ABSTRACT
Long-term and short-term memories differ primarily in the duration of their retention. At a molecular level, long-term memory (LTM) is distinguished from short-term memory (STM) by its requirement for new gene expression. In addition to transcription (nuclear gene expression) the translation of stored mRNAs is necessary for LTM formation. The mechanisms and functions for temporal and spatial regulation of mRNAs required for LTM is a major contemporary problem, of interest from molecular, cell biological, neurobiological and clinical perspectives. This review discusses primary evidence in support for translational regulatory events involved in LTM and a model in which different phases of translation underlie distinct phases of consolidation of memories. However, it focuses largely on mechanisms of memory persistence and the role of prion-like domains in this defining aspect of long-term memory. We consider primary evidence for the concept that Cytoplasmic Polyadenylation Element Binding (CPEB) protein enables the persistence of formed memories by transforming in prion-like manner from a soluble monomeric state to a self-perpetuating and persistent polymeric translationally active state required for maintaining persistent synaptic plasticity. We further discuss prion-like domains prevalent on several other RNA-binding proteins involved in neuronal translational control underlying LTM. Growing evidence indicates that such RNA regulatory proteins are components of mRNP (RiboNucleoProtein) granules. In these proteins, prion-like domains, being intrinsically disordered, could mediate weak transient interactions that allow the assembly of RNP granules, a source of silenced mRNAs whose translation is necessary for LTM. We consider the structural bases for RNA granules formation as well as functions of disordered domains and discuss how these complicate the interpretation of existing experimental data relevant to general mechanisms by which prion-domain containing RBPs function in synapse specific plasticity underlying LTM.
Introduction
The formation of stable memories allows organisms to encode and retrieve information about facts, events, people, places, past experiences or learned skills and is of fundamental importance for guiding behavior. In experimental systems used to study long-term memory, the process could most simply manifest as a change in aversion or attraction to a particular stimulus based on previous experience. Enduring memories are thought to be achieved through long-lasting functional and structural plasticity of synapses that encode behavior. Understanding how neurons form and retain stable memories is one of the most intriguing and intensively studied aspects of neuroscience.
One critical difference between transient (short-term) and stable (long-term) memories is that formation of the latter requires new gene expression. This involves not only transcription of new mRNAs in memory encoding neurons, but also experience-induced translation of quiescent mRNAs stored at synapses that are activated by the experience. Thus, the consolidation of long-term memories requires both cell-wide and local mRNA translation, the latter enabling spatio-temporal regulation of gene expression in neurons.Citation1,2 For this reason, understanding regulation of mRNA translation remains an important goal of research on the cellular and molecular features of long-term memory.
The translational control of mRNAs requires RNA localization in dendrites as well as activity-induced translation at specific synapses. mRNA-associated regulatory proteins along with non-coding RNAs like microRNAs enable both basal repression and activity-dependent induction of mRNA translation. In neurons, translationally repressed mRNAs are held in assemblies of neuronal RiboNucleoProtein (RNP) particles or granules, cytoplasmic foci that mediate specific aspects of translational control.Citation3,4 The assembly and disassembly of these granules is thought to be mediated by protein domains with low sequence complexity (LC domains) including prion-like domains, which are found frequently on RNA binding proteins.Citation5-8 In one model, these LC domains serve to hold groups of individual mRNPs with repressed mRNAs together and make them available for translational activation at sites to which the assemblies are localized. In this simple model, LC domains function predominantly to gate mRNA translation that is required for the induction of long-term plasticity and memory. However, prion-like domains also classically undergo transitions into β-sheet rich conformations that nucleate and support the formation of self-sustaining and stable amyloid-like fibers. Such activity-induced transitions in RNA regulatory proteins, if they occurred, could potentially provide a mechanism for persistent and long-lasting changes in synaptic translation that may underlie long-term synaptic plasticity and memory. Such a model has been proposed and is supported by several observations of the prion-domain containing RNA regulatory protein CPEB.Citation9,10
This review will introduce diverse aspects of molecular biology of long-term memory, discuss the role of translational control mechanisms in long-term synaptic plasticity with a particular focus on the role and potential mechanisms by which prion-domain containing RNA-binding proteins enable long-term memory formation and retention.
