1 deletions using a new strategy for identification of SMN1 subtle mutations. Hum Mutat 2004; 24:417-27; PMID:15459957; https://doi.org/10.1002/humu.20092
- Wirth B, Herz M, Wetter A, Moskau S, Hahnen E, Rudnik-Schöneborn S, Wienker T, Zerres K, Rudnik-Schoneborn S, Wienker T, et al. Quantitative analysis of survival motor neuron copies: identification of subtle SMN1 mutations in patients with spinal muscular atrophy, genotype-phenotype correlation, and implications for genetic counseling. Am J Hum Genet 1999; 64:1340-56; PMID:10205265; https://doi.org/10.1086/302369
- Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci U S A 1999; 96:11167-72; PMID:10500148; https://doi.org/10.1073/pnas.96.20.11167
- Workman E, Saieva L, Carrel TL, Crawford TO, Liu D, Lutz C, Beattie CE, Pellizzoni L, Burghes AHM. A SMN missense mutation complements SMN2 restoring snRNPs and rescuing SMA mice. Hum Mol Genet 2009; 18:2215-29; PMID:19329542; https://doi.org/10.1093/hmg/ddp157
- Sarachan KL, Valentine KG, Gupta K, Moorman VR, Gledhill JM, Bernens M, Tommos C, Wand AJ, Van Duyne GD. Solution structure of the core SMN–Gemin2 complex. Biochem J 2012; 445:361-70; PMID:22607171; https://doi.org/10.1042/BJ20120241
- Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol 2002; 14:305-12; PMID:12067652; https://doi.org/10.1016/S0955-0674(02)00332-0
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bähler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 2012; 151:671-83; PMID:23101633; https://doi.org/10.1016/j.cell.2012.09.019
- Paushkin S, Charroux B, Abel L, Perkinson RA, Pellizzoni L, Dreyfuss G. The survival motor neuron protein of Schizosacharomyces pombe: Conservation of survival motor neuron interaction domains in divergent organisms. J Biol Chem 2000; 275:23841-6; PMID:10816558; https://doi.org/10.1074/jbc.M001441200
- Gao L, Frey MR, Matera AG. Human genes encoding U3 snRNA associate with coiled bodies in interphase cells and are clustered on chromosome 17p11.2 in a complex inverted repeat structure. Nucleic Acids Res 1997; 25:4740-7; PMID:9365252; https://doi.org/10.1093/nar/25.23.4740
- Jacobs EY, Frey MR, Wu W, Ingledue TC, Gebuhr TC, Gao L, Marzluff WF, Matera AG. Coiled bodies preferentially associate with U4, U11, and U12 small nuclear RNA genes in interphase HeLa cells but not with U6 and U7 genes. Mol Biol Cell 1999; 10:1653-63; PMID:10233169; https://doi.org/10.1091/mbc.10.5.1653
- Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G, Hakim O, Baek S, Hager GL, Dundr M. Cajal bodies are linked to genome conformation. Nat Commun 2016; 7:10966; PMID:26997247; https://doi.org/10.1038/ncomms10966
- Bäumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy. PLoS Genet 2009; 5:e1000773; https://doi.org/10.1371/journal.pgen.1000773
- Lotti F, Imlach WL, Saieva L, Beck ES, Hao LT, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell 2012; 151:440-54; PMID:23063131; https://doi.org/10.1016/j.cell.2012.09.012
- Luchetti A, Ciafrè S, Murdocca M, Malgieri A, Masotti A, Sanchez M, Farace M, Novelli G, Sangiuolo F. A perturbed MicroRNA expression pattern characterizes embryonic neural stem cells derived from a severe mouse model of spinal muscular atrophy (SMA). Int J Mol Sci 2015; 16:18312-27; PMID:26258776; https://doi.org/10.3390/ijms160818312
- Maeda M, Harris AW, Kingham BF, Lumpkin CJ, Opdenaker LM, McCahan SM, Wang W, Butchbach MER. Transcriptome profiling of spinal muscular atrophy motor neurons derived from mouse embryonic stem cells. PLoS One 2014; 9:e106818; PMID:25191843; https://doi.org/10.1371/journal.pone.0106818
- Murray LM, Beauvais A, Gibeault S, Courtney NL, Kothary R. Transcriptional profiling of differentially vulnerable motor neurons at pre-symptomatic stage in the Smn 2b/- mouse model of spinal muscular atrophy. Acta Neuropathol Commun 2015; 3:55; PMID:26374403; https://doi.org/10.1186/s40478-015-0231-1
- Ng S-Y, Soh BS, Rodriguez-Muela N, Hendrickson DG, Price F, Rinn JL, Rubin LL. Genome-wide RNA-Seq of human motor neurons implicates selective ER stress activation in spinal muscular atrophy. Cell Stem Cell 2015; 17(5):569-84
