ABSTRACT
The biogenesis of small nuclear ribonucleoproteins (snRNPs), small Cajal body-specific RNPs (scaRNPs), small nucleolar RNPs (snoRNPs) and the telomerase RNP involves Cajal bodies (CBs). Although many components enriched in the CB contain post-translational modifications (PTMs), little is known about how these modifications impact individual protein function within the CB and, in concert with other modified factors, collectively regulate CB activity. Since all components of the CB also reside in other cellular locations, it is also important that we understand how PTMs affect the subcellular localization of CB components. In this review, we explore the current knowledge of PTMs on the activity of proteins known to enrich in CBs in an effort to highlight current progress as well as illuminate paths for future investigation.
Introduction
There are different types of ribonucleoproteins (RNPs), which are comprised of non-coding RNA and associated proteins. RNPs take part in fundamental cellular activities such as translation and pre-mRNA splicing. While the nucleolus serves as the initial hub for the generation of RNPs that take part in translation (ribosomes), the subnuclear domain known as the Cajal body (CB) contributes to the biogenesis of small nuclear RNPs (snRNPs), which are involved in pre-mRNA splicing.Citation1 In addition to snRNPs, CBs also contain another type of RNP: small Cajal body-specific RNPs (scaRNPs). The presence of scaRNPs in the CB serves to modify the snRNA component of snRNPs. The modification of snRNAs allows for the full-functionality of the snRNP. Base paring between the scaRNA of the scaRNP with the snRNA of the snRNP guides the modification of target sites within the snRNA by ribose methylation (for box C/D scaRNPs) or pseudouridylation (for box H/ACA scaRNPs).Citation2 CBs also take part in the biogenesis of small nucleolar RNPs (snoRNPs). Like scaRNPs, there are 2 classes of snoRNPs: box H/ACA and box C/D. The target sites for snoRNPs are rRNA and the RNA polymerase III transcribed, nucleolar-trafficked, U6 snRNA.Citation2 In addition to snRNPs, scaRNPs and snoRNPs, CBs also contain the RNP responsible for the formation of telomeres, telomerase.Citation3 The RNA component of telomerase, known as TERC or TR (hTR if referring to that in human) is an H/ACA class of small non-coding RNA and is considered to be a scaRNA. Mature telomerase contains hTR in a complex with core proteins and the telomerase reverse transcriptase (hTERT).Citation4 The CB is thought to be the assembly point for the incorporation of hTERT into the nascent telomerase complex. Along with snRNPs, scaRNPs and telomerase, the CB is enriched for many proteins. All of these proteins localize to other cellular locales in addition to the CB. The functions of these proteins in the CB is diverse, but collectively are thought to contribute to the RNP biogenesis mission of the CB. Like other cellular processes, RNP biogenesis, and thus CB activity, is regulated but an understanding of this regulation is far from complete. For example, many of the proteins enriched within the CB are post-translationally modified, but the functional consequences of these modifications are, for the most part, unclear. In this review, we will summarize what is known about the regulation of CB activity by exploring the current literature in regards to the post-translational modification of proteins enriched in the CB. Specifically, we will detail the (putative) function of proteins within the CB and describe any evidence as to if post-translational modification impacts these functions. The components that conduct these modifications, if they are known, will also be examined. We will also discuss the signals that govern these modifications in order to understand the local and global controls that influence the RNP biogenesis activity of CBs.
We will begin our analysis with 3 major components of the CB: coilin, SMN and WRAP53. We will then discuss other proteins that accumulate in CBs and contribute to snRNP, telomerase, scaRNP, or snoRNP formation. lists the proteins we will examine, along with known modifications of these proteins. The modification information was obtained from UniProt and PhosphoSitePlus websites and includes evidence obtained from large-scale mass spectrometric approaches and directed studies. As stated above, although all of the proteins listed in localize to other regions of the cell in addition to CBs, we will focus our discussion on the activity of these proteins in CBs. However, we will also provide information about non-CB functions when appropriate to give an indication as to how post-translational modification influences the function of a given protein.
Table 1. Modified CB proteinsFootnote*.
Coilin
What does coilin do in the CB?
Human coilin (UniProt # P38432) contains numerous modifications () and is considered the CB marker protein.Citation5,6 Given coilin's interaction with other proteins and RNAs in the CB, it likely plays a structural role in CB formation.Citation7-13 Reduction or depletion of coilin abolishes canonical CBs.Citation14,15 Coilin has been shown to have RNA degradation/processing activity.Citation16 Although the functional consequence of this activity is not fully understood, it is possible that coilin may directly contribute to snRNA, scaRNA and hTR processing and/or incorporation of these RNA into RNPs.
How does modification impact coilin's CB activity?
Phosphorylation
The first report of coilin modification took place in 1993, when Lamond and colleagues observed that coilin is phosphorylated, and this phosphorylation increases during mitosis when CBs disassemble.Citation17 Work by this same group demonstrated that one potential phosphorylation site within coilin, S202, mis-localizes coilin to the nucleolus when this residue is changed to mimic a constitutively phosphorylated residue (S202D).Citation18 The Lamond lab also showed that phosphatase inhibition by okadaic acid treatment disrupts CBs and results in the mis-localization of coilin to the nucleolus.Citation19 This finding clearly indicates that phosphorylation impacts CB formation. Our studies have shown that coilin is a self-interacting protein, and the extent of this self-interaction decreases during mitosis when coilin is hyperphosphorylated and CBs disassemble.Citation7 Hyperphosphorylation of coilin, therefore, may trigger CB disassembly. We have also observed that coilin in cell lines with CBs (such as HeLa) is less phosphorylated than that found in some primary cell lines (such as WI-38), which have few CBs.Citation20 In fact, coilin phosphorylation in interphase primary cells is apparently about the same as that found in mitotic HeLa cells. Point mutations of known phosphorylation sites further demonstrate that phosphorylation impacts coilin function, and thus CB activity, as well as proliferation rate.Citation21 Specifically, one change in coilin (S184A) results in the mis-localization of coilin to the nucleolusCitation7 while another change (S489D) decreases the in vitro RNA processing activity of coilin.Citation22 Besides coilin self-association, phosphorylation also appears to impact coilin association with other proteins and RNA. For example, phosphorylation of the C-terminus of coilin may help disengage the SMN complex from CBs and allow for snRNP accumulation in this subnuclear domain.Citation11 Additionally, the interaction profile of RNAs associated with coilin changes in response to nocodazole, which arrests cells in mitosis and generates hyperphosphorylated coilin.Citation12 Specifically, nocodazole treatment decreases the level the box C/D scaRNA 2 and 9 in the endogenous coilin immunoprecipitation complex approximately 4-fold compared to that present in untreated cells. Since another study has observed that ectopically expressed coilin-GFP associates with hundreds of non-coding RNAs,Citation13 it is likely, therefore, that coilin phosphorylation impacts CB activity by influencing the composition of the CB both in terms of proteins and RNAs. In addition, coilin may play a more direct role in snRNP, scaRNP, snoRNP and telomerase biogenesis than previously appreciated, and this activity is also likely to be regulated by phosphorylation. Recent work has shown that vaccinia-related kinase (VRK1) may play an important role in regulating CB dynamics in a cell cycle dependent manner by phosphorylating coilin (at S184) and protecting coilin from ubiquitination and proteasomal degradation.Citation23 More studies of this kind will be necessary to understand the role that coilin phosphorylation has on CB integrity and activity.
Methylation
Coilin has been shown to contain symmetrical dimethyl arginine (sDMA) in a region of the protein known as the RG (arginine/glycine) box.Citation9,24 Coilin interaction with SMN is contingent upon the presence of sDMAs in coilin.Citation8,9,24 Mutations in the coilin RG-box decrease interaction with SMN and result in coilin foci that do not enrich for SMN. Instead, nuclear SMN in cells expressing coilin with a mutated RG box form foci known as Gems.Citation8 In agreement with these results, hypomethylated coilin is correlated with decreased SMN interaction and increased Gem frequency.Citation9,24,25 Other work has shown that coilin localization is dependent upon its modification: hypomethylated coilin is enriched in the nucleolus while methylated coilin localizes to CBs.Citation26 Interestingly, hypomethylated coilin in the nucleolus is not associated with SMN or snRNPs,Citation26 clearly demonstrating that sDMA modification of coilin can affect its localization and association with other proteins.
Acetylation, sumoylation and ubiquitination
Large-scale mass spectrometric work has shown that coilin is modified by acetylation (at residues K204, K405, K496), sumoylation (K127, K204, K281) and ubiquitination (K476, K496, K555). Unfortunately, the functional consequences of these modifications on coilin activity are unknown. We have previously reportedCitation27 that coilin interacts with PIASy, which is a SUMO E3 ligase enriched within another nuclear domain known as the PML body. It is not known if coilin is a substrate for PIASy-mediated sumoylation, but it is known that SUMO-1 can be detected in CBs.Citation28 The interaction between coilin and PIASy may thus provide an avenue for the sumoylation of proteins enriched within the CB.
SMN
What does SMN do in the CB?
Human SMN (UniProt # Q16637) is extensively modified () and very well studied, in part because this protein is mutated in most cases of the neurodegenerative disease spinal muscular atrophy (SMA). The best-characterized activity of SMN actually takes place in the cytoplasm, not the CB.Citation29-34 In the cytoplasm, SMN plays crucial roles in ensuring that Sm proteins assemble properly onto U1, U2, U4 and U5 spliceosomal snRNAs during snRNP biogenesis, and the import of these nascent snRNPs to the nucleus, where they first target to CBs.Citation32,35-38 The exact function of SMN in the CB is unclear, but likely involves more than being a delivery vehicle for newly assembled, yet not mature, snRNPs to the CB. Before snRNPs are ready for splicing, additional snRNP-specific proteins are assembled onto the RNA. Additionally, the snRNA component of the snRNP contains modifications (ribose methylations and pseudouridylations) that are required for full functionality of the snRNP.Citation39 These snRNA modifications take place in the CB via scaRNPs.Citation2 Since scaRNPs are similar to snRNPs in that they are comprised of an RNA (scaRNA) and associated proteins, it is possible the SMN in the CB helps assemble scaRNPs in a similar manner as that found for snRNPs. Furthermore, SMN in the CB may also contribute toward telomerase holoenzyme assembly, which is known to take place in the CB. Another idea as to what SMN may be doing in the CB involves the recycling or regeneration of snRNPs after a splicing reaction takes place.Citation10,40 Reduction of SMN by RNAi disrupts CBs,Citation15 clearly indicating that this protein is vital for proper CB formation and composition. Regarding the function of SMN that when disrupted causes SMA, there are some opposing ideas as to if alterations in the snRNP biogenesis-promoting role of SMN is the main culprit.Citation41,42,43 The disruption of other functions for SMN, such as those involved in muscle formation and neuromuscular junctions, as well as in the afferent nerves, may also contribute to the SMA phenotype.Citation44,45 Recent work has shown that SMN takes part in the formation of messenger RNPs which are comprised of mRNA and mRNA binding proteins.Citation46 Therefore, reduction in the level of functional SMN may cause disruptions in many cellular processes, some of which take place in the CB, but all of which result in SMA.
How does modification impact SMN's CB activity?