RNA regulation is central to long-term memory formation
Memories were originally classified as short-term (STM) or long-term (LTM) based on the duration of memory retention and training regimens required for their acquisition. Short-term memories last from minutes to hours, while long-term memories last from days to weeks or sometimes for a lifetime. Subsequent studies showed that STM and LTM could be more precisely discriminated based on their need for new gene expression: STM can be induced without the need for new gene expression; in contrast the establishment of LTM is absolutely dependent on new transcription and translation.Citation2,11,12
Because perception and behavior derive from activity in the underlying connected systems of neurons, STM and LTM are respectively associated with short-term (transient) and long-term (persistent) changes of synaptic and neuronal properties in underlying behavioral circuits. Further, like STM and LTM, short-term and long-term plasticity also differ in terms of their respective dependence on new gene expression.Citation13-15
Studies of the molecular mechanisms of memory have extensively studied mechanisms of synaptic plasticity in reduced neuronal preparations, focusing either on Long Term Potentiation (LTP)/ Long-term Depression (LTD) in the mammalian brain or on synaptic facilitation in invertebrate models like Drosophila and Aplysia.Citation16-19 Both LTP and facilitation exist in 2 phases. For LTP: Early (E-LTP) that is short-term and late LTP (L-LTP) that is long-lasting; for facilitation, short term facilitation (STF) that is transient and long-term facilitation (LTF) that can last for several days.Citation15,20 Several studies of mammalian LTP and Aplysia synaptic facilitation have now established that gene expression is necessary only for the respective long-term forms of synaptic plasticity and that this is mediated in part through local protein synthesis at synapses.Citation21-23
During behavioral sensitization in Aplysia, the animals learn to strengthen a reflexive response to a previously mild stimulus (touch) if this is presented after a strongly aversive, “sensitizing” stimulus (a electrical shock). Single training trials lead to short-term sensitization lasting for hours and multiple spaced trials lead to long-term sensitization that last for days.Citation24 Formation of short term or long term memory is mirrored by short-term or long-term facilitation (increased neurotransmitter release) from sensory neurons that mediate touch onto motor neuron synapses that mediate a withdrawal response.Citation15,25 This facilitation of neurotransmitter release is mediated by shock-triggered serotonin that induces cAMP signaling and resultant PKA activation in presynaptic endings of sensory neurons.Citation26
Both short-term (STF) and long-term facilitation (LTF) can be reconstituted in synapses formed in primary cultures composed exclusively of dissected sensory and motor neurons.Citation20,27-29 In these cultured sensorimotor synapses, spaced application of 5 pulses of serotonin (5X5HT) leads to LTF that lasts for either 24 or 72 hours depending on the exact stimulation regimen.Citation30 Both 24 hour and 72 hour LTF are dependent on transcription and translation of mRNAs and 72 hour LTF is associated with the formation of stable new synaptic connections.Citation31
5X5HT triggered LTF activates protein kinase A and MAPK and induces the translocation of these kinases into the nucleus.Citation32 In the nucleus, through phosphorylation, the kinases turn on a transcriptional activator CREB1 (cAMP response element binding protein-1) while inactivating a transcriptional repressor CREB2.Citation30,33,34 This initiates a learning-associated transcriptional cascade of activity-dependent mRNAs whose translation is required for LTM.
In addition, LTM-producing stimuli activate local translation of mRNA stored at synapses. The concept of local translation at synapses was first suggested by the discovery of synapse-associated polyribosomes and other components of the translational machinery in dendrites, indicating that synapses are capable of acting as autonomous compartments for neuronal protein synthesis.Citation35-39 Further, several dendritically localized mRNAs were identified and activity induced transport of synaptic mRNAs to dendrites were reported.Citation40-42 Subsequently, enhanced accumulation in levels of specific mRNAs like CaMKII in dendritic spines following neuronal activity and increase in the polyadenylation and translation of stored, partially deadenylated CaMKII mRNA in mammalian synaptosomal preparations, following glutamate stimulation were observed.Citation43-45 Further studies in Aplysia,Citation46-48 DrosophilaCitation49 and mammalian systemsCitation50,51 showed that protein synthesis at synapses was induced by synaptic activity and provided evidence consistent with it being is required for long-term synaptic plasticity.
In Aplysia a key observation is that local application of a translational inhibitor to the sensorimotor synapse subjected to 5X5HT stimulation prevents the formation of LTF at this synapses.Citation30 Taken together, the data indicate that 5X5HT applied directly to the sensorimotor synapse triggers rapid local translation of synaptic mRNAs that is required for a retrograde (presynapse to cell body) signaling pathway necessary for synaptically-driven induction of LTF.Citation52 These findings were initially viewed as definitive evidence for local translation being necessary for long-term plasticity. However, the recent observation that postsynaptic translation in motor neurons is also required for LTF has complicated the interpretation of these findings.Citation46,53,54 It remains theoretically possible that while local translation occurs, the effect of synaptically applied translational inhibitorsCitation30 may have been due to its effects on cell-wide translation in the postsynaptic motor neuron.
Synapse specificity of LTM and 2 functions for local mRNA translation
How nuclear transcription can strengthen only a small subset of the thousands of synapse made by a single neuron remains an important question. A major initial assumption was that although transcription occurs in the nucleus and mRNA translation largely in the soma, protein products encoded by activity-regulated mRNAs can only induce synapse strengthening at synapses that are, in some way, marked or “tagged” by recent synaptic activity. The requirement for a local tag would restrict long-term plasticity to active synapses thereby ensuring appropriate selectivity and specificity.Citation55,56 This idea was formulated into the synaptic tagging theory, which postulated that even weak stimulation is capable of laying down a local, synaptic tag which renders synapses transiently receptive to long-term synaptic changes via proteins induced by strong stimulation of a different synapse on the same cell.Citation57
The phenomenon of synaptic tagging was experimentally demonstrated in hippocampal slices where it was found that weak tetanic stimulation that normally leads to only E-LTP can instead induce L-LTP at a synapse if multiple spaced tetani have recently been applied to other input neurons in the same population. The ability of the synaptic tag to enable capture is retained for 3 hours.Citation58,59
The potential role of local protein-synthesis in laying down this tag remained confusing for 2 reasons: first, strong stimulation was thought to be required for local protein translation; and second, early experiments in rat hippocampal cultures suggested that local translation was not required for synaptic tagging. Thus, local application of translational inhibitors to weakly stimulated hippocampal synapses did not affect their ability to “capture” L-LTP induced through a different set of synapses.Citation55,58,60
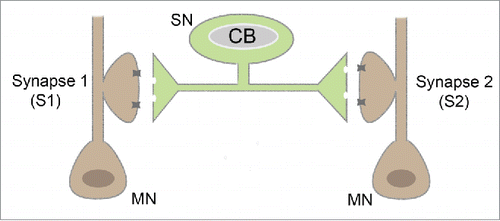
Experiments to address this issue performed in the Aplysia sensorimotor cultures confirmed but also curiously extended models for the nature and function of synaptic tagging and the role of local translation. These experiments to study synapse-specificity of LTM in built on one crucial technical advance. In primary cultures of Aplysia sensory and motor neurons, a single sensory neuron could be induced to form synapses with 2 different motor neurons in culture. In these “bifurcated” sensory neurons, it is possible to isolate and independently study LTF formation at each of 2 synapses made by the same sensory neuron Citation30 ().