- Wertz MH, Winden K, Neveu P, Ng SY, Ercan E, Sahin M. Cell-type-specific miR-431 dysregulation in a motor neuron model of spinal muscular atrophy. Hum Mol Genet 2016; 0:ddw084; PMID:27005422
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 2008; 133:585-600; PMID:18485868; https://doi.org/10.1016/j.cell.2008.03.031
- Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 2015; 529:48-53; PMID:26700805; https://doi.org/; https://doi.org/10.1038/nature16469
- Sims RJ, Rojas LA, Beck D, Bonasio R, Schüller R, Drury WJ, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science 2011; 332:99-103; PMID:21454787; https://doi.org/10.1126/science.1202663
- Suraweera A, Lim YC, Woods R, Birrell GW, Nasim T, Becherel OJ, Lavin MF. Functional role for senataxin, defective in ataxia oculomotor apraxia type 2, in transcriptional regulation. Hum Mol Genet 2009; 18:3384-96; PMID:19515850; https://doi.org/10.1093/hmg/ddp278
- Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell 2011; 42:794-805; PMID:21700224; https://doi.org/10.1016/j.molcel.2011.04.026
- Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev 2014; 28:1384-96; PMID:24990962; https://doi.org/10.1101/gad.242990.114
- Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet 2015; 16:583-97; PMID:26370899; https://doi.org/10.1038/nrg3961
- Smith KP, Lawrence JB. Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies. Mol Biol Cell 2000; 11:2987-98; PMID:10982395; https://doi.org/10.1091/mbc.11.9.2987
- Suzuki T, Izumi H, Ohno M. Cajal body surveillance of U snRNA export complex assembly. J Cell Biol 2010; 190:603-12; PMID:20733056; https://doi.org/10.1083/jcb.201004109
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 2004; 16:777-87; PMID:15574332; https://doi.org/10.1016/j.molcel.2004.11.013
- Matera AG. Of coiled bodies, gems, and salmon. J Cell Biochem 1998; 70:181-92; PMID:9671224; https://doi.org/10.1002/(SICI)1097-4644(19980801)70:2%3c181::AID-JCB4%3e3.0.CO;2-K
- Matera AG, Frey MR. Coiled bodies and gems: Janus or Gemini? Am J Hum Genet 1998; 63:317-21; PMID:9683623; https://doi.org/10.1086/301992
- Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, Smn. J Cell Biol 1999; 147:715-28; PMID:10562276; https://doi.org/10.1083/jcb.147.4.715
- Hua Y, Zhou J. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett 2004; 572:69-74; PMID:15304326; https://doi.org/10.1016/j.febslet.2004.07.010
- Anderson P, Kedersha N. Stress granules: the Tao of RNA triage. Trends Biochem Sci 2008; 33:141-50; PMID:18291657; https://doi.org/10.1016/j.tibs.2007.12.003
- Zou T, Yang X, Pan D, Huang J, Sahin M, Zhou J. SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell Mol Neurobiol 2011; 31:541-50; PMID:21234798; https://doi.org/10.1007/s10571-011-9647-8
- Wu CY, Whye D, Glazewski L, Choe L, Kerr D, Lee KH, Mason RW, Wang W. Proteomic assessment of a cell model of spinal muscular atrophy. BMC Neurosci 2011; 12:25; PMID:21385431; https://doi.org/10.1186/1471-2202-12-25
- Garcia EL, Lu Z, Meers MP, Praveen K, Matera AG. Developmental arrest of Drosophila survival motor neuron (Smn) mutants accounts for differences in expression of minor intron-containing genes. RNA 2013; 19:1510-6; PMID:24006466; https://doi.org/10.1261/rna.038919.113
- Genabai NK, Ahmad S, Zhang Z, Jiang X, Gabaldon CA, Gangwani L. Genetic inhibition of JNK3 ameliorates spinal muscular atrophy. Hum Mol Genet 2015; 24:6986-7004; PMID:26423457
- Garcia EL, Wen Y, Praveen K, Matera AG. Transcriptomic comparison of Drosophila snRNP biogenesis mutants reveals mutant-specific changes in pre-mRNA processing: implications for spinal muscular atrophy. RNA 2016; 22(8):1215-27
- Deshpande M, Rodal AA. The crossroads of synaptic growth signaling, membrane traffic and neurological disease: Insights from Drosophila. Traffic 2016; 17:87-101; PMID:26538429; https://doi.org/10.1111/tra.12345
- Patitucci TN, Ebert AD. SMN deficiency does not induce oxidative stress in SMA iPSC-derived astrocytes or motor neurons. Hum Mol Genet 2016; 25:514-23; PMID:26643950; https://doi.org/10.1093/hmg/ddv489