Phosphorylation
Somewhat surprisingly, the first report of SMN post-translational modification (phosphorylation) was published in 2004,Citation47 9 y after the identification of SMN, which took place in 1995.Citation48 A functional consequence of SMN phosphorylation was soon identified, and centered upon the differential phosphorylation of cytoplasmic and nuclear SMN.Citation49 Briefly, SMN was shown to be phosphorylated on serines 28 and 31, and these modifications were more enriched in cytoplasmic SMN compared to nuclear SMN. This group then examined the activity of cytoplasmic SMN versus nuclear SMN using an snRNP assembly assay and observed that cytoplasmic SMN, which is phosphorylated on S28 and S31, is more active than nuclear SMN which is hypophosphorylated relative to cytoplasmic SMN. Hence phosphorylation of cytoplasmic SMN is an activator of the SMN complex.Citation49 Other studies have shown that dephosphorylation of SMN, which occurs via the action of the PPM1G phosphatase, is necessary for SMN localization to CBs.Citation50 Another phosphatase, PP1γ, likely also acts upon SMN (or other members of the SMN complex).Citation51 It is not known if PP1γ targets the same residues as PPM1G, but reduction of these 2 phosphatases results in different affects on SMN localization: PPM1G reduction decreases SMN enrichment in CBsCitation50 while PP1γ reduction increases SMN CB localization.Citation51 Other work, however, has shown that, like PPM1G, PP1γ reduction decreases SMN localization in CBs.Citation52 Phospho-null mutants of SMN (S28A/S31A) fail to show large cytoplasmic aggregates like those formed by WT SMN when overexpressed in cell lines.Citation53 This finding suggests that the phosphorylation of SMN may promote self-association and SMN complex formation. By forming these complexes, it is hypothesized that phosphorylated SMN is more stable and less subjected to the ubiquitin proteasome system.Citation54 Additional insight into SMN phosphorylation has come from work conducted by Gruss and colleagues, who have demonstrated that 7 out of the 9 components of the SMN complex are phosphorylated.Citation55 SMN contains serine and threonine phosphorylations, and mutation of these sites disrupts localization to CBs and association with coilin.Citation55 Three tyrosine residues have also been shown to be phosphorylated in SMN, and simultaneous mutation of these 3 tyrosine sites to phenylalanine (Y109F, Y127F, Y130F; demarcated in paper as SMN_Y) greatly reduces enrichment in CBs and nearly abolishes interaction with coilin.Citation55 Collectively, this work clearly demonstrates that SMN, and the SMN complex, is regulated by phosphorylation and future work clearly elucidating the kinases and phosphatases responsible for this regulation is of great importance. This work has already started in earnest.Citation52
Methylation
SMN is methylated on arginine 204.Citation56 The functional consequence of this modification on SMN activity is unknown. SMN has been shown to have preferential interaction with proteins containing symmetrical dimethyl arginine (sDMA). Proteins with sDMAs that have been shown to increase interaction with SMN include coilin,Citation9,24 SmD1 and SmD3,Citation57 SmB/B',Citation57,58 Lsm4,Citation58 Epstein-Barr Virus Nuclear Antigen 2 (EBNA2)Citation59; FGF-223Citation60 and the C-terminal domain of RNA polymerase II.Citation61
Acetylation, sumoylation and ubiquitination
SMN is acetylated at 3 residues (), but nothing is known about how this modification impacts its activity. Recent work has shown that SMN is modified by SUMO1 at residue K119.Citation62 Sumoylated SMN is not enriched in the CB, which contains the desumoylation enzyme USPL1. This suggests that desumoylation of SMN takes place in the CB. This work has also identified a SUMO-interacting motif (SIM)-like motif in close proximity to the sumoylated K119 residue, both of which reside within the Tudor domain. The authors' propose that SMN sumoylation and interaction with proteins via the SIM-like motif are important for CB formation and/or composition.Citation62 Regarding ubiquitination, mass spectrometric approaches have shown that SMN is ubiquitinated on residues K41, K51, K184, K186 and K209. The first report of ubiquitinated SMN showed that SMN protein levels could be increased in SMA patient cells upon treatment with the proteasome inhibitor MG132.Citation63 As mentioned above, the impact of ubiquitination and phosphorylation on SMN stability and complex formation was also examined.Citation54 The protein UCHL1 (ubiquitin C-terminal hydrolase L1) was found to be increased in SMA patient cells, and overexpression of UCHL1 decreased SMN levels.Citation64 Moreover, UCHL1 can ubiquitinate SMN, likely facilitating SMN degradation. Inhibition of UCHL1 activity increased SMN levels, suggesting that UCHL1 could be a therapeutic target for the treatment of SMA. However, another report has found that inhibition of UCHL1 actually exacerbates SMA in a mouse model.Citation65 The effect of monoubiquitination of SMN has also been explored.Citation66 The ubiquitin ligase Itch interacts with SMN and monoubiquitinates it at residues K179 and K209. Monoubiquitination of SMN affects its cellular localization. Ubiquitination-incompetent SMN mutant is capable of forming SMN oligomers and the SMN complex, but this mutant accumulates primarily in many nuclear foci with some nucleolar accumulations. In contrast, a SMN mutant fused with ubiquitin to mimic monoubiquitination primarily localizes to the cytoplasm. Nuclear accumulations with this mutant are present but appear to be in nucleoli. Hence the monoubiquitination of SMN may be a signal for nuclear export or inhibition of nuclear import. Both SMN mutants (ubiquitination-incompetent and monoubiquitination mimic) disrupt canonical CBs.Citation66 More broadly speaking, the disruption of ubiquitin homeostasis, via SMN interaction with the ubiquitin-like modifier activating enzyme 1 (UBA1), may, along with alterations in the β-catenin signaling, promote SMA.Citation65,67
WRAP53
What does WRAP53 do in the CB?
Unlike coilin and SMN, Human WRAP53 (UniProt # Q9BUR4) is not extensively modified, containing only phosphorylated and ubiquitinated residues (). WRAP53 (also known as TCAB1 or WDR79) was first identified in 2009 as the protein that binds the CAB motif present in box H/ACA scaRNAs and telomerase RNA, and targets these RNAs to the CB.Citation68,69 Like SMN, WRAP53 localizes to the cytoplasm and CB. Reduction of WRAP53 by RNAi disrupts CBs and mis-localizes coilin to the nucleolus,Citation70 demonstrating that this protein plays an important role in CB integrity and nuclear organization. Mutations in WRAP53 are found in patients with dyskeratosis congenita, a disease caused by insufficient levels of telomerase.Citation71 These findings show that WRAP53 is a crucial player in the trafficking and formation of the telomerase RNP. Given its interaction with the CAB motif present in box H/ACA scaRNAs, WRAP53 is also an important factor in that class of scaRNP biogenesis. It is important to note that human box C/D scaRNAs lack a CAB motif and WRAP53 interaction with these RNAs is far lower that that observed for box H/ACA scaRNAs.Citation72 This finding brings into question how box C/D scaRNAs are targeted to the CB for incorporation into a scaRNP. For some box C/D scaRNAs (those that are intron encoded), it appears that a motif known as the G.U/U.G wobble stem associates with WRAP53, allowing for the accumulation of these scaRNAs in the CB.Citation72 We have provided evidence that coilin, which highly associates with the box C/D scaRNAs 2, 9 and 17, may also play a role in the targeting of these scaRNAs to the CB.Citation12 Apart from the role of WRAP53 in CB-centered telomerase and scaRNP biogenesis, other work has shown that this protein is an essential player in the DNA double strand break repair pathway.Citation73
How does modification impact WRAP53s CB activity?
Phosphorylation and ubiquitination
No studies have specifically investigated how post-translational modification of WRAP53 influences its activities. Clearly, therefore, this is an open area of investigation ripe for the picking. WRAP53 contains 20 residues that are phosphorylated (), 15 of which reside in the N-terminal 150 amino acids of the protein. It is possible that WRAP53 interactions through its WD40 domain are regulated by phosphorylation of the N-terminus. Additionally, phosphorylation of WRAP53 may regulate its association with scaRNAs and telomerase RNA as well as its trafficking and localization to CBs. Regarding ubiquitination, WRAP53 residues K176 and K388 are modified in this manner. Like phosphorylation, however, it is not known how these modifications impact WRAP53 activity, localization or stability.
CRM1
What does CRM1 do in the CB?
CRM1 (UniProt # O14980) localizes to the nucleoplasm and CB.Citation74,75 CRM1 is part of the export complex that takes newly transcribed snRNAs from the nucleus to the cytoplasm for further maturation steps.Citation76 Before the journey to the cytoplasm, the export complex likely transits through the CB for remodeling steps which include the removal of the integrator complex responsible for the 3´-end processing of snRNA.Citation1,77-79 CRM1 also plays a role in the targeting of snoRNPs to the nucleolus, which, like snRNPs, also appear to transit through the CB during their biogenesis.Citation75,80-82 Inhibition of CRM1, and hence export of nascent spliceosomal snRNAs to the cytoplasm, disrupts CB composition and dynamics,Citation75,83 thereby reinforcing the idea that the CB is a dynamic nuclear structure responsive to the RNP demands of the cell. Moreover, selective inhibitors of nuclear export (SINE) that target CRM1 are current being developed to treat triple negative breast cancer,Citation84 implicating CRM1 RNP biogenesis activity as a potential target to treat cancer.
How does modification impact CRM1s CB activity?
Phosphorylation, acetylation and ubiquitination
CRM1 is modified by phosphorylation, acetylation and ubiquitination (). Given its important role in nuclear export, it is not surprising that many reports regarding the impact of PTMs do not necessarily directly involve modifications of CRM1. Rather, most of these reports discuss how the modification of cargo proteins influences their export by the CRM1 complex. One exception is the finding that CRM1 S391 phosphorylation by CDK1/cyclinB during mitosis controls CRM1 interactions, localization and mitotic functions.Citation85 Considering that CRM1 is phosphorylated on 20 residues, including S391, it will be of great interest to decipher how these PTMs regulate the RNP biogenesis-promoting activities of the CRM1 complex.
DAXX
What does DAXX do in the CB?
DAXX (UniProt # Q9UER7) is a well-studied transcriptional co-repressor protein that is involved in many pathways and interacts with many factors.Citation86 DAXX localizes to the cytoplasm and the nucleus, and in the nucleus DAXX is sequestered into PML bodies in order to regulate its activity.Citation86 Recent work has shown that DAXX can localize to CB in a cell cycle-dependent manner, with maximal enrichment in CBs taking place in early to mid S phase.Citation87 Since telomerase reverse transcriptase (TERT) accumulation with CBs also peaks during this phase of the cell cycle, coinciding with the assembly of the telomerase holoenzyme,Citation88-90 it is possible that the presence of DAXX in CBs promotes telomerase assembly (via interactions with telomerase subunits) and targeting of telomerase to telomeres.Citation87
How does modification impact DAXX's CB activity?
Phosphorylation, acetylation, sumoylation and ubiquitination
DAXX is extensively modified (), and the impact of these modifications on non-CB activities of DAXX has been well investigated.Citation91 Since the report about DAXX's involvement in telomerase biogenesis is relatively recent (2015), it is not surprising that the impact of post-translational modifications on this facet of DAXX function has not been examined. However, given that DAXX is a protein of 740 amino acids that contains 50 phosphorylated residues, it is likely that the telomerase-promoting activity of DAXX is regulated by phosphorylation.
Dyskerin
What does dyskerin do in the CB?
Dyskerin (UniProt # O60832) localizes to the nucleolus and CB.Citation92 Mutations in dyskerin cause X-linked dyskeratosis congenita,Citation93,94 demonstrating the importance of this protein in telomerase biogenesis. The co-transcriptional association of dyskerin and the proteins NHP2 and NOP10 with hTR is an early step in telomerase biogenesis.Citation3 This early complex also contains NAF1, which is replaced by GAR1 in the mature complex. Dyskerin is also the enzymatic component of box H/ACA small nucleolar RNPs (snoRNPs) and scaRNPs, which are responsible for the pseudouridylation of rRNA and snRNA, respectively.Citation95 Because hTR lacks any complementary of its hairpin pockets to any known target sequence, telomerase lacks the dyskerin-mediated pseudouridylase activity found in other H/ACA RNPs.Citation3 We have recently identified novel interactions between coilin and SMN with the telomerase components dyskerin and NAF1.Citation96 We have also found that alterations in the level of coilin and SMN impact the composition of the dyskerin complex and alter the association of dyskerin with hTR.Citation96 Hence dyskerin incorporation into both telomerase and box H/ACA scaRNPs/snoRNPs may be regulated by associations with CB proteins.
How does modification impact dyskerin's CB activity?
Phosphorylation and acetylation
Although dyskerin contains 17 phosphorylated sites (), there is no published information about how this modification impacts telomerase, scaRNP or snoRNP biogenesis. Nor is there information about if phosphorylation regulates the pseudouridylase activity of dyskerin. In mouse dyskerin, S121 is phosphorylated (, PhosphoSitePlus). Interestingly, this site is conserved in human dyskerin and a mutation of this residue (S121G) is associated with dyskeratosis congenita (DC).Citation97 It is possible, therefore, that some cases of DC are the result of altered dyskerin activity due to insufficient phosphorylation. In terms of acetylation, the influence of this modification on the activity of dyskerin activity is unknown.
Sumoylation and ubiquitination
Many mutations in dyskerin associated with DC are clustered in 2 regions: amino acid 31-72 and amino acid 314-420.Citation98 The 31-72 region contains a sumoylated residue (K39), that is mutated (K39E) in DC.Citation99 The 31-72 region also contains a hydrophobic cluster sumoylation motif that encompasses K39, and each residue of this motif is mutated in DC.Citation99 This group also found that mutation of K39 that prevents its sumoylation (K39R) results in decreased hTR levels as well as reduced telomerase activity and telomere maintenance.Citation99 The sumoylation of K39 was also shown to increase dyskerin protein stability, while the K39R mutation did not show this phenotype.Citation99 Since K39 is also likely to be ubiquitinated (),Citation100,101 modification of this residue by sumoylation or ubiquitination most likely represents a crucial regulatory control that impacts overall dyskerin levels in the cell.
Fam118B
What does Fam118B do in the CB?
Fam118B (UniProt # Q9BPY3) was identified in a screen for coilin interacting proteins.Citation102 Alteration of Fam118B expression levels (either by reduction or overexpression) disrupts canonical CB structure and composition. Depletion of Fam118B was also shown to decrease splicing capacity, inhibit cell proliferation and reduce the amount of symmetrical dimethyl arginine present in SmD1.Citation102 Consequently, Fam118B is an important player not only in the formation of CBs, but also in the interaction of SMN with Sm proteins which is required for snRNP biogenesis.
How does modification impact Fam118B's CB activity?
Phosphorylation, acetylation, ubiquitination
Fam118B contains 5 phosphorylation sites, 2 of which take place on tyrosines (). Currently, there is no information about the role of phosphorylation on Fam118B activity. This protein is also acetylated and ubiquitinated, but, like phosphorylation, the role that these PTMs have on Fam118B activity, and by extension CB activity, is unknown at present.
Fibrillarin
What does fibrillarin do in the CB?
Fibrillarin (UniProt # P22087) localizes to the nucleolus and CBCitation103,104 and is an essential component of box C/D scaRNPs and snoRNPs which are responsible for the modification of snRNA and rRNA, respectively, by methylation of ribose. Fibrillarin is not only a core component of box C/D scaRNPs and snoRNPs, but is also an RNA binding protein that interacts with scaRNAs and snoRNAs, and is the methyltransferase which modifies target sites within snRNA and rRNA.Citation105-107 Fibrillarin has also been shown to interact with SMN via a glycine/arginine rich (GAR) domain present within fibrillarin,Citation108,109 suggesting that SMN may contribute to the formation of box C/D scaRNPs and snoRNPs. In Drosophila, fibrillarin recruitment to CBs has been shown to be contingent upon poly(ADP-ribose) polymerase (PARP1), a well-studied modifier of nuclear proteins by ADP-ribosylation.Citation110
How does modification impact fibrillarin's CB activity?