As expected, LTF induced by 5X5HT stimulation of one synapse of a bifurcated neuron is restricted to that synapse. However, it can be captured by the second synapse contemporaneously tagged by a single 5HT pulse.Citation52 Consistent with observations in hippocampal neurons, inhibiting protein synthesis at the synapse tagged by 1X5HT, does not affects its ability to capture LTH if this was assessed 24 hours after 5X5HT stimulation. Thus, local protein synthesis is not required for the initial synaptic tag. However, and curiously, if protein synthesis is inhibited, then the captured LTH decays over 72 hours.Citation30,52
Two temporally distinct phases, functions and mechanisms for translational control of synaptic mRNAs are indicated by studies at the Aplysia sensorimotor synapse. There are 2 main arguments for this. First, local translation is required for LTF induction at the sensorimotor synapse subjected to 5X5HT stimulation. Thus, it plays an early role in LTF induction. Second, it is also required at a second, tagged synapse that captures LTF, not for the early capture event itself, but for the persistence of LTF for 72 hours or beyond. Thus, local translation appears to be required for both immediate (plasticity induction) and late (plasticity persistence) events that mediate LTF consolidation.
Additional experiments throw further light on the nature of the latter slow process. In the bifurcated neuron system, 5X5HT pulses applied directly on the soma of the sensory neuron were sufficient to induce nuclear translocation of MAPK, CREB-dependent transcription and, remarkably, cell-wide 24 hour LTF. This cell-wide facilitation did not require local translation at synapses. However, following cell-wide facilitation, a single pulse of serotonin applied to a synapse was sufficient to stabilize LTF for 72 hours. This occurred along with the growth of new stable contacts sites at the 1X5HT-tagged synapse.Citation30,33,52 Summary of various 5 –HT stimulation protocols and associated synapse specific, cell wide or capture based LTF given in .
Table 1. Stimulation protocols and associated LTF in the Aplysia bifurcated culture system.
Taken together the data point to the following scenario. (1) 5X5HT acts to induce translation of pre-existing mRNAs stored in translationally repressed form at synapses. This contributes to a retrograde signaling pathway that results in CREB-dependent transcription of activity induced mRNAS (2) 1X 5HT tags synapses in a manner that somehow allows CREB-dependent mRNAs to be transported toward and/or specifically stabilized in the vicinity of the tag. (3) Over a period of days, mRNAs translated close to tagged synapses express proteins that enable synaptic growth and the local formation of new stable synaptic contacts. These encode long-lasting memory. Importantly, a recent study using Aplysia sensor-motor neuron culture has shown that following stimulation by local application of neurotransmitter, mRNAs are trafficked cell wide rather than directed traffic to only stimulated synapses. However, enhanced translation of the localized mRNAs occurred only at activated synapses, in a Ca2+ dependent manner.Citation61
Mechanisms and functions of regulated translation in LTM
mRNAs are transported and localized to synapses in a translationally repressed state. How they are repressed, packaged, transported and subsequently regulated by local neuronal activity is a focus of contemporary studies. Several RNA binding proteins (RBPs) have been implicated in transport, localization and/or translational control of synaptic mRNAs: for example, ZBP1, FMRP, CPEB1, Staufen and the Exon-Junction Complex.Citation62-65
RBPs typically interact with specific regulatory elements on the 3′ UTR of mRNAs to enable their localization or regulation. Dendritic targeting elements are found on many dendritically localized mRNAs like MAP2 and CaMKII.Citation66 Individual RiboNucleoprotein (mRNP) particles formed by association of RNA and binding proteins assemble into larger RNA granules and, in such large complexes, are transported along cytoskeletal tracks by motor proteins enabling their localization.Citation4,67β- actin mRNA, which plays an important role in memory associated synaptic plasticity, is present in masked form in resting neurons. Activity induces mobility and unmasking of mRNA leading to protein synthesis.Citation68 Thus many lines of evidence indicate that during neuritic transport and localization, mRNAs are held in translationally repressed states, masked from the translational machinery. Once localized, they may be activated by one of several alternative mechanisms.Citation69-72
Translation of mRNAs that are packaged into RNPs are regulated at multiple stages of their translation. However the most common mechanism involve regulation of initiation step of mRNA translation. Cap dependent translation initiation is regulated by a multitude of of 5′- 3′ regulatory elements on mRNAs and various RBPs that bind and suppress initiation by interaction with elongation factors (eIFs). However, in addition to initiation step, translation of mRNAs housed in RNPs can be regulated at multiple stages using other mechanisms.Citation72,73 The most studied, canonical mechanism of translational activation involves the Cytoplasmic Polyadenylation Element Binding (CPEB1) protein and its regulation of CaMKII mRNA.Citation43,44,74,75
Activity induced translational regulation has been extensively studied in the case of CaMKII mRNA which is one of the most abundant protein in neurons especially enriched in the post synaptic density (PSD). NMDA receptor activation mediated Ca2+ entry leads to phosphorylation of the T286 residue of the pseudosubstrate domain of the α subunit (T287 of β) leading to a Ca2+ independent autonomous state of activity by autophosphorylation. Constant exchange of subunits between the holoenzymes enable the spread of the active state between CaMKII enzyme assemblies.Citation76-78 These features allow the enzyme to be active even after the cessation of neuronal stimulation, a feature attributed to its role in long term memory storage.