Phosphorylation, methylation, acetylation and ubiquitination
Nothing is known about how acetylation and ubiquitination regulate fibrillarin incorporation into scaRNPs/snoRNPs, interaction with scaRNAs/snoRNAs, methyltransferase activity, or localization to CBs. Likewise, although it is known that fibrillarin is phosphorylated on 8 residues, the impact of this PTM on fibrillarin function and localization is unknown. The most well-studied PTM of fibrillarin is methylation. Arginines within the GAR domain of fibrillarin are dimethylated,Citation103,111-114 indicating that this modification may serve to regulate the interaction between fibrillarin and SMN. It should be pointed out that the type of methylation present in fibrillarin, asymmetric dimethylarginine (aDMA),Citation115 is more common than the symmetric dimethylarginine (sDMA) modification present within some Sm proteins and coilin, which strengthens their interaction with SMN.Citation9,24,58,116 Unlike sDMA, the aDMA modification present within the GAR domain of fibrillarin does not influence its interaction with SMN.Citation117 It is not clear if aDMA modifications in fibrillarin impact other aspects of its function, including localization, assembly into box C/D scaRNPs/snoRNPs, RNA binding and interaction with other proteins.
GAR1
What does GAR1 do in the CB?
Like fibrillarin, GAR1 (UniProt # Q9NY12) localizes to the nucleolus and CB.Citation118 GAR1 replaces NAF1 in telomerase biogenesis, and is present in the mature telomerase RNP.Citation3 GAR1 is also a core component of box H/ACA scaRNPs and snoRNPs, which are involved in the pseudouridylation of target sites within snRNA and rRNA, respectively.Citation119 GAR1 is so named because it contains 2 glycine/arginine rich (GAR) domains,Citation119 which contain aDMA modifications.Citation117,120 GAR1 interacts with SMN via either of its GAR domains, one of which is located at the N-terminus and the other is found at the C-terminus.Citation108,117 The exchange of NAF1 for GAR1 in both telomerase RNP and box H/ACA snoRNP (and presumably H/ACA scaRNPs) is thought to take place in the CB.Citation121
How does modification impact GAR1s CB activity?
Phosphorylation, methylation, acetylation and ubiquitination
GAR1 contains PTMs (), but little is known about how/if these modifications regulate GAR1 activity. GAR1 contains 5 phosphorylated residues, 4 of which are tyrosines. None of the phosphorylation sites lie within the GAR domains present at the N- and C-terminus. The GAR domain does contain aDMA,Citation117,120 but this modification does not alter GAR1 interaction with SMN.Citation117 It is possible that the aDMA modifications found within fibrillarin and GAR1 do in fact alter interaction with SMN in cells, but the experimental techniques used to assess the SMN/fibrillarin, SMN/GAR1 interaction may lack the resolution to detect such aDMA-mediated alterations. As with fibrillarin, the impact of aDMA on the activity of GAR1 as a core component of telomerase and box H/ACA snoRNPs and scaRNPs is unknown. It is possible that this modification regulates the exchange with NAF1 in the formation of the mature RNPs that takes place in the CB. We have recently observed that NAF1 is more highly recovered in SMN immunoprecipitation (IP) complexes than that found in coilin IP complexes.Citation96 Hence the interplay between SMN, coilin, GAR1, NAF1 and fibrillarin, which is likely regulated by PTMs, may contribute toward the formation of mature telomerase and H/ACA RNPs.
Nopp140
What does Nopp140 do in the CB?
Nopp140 (UniProt # Q14978) is enriched in the CB and nucleolusCitation92,122 and plays a crucial role in the formation of the ribosome.Citation123 It forms a complex with dyskerin, which also localizes to the CB and nucleolus.Citation92 Additionally, Nopp140 has been shown to interact with coilinCitation124 and both classes (box H/ACA and box C/D) of snoRNPs,Citation118 giving rise to the hypothesis that Nopp140 serves as a snoRNP chaperone and the link between the CB and nucleolus.Citation118 Given these characteristics, it is possible that Nopp140 also contributes to scaRNP biogenesis in the CB. Intriguingly, studies using SMA cell lines have revealed a link between SMN and Nopp140 in the CB: the loss of Nopp140 from CBs is correlated with the severity of SMA.Citation125 In other words, lines derived from SMA type I (the most severe) patients had the lowest proportion of CBs with Nopp140 compared to lines derived from SMA type II or type III patients.Citation125 These findings indicate that SMN may contribute to Nopp140 functions in the CB, possibly related to H/ACA snoRNP or scaRNP biogenesis.Citation125
How does modification impact Nopp140s CB activity?
Phosphorylation, methylation, acetylation and ubiquitination
The role of methylation, acetylation and ubiquitination on Nopp140 activity is largely unexplored. In contrast, Nopp140 phosphorylation has been investigated. Nopp140 is one of the most highly phosphorylated proteins in the cell,Citation126,127 and is a substrate for casein kinase II (CK2)Citation127 and protein kinase A.Citation128 Phosphatases acting upon Nopp140 in the cell are at present unknown. Using an in vitro assay, the phosphorylation of Nopp140 was found to be required for the association of Nopp140 with snoRNPs.Citation129 Therefore, kinases and phosphatases likely regulate the snoRNP chaperone activity of Nopp140. Nopp140 has been shown to interact with CK2,Citation130 and it is likely that this interaction inhibits the activity of CK2.Citation131 In particular, the phosphorylation of Nopp140 S574 greatly increases its affinity for CK2, providing a molecular switch that inactivates CK2. CK2 repression by Nopp140 is antagonized by inositol hexakisphosphate (IP6), which reactivates CK2 by competing with Nopp140 for binding at the CK2 substrate binding site.Citation131
PA28γ
What does PA28γ do in the CB?
PA28γ (UniProt # P61289) is a very well studied proteasome activator.Citation132 In conditions of stress, such as that induced by UV-C exposure, CBs are disrupted and PA28γ co-localization with coilin is increased.Citation133 It is important to note that PA28γ is not present in CBs in untreated cells, but rather is dispersed uniformly in the nucleoplasm.Citation133 Interestingly, reduction of PA28γ by RNAi attenuates the impact of UV-C-mediated disruption on CBs, while overexpression of PA28γ promotes CB disassembly, verifying that PA28γ contributes to CB integrity.Citation133 In support of the idea that stress results in the increased association of coilin with PA28γ, Lafarga and colleagues have observed that motor neurons from patients with type I SMA have increased co-localization of coilin and PA28γ both in CBs and nucleoli compared to control cells.Citation134 These findings strongly indicate that PA28γ is responsive to various stresses and contributes to the reorganization of CB components.
How does modification impact PA28γ's CB activity?
Phosphorylation, methylation, acetylation, sumoylation and ubiquitination
PA28γ contains several PTMs (). Specific information about how these modifications impact the function of PA28γ in CB integrity is lacking. Regarding the UV-C-induced disruption of CBs, it was observed that PA28γ levels do not change significantly after this treatment. Consequently, one idea as to how the association of PA28γ with coilin is regulated is by phosphorylation of coilin or PA28γ. The kinase MEKK3 may participate in this process by modifying PA28γ133, but this has not been proven as of yet.
PHAX
What does PHAX do in the CB?
Like CRM1, PHAX (UniProt # Q9H814) takes part in the export of spliceosomal snRNACitation1,135 and is localized to the CB and nucleoplasm. Along with the cap binding complex (CBC) and ARS2, PHAX interacts with the 7-methylguanosine cap of nascent snRNA to form an export complex that also contains CRM1.Citation1,135,136 After remodeling steps that take place in the CB, the export complex transits to the cytoplasm.Citation1,78,82 The directionality of the nuclear export transport is conferred by CRM1-associated RAN bound to GTP. Once in the cytoplasm, GTP is hydrolyzed resulting in RAN-GDP and disassembly of the export complex. As detailed below, phosphorylation of PHAX also regulates the formation of the export complex and influences its directionality. Reduction of PHAX by RNAi disrupts CBs,Citation15 demonstrating that snRNP biogenesis is required for CB formation.
How does modification impact PHAX's CB activity?
Phosphorylation, methylation and acetylation
PHAX contains several modifications () but the PTM of PHAX that has been most investigated is phosphorylation. Indeed, PHAX stands for phosphorylated adapter for RNA export.Citation135 Nuclear PHAX is phosphorylated, and this modification is essential to form the export complex.Citation135 After cargo is delivered to the cytoplasm, PHAX is dephosphorylated and this, along with GTP hydrolysis by CRM1-associated RAN, disassembles the complex allowing for the recycling of PHAX and the other export components back into the nucleus.Citation135 Since, like CRM1, PHAX is also involved in the intranuclear transport of U3, U8 and U13 snoRNAs as well as telomerase RNA,Citation82,137 it is possible that phosphorylation also regulates this aspect of PHAX activity. In follow up work, Ohno and colleagues identified the sites within mouse PHAX that are phosphorylated: S14, S16, S18, S56, S57, S60 and S63.Citation138 Of these sites, S56, S57, S60 and S63 are the most important for export complex formation. The kinase responsible for PHAX phosphorylation is the nucleus-enriched casein kinase 2 and the cytoplasmic phosphatase that acts upon PHAX is the protein phosphatase 2A (PP2A).Citation138 It will be of interest in future studies to investigate how phosphorylation of these sites within PHAX regulates the intranuclear transport of U3, U8 and U13 snoRNAs, as well as telomerase RNA and scaRNAs.
SART3
What does SART3 do in the CB?
SART3 (UniProt # Q15020) is an snRNP assembly factor that interacts with U6 snRNA and accumulates in CBs.Citation139 It is suspected that SART3 association with the U6 snRNP promotes annealing with the U4 snRNP, forming the U4/U6 di-snRNP, and this activity takes place in the CB.Citation139 This annealing of U4 and U6 snRNPs is hypothesized to take place upon the synthesis of new snRNPs as well as after a splicing reaction takes place.Citation139 Building upon this work, Stanek and colleagues observed that inhibition of snRNP assembly induces CBs in primary cell lines (WI-38) that normally have very few CBs.Citation140 SART3, which is a coilin interacting protein, is required for the induction of CBs and the accumulation of incomplete snRNPs with coilin in CBs. These findings suggest that the CB is formed in response to an increased demand for snRNPs that outpaces the capacity of the nucleoplasm to accurately form these snRNPs. Hence the CB is involved in the quality control of snRNP assembly.Citation140
How does modification impact SART3s CB activity?
Phosphorylation, methylation, acetylation and ubiquitination
SART3 is extensively modified, containing ubiquitinated, acetylated, methylated and phosphorylated residues (). Somewhat surprisingly, there is no published information regarding the impact of these PTMs on the snRNP assembly activity of CBs. Nor is there any information about if the PTMs in SART3 affect CB induction in primary cell lines upon inhibition of snRNP assembly. Since SART3 is phosphorylated on 20 residues, it will be very interesting to ascertain if the modification of these residues is part of the regulatory system that governs CB formation, number and size.
SmD1
What does SmD1 do in the CB?
SmD1 (UniProt # P62314) is a core component of snRNPs. We are using SmD1 as representative of all Sm proteins, although not all Sm proteins share the same PTMs as found on SmD1. Along with SmB/B´, SmD2, SmD3, SmE, SmF and SmG, SmD1 is loaded onto the Sm motif cis element present within spliceosomal snRNAs.Citation1,2 This Sm ring assembly step of snRNP formation takes place in the cytoplasm, under the control of the SMN complex.Citation1,2 After additional cytoplasmic modifications of the snRNA, including 3´-end processing and trimethylguanosine cap formation,Citation1,2 the nascent snRNP is imported into the nucleus with the help of the SMN complex and other nuclear import factors,Citation1,141 where it first localizes to CB.Citation36 In addition to Sm proteins, the newly imported snRNP acquires additional snRNP-specific proteins, and it possible that this takes place in the CB.Citation2
The CB is also the site wherein the snRNA component of the snRNP is modified by pseudouridylation (by box H/ACA scaRNPs) or methylation (by box C/D scaRNPs).Citation81 These modifications are guided by complementary base pairing of the scaRNA component of the scaRNP with target sites within snRNA of the snRNP.Citation106,142-145 After all these biogenesis steps, snRNPs localize to active sites of transcription where splicing takes place or are stored in speckles.Citation1,2 Coilin interaction with Sm proteins and SMN at the CB may disengage snRNPs from the SMN complex.Citation10,11
How does modification impact SmD1s CB activity?
Phosphorylation, methylation and ubiquitination
Although SmD1 is phosphorylated and ubiquitinated (), the most extensively studied PTM of SmD1 is methylation. Specifically, SmD1 contains sDMAs, which increases its interaction with SMN.Citation116 The sDMA modification within SmD1 facilitates interaction with the SMN complex in the cytoplasm so that Sm ring assembly takes place efficiently and on the proper site of the snRNA, thereby preventing Sm binding to other RNAs and forming a deleterious complex. It is not known if methylation or phosphorylation of SmD1, or PTMs found on other Sm proteins, alters their localization in CBs or interactions with CB components such as coilin. We have found, however, that one Sm protein (SmB´) preferentially interacts with a phosphomimetic coilin fragment but SMN prefers binding to a phospho-null coilin fragment.Citation11,146 This finding clearly shows that PTMs likely regulate CB protein dynamics and influence RNP biogenesis within this subnuclear organelle. The major methyltransferase that generates sDMAs on Sm proteins and Lsm proteins (which are part of the U6 snRNP), as well as on coilin is PRMT5.Citation24,57,58,111,116,147 Recent work has shown a functional interaction between PRMT5 and huntingtin (Htt), and observed that sDMA modification of known PRMT5 substrates, such as coilin, is reduced in Huntington disease.Citation148 Thus, disrupted modification of PRMT5 target proteins may contribute to Huntington disease pathology. In addition to PRMT5, another methyltransferase, PRMT7, also contributes to sDMA modifications found in Sm proteins and, like PRMT5, is required for snRNP assembly.Citation149
TERT
What does TERT do in the CB?
TERT (UniProt # O14746) is the telomerase reverse transcriptase. This subunit of telomerase is responsible for adding the repeat sequence TTAGGG to the ends of chromosomes using telomerase RNA as the template.Citation3 Telomerase holoenzyme assembly is suspected to take place in the CB,Citation150 which may then deliver telomerase to telomeres.Citation151
How does modification impact TERT's CB activity?
Phosphorylation, acetylation and ubiquitination
TERT contains several PTMs (). Most notably, TERT contains 11 phosphorylated residues. There has been some effort at understanding how PTMs regulate TERT function. Specifically, TERT has been shown to be a substrate for the E3 ubiquitin ligase Hdm2, thereby negatively regulating telomerase activity.Citation152 Also, several studies have shown that phosphorylation affects TERT: the Akt kinase-mediated phosphorylation of TERT increases telomerase activity,Citation153 but the c-Abl tyrosine kinase, which is activated by double stranded DNA breaks, inhibits TERT activity upon phosphorylation.Citation154 Protein phosphatase 2A (PP2A) has been shown to inhibit telomerase activity.Citation155 Furthermore, phosphorylation of TERT, especially by Akt at S227, has been shown to be important for the nuclear localization of TERT.Citation156,157 Despite these efforts, no studies have investigated how TERT PTMs regulate its association with the CB, or the delivery of telomerase by CBs to telomeres.
TGS1
What does TGS1 do in the CB?
The localization of TGS1, trimethylguanosine synthase I (UniProt # Q96RS0), is similar to SMN in that it is found in the cytoplasm and CBs.Citation158 Additionally, TGS1 directly interacts with SMN and forms the 5′ cap of spliceosomal snRNAs in the cytoplasm.Citation158 Trimethylguanosine cap formation of snoRNAs, which takes place in the CB, is catalyzed by a shorter TGS1 isoform compared to the full-length isoform which acts upon snRNAs in the cytoplasm.Citation159
How does modification impact TGS1s CB activity?
Phosphorylation and ubiquitination
Proteasomal processing of TGS1, resulting in the generation of a shorter nuclear isoform, may very well be regulated by the phosphorylation status of TGS1, which contains 30 phosphorylated residues. Unfortunately, no studies regarding the impact of phosphorylation on the trimethylguanosine synthase activity of TGS1, its interaction with the SMN complex, its localization, or its processing have been reported.
TOE1
What does TOE1 do in the CB?
TOE1 is a target of the immediate early transcription factor Egr1, and plays a role as an inhibitor of cell growth by influencing the level of the cyclin-dependent kinase inhibitor p21.Citation160 Curiously, TOE1 (also known as hCaf1z) localizes to the nucleus and CB.Citation161 Additionally, TOE1 forms a complex with hCcr4d (which is also found in the CB) and has deadenylation activity.Citation161 Both coilin and SMN interact with TOE1, and reduction of TOE1 disrupts CBs, decreases pre-mRNA splicing and inhibits cellular proliferation.Citation162 A possible function for TOE1 (and the TOE1/hCcr4d complex) in the CB is not entirely obvious considering that RNA with polyA tails do not significantly accumulate in the CB. Indeed, the RNAs that accumulate in CBs (i.e. snRNAs, snoRNAs, scaRNAs) are not polyadenylated. However, fairly recent work has identified a polyadenylation pathway for the maturation of hTR.Citation163,164 These authors posit that much of this process occurs in the nucleolus, but leave open the possibility that final trimming of hTR may occur in CBs. If so, it stands to reason that TOE1, a 3′ to 5′ exonuclease, may contribute to hTR processing. TOE1 may also contribute to the 3′ processing of other RNAs that accumulate in CBs.Citation161
How does modification impact TOE1s CB activity?
Phosphorylation
Human TOE1 is phosphorylated on 7 residues, 6 serines and 1 threonine. At present, there is no information regarding the kinases and phosphatases that modify TOE1. Also lacking are any functional studies that indicate how phosphorylation of TOE1 impacts its function, interactions, or localization to CBs.
USPL1
What does USPL1 do in the CB?
USPL1, ubiquitin-specific protease-like 1 (UniProt # Q5W0Q7), was recently identified as a component of CBs that is essential for proliferation and canonical CB formation.Citation165 USPL1 is the third type of SUMO protease reported in the literature, but this activity does not underlie the alterations in CB formation and impaired proliferation observed upon reduction of USPL1.Citation165 Additional work on USPL1 has shown that snRNA transcription is reduced upon depletion of USPL1 by RNAi.Citation166 Moreover, snRNP assembly and pre-mRNA splicing are negatively impacted upon reduction of USPL1.Citation166 USPL1 also associates with U1 and U2 snRNA gene loci and interacts with components of the little elongation complex, which is involved in the regulation of snRNA gene transcription.Citation166 Together, these data support the idea that USPL1 plays an important role in the transcription of snRNA genes by RNA polymerase II.Citation166
How does modification impact USPL1s CB activity?
Phosphorylation
Not surprisingly considering that USPL1 was only recently identified, no studies regarding the regulatory role of PTMs on USPL1 activities in the CB have been reported. Considering that USPL1 has 12 phosphorylated residues, it is likely that this post-translational modification contributes to USPL1 function and, by extension, CB formation and activity.
What signals promote modification of CB proteins?
A variety of conditions have been shown to impact CBs.Citation146,167 In regards to CB formation, a parallel can be drawn between the biogenesis of nucleoli at sites of rDNA transcription and the nucleation of CBs at sites of snRNA gene transcription (such as RNU1 and RNU2).Citation146,167 Astonishingly, recent work has found that CB association with certain gene loci contributes to genome organization, especially of chromosome 1, and CBs impact the expression of many genes and RNA splicing.Citation168 Inhibition of transcription disrupts canonical CBs,Citation146,167 clearly indicating that the CB is a dynamic subnuclear structure that is responsive to the overall snRNA levels in the cell. It is likely that cellular cues regulating snRNA transcription levels reach the CB via signal transduction cascades that culminate in the post-translational modification of proteins enriched within the CB, but a clear elucidation of such pathways awaits further investigation. Interestingly, reduction of CB components, such as WRAP53, resulting in CB disruption appears to induce a feedback control mechanism designed to increase important CB proteins, such as coilin, by stimulating the transcription of genes, such as COIL, that encode these proteins.Citation168 Since coilin is extensively modified, this putative compensatory mechanism must also include the regulation of enzymes that properly configure coilin.
Although not extensively discussed here, it is also highly likely that the activity of the SNAPc, little elongation and integrator complexes, all of which contribute to snRNA gene transcription, are regulated by PTMs. One example, ICE1, which is part of the little elongation complex, contains (according to PhosPhoSitePlus) 72 phosphorylated residues. Unfortunately, a PubMed search for ICE1 and phosphorylation yields zero results. Additionally, INTS1, a component of the integrator complex, is phosphorylated on 37 residues but no functional information regarding these modifications is present. Regarding the SNAPc complex, a bit more information regarding PTMs and the function of this complex is known compared to the little elongation or integrator complexes. For example, it has been shown that CK2 phosphorylates SNAP190, which in turn regulates SNAPc complex binding and transcription of the U6 snRNA gene by RNA polymerase III.Citation169 Clearly, therefore, the post-translational regulation of these 3 transcription complexes is largely unchartered territory and fertile ground for investigation.
Additional, but unknown at present, signaling pathways likely also impact the modification of CB proteins during the cell cycle and development. For example, CBs are disassembled during mitosisCitation170,171 and the composition and number of CBs changes during development.Citation172,173 Additionally, immortalization and transformation both increase the frequency and size of CBs,Citation174 while viral infection by adenovirus and herpes virus disrupts CBs and alters the localization of some components, such as coilin.Citation175-177 Finally, serum starvation, DNA damage and inhibition of translation, nuclear export and phosphatases all disrupt canonical CBs.Citation146 In regards to serum starvation, 2 proteins that respond to mitogenic stimuli and impact CBs are FGF-223 and ZPR1. FGF-223, as mentioned above, interacts with SMN and contains sDMAs.Citation60 Moreover, FGF-223 is part of the INFS (integrative nuclear FGR1 signaling) pathway and thus may transmit mitogenic signals to SMN and CBs.Citation178,179 A connection between the epidermal growth factor receptor (EGFR) and CBs is made via ZPR1. ZPR1 binds the cytoplasmic domain of EGFR and, upon EGF binding to EGFR, ZPR1 is released and can then interact with elongation factor 1α and SMN. The interaction between ZPR1 and SMN increases the localization of SMN in CBs.Citation180 Together with TOE1, therefore, ZPR1 and FGF-223 may be mediators of proliferative signals to CBs, possibly resulting in the modification of CB-enriched proteins. Another signaling protein found in CBs is peroxiredoxin V, which is responsive to reactive oxygen species.Citation181 It is possible that peroxiredoxin V accumulation in CBs is an indicator of a stress response pathway that may alter the composition of protein modification within the CB.
Although the conditions listed above (cell cycle, development, etc.) most likely trigger signal transduction cascades that result in the modification of CB proteins, the identification of the modifying enzymes and the functional consequence of such PTMs on CB proteins and CB function is largely unknown. Known modification factors that act upon the CB proteins listed in are shown in . As an example of how we need more clarification regarding PTMs and the CB, it is well established that CBs disassemble during mitosis, which is when coilin is hyperphosphorylated.Citation17 Since the self-association of coilin is reduced upon hyperphosphorylation,Citation7 a model can be developed in which the phosphorylation state of coilin impacts CB formation. In order for this model to be complete, however, one needs to identify the kinases and phosphatases that modify coilin in a cell cycle-dependent manner. Recent work has shown that VRK1 may be one such kinase.Citation23
Table 2. Modifiers of CB proteins.
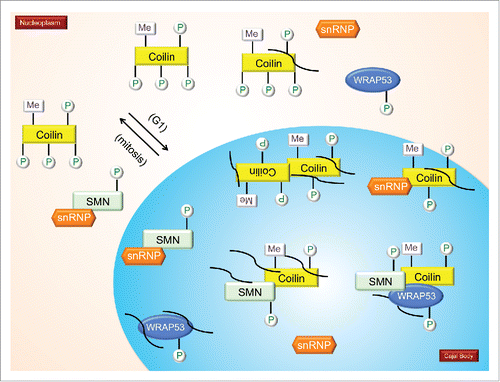
Several figures have been provided which depict the current knowledge about the PTM of CB-enriched proteins. In , we show the impact of PTMs on snRNP biogenesis, with specific emphasis on SMN modifications as well as the snRNA export pathway. focuses on coilin modifications and modifiers. specifically examines the impact of PTMs on the interactions and localization of SMN and coilin.
Figure 1. PTMs and snRNP biogenesis. A model showing known modifications of proteins involved in snRNP biogenesis, including the snRNA export pathway. Enzymes that modify these proteins are shown. Phosphorylated, methylated, ubiquitinated and sumoylated proteins are indicated. Processes marked by question marks indicate that there is no information on how PTMs affect localization or interaction; or where the PTM is known, but the enzyme(s) responsible for the modification are not known. Proteins that are modified, such as TGS1, but the functional consequence of this modification and the modifying enzymes are unknown, are indicated in the figure but lack denoted modification. Also not shown in the model are proteins with combinations of PTMs. (P = Phosphorylation; Me = Methylation; So = Sumoylation; Ub = Ubiquitination).
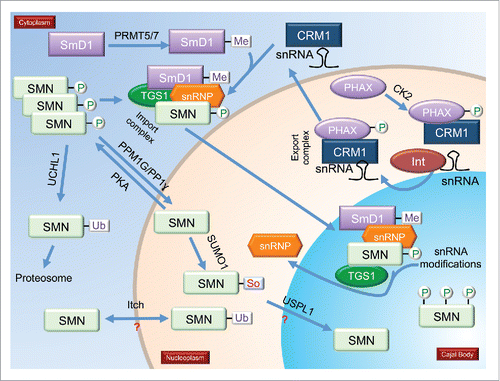
Figure 2. Modifications and modifiers of coilin, the CB marker protein. Enzymes known to modify coilin are shown. Not shown are coilin proteins with multiple different PTMs. During mitosis, coilin is hyperphosphorylated (coilin with 6 phosphorylations), which correlates with decreased coilin self-association and disassembled CBs. Hypomethylated coilin, which is enriched within the nucleolus, is indicated. Also shown in the nucleolus is a coilin phosphomutant (S184A). More details in text.