Neuronal stimulation leads to recruitment of CaMKII mRNA into cytoplasmic granules that are trafficked to dendrites. Cis regulatory elements on the 3′UTR that interact with RNP components enable its localization and targeting to the dendrites.Citation79,80 Disruption of its dendritic targeting leads to impairment of the late phase of LTP and consolidation of memory.Citation81 Following tetanic stimulation, CaMKII levels increase in the memory associated CA1 region of the brain and LTP induction stimulates CaMKII activity in single spines.Citation45,82 Several studies have shown that memory associated LTP is associated with CaMKII activation and phosphorylation. Consistently inhibitors of CaMKII and mutation in the T286 residue blocks specifically the induction of LTP.Citation83,84 Consistent with these results RNP components are also found to associate with and regulate the translation of CaMKII in a manner required for LTM.Citation74,85,86
In addition to RBPs, neuronal mRNA translation is regulated by non coding RNAs such as microRNAs (miRNAs) that are abundant in brain tissue.Citation87 miRNA association with mRNAs by virtue of miRNA binding sites on 3-UTR can induce translational repression as well as promote the localization of RNAs to processing bodies. For example, the miRNA 134 regulates the repression of Limk1 encoding mRNAs in dendritic spines of hippocampal neurons, until its translation and Limk1 protein expression is induced by BDNF treatment.Citation88 miRNAs interact directly or indirectly with RBPs including RISC ( RNA Induced Silencing Complex). For example FMRP interacts with miRNA and RISC components to regulate expression of bound mRNAs.Citation89 Similarly, the interaction of GW182 with other core RISC proteins such as Argonaute (Ago1) is required for miRNA mediated repression.Citation90 Other prominent examples are of Armitage, whose degradation releases translational repression of CaMKII at Drosophila synapses and MOV10, a RISC protein under activity dependent degradative control by proteosomal machinery, that regulates mRNAs like CaMKII and Lypia1.Citation49,91
Since the discovery of dendritic translation, understanding the cellular and molecular mechanisms underlying regulation of mRNA transport, localization and local translation has been the primary focus of studies on memory associated synaptic plasticity. Over the past 12 y, a series of papers point to a model in which a component of earlier discussed synaptic tag includes local, self-sustaining switch in conformation of CPEB from one that is predominantly repressive to one that is translationally active. This self-sustaining switch is proposed to play a key role in the persistence of long-term plasticity and memory which is discussed in detail later in this review. Before that we discuss briefly, the models proposed for persistence of long-term memory.
Biochemical models for LTM maintenance
How memories outlast molecular turnover is a question that has always baffled neurobiologists. Formation of long term memory has been attributed to increased (or altered) a) release of neurotransmitters, b) receptor densities on synaptic membranes and c) synaptic contact number, each of which could be dependent on increased protein translation.Citation92 However, any biochemical model proposed for ‘maintenance’ of LTM primarily had to explain how synapse-associated protein synthesis could account for memory that lasts for days or years given the decay of most proteins would occur in span of hours. Therefore, theories put forth for persistence of LTM mostly centered around bistable autocatalytic loops that would ensure the sustained altered state of a synapse without need for continuous neuronal stimulation.
Francis Crick in 1984 raised the possibility that memories are stored as sustained secondary modifications to DNA and protein that alter synaptic strength. To overcome the limitation of molecular turnover he modeled enzymatic reactions based on modifications of symmetric molecules akin to that of methylated DNA cytosine residues.Citation93,94 Later, Lisman proposed and formalized the theory that autophosphorylation of CaMKII could underlie persistence of memory. The model proposed that autophosphorylation mediated ‘on’ state of the bistable switch involving CaMKII would render the system immune to dephosphorylation and protein degradation enabling information storage indefinitely.Citation83,95 PKMζ is another enzyme implicated in long-term memory storage. It is an atypical PKC enzyme with the ability to be autonomously active once synthesized, thereby stabilizing lasting changes required for sustained LTP.Citation96-99 However more recent studies show that PKMζ, is dispensable for LTP associated with memory.Citation100-102 In 1998, Tompa and Friedrich put forth the prion theory of memory wherein they suggested that propagation of a non-toxic conformational state of prions could underlie cellular functions like memory storage.Citation103 The theory suggested that certain conformational states adopted by prion proteins enable them to retain an self-sustaining altered state indefinitely, analogous to the bistable switch models earlier proposed by Crick and Lisman, thereby rendering memories immune to molecular turnover. Potential prion-like mechanisms underlying memory persistence were later dramatically proposed and experimentally argued in the context of CPEB protein, which contains an N-terminal prion-like domain. We consider the background and status of the ‘prion hypothesis’ for LTM storage in more detail below.
Prions – as basis for long term memory
Discovery and characterization of prion protein
Prions are self-propagating, infectious protein particles first described by Prusiner as the causative factor behind Creutzfeldt – Jakob disease (CJD).Citation104 The prion protein PrP exists in 2 different states, the normal cellular isoform PrPC and the toxic isoform PrPSc. PrPSc is capable of self-propagation by converting the PrPC isoform to PrPSc isoform in a self-templating manner.Citation105,106 PrPSc forms insoluble aggregates called amyloid fibers that are resistant to proteolytic digestion and the accumulation of these in neurons leads to neurodegeneration.Citation107-109 These two isoforms differ only in their conformation: PrPSc is characterized by β sheets running parallel to the axis of the fiber (cross β sheet) whereas PrPC is rich in α helical content.Citation110,111 The self-templating mode of propagation and conformational switching are features of prions that are proposed to be important for memory persistence.Citation14 The prions that underlie mammalian Transmissible Spongiform Encephalopathies (TSEs) form amyloids that are extremely stable and irreversible to the non-prion conformation.Citation105,112 Recent studies do indicate that interaction of PrP with RNA could drive its conformational switching.Citation113
Characterization of prions and prion domains
Since the discovery of infectious prions, several prion-domain proteins that confer adaptive advantage have been identified in fungi. Prion-mechanisms, that explain how proteins can act as self-replicating epigenetic units of inheritance enabled scientists to understand non mendelian, dominant (“super-suppressor”) inheritance patterns observed in yeast.Citation114,115 [PSI+] element, which is the prion like form of the cellular protein Sup35 which is a translation termination factor, is the best characterized prion-like protein in yeast.Citation116-118 Since then other prion-like proteins with distinct cellular functions such as [URE3] [RNQ+] and [SWI+] that are prion-activated isoforms of Ure2p, Rnq1p, and Swi1 respectively, have been identified in yeast.Citation119
Further characterization of identified yeast prions revealed that conformational switching of normal cellular proteins to prion-like isoforms are dependent on “prion domains” (PrD) that are enriched with Glutamine (Q) and Asparagine (N) residues and are largely devoid of charged residues.Citation119,120 These domains are structurally autonomous and can impart prion-forming ability to novel proteins.