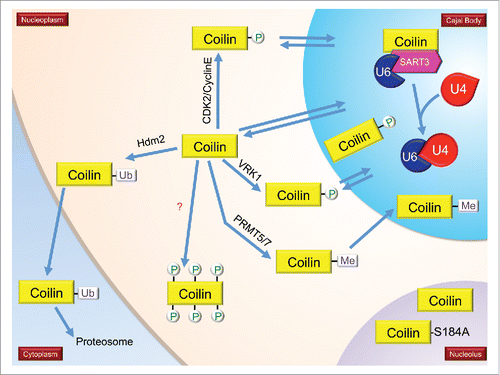
Figure 3. The impact of PTMs on the interactions and localizations of SMN and coilin. It is hypothesized that coilin in the nucleoplasm is hyperphosphorylated compared to coilin in CBs. During mitosis, coilin is hyperphosphorylated and CBs disassemble. CBs reform early to mid G1 (indicated in model). Mitotic coilin is reduced in its self-association compared to interphase coilin (paired coilin in the CB). Coilin interacts with SMN, Sm proteins, sRNPs and WRAP53. Methylation of coilin (and certain SM proteins) increases it interaction with SMN. Phosphorylation of coilin differentially affects its interaction with SMN and Sm proteins, with SMN preferentially binding to coilin that is less phosphorylated than that preferred by Sm proteins. The differential interaction of coilin with SMN and Sm proteins may disengage the nascent snRNP from the SMN complex in the CB, allowing for subsequent snRNP biogenesis steps, including snRNA modification and snRNP-specific protein association. Many non-coding RNAs, such as scaRNAs, snRNAs and snoRNAs are enriched in the CB and coilin complex (curved lines). Coilin interaction with RNAs decreases when coilin is hyperphosphorylated.Citation12
Future directions
Moving forward, a more complete understanding as to how PTMs impact the function of proteins enriched within the CB will be crucial in order to decipher how this subnuclear domain is formed and its activity regulated. At present, there is little information that details protein modification within the CB vs the modification status of these same proteins in other cellular locales. This type of information is known for SMN in that cytoplasmic SMN is more likely to be phosphorylated at serine 28 and 31 compared to nuclear SMNCitation49 and coilin since it has been shown that hypomethylated coilin localizes to the nucleolus.Citation26 Ideally, the modification of all CB proteins shown in could be determined for these proteins in all the cellular localizations they accumulate, be it the CB, nucleolus, nucleoplasm or cytoplasm. In so doing, the critical modifications that impact the localization and function of these proteins could be more easily ascertained. This information may also help identify the cellular location of the enzymes that modify these CB-enriched proteins. Along these lines, the cellular cascades that trigger CB formation and govern their numbers also need to be more fully elucidated. Strikingly, all 18 of the proteins we evaluated here are phosphorylated, but astonishingly little has been done to evaluate how this modification impacts their activity, either in the CB or other cellular locations. It is our hope that the presentation of these gaps in our knowledge will spur the research community to make a concerted effort culminating in an understanding of regulated Cajal body activity.
Since the CB plays an important role in the biogenesis of snRNP, scaRNP, snoRNP and telomerase, all of which are required for efficient and rapid cell proliferation, identification of signaling pathways that could be inhibited should be of therapeutic value in the treatment of cancer. In support of this hypothesis, reduction of many different inhabitants of the CB, as detailed above, results in decreased proliferation of cancer cell lines. Additionally, SMA and DC, both of which are diseases that have alterations in RNPs, may in fact be the result of more widespread disruptions in CB functions related to the biogenesis of other RNPs besides snRNPs for SMA and telomerase for DC. Studies that seek to clarify how PTMs regulate the formation of these RNPs, therefore, may shed light on the SMA and DC disease states.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- Praveen K, Wen Y, Gray KM, Noto JJ, Patlolla AR, Van Duyne GD, Matera AG. SMA-causing missense mutations in survival motor neuron (Smn) display a wide range of phenotypes when modeled in Drosophila. PLoS Genet 2014; 10:e1004489; PMID:25144193; https://doi.org/10.1371/journal.pgen.1004489
- Kiss T. Biogenesis of small nuclear RNPs. J Cell Sci 2004; 117:5949-51; PMID:15564372; https://doi.org/10.1242/jcs.01487
- Egan ED, Collins K. Biogenesis of telomerase ribonucleoproteins. Rna 2012; 18:1747-59; PMID:22875809; https://doi.org/10.1261/rna.034629.112
- Trahan C, Dragon F. Dyskeratosis congenita mutations in the H/ACA domain of human telomerase RNA affect its assembly into a pre-RNP. Rna 2009; 15:235-43; PMID:19095616; https://doi.org/10.1261/rna.1354009
- Raska I, Andrade LEC, Ochs RL, Chan EKL, Chang C-M, Roos G, Tan EM. Immunological and ultrastructural studies of the nuclear coiled body with autoimmune antibodies. Exp Cell Res 1991; 195:27-37; PMID:2055273; https://doi.org/10.1016/0014-4827(91)90496-H
- Andrade LEC, Chan EKL, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: Immunological characterization and cDNA cloning of p80 coilin. J Exp Med 1991; 173:1407-19; PMID:2033369; https://doi.org/10.1084/jem.173.6.1407
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell 2000; 11:4159-71; PMID:11102515; https://doi.org/10.1091/mbc.11.12.4159
- Hebert MD, Szymczyk PW, Shpargel KB, Matera AG. Coilin forms the bridge between Cajal bodies and SMN, the spinal muscular atrophy protein. Genes Dev 2001; 15:2720-9; PMID:11641277; https://doi.org/10.1101/gad.908401
- Hebert MD, Shpargel KB, Ospina JK, Tucker KE, Matera AG. Coilin methylation regulates nuclear body formation. Dev Cell 2002; 3:329-37; PMID:12361597; https://doi.org/10.1016/S1534-5807(02)00222-8
- Xu H, Pillai RS, Azzouz TN, Shpargel KB, Kambach C, Hebert MD, Schumperli D, Matera AG. The C-terminal domain of coilin interacts with Sm proteins and U snRNPs. Chromosoma 2005; 114:155-66; PMID:16003501; https://doi.org/10.1007/s00412-005-0003-y
- Toyota CG, Davis MD, Cosman AM, Hebert MD. Coilin phosphorylation mediates interaction with SMN and SmB'. Chromosoma 2010; 119:205-15; PMID:19997741; https://doi.org/10.1007/s00412-009-0249-x
- Enwerem II, Velma V, Broome HJ, Kuna M, Begum RA, Hebert MD. Coilin association with Box C/D scaRNA suggests a direct role for the Cajal body marker protein in scaRNP biogenesis. Biol Open 2014; 3:240-9; PMID:24659245; https://doi.org/10.1242/bio.20147443
- Machyna M, Kehr S, Straube K, Kappei D, Buchholz F, Butter F, Ule J, Hertel J, Stadler PF, Neugebauer KM. The coilin interactome identifies hundreds of small noncoding RNAs that traffic through Cajal bodies. Mol Cell 2014; 56:389-99; PMID:25514182; https://doi.org/10.1016/j.molcel.2014.10.004
- Tucker KE, Berciano MT, Jacobs EY, LePage DF, Shpargel KB, Rossire JJ, Chan EK, Lafarga M, Conlon RA, Matera AG. Residual Cajal bodies in coilin knockout mice fail to recruit Sm snRNPs and SMN, the spinal muscular atrophy gene product. J Cell Biol 2001; 154:293-307; PMID:11470819; https://doi.org/10.1083/jcb.200104083
- Lemm I, Girard C, Kuhn AN, Watkins NJ, Schneider M, Bordonne R, Luhrmann R. Ongoing U snRNP biogenesis is required for the integrity of Cajal bodies. Mol Biol Cell 2006; 17:3221-31; PMID:16687569; https://doi.org/10.1091/mbc.E06-03-0247
- Broome HJ, Hebert MD. In vitro RNase and nucleic acid binding activities implicate coilin in U snRNA processing. PLoS One 2012; 7:e36300; PMID:22558428; https://doi.org/10.1371/journal.pone.0036300
- Carmo-Fonseca M, Ferreira J, Lamond AI. Assembly of snRNP-containing coiled bodies is regulated in interphase and mitosis - Evidence that the coiled body is a kinetic nuclear structure. J Cell Biol 1993; 120:841-52; PMID:7679389; https://doi.org/10.1083/jcb.120.4.841
- Lyon CE, Bohmann K, Sleeman J, Lamond AI. Inhibition of protein dephosphorylation results in the accumulation of splicing snRNPs and coiled bodies within the nucleolus. Exp Cell Res 1997; 230:84-93; PMID:9013710; https://doi.org/10.1006/excr.1996.3380
- Sleeman J, Lyon CE, Platani M, Kreivi J-P, Lamond AI. Dynamic interactions between splicing snRNPs, coiled bodies and nucleoli revealed using snRNP protein fusions to the green fluorescent protein. Exp Cell Res 1998; 243:290-304; PMID:9743589; https://doi.org/10.1006/excr.1998.4135
- Hearst SM, Gilder AS, Negi SS, Davis MD, George EM, Whittom AA, Toyota CG, Husedzinovic A, Gruss OJ, Hebert MD. Cajal-body formation correlates with differential coilin phosphorylation in primary and transformed cell lines. J Cell Sci 2009; 122:1872-81; PMID:19435804; https://doi.org/10.1242/jcs.044040
- Carrero ZI, Velma V, Douglas HE, Hebert MD. Coilin phosphomutants disrupt cajal body formation, reduce cell proliferation and produce a distinct coilin degradation product. PLoS One 2011; 6:e25743; PMID:21991343; https://doi.org/10.1371/journal.pone.0025743
- Broome HJ, Carrero ZI, Douglas HE, Hebert MD. Phosphorylation regulates coilin activity and RNA association. Biol Open 2013; 2:407-15; PMID:23616925; https://doi.org/10.1242/bio.20133863
- Cantarero L, Sanz-Garcia M, Vinograd-Byk H, Renbaum P, Levy-Lahad E, Lazo PA. VRK1 regulates Cajal body dynamics and protects coilin from proteasomal degradation in cell cycle. Sci Rep 2015; 5:10543; PMID:26068304; https://doi.org/10.1038/srep10543
- Boisvert FM, Cote J, Boulanger MC, Cleroux P, Bachand F, Autexier C, Richard S. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J Cell Biol 2002; 159:957-69; PMID:12486110; https://doi.org/10.1083/jcb.200207028
- Clelland AK, Kinnear NP, Oram L, Burza J, Sleeman JE. The SMN protein is a key regulator of nuclear architecture in differentiating neuroblastoma cells. Traffic 2009; 10:1585-98; PMID:19735367; https://doi.org/10.1111/j.1600-0854.2009.00972.x
- Tapia O, Bengoechea R, Berciano MT, Lafarga M. Nucleolar targeting of coilin is regulated by its hypomethylation state. Chromosoma 2010; 119:527-40; PMID:20449600; https://doi.org/10.1007/s00412-010-0276-7
- Sun J, Xu H, Subramony SH, Hebert MD. Interactions between Coilin and PIASy partially link Cajal bodies to PML bodies. J Cell Sci 2005; 118:4995-5003; PMID:16219678; https://doi.org/10.1242/jcs.02613
- Navascues J, Bengoechea R, Tapia O, Casafont I, Berciano MT, Lafarga M. SUMO-1 transiently localizes to Cajal bodies in mammalian neurons. J Struct Biol 2008; 163:137-46; PMID:18571432; https://doi.org/10.1016/j.jsb.2008.04.013
- Fischer U, Liu Q, Dreyfuss G. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 1997; 90:1023-9; PMID:9323130; https://doi.org/10.1016/S0092-8674(00)80368-2
- Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci U S A 1999; 96:11167-72; PMID:10500148; https://doi.org/10.1073/pnas.96.20.11167
- Pellizzoni L, Yong J, Dreyfuss G. Essential Role for the SMN Complex in the Specificity of snRNP Assembly. Science 2002; 298:1775-9; PMID:12459587; https://doi.org/10.1126/science.1074962
- Meister G, Eggert C, Fischer U. SMN-mediated assembly of RNPs: a complex story. Trends Cell Biol 2002; 12:472-8; PMID:12441251; https://doi.org/10.1016/S0962-8924(02)02371-1
- Paushkin S, Gubitz AK, Massenet S, Dreyfuss G. The SMN complex, an assemblyosome of ribonucleoproteins. Curr Opin Cell Biol 2002; 14:305-12; PMID:12067652; https://doi.org/10.1016/S0955-0674(02)00332-0
- Coady TH, Lorson CL. SMN in spinal muscular atrophy and snRNP biogenesis. Wiley Interdisciplinary Rev RNA 2011; 2:546-64; PMID:21957043; https://doi.org/10.1002/wrna.76
- Massenet S, Pellizzoni L, Paushkin S, Mattaj IW, Dreyfuss G. The SMN complex is associated with snRNPs throughout their cytoplasmic assembly pathway. Mol Cell Biol 2002; 22:6533-41; PMID:12192051; https://doi.org/10.1128/MCB.22.18.6533-6541.2002