A genome-wide screen performed in S. cerevisiae identified ∼200 proteins with candidate prion domains of which 24 were found capable of amyloid formation based on cell biological and biochemical assays. Notably, when classified according to cellular functions a statistically startling over-representation was seen for proteins involved in RNA binding, transport and processing in the collection of prion-domain containing proteins.Citation121,122
The unusual composition of the PrDs enables the ensemble of multiple cross β sheets that act as a nucleation point for aggregation of prion proteins. Short oligopeptide repeat sequences that reside within the PrDs also have role in the conformational plasticity.Citation123-125 Shuffled sequences of aminoacids from yeast prion domains remain capable of forming amyloids in vitro, indicating that the amino acid composition of prion domains, rather than their sequence alone, contributes importantly to amyloid assembly.Citation126 Recently synthetic prions have been made de novo enabled by the development of computer algorithms like PAPA (Prion Aggregation Prediction Algorithm) that uses amino acid composition to predict prion propensities of intrinsically disordered protein domains.Citation127 However, these analyses likely underestimate functional prion domains in the genome because several known prions are not enriched in Q/N residues. For instance, the yeast prion protein Mod5 lacks Q/N rich domains although it forms amyloid like fibers.Citation128
The characteristic “cross-β sheet” structure of amyloids, resistance to detergents, proteases or chaotropic agents, binding to dyes like Congo red or Thioflavin, ability to induce oligomerization of naïve proteins in vitro and stable propagation and self replication in-vivo are now considered standard attributes of prion filaments, which are enabled by PrDs. PrDs are considered as intrinsically disordered domains, which by sampling multiple conformational states, can occasionally achieve β-sheet rich conformations that support amyloid fiber assembly.
Studies of prions in yeast have led to the proposal that prions ensure heritable phenotypic diversification in response to environmental signals.Citation119 The discovery of non-toxic yeast prions in the yeast genome prompted speculation and further research into functional prions in metazoan organisms. Indeed, this led to the original speculation that prion-like self-sustaining transitions in molecular structures could contribute to the persistence of memory.Citation103
Insights from CPEB / Orb2
Prion-like, self-propagating conformational switches could potentially explain how memories endure in a manner that is resistant to molecular turnover. The discovery that in addition to its canonical form Cytoplasmic Polyadenylation Element Binding (CPEB) protein was capable of existing in prion like conformations in vitro and are required for long-term memory storage in Aplysia provided the first evidence in support of such a possibility.
In neurons, CPEB is localized to synapses and is enriched in the post-synaptic density. In the PSD, in response to neuronal stimulation, CPEB likely activates translation of CaMKII and similar mRNAs that possess CPEs in their UTRs.Citation43 It also associates with motor proteins for mRNA transport and has a role in packaging of bound mRNAs to RNP complexes.Citation129 CPEB initiates polyadenylation induced translation of dormant mRNAs during Xenopus oocyte maturation.Citation130 In developing oocytes, following nuclear export, CPE containing mRNAs are bound by CPEB1 as well as other interacting proteins like PARN (poly A ribonuclease) and Gld2 (polyA polymerase), leading to removal of polyA tail of mRNAs, as PARN overrides Gld2 activity. This leads to translational suppression of mRNAs. However upon activity-induced phosphorylation of CPEB, PARN dissociates from the complex and the mRNA is polyadenylated leading to translation.Citation131
In neurons, CPEB possibly functions in close association with Maskin or other 4E binding proteins like neurogidin to regulate translation.Citation132 In its basal state, 4E-BPs interacts with CPEB as well as the translation initiation factor eIF4E, inhibiting eIF4E interaction with eIF4G and thereby preventing translation (). Upon NMDA mediated neuronal activation, CPEB phosphorylation leads to dissociation of 4E-BP from eIF4E, resulting in the recruitment of CPSF and Gld-2 dependent polyadenylation of mRNAs leading to translation.Citation44,133 Proteins expressed in this manner could (a) effect local synaptic changes; (b) initiate signaling processes to the nucleus; (c) tag synapses such that they are capable of being altered by nuclear-gene expression.
Figure 2. Prion mechanism of CPEB in memory (A) Mechanism of CPEB function in translational regulation: In the basal state CPEB interacts with Maskin or similar 4E-BPs which further interacts with eIF4E and prevents eIF4E interaction with eIF4G. Upon neuronal activity, CPEB gets phosphorylated and polyadenylation enables PABP association with eIF4G, which then disrupts 4E-BP-eIF4E interaction thereby permitting eIF4E-eIF4G interaction and resultant translation. (B) In the basal state both prionogenic and non-prion forms of CPEB exists mostly as monomers and can bind RNA and keep it in a translationally repressed state. Upon activity, the prionogenic form induces aggregation and the prion like nature induces self assembly and propagation of the aggregates where the RNA gets translated (C) CPEB monomers in the resting synapses and Q/N domain mediated amyloid like assemblies in active synapses.