- Sleeman JE, Lamond AI. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr Biol 1999; 9:1065-74; PMID:10531003; https://doi.org/10.1016/S0960-9822(99)80475-8
- Narayanan U, Ospina JK, Frey MR, Hebert MD, Matera AG. SMN, the spinal muscular atrophy protein, forms a pre-import snRNP complex with snurportin1 and importin β. Hum Mol Genet 2002; 11:1785-95; PMID:12095920; https://doi.org/10.1093/hmg/11.15.1785
- Narayanan U, Achsel T, Luhrmann R, Matera AG. Coupled in vitro import of U snRNPs and SMN, the spinal muscular atrophy protein. Mol Cell 2004; 16:223-34; PMID:15494309; https://doi.org/10.1016/j.molcel.2004.09.024
- Yu YT, Shu MD, Steitz JA. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. Embo J 1998; 17:5783-95; PMID:9755178; https://doi.org/10.1093/emboj/17.19.5783
- Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell 1998; 95:615-24; PMID:9845364; https://doi.org/10.1016/S0092-8674(00)81632-3
- Lotti F, Imlach WL, Saieva L, Beck ES, Hao le T, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, et al. An SMN-dependent U12 splicing event essential for motor circuit function. Cell 2012; 151:440-54; PMID:23063131; https://doi.org/10.1016/j.cell.2012.09.012
- Praveen K, Wen Y, Matera AG. A Drosophila model of spinal muscular atrophy uncouples snRNP biogenesis functions of survival motor neuron from locomotion and viability defects. Cell Reports 2012; 1:624-31; PMID:22813737; https://doi.org/10.1016/j.celrep.2012.05.014
- Garcia EL, Lu Z, Meers MP, Praveen K, Matera AG. Developmental arrest of Drosophila survival motor neuron (Smn) mutants accounts for differences in expression of minor intron-containing genes. Rna 2013; 19:1510-6; PMID:24006466; https://doi.org/10.1261/rna.038919.113
- Goulet BB, Kothary R, Parks RJ. At the “junction” of spinal muscular atrophy pathogenesis: the role of neuromuscular junction dysfunction in SMA disease progression. Curr Mol Med 2013; 13:1160-74; PMID:23514457; https://doi.org/10.2174/15665240113139990044
- Mentis GZ, Blivis D, Liu W, Drobac E, Crowder ME, Kong L, Alvarez FJ, Sumner CJ, O'Donovan MJ. Early functional impairment of sensory-motor connectivity in a mouse model of spinal muscular atrophy. Neuron 2011; 69:453-67; PMID:21315257; https://doi.org/10.1016/j.neuron.2010.12.032
- Donlin-Asp PG, Bassell GJ, Rossoll W. A role for the survival of motor neuron protein in mRNP assembly and transport. Curr Opin Neurobiol 2016; 39:53-61; PMID:27131421; https://doi.org/10.1016/j.conb.2016.04.004
- La Bella V, Kallenbach S, Pettmann B. Post-translational modifications in the survival motor neuron protein. Biochem Biophys Res Commun 2004; 324:288-93; PMID:15465016; https://doi.org/10.1016/j.bbrc.2004.09.057
- Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell 1995; 80:155-65; PMID:7813012; https://doi.org/10.1016/0092-8674(95)90460-3
- Grimmler M, Bauer L, Nousiainen M, Korner R, Meister G, Fischer U. Phosphorylation regulates the activity of the SMN complex during assembly of spliceosomal U snRNPs. EMBO Rep 2005; 6:70-6; PMID:15592453; https://doi.org/10.1038/sj.embor.7400301
- Petri S, Grimmler M, Over S, Fischer U, Gruss OJ. Dephosphorylation of survival motor neurons (SMN) by PPM1G/PP2Cgamma governs Cajal body localization and stability of the SMN complex. J Cell Biol 2007; 179:451-65; PMID:17984321; https://doi.org/10.1083/jcb.200704163
- Renvoise B, Querol G, Verrier ER, Burlet P, Lefebvre S. A role for protein phosphatase PP1gamma in SMN complex formation and subnuclear localization to Cajal bodies. J Cell Sci 2012; 125:2862-74; PMID:22454514; https://doi.org/10.1242/jcs.096255
- Husedzinovic A, Neumann B, Reymann J, Draeger-Meurer S, Chari A, Erfle H, Fischer U, Gruss OJ. The catalytically inactive tyrosine phosphatase HD-PTP/PTPN23 is a novel regulator of SMN complex localization. Mol Biol Cell 2015; 26:161-71; PMID:25392300; https://doi.org/10.1091/mbc.E14-06-1151
- Aoki Y, Fukao T, Zhang G, Ohnishi H, Kondo N. Mutation in the Q28SDD31SD site, but not in the two SQ sites of the survival of motor neuron protein, affects its foci formation. Int J Mol Med 2010; 26:667-71; PMID:20878088; https://doi.org/10.3892/ijmm_00000512
- Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol 2009; 29:1107-15; PMID:19103745; https://doi.org/10.1128/MCB.01262-08
- Husedzinovic A, Oppermann F, Draeger-Meurer S, Chari A, Fischer U, Daub H, Gruss OJ. Phosphoregulation of the human SMN complex. Eur J Cell Biol 2014; 93:106-17; PMID:24602413; https://doi.org/10.1016/j.ejcb.2014.01.006
- Guo A, Gu H, Zhou J, Mulhern D, Wang Y, Lee KA, Yang V, Aguiar M, Kornhauser J, Jia X, et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteomics 2014; 13:372-87; PMID:24129315; https://doi.org/10.1074/mcp.O113.027870
- Friesen WJ, Paushkin S, Wyce A, Massenet S, Pesiridis GS, Van Duyne G, Rappsilber J, Mann M, Dreyfuss G. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol 2001; 21:8289-300; PMID:11713266; https://doi.org/10.1128/MCB.21.24.8289-8300.2001
- Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 2001; 7:1531-42; PMID:11720283; https://doi.org/10.1017/S135583820101442X
- Barth S, Liss M, Voss MD, Dobner T, Fischer U, Meister G, Grasser FA. Epstein-Barr virus nuclear antigen 2 binds via its methylated arginine-glycine repeat to the survival motor neuron protein. J Virol 2003; 77:5008-13; PMID:12663808; https://doi.org/10.1128/JVI.77.8.5008-5013.2003
- Bruns AF, Grothe C, Claus P. Fibroblast growth factor 2 (FGF-2) is a novel substrate for arginine methylation by PRMT5. Biol Chem 2009; 390:59-65; PMID:19086919; https://doi.org/10.1515/BC.2009.001
- Zhao DY, Gish G, Braunschweig U, Li Y, Ni Z, Schmitges FW, Zhong G, Liu K, Li W, Moffat J, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature 2016; 529:48-53; PMID:26700805; https://doi.org/10.1038/nature16469
- Tapia O, Lafarga V, Bengoechea R, Palanca A, Lafarga M, Berciano MT. The SMN Tudor SIM-like domain is key to SmD1 and coilin interactions and to Cajal body biogenesis. J Cell Sci 2014; 127:939-46; PMID:24413165; https://doi.org/10.1242/jcs.138537
- Chang HC, Hung WC, Chuang YJ, Jong YJ. Degradation of survival motor neuron (SMN) protein is mediated via the ubiquitin/proteasome pathway. Neurochem Int 2004; 45:1107-12; PMID:15337310; https://doi.org/10.1016/j.neuint.2004.04.005
- Hsu SH, Lai MC, Er TK, Yang SN, Hung CH, Tsai HH, Lin YC, Chang JG, Lo YC, Jong YJ. Ubiquitin carboxyl-terminal hydrolase L1 (UCHL1) regulates the level of SMN expression through ubiquitination in primary spinal muscular atrophy fibroblasts. Clin Chim Acta 2010; 411:1920-8; PMID:20713032; https://doi.org/10.1016/j.cca.2010.07.035
- Powis RA, Mutsaers CA, Wishart TM, Hunter G, Wirth B, Gillingwater TH. Increased levels of UCHL1 are a compensatory response to disrupted ubiquitin homeostasis in spinal muscular atrophy and do not represent a viable therapeutic target. Neuropathol Appl Neurobiol 2014; 40:873-87; PMID:25041530; https://doi.org/10.1111/nan.12168
- Han KJ, Foster D, Harhaj EW, Dzieciatkowska M, Hansen K, Liu CW. Monoubiquitination of survival motor neuron regulates its cellular localization and Cajal body integrity. Hum Mol Genet 2016; 25:1392-405; PMID:26908624; https://doi.org/10.1093/hmg/ddw021
- Aghamaleky Sarvestany A, Hunter G, Tavendale A, Lamont DJ, Llavero Hurtado M, Graham LC, Wishart TM, Gillingwater TH. Label-free quantitative proteomic profiling identifies disruption of ubiquitin homeostasis as a key driver of Schwann cell defects in spinal muscular atrophy. J Proteome Res 2014; 13:4546-57; PMID:25151848; https://doi.org/10.1021/pr500492j
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA. A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 2009; 34:47-57; PMID:19285445; https://doi.org/10.1016/j.molcel.2009.02.020
- Venteicher AS, Abreu EB, Meng Z, McCann KE, Terns RM, Veenstra TD, Terns MP, Artandi SE. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009; 323:644-8; PMID:19179534; https://doi.org/10.1126/science.1165357
- Mahmoudi S, Henriksson S, Weibrecht I, Smith S, Soderberg O, Stromblad S, Wiman KG, Farnebo M. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLoS Biol 2010; 8:e1000521; PMID:21072240; https://doi.org/10.1371/journal.pbio.1000521
- Zhong F, Savage SA, Shkreli M, Giri N, Jessop L, Myers T, Chen R, Alter BP, Artandi SE. Disruption of telomerase trafficking by TCAB1 mutation causes dyskeratosis congenita. Genes Dev 2011; 25:11-6; PMID:21205863; https://doi.org/10.1101/gad.2006411
- Marnef A, Richard P, Pinzon N, Kiss T. Targeting vertebrate intron-encoded box C/D 2′-O-methylation guide RNAs into the Cajal body. Nucleic Acids Res 2014; 42:6616-29; PMID:24753405; https://doi.org/10.1093/nar/gku287
- Henriksson S, Rassoolzadeh H, Hedstrom E, Coucoravas C, Julner A, Goldstein M, Imreh G, Zhivotovsky B, Kastan MB, Helleday T, et al. The scaffold protein WRAP53beta orchestrates the ubiquitin response critical for DNA double-strand break repair. Genes Dev 2014; 28:2726-38; PMID:25512560; https://doi.org/10.1101/gad.246546.114
- Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. Molecular cloning and cell cycle-dependent expression of mammalian CRM1, a protein involved in nuclear export of proteins. J Biol Chem 1997; 272:29742-51; PMID:9368044; https://doi.org/10.1074/jbc.272.47.29742
- Sleeman J. A regulatory role for CRM1 in the multi-directional trafficking of splicing snRNPs in the mammalian nucleus. J Cell Sci 2007; 120:1540-50; PMID:17405816; https://doi.org/10.1242/jcs.001529
- Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997; 90:1051-60; PMID:9323133; https://doi.org/10.1016/S0092-8674(00)80371-2
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 2005; 123:265-76; PMID:16239144; https://doi.org/10.1016/j.cell.2005.08.019
- Suzuki T, Izumi H, Ohno M. Cajal body surveillance of U snRNA export complex assembly. J Cell Biol 2010; 190:603-12; PMID:20733056; https://doi.org/10.1083/jcb.201004109
- Takata H, Nishijima H, Maeshima K, Shibahara K. The integrator complex is required for integrity of Cajal bodies. J Cell Sci 2012; 125:166-75; PMID:22250197; https://doi.org/10.1242/jcs.090837
- Verheggen C, Lafontaine DL, Samarsky D, Mouaikel J, Blanchard JM, Bordonne R, Bertrand E. Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments. Embo J 2002; 21:2736-45; PMID:12032086; https://doi.org/10.1093/emboj/21.11.2736
- Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. Embo J 2003; 22:1878-88; PMID:12682020; https://doi.org/10.1093/emboj/cdg187
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 2004; 16:777-87; PMID:15574332; https://doi.org/10.1016/j.molcel.2004.11.013
- Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol 1999; 147:715-28; PMID:10562276; https://doi.org/10.1083/jcb.147.4.715
- Schroder J, Cheng B, Ebinger M, Kohrmann M, Wu O, Kang DW, Liebeskind DS, Tourdias T, Singer OC, Christensen S, et al. Validity of acute stroke lesion volume estimation by diffusion-weighted imaging-Alberta Stroke Program Early Computed Tomographic Score depends on lesion location in 496 patients with middle cerebral artery stroke. Stroke 2014; 45:3583-8; PMID:25316278; https://doi.org/10.1161/STROKEAHA.114.006694
- Wu Z, Jiang Q, Clarke PR, Zhang C. Phosphorylation of Crm1 by CDK1-cyclin-B promotes Ran-dependent mitotic spindle assembly. J Cell Sci 2013; 126:3417-28; PMID:23729730; https://doi.org/10.1242/jcs.126854
- Lindsay CR, Morozov VM, Ishov AM. PML NBs (ND10) and Daxx: from nuclear structure to protein function. Front Biosci 2008; 13:7132-42; PMID:18508722; https://doi.org/10.2741/3216
- Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, Lawrenson K, McGuffog L, Healey S, Lee JM, et al. Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet 2015; 47:164-71; PMID:25581431; https://doi.org/10.1038/ng.3185
- Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol 2004; 164:647-52; PMID:14981093; https://doi.org/10.1083/jcb.200310138
- Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell 2004; 15:81-90; PMID:14528011; https://doi.org/10.1091/mbc.E03-07-0525
- Cristofari G, Adolf E, Reichenbach P, Sikora K, Terns RM, Terns MP, Lingner J. Human telomerase RNA accumulation in Cajal bodies facilitates telomerase recruitment to telomeres and telomere elongation. Mol Cell 2007; 27:882-9; PMID:17889662; https://doi.org/10.1016/j.molcel.2007.07.020
- Tang J, Agrawal T, Cheng Q, Qu L, Brewer MD, Chen J, Yang X. Phosphorylation of Daxx by ATM contributes to DNA damage-induced p53 activation. PLoS One 2013; 8:e55813; PMID:23405218; https://doi.org/10.1371/journal.pone.0055813
- Meier UT, Blobel G. NAP57, a mammalian nucleolar protein with a putative homolog in yeast and bacteria. J Cell Biol 1994; 127:1505-14; PMID:7798307; https://doi.org/10.1083/jcb.127.6.1505
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nature Genet 1998; 19:32-8; PMID:9590285; https://doi.org/10.1038/ng0598-32
- Mitchell JR, Wood E, Collins K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 1999; 402:551-5; PMID:10591218; https://doi.org/10.1038/990141
- Lafontaine DLJ, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D. The box H + ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev 1998; 12:527-37; PMID:9472021; https://doi.org/10.1101/gad.12.4.527
- Poole AR, Hebert MD. SMN and coilin negatively regulate dyskerin association with telomerase RNA. Biol Open 2016; 5(6):726-35; PMID:27215323; https://doi.org/10.1242/bio.018804
- Grozdanov PN, Fernandez-Fuentes N, Fiser A, Meier UT. Pathogenic NAP57 mutations decrease ribonucleoprotein assembly in dyskeratosis congenita. Hum Mol Genet 2009; 18:4546-51; PMID:19734544; https://doi.org/10.1093/hmg/ddp416
- Marrone A, Walne A, Dokal I. Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr Opin Genet Dev 2005; 15:249-57; PMID:15917199; https://doi.org/10.1016/j.gde.2005.04.004
- Brault ME, Lauzon C, Autexier C. Dyskeratosis congenita mutations in dyskerin SUMOylation consensus sites lead to impaired telomerase RNA accumulation and telomere defects. Hum Mol Genet 2013; 22:3498-507; PMID:23660516; https://doi.org/10.1093/hmg/ddt204
- Thomalla G, Cheng B, Ebinger M, Hao Q, Tourdias T, Wu O, Kim JS, Breuer L, Singer OC, Warach S, et al. DWI-FLAIR mismatch for the identification of patients with acute ischaemic stroke within 4.5 h of symptom onset (PRE-FLAIR): a multicentre observational study. Lancet Neurol 2011; 10:978-86; PMID:21978972; https://doi.org/10.1016/S1474-4422(11)70192-2
- Mertins P, Qiao JW, Patel J, Udeshi ND, Clauser KR, Mani DR, Burgess MW, Gillette MA, Jaffe JD, Carr SA. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nat Methods 2013; 10:634-7; PMID:23749302; https://doi.org/10.1038/nmeth.2518
- Li Y, Fong KW, Tang M, Han X, Gong Z, Ma W, Hebert M, Songyang Z, Chen J. Fam118B, a newly identified component of Cajal bodies, is required for Cajal body formation, snRNP biogenesis and cell viability. J Cell Sci 2014; 127:2029-39; PMID:24569877; https://doi.org/10.1242/jcs.143453
- Ochs RL, Lischwe MA, Spohn WH, Busch H. Fibrillarin: a new protein of the nucleolus identified by autoimmune sera. Biol Cell 1985; 54:123-33; PMID:2933102; https://doi.org/10.1111/j.1768-322X.1985.tb00387.x
- Raska I, Ochs RL, Andrade LEC, Chan EKL, Burlingame R, Peebles C, Gruol D, Tan EM. Association between the nucleolus and the coiled body. J Struct Biol 1990; 104:120-7; PMID:2088441; https://doi.org/10.1016/1047-8477(90)90066-L
- Schimmang T, Tollervey D, Kern H, Frank R, Hurt EC. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. Embo J 1989; 8:4015-24; PMID:2686980
- Kiss-Laszlo Z, Henry Y, Bachellerie JP, Caizergues-Ferrer M, Kiss T. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 1996; 85:1077-88; PMID:8674114; https://doi.org/10.1016/S0092-8674(00)81308-2
- Wang H, Boisvert D, Kim KK, Kim R, Kim SH. Crystal structure of a fibrillarin homologue from Methanococcus jannaschii, a hyperthermophile, at 1.6 A resolution. EMBO J 2000; 19:317-23; PMID:10654930; https://doi.org/10.1093/emboj/19.3.317
- Pellizzoni L, Baccon J, Charroux B, Dreyfuss G. The survival of motor neurons (SMN) protein interacts with the snoRNP proteins fibrillarin and GAR1. Curr Biol 2001; 11:1079-88; PMID:11509230; https://doi.org/10.1016/S0960-9822(01)00316-5
- Jones KW, Gorzynski K, Hales CM, Fischer U, Badbanchi F, Terns RM, Terns MP. Direct interaction of the spinal muscular atrophy disease protein SMN with the small nucleolar RNA-associated protein fibrillarin. J Biol Chem 2001; 276:38645-51; PMID:11509571; https://doi.org/10.1074/jbc.M106161200
- Kotova E, Jarnik M, Tulin AV. Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet 2009; 5:e1000387; PMID:19229318; https://doi.org/10.1371/journal.pgen.1000387
- Branscombe TL, Frankel A, Lee JH, Cook JR, Yang Z, Pestka S, Clarke S. PRMT5 (Janus kinase-binding protein 1) catalyzes the formation of symmetric dimethylarginine residues in proteins. J Biol Chem 2001; 276:32971-6; PMID:11413150; https://doi.org/10.1074/jbc.M105412200
- Christensen ME, Fuxa KP. The nucleolar protein, B-36, contains a glycine and dimethylarginine-rich sequence conserved in several other nuclear RNA-binding proteins. Biochem Biophys Res Commun 1988; 155:1278-83; PMID:3140806; https://doi.org/10.1016/S0006-291X(88)81279-8
- Brahms H, Raymackers J, Union A, de Keyser F, Meheus L, Luhrmann R. The C-terminal RG dipeptide repeats of the spliceosomal Sm proteins D1 and D3 contain symmetrical dimethylarginines, which form a major B-cell epitope for anti-Sm autoantibodies. J Biol Chem 2000; 275:17122-9; PMID:10747894; https://doi.org/10.1074/jbc.M000300200
- Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol 1995; 15:2800-8; PMID:7739561; https://doi.org/10.1128/MCB.15.5.2800
- Gary JD, Clarke S. RNA and protein interactions modulated by protein arginine methylation. Prog Nucleic Acid Res Mol Biol 1998; 61:65-131; PMID:9752719; https://doi.org/10.1016/S0079-6603(08)60825-9
- Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell 2001; 7:1111-7; PMID:11389857; https://doi.org/10.1016/S1097-2765(01)00244-1
- Whitehead SE, Jones KW, Zhang X, Cheng X, Terns RM, Terns MP. Determinants of the interaction of the spinal muscular atrophy disease protein SMN with the dimethylarginine-modified box H/ACA small nucleolar ribonucleoprotein GAR1. J Biol Chem 2002; 277:48087-93; PMID:12244096; https://doi.org/10.1074/jbc.M204551200
- Yang Y, Isaac C, Wang C, Dragon F, Pogacic V, Meier UT. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol Biol Cell 2000; 11:567-77; PMID:10679015; https://doi.org/10.1091/mbc.11.2.567
- Girard JP, Lehtonen H, Caizergues-Ferrer M, Amalric F, Tollervey D, Lapeyre B. GAR1 is an essential small nucleolar RNP protein required for pre-rRNA processing in yeast. Embo J 1992; 11:673-82; PMID:1531632
- Frankel A, Clarke S. RNase treatment of yeast and mammalian cell extracts affects in vitro substrate methylation by type I protein arginine N-methyltransferases. Biochem Biophys Res Commun 1999; 259:391-400; PMID:10362520; https://doi.org/10.1006/bbrc.1999.0779
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 2006; 173:207-18; PMID:16618814; https://doi.org/10.1083/jcb.200601105
- Meier UT, Blobel G. A nuclear localization signal binding protein in the nucleolus. J Cell Biol 1990; 111:2235-45; PMID:2177472; https://doi.org/10.1083/jcb.111.6.2235
- He F, James A, Raje H, Ghaffari H, DiMario P. Deletion of Drosophila Nopp140 induces subcellular ribosomopathies. Chromosoma 2015; 124:191-208; PMID:25384888; https://doi.org/10.1007/s00412-014-0490-9
- Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol 1998; 142:407-17; https://doi.org/10.1083/jcb.142.2.319
- Renvoise B, Colasse S, Burlet P, Viollet L, Meier UT, Lefebvre S. The loss of the snoRNP chaperone Nopp140 from Cajal bodies of patient fibroblasts correlates with the severity of spinal muscular atrophy. Hum Mol Genet 2009; 18:1181-9; PMID:19129172; https://doi.org/10.1093/hmg/ddp009
- Meier UT, Blobel G. Nopp140 shuttles on tracks between nucleolus and cytoplasm. Cell 1992; 70:127-38; PMID:1623516; https://doi.org/10.1016/0092-8674(92)90539-O
- Meier UT. Comparison of the rat nucleolar protein nopp140 with its yeast homolog SRP40. Differential phosphorylation in vertebrates and yeast. J Biol Chem 1996; 271:19376-84; PMID:8702624
- Chiu CM, Tsay YG, Chang CJ, Lee SC. Nopp140 is a mediator of the protein kinase A signaling pathway that activates the acute phase response alpha1-acid glycoprotein gene. J Biol Chem 2002; 277:39102-11; PMID:12167624; https://doi.org/10.1074/jbc.M205915200
- Wang C, Query CC, Meier UT. Immunopurified small nucleolar ribonucleoprotein particles pseudouridylate rRNA independently of their association with phosphorylated Nopp140. Mol Cell Biol 2002; 22:8457-66; PMID:12446766; https://doi.org/10.1128/MCB.22.24.8457-8466.2002
- Li D, Meier UT, Dobrowolska G, Krebs EG. Specific interaction between casein kinase 2 and the nucleolar protein Nopp140. J Biol Chem 1997; 272:3773-9; PMID:9013635; https://doi.org/10.1074/jbc.272.6.3773
- Bojesen SE, Pooley KA, Johnatty SE, Beesley J, Michailidou K, Tyrer JP, Edwards SL, Pickett HA, Shen HC, Smart CE, et al. Multiple independent variants at the TERT locus are associated with telomere length and risks of breast and ovarian cancer. Nat Genet 2013; 45:371-84, 84e1-2; PMID:23535731; https://doi.org/10.1038/ng.2566
- Realini C, Jensen CC, Zhang Z, Johnston SC, Knowlton JR, Hill CP, Rechsteiner M. Characterization of recombinant REGalpha, REGbeta, and REGgamma proteasome activators. J Biol Chem 1997; 272:25483-92; PMID:9325261; https://doi.org/10.1074/jbc.272.41.25483
- Cioce M, Boulon S, Matera AG, Lamond AI. UV-induced fragmentation of Cajal bodies. J Cell Biol 2006; 175:401-13; PMID:17088425; https://doi.org/10.1083/jcb.200604099
- Tapia O, Bengoechea R, Palanca A, Arteaga R, Val-Bernal JF, Tizzano EF, Berciano MT, Lafarga M. Reorganization of Cajal bodies and nucleolar targeting of coilin in motor neurons of type I spinal muscular atrophy. Histochem Cell Biol 2012; 137:657-67; PMID:22302308; https://doi.org/10.1007/s00418-012-0921-8
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 2000; 101:187-98; PMID:10786834; https://doi.org/10.1016/S0092-8674(00)80829-6
- Hallais M, Pontvianne F, Andersen PR, Clerici M, Lener D, Benbahouche Nel H, Gostan T, Vandermoere F, Robert MC, Cusack S, et al. CBC-ARS2 stimulates 3′-end maturation of multiple RNA families and favors cap-proximal processing. Nat Struct Mol Biol 2013; 20:1358-66; PMID:24270878; https://doi.org/10.1038/nsmb.2720
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell 2004; 16:789-98; PMID:15574333; https://doi.org/10.1016/j.molcel.2004.11.012
- Kitao S, Segref A, Kast J, Wilm M, Mattaj IW, Ohno M. A compartmentalized phosphorylation/dephosphorylation system that regulates U snRNA export from the nucleus. Mol Cell Biol 2008; 28:487-97; PMID:17967890; https://doi.org/10.1128/MCB.01189-07
- Stanek D, Rader SD, Klingauf M, Neugebauer KM. Targeting of U4/U6 small nuclear RNP assembly factor SART3/p110 to Cajal bodies. J Cell Biol 2003; 160:505-16; PMID:12578909; https://doi.org/10.1083/jcb.200210087