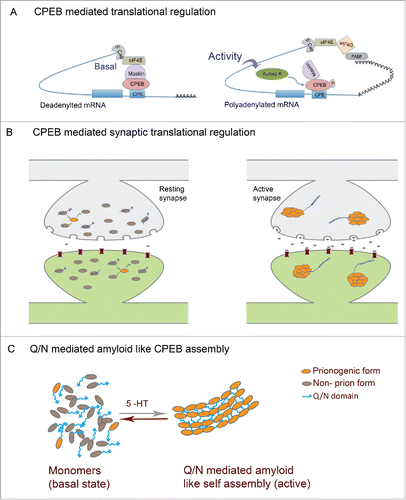
Several lines of evidence support a model in which prion-like characteristics of the CPEB protein enable activity-induced conformational switching from soluble monomer to a self-propagating oligomeric “prion” form to facilitate the persistence of long-term plasticity of specific synapses. These experiments are reviewed below while alternative and/or additional roles for the CPEB and other prion-like domains are also proposed.
What would constitute decisive proof for the CPEB prion-model for LTM
As proving a biological hypothesis can be close to impossible, the absence of conclusive proof is not a particularly severe criticism of the prion model for long-term memory. That being stated, it is useful to outline key requirements for establishing a model, in order to have an objective yardstick for assessing the conclusiveness of current data, which have been discussed and/or reviewed in recent articles.Citation9,10
| (a) | Synaptic activity that triggers long-term memory should be demonstrated to result in conformational switch of CPEB from a monomeric to oligomeric prion-like form at active but not at inactive synapses. | ||||
| (b) | At the activated synapse, the new oligomeric CPEB should be capable of self-renewal, recruiting monomeric CPEB into oligomers over the extended period for which memory is retained. | ||||
| (c) | The self-sustaining prion form should have an activity distinct from the soluble monomeric form and this new activity should be necessary for the persistence of long-term memory in vivo, and sufficient for CPEB's function in memory persistence. | ||||
Evidence from experiments in Aplysia and Drosophila summarized below provide substantial support for this model. However, considerable additional evidence remains necessary to definitively meet each of the major requirements for establishing the prion model.
Molecular properties of aplysia CPEB in vitro and in yeast
Aplysia CPEB1 belongs to the CPEB1 subfamily and is involved in translational regulation in a CPE dependent manner.Citation43,130 The N-terminal domain of ApCPEB has a high Q/N content and lacks an ordered secondary structure, similar to yeast prion domains. Consistent with this, ApCPEB N-terminal domain shows prion like behavior. In yeast cells, ApCPEB N-terminal fused to GFP, shows punctate expression suggestive of heritable aggregates. Consistent with this, the N-terminus of Aplysia CPEB (as well as related domains from Drosophila and mouse orthologs) can confer prion-like behavior to heterologous fusion proteins.Citation134-136
For example, when ApCPEB N-terminus fusions with a constitutively active Glucocorticoid Receptor (GR) are expressed in yeast cells carrying a GR-responsive, β-gal reporter, it is possible to easily read out the activity state of the GR-fusion protein from the color of colonies (white when inactive and blue when active). In this assay the fusion proteins produces metastable blue and white colonies, switch from blue (active) to white (inactive) colonies at rates consistent with prion-like conformational transition. This inactive state is heritable, although it shows occasional reversion. The functional state depicted by GR activity also corresponds with specific physical states of the protein as seen in sucrose gradient assay, where extracts from white colonies settle at the bottom. Thus the Q/N rich domain of ApCPEB confers on a fusion protein, distinct functional and physical properties in a manner that is heritable, transmissible and interconvertible and thereby show fundamental prion like behavior.Citation134 Si et al., also provide interesting data in yeast cells, which are consistent with a model in which only the aggregated form supports translational activation. However, additional direct mechanistic support for this proposal is required. Current data do not completely exclude alternative models E.g. :(a) that aggregation induces translation by blocking a translational repressor activity of the soluble form; and (b) that small oligomers, rather than proposed self-perpetuating amyloids, mediate translational activation in vivo. Recombinant CPEB-Q also demonstrates amyloid fiber formation and purified CPEB amyloid fibers can induce changes in CPEB activity state in a self-perpetuating manner. These results are indicative of a protein-only mechanism that could be used by ApCPEB to create long lasting biochemical traces required for memory persistence.Citation137
CPEB functions and properties in aplysia synapses
In Aplysia sensorimotor cultures, 1 hour after initiation of prolonged 5-HT application, ApCPEB increases 4-5-fold in a protein synthesis dependent manner. Interestingly, single application of 5-HT also induces ApCPEB locally in neurites, as evident from increased immunoreactivity at stimulated synapses. This occurs in neurites devoid of cell bodies as well, confirming local translation of CPEB. However, whether this increased CPEB levels correlates with aggregation of CPEB is unclear. Application of antisense oligos against CPEB at the cell body, result in the premature decay of LTF after 24 hours compared to more than 72 hours for controls. Thus, ApCPEB1 is necessary for the persistent phase of LTF and is either not required, or only required at lower levels, for LTF induction.Citation138
Prion like physical and functional properties of ApCPEB and its role in sustenance of LTF was shown in a subsequent study in 2010. Overexpression of ApCPEB fused to EGFP showed punctate expression in sensory neurons. An antibody (Ab 464) raised against the N-terminal sequences stains puncta observed in sensory neurons and IP using this antibody yields multimeric ApCPEB resistant to detergents. Notably, the punctae formed by ApCPEB do not colocalize with P body markers like Dcp1, suggesting they are distinct from known endogenous RNP granules seen in neurons. Fluorescence reconstitution assays show that the multimers of ApCPEB found in sensory neurons are formed by self assembly and are homotypic. Consistent with prion-like nature, recombinant ApCPEB shows Thioflavin-T reactivity in vitro and the dye binds punctate ApCPEB in sensory neurons suggestive of high amyloid content. Assays using photoactivable GFP to track new protein synthesis, show that the existing aggregates start showing presence of new ApCPEB proteins (green to red transformation) within 4 hours, indicating the self sustaining nature of ApCPEB assembly into these puncta. Importantly, 5x 5HT application onto sensory neurons increases ApCPEB puncta in neurons and 5HT treated sensory neurons show increased ApCPEB multimer formation. In sensory motor neuron cultures, 5× 5HT application leads to LTF that lasts beyond 48 hours. However overexpression of ApCPEB that leads to multimerization in the synaptic as well as extra synaptic areas blocks sustenance of LTF beyond upto 48 hours, an effect dependent on mRNA binding suggesting that the block in 48 hour LTF may arise from key mRNAs being sequestered in ApCPEB aggregates.Citation139
In bifurcated sensory-motor neuron cultures, 5 × 5HT application to one branch normally leads to LTF lasting beyond 72 hours. However, sustained LTF is not seen when protein synthesis is locally inhibited 24 or 48 hours after 5x 5HT application. Indeed, this leads to retraction of newly formed synaptic varicosities as well. Interestingly, blocking ApCPEB using antisense oligonucleotides 24 hours after 5x 5HT treatment also impairs stable maintenance of LTF and associated growth beyond 72 hours. Taking into consideration the conventional role of CPEB in mRNA regulation, these data indicate that ApCPEB regulates translation of synaptic mRNAs during the temporal phase where local protein synthesis has a key role in stabilizing the plasticity of activated synapses, thereby facilitating persistence of memory.Citation140 Together, the yeast experiments using ApCPEB and the in vivo studies in Aplysia sensory neuron cultures are consistent with ApCPEB functioning like a prion by virtue of its N-terminal domain and that ApCPEB mediated prion mechanisms are involved in long lasting LTF of Aplysia sensory motor synapses. The general idea here is that in the resting state of the neurons CPEB acts as a translational repressor of synaptically localized mRNAs. Upon neuronal stimulation the conversion to an active prion like form is induced which then permits translation and the existence of an altered translational state of specific synapses that subsequently facilitates long lasting synaptic changes ().
CPEB1 in Drosophila – Orb2
Drosophila Orb2 belongs to the CPEB2 subfamily of proteins that regulate mRNA translation in a CPE independent manner unlike the homologous “Orb” protein of the CPEB1 subfamily that regulates translation in a CPE dependent manner.Citation141,142 However, similar to ApCPEB1, Drosophila Orb2 contains prion-like Q/N rich domains and appear capable of supporting prion-like assembly. Work in Drosophila, where genetic perturbations can be combined with analyses of long- and short-term memory, have made important contributions to current knowledge of CPEB/ Orb2 function in vivo.
Single or massed olfactory aversive conditioning trials in Drosophila lead different types of short-term memory (STM) that last from few to 24 hours In contrast, spaced training with intervals between training sessions, leads additionally to the formation of protein-synthesis dependent LTM that can persist beyond 7 d.Citation13 In yet another assay (courtship conditioning), young male flies that have suffered multiple rejections from a mated female over a 5 hour training session, show a prolonged suppression of courtship behavior that lasts for well over 24 hours.Citation143
In the fly brain, Orb2 is highly expressed in the neurons and predominantly in the mushroom bodies, a neuropil region previously associated with courtship conditioning and olfactory memory formation.Citation144,145 Drosophila Orb2 mutants engineered to lack the N-terminal prion-like domain show defects in long-term courtship suppression. They are unable to recall rejection for more than 9 hours, after which they court with the enthusiasm of naive flies. Significantly, learning, short-term memory and immediate recall are intact in Orb2 mutants. Additional experiments reveal that Orb2 is necessary during or shortly after the training for its function in long-term 24-hour memory. Consistent with expression pattern, transgenic expression of Orb2 in adult mushroom-body neurons rescues the LTM defect observed in Orb2 mutants deleted for the prion-like domain.Citation146
Drosophila Orb2 has 2 isoforms, Orb2A and Orb2B. The Orb2B isoform is very abundant in fly brain whereas Orb2A is normally nearly undetectable in the fly brain. The two isoforms have similar C-terminal and RNA-binding (RRM) domains but differ in the sequences preceding their N-termini. The Orb2A has a short 8 amino acid stretch whereas in Orb2B isoform it is 162 amino acids long.Citation147,148 Both Orb2A and Orb2B interact with transcripts of Tequila, DaPKC and Murashka, which are required for LTM formation. Removal of the Zn finger RNA-binding domain from the Orb2B isoform abolishes its association with these target mRNA suggestive of direct interaction with target RNAs. The translation of target mRNAs is suppressed when either of the 2 isoforms are expressed in S2 cells that lack endogenous Orb2, clearly indicating a role for Orb2 in mRNA repression.Citation149 Taken together, the results indicate that Orb2 is required for the maintenance of long-term memory through a mechanism that requires its N-terminal domain as well as its ability to regulate translation of mRNAs required for stable alteration of activated synapses.