- Novotny I, Malinova A, Stejskalova E, Mateju D, Klimesova K, Roithova A, Sveda M, Knejzlik Z, Stanek D. SART3–dependent accumulation of incomplete spliceosomal snRNPs in cajal bodies. Cell Reports 2015; 10:429–40; PMID:25600876; https://doi.org/10.1016/j.celrep.2014.12.030
- Mates L, Nicolae C, Morgelin M, Deak F, Kiss I, Aszodi A. Mice lacking the extracellular matrix adaptor protein matrilin-2 develop without obvious abnormalities. Matrix Biol 2004; 23:195-204; PMID:15296947; https://doi.org/10.1016/j.matbio.2004.05.003
- Ganot P, Bortolin ML, Kiss T. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 1997; 89:799-809; PMID:9182768; https://doi.org/10.1016/S0092-8674(00)80263-9
- Ni J, Tien AL, Fournier MJ. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 1997; 89:565-73; PMID:9160748; https://doi.org/10.1016/S0092-8674(00)80238-X
- Tycowski KT, You ZH, Graham PJ, Steitz JA. Modification of U6 spliceosomal RNA is guided by other small RNAs. Mol Cell 1998; 2:629-38; PMID:9844635; https://doi.org/10.1016/S1097-2765(00)80161-6
- Darzacq X, Jady BE, Verheggen C, Kiss AM, Bertrand E, Kiss T. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. Embo J 2002; 21:2746-56; PMID:12032087; https://doi.org/10.1093/emboj/21.11.2746
- Hebert MD. Phosphorylation and the Cajal body: modification in search of function. Arch Biochem Biophys 2010; 496:69-76; PMID:20193656; https://doi.org/10.1016/j.abb.2010.02.012
- Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol 2001; 11:1990-4; PMID:11747828; https://doi.org/10.1016/S0960-9822(01)00592-9
- Ratovitski T, Arbez N, Stewart JC, Chighladze E, Ross CA. PRMT5- mediated symmetric arginine dimethylation is attenuated by mutant huntingtin and is impaired in Huntington's disease (HD). Cell Cycle 2015; 14:1716-29; PMID:25927346; https://doi.org/10.1080/15384101.2015.1033595
- Gonsalvez GB, Tian L, Ospina JK, Boisvert FM, Lamond AI, Matera AG. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J Cell Biol 2007; 178:733-40; PMID:17709427; https://doi.org/10.1083/jcb.200702147
- Tomlinson RL, Ziegler TD, Supakorndej T, Terns RM, Terns MP. Cell Cycle-regulated Trafficking of Human Telomerase to Telomeres. Mol Biol Cell 2006; 17:955-65; PMID:16339074; https://doi.org/10.1091/mbc.E05-09-0903
- Jady BE, Richard P, Bertrand E, Kiss T. Cell Cycle-dependent Recruitment of Telomerase RNA and Cajal Bodies to Human Telomeres. Mol Biol Cell 2006; 17:944-54; PMID:16319170; https://doi.org/10.1091/mbc.E05-09-0904
- Oh W, Lee EW, Lee D, Yang MR, Ko A, Yoon CH, Lee HW, Bae YS, Choi CY, Song J. Hdm2 negatively regulates telomerase activity by functioning as an E3 ligase of hTERT. Oncogene 2010; 29:4101-12; PMID:20453884; https://doi.org/10.1038/onc.2010.160
- Kang SS, Kwon T, Kwon DY, Do SI. Akt protein kinase enhances human telomerase activity through phosphorylation of telomerase reverse transcriptase subunit. J Biol Chem 1999; 274:13085-90; PMID:10224060; https://doi.org/10.1074/jbc.274.19.13085
- Kharbanda S, Kumar V, Dhar S, Pandey P, Chen C, Majumder P, Yuan ZM, Whang Y, Strauss W, Pandita TK, et al. Regulation of the hTERT telomerase catalytic subunit by the c-Abl tyrosine kinase. Curr Biol 2000; 10:568-75; PMID:10837221; https://doi.org/10.1016/S0960-9822(00)00483-8
- Li H, Zhao LL, Funder JW, Liu JP. Protein phosphatase 2A inhibits nuclear telomerase activity in human breast cancer cells. J Biol Chem 1997; 272:16729-32; PMID:9201974; https://doi.org/10.1074/jbc.272.27.16729
- Chung J, Khadka P, Chung IK. Nuclear import of hTERT requires a bipartite nuclear localization signal and Akt-mediated phosphorylation. J Cell Sci 2012; 125:2684-97; PMID:22366458; https://doi.org/10.1242/jcs.099267
- Jeong SA, Kim K, Lee JH, Cha JS, Khadka P, Cho HS, Chung IK. Akt-mediated phosphorylation increases the binding affinity of hTERT for importin α to promote nuclear translocation. J Cell Sci 2015; 128:2287-301; PMID:25999477; https://doi.org/10.1242/jcs.166132
- Mouaikel J, Narayanan U, Verheggen C, Matera AG, Bertrand E, Tazi J, Bordonne R. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep 2003; 4:616-22; PMID:12776181; https://doi.org/10.1038/sj.embor.embor863
- Girard C, Verheggen C, Neel H, Cammas A, Vagner S, Soret J, Bertrand E, Bordonne R. Characterization of a short isoform of human Tgs1 hypermethylase associating with small nucleolar ribonucleoprotein core proteins and produced by limited proteolytic processing. J Biol Chem 2008; 283:2060-9; PMID:18039666; https://doi.org/10.1074/jbc.M704209200
- De Belle I, Wu JX, Sperandio S, Mercola D, Adamson ED. In vivo cloning and characterization of a new growth suppressor protein TOE1 as a direct target gene of Egr1. J Biol Chem 2003; 278:14306-12; PMID:12562764; https://doi.org/10.1074/jbc.M210502200
- Wagner E, Clement SL, Lykke-Andersen J. An unconventional human Ccr4-Caf1 deadenylase complex in nuclear cajal bodies. Mol Cell Biol 2007; 27:1686-95; PMID:17178830; https://doi.org/10.1128/MCB.01483-06
- Fong KW, Li Y, Wang W, Ma W, Li K, Qi RZ, Liu D, Songyang Z, Chen J. Whole-genome screening identifies proteins localized to distinct nuclear bodies. J Cell Biol 2013; 203:149-64; PMID:24127217; https://doi.org/10.1083/jcb.201303145
- Moon DH, Segal M, Boyraz B, Guinan E, Hofmann I, Cahan P, Tai AK, Agarwal S. Poly(A)-specific ribonuclease (PARN) mediates 3′-end maturation of the telomerase RNA component. Nat Genet 2015; 47:1482-8; PMID:26482878; https://doi.org/10.1038/ng.3423
- Nguyen D, Grenier St-Sauveur V, Bergeron D, Dupuis-Sandoval F, Scott MS, Bachand F. A Polyadenylation-Dependent 3′ End Maturation pathway is required for the synthesis of the human telomerase RNA. Cell Reports 2015; 13:2244-57; PMID:26628368; https://doi.org/10.1016/j.celrep.2015.11.003
- Schulz S, Chachami G, Kozaczkiewicz L, Winter U, Stankovic-Valentin N, Haas P, Hofmann K, Urlaub H, Ovaa H, Wittbrodt J, et al. Ubiquitin-specific protease-like 1 (USPL1) is a SUMO isopeptidase with essential, non-catalytic functions. EMBO Rep 2012; 13:930-8; PMID:22878415; https://doi.org/10.1038/embor.2012.125
- Hutten S, Chachami G, Winter U, Melchior F, Lamond AI. A role for the Cajal-body-associated SUMO isopeptidase USPL1 in snRNA transcription mediated by RNA polymerase II. J Cell Sci 2014; 127:1065-78; PMID:24413172; https://doi.org/10.1242/jcs.141788
- Hebert MD. Signals controlling Cajal body assembly and function. Int J Biochem Cell Biol 2013; 45:1314-7; PMID:23583661; https://doi.org/10.1016/j.biocel.2013.03.019
- Wang Q, Sawyer IA, Sung MH, Sturgill D, Shevtsov SP, Pegoraro G, Hakim O, Baek S, Hager GL, Dundr M. Cajal bodies are linked to genome conformation. Nat Commun 2016; 7:10966; PMID:26997247; https://doi.org/10.1038/ncomms10966
- Gu L, Husain-Ponnampalam R, Hoffmann-Benning S, Henry RW. The protein kinase CK2 phosphorylates SNAP190 to negatively regulate SNAPC DNA binding and human U6 transcription by RNA polymerase III. J Biol Chem 2007; 282:27887-96; PMID:17670747; https://doi.org/10.1074/jbc.M702269200
- Andrade LEC, Tan EM, Chan EKL. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA 1993; 90:1947-51; PMID:8446613; https://doi.org/10.1073/pnas.90.5.1947
- Antoniou M, Carmo-Fonseca M, Ferreira J, Lamond AI. Nuclear organization of splicing snRNPs during differentiation of murine erythroleukemia cells in vitro. J Cell Biol 1993; 123:1055-68; PMID:8245117; https://doi.org/10.1083/jcb.123.5.1055
- Young PJ, Le TT, Dunckley M, Nguyen TM, Burghes AH, Morris GE. Nuclear gems and Cajal (coiled) bodies in fetal tissues: nucleolar distribution of the spinal muscular atrophy protein, SMN. Exp Cell Res 2001; 265:252-61; PMID:11302690; https://doi.org/10.1006/excr.2001.5186
- Strzelecka M, Oates AC, Neugebauer KM. Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 2010; 1:96-108; PMID:21327108; https://doi.org/10.4161/nucl.1.1.10680
- Spector DL, Lark G, Huang S. Differences in snRNP localization between transformed and nontransformed cells. Mol Biol Cell 1992; 3:555-69; PMID:1535243; https://doi.org/10.1091/mbc.3.5.555
- Rebelo L, Almeida F, Ramos C, Bohmann K, Lamond AI, Carmo-Fonseca M. The dynamics of coiled bodies in the nucleus of adenovirus-infected cells. Mol Biol Cell 1996; 7:1137-51; PMID:8862526; https://doi.org/10.1091/mbc.7.7.1137
- Rodrigues SH, Silva NP, Delicio LR, Granato C, Andrade LE. The behavior of the coiled body in cells infected with adenovirus in vitro. Mol Biol Rep 1996; 23:183-9; PMID:9112227; https://doi.org/10.1007/BF00351167
- Morency E, Sabra M, Catez F, Texier P, Lomonte P. A novel cell response triggered by interphase centromere structural instability. J Cell Biol 2007; 177:757-68; PMID:17548509; https://doi.org/10.1083/jcb.200612107
- Claus P, Doring F, Gringel S, Muller-Ostermeyer F, Fuhlrott J, Kraft T, Grothe C. Differential intranuclear localization of fibroblast growth factor-2 isoforms and specific interaction with the survival of motoneuron protein. J Biol Chem 2003; 278:479-85; PMID:12397076; https://doi.org/10.1074/jbc.M206056200
- Bruns AF, van Bergeijk J, Lorbeer C, Nolle A, Jungnickel J, Grothe C, Claus P. Fibroblast growth factor-2 regulates the stability of nuclear bodies. Proc Natl Acad Sci U S A 2009; 106:12747-52; PMID:19617559; https://doi.org/10.1073/pnas.0900122106
- Gangwani L, Mikrut M, Theroux S, Sharma M, Davis RJ. Spinal muscular atrophy disrupts the interaction of ZPR1 with the SMN protein. Nat Cell Biol 2001; 3:376-83; PMID:11283611; https://doi.org/10.1038/35070059
- Kropotov AV, Grudinkin PS, Pleskach NM, Gavrilov BA, Tomilin NV, Zhivotovsky B. Downregulation of peroxiredoxin V stimulates formation of etoposide-induced double-strand DNA breaks. FEBS Lett 2004; 572:75-9; PMID:15304327; https://doi.org/10.1016/j.febslet.2004.07.011
- Liu J, Hebert MD, Ye Y, Templeton DJ, Kung H, Matera AG. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J Cell Sci 2000; 113(Pt 9):1543-52; PMID:10751146
- Sanz-Garcia M, Vazquez-Cedeira M, Kellerman E, Renbaum P, Levy-Lahad E, Lazo PA. Substrate profiling of human vaccinia-related kinases identifies coilin, a Cajal body nuclear protein, as a phosphorylation target with neurological implications. J Proteomics 2011; 75:548-60; PMID:21920476; https://doi.org/10.1016/j.jprot.2011.08.019
- Magni M, Ruscica V, Buscemi G, Kim JE, Nachimuthu BT, Fontanella E, Delia D, Zannini L. Chk2 and REGgamma-dependent DBC1 regulation in DNA damage induced apoptosis. Nucleic Acids Res 2014; 42:13150-60; PMID:25361978; https://doi.org/10.1093/nar/gku1065
- Wu CY, Curtis A, Choi YS, Maeda M, Xu MJ, Berg A, Joneja U, Mason RW, Lee KH, Wang W. Identification of the phosphorylation sites in the survival motor neuron protein by protein kinase A. Biochim Biophys Acta 2011; 1814:1134-9; PMID:21609790; https://doi.org/10.1016/j.bbapap.2011.04.015
- Carnegie GK, Sleeman JE, Morrice N, Hastie CJ, Peggie MW, Philp A, Lamond AI, Cohen PT. Protein phosphatase 4 interacts with the Survival of Motor Neurons complex and enhances the temporal localisation of snRNPs. J Cell Sci 2003; 116:1905-13; PMID:12668731; https://doi.org/10.1242/jcs.00409
- Kwon DY, Dimitriadi M, Terzic B, Cable C, Hart AC, Chitnis A, Fischbeck KH, Burnett BG. The E3 ubiquitin ligase mind bomb 1 ubiquitinates and promotes the degradation of survival of motor neuron protein. Mol Biol Cell 2013; 24:1863-71; PMID:23615451; https://doi.org/10.1091/mbc.E13-01-0042
- Han KJ, Foster DG, Zhang NY, Kanisha K, Dzieciatkowska M, Sclafani RA, Hansen KC, Peng J, Liu CW. Ubiquitin-specific protease 9x deubiquitinates and stabilizes the spinal muscular atrophy protein-survival motor neuron. J Biol Chem 2012; 287:43741-52; PMID:23112048; https://doi.org/10.1074/jbc.M112.372318