Two subsequent studies show how each of the 2 Orb2 isoforms contribute to mechanistically distinct Orb2 functions required for LTM persistence. Several observations indicate that Orb2A oligomerise better than Orb2B. Direct biochemical studies show that recombinant Orb2A form SDS resistant oligomers more efficiently. IP from S2 cells yield monomers as well as oligomers and absence of the Q/N domain leads to a loss of oligomers. Synaptosome preparations from heads of tyramine or dopamine stimulated/fed flies show enrichment of oligomeric Orb2, resistant to SDS and other denaturants, which persist for around 20 hours after feeding. Immunoprecipitates form fly brains expressing only Orb2B isoform do not show any oligomers, consistent with a role for Orb2A in aggregation. However this oligomerization is absent in Q/N domain mutants as well as flies lacking Q domain just in the Orb2A.Citation147,148 The 8 amino acids that confer the specificity on Orb2A isoform do not oligomerise on their own indicating that they only act as catalysts for aggregation in S2 cells. Surprisingly, a single point mutation of a conserved phenyl alanine residue to tyrosine (Orb2AF5Y ) in the 8 amino acid stretch of the 2A isoform, dramatically reduces its aggregation capability. These observations show that Orb2A has a role in potentially amyloid-like aggregation of Orb2. Considering the level of expression of Orb2A is very low compared to Orb2B, it is very likely that the Orb2A isoform acts as a nucleation point for oligomerization and recruits Orb2B as well into the complex.Citation148
Strikingly, the Orb2AF5Y mutants defective in oligomer formation show defective persistence of memory associated with, courtship conditioning (early decay, starting at 36 hrs) as well as appetitive conditioning (intact at 24 hrs, but not 48hrs), which is suggestive for a role of Orb2A oligomerization in the persistence of memory.Citation148 These studies thus put forth a possible mechanism of Orb2 function in LTM persistence, wherein following neurotransmitter stimulation, Orb2A induces aggregation by virtue of its Q/N domain recruiting Orb2B also into the same complexes. The Orb2B isoform function more as a conventional CPEB by virtue of its RNA binding domain to translationally regulate mRNAs involved in memory formation. This enables sustenance of alterations in the stimulated synapses required for endurance of formed memories.
The formation of Orb2 oligomers is under control of a protein network that appears to act mainly to regulate the levels, stability and properties of Orb2A. This network including of protein phosphatase 2A (PP2A), Lim Kinase and Transducer of Erb (Tob) ensure formation of Orb2 complexes in a spatially restricted manner.Citation150 Importantly, although Orb2 oligomeric complexes share biochemical features with pathological amyloids, the proposed Orb2 amyloids appear uniquely transient in nature in contrast to classical irreversible and pathological amyloid assemblies.Citation135
The inherent differences in the properties of the 2 isoforms also impinge on the domains of the isoforms specifically required for maintenance of LTM. Flies lacking the Q/N domain in Orb2A but not Orb2B show severe LTM impairment. However, RNA-binding domains in Orb2B but not the Orb2A isoform is necessary for long term courtship memory. Consistent with this a trans heterozygous combination of a Q/N-deleted Orb2B and an RBD-deleted Orb2A show normal LTM as well as presence of Orb2 oligomers in the brain.Citation147 These results show that the oligomerization as well as mRNA regulation is crucial for persistence of LTM but additionally indicate a possible mechanism of Orb2 function in LTM.
Recent studies have shown further distinctive properties of the Orb2 isoforms pertinent to LTM formation. The Orb2 monomer and the oligomer formed by self assembly have different roles in mRNA regulation. The monomer enhances deadenylation of bound mRNAs and thus makes them more susceptible to degradation whereas the oligomer increases polyA length, which possibly stabilizes the mRNA and enhances translation. Deadenylation by the monomer is mediated by CG13928 and the translational activity of oligomeric Orb2 is mediated by CG4612, which is also required for LTM formation.Citation151 These observations suggest that in the basal state Orb2 binds mRNA and keeps them in a repressed state with the help of the deadenylation complex, and upon activation oligomerization initiates translation of mRNAs that were previously sequestered in repressed state.
Additional very interesting but yet puzzling observations lead to the suggestion that Orb2A functions during the acquisition phase of LTM to tags active synapses which are later reactivated during delayed memory replay required for LTM consolidation.Citation152 During delayed memory replay, a second burst of dopamine release drives Orb2A-Orb2B oligomerization that promotes translation required for LTM.Citation152 While the observations appear robust, the nature of the Orb2A dependent early “tag” is mysterious given that this does not seem to require RNA-binding. Similarly, the effect of early Orb2A activity on Orb2 functions induced during memory replay remains unclear as also as the identity and function of proteins synthesized in the delayed phase of LTM consolidation.
Thus, while great progress has been made in identifying various distinctive functions and properties of Orb2 domains required for the persistence of LTM, these have yet to provide a clear and integrated picture of how and when Orb2 and its prion-like domain function in activity-related translational control necessary for LTM.
Mouse CPEB
Mouse CPEB1 belongs to the CPEB1 subfamily, which also contains the Aplysia CPEB1, Xenopus CPEB and the Drosophila Orb proteins. It is localized to synaptic regions in the mouse brain and is implicated in selective modulation of LTP and LTD in hippocampal neurons.Citation153,154 CPEB1 knockout mouse also display moderate defects in synaptic efficacy and extinction of hippocampal memories.Citation155 Mouse CPEB2-4 belongs to the CPEB2 subfamily and regulates translation in CPE independent manner.Citation156 In mouse hippocampal neurons CPEB3 is activated and ubiquitinated by Neuralized1, an E3 ubiquitin ligase that is implicated in memory formation and synaptic plasticity. Neuralized overexpression leads to structural plasticity of dendritic spines and upregulation of AMPA receptors GluA1 and GluA2 that are regulatory targets of CPEB3.Citation157 Like CPEB1 and Orb2 in Drosophila, CPEB3 forms aggregates, which are self-propagating and heritable in yeast, in a N terminal subdomain dependent manner. CPEB3 aggregation is reported to occur in mouse brain upon appropriate neuronal stimulation. In synapses, CPEB3 physically interacts with actin and its aggregation is dependent on the actin framework. Consistent with this, actin expression is regulated by CPEB3, in response to neuronal activity.Citation136 The interaction with actin framework is suggested to play a role in maintaining an open conformation, which can more efficiently seed aggregation of prions. CPEB3 knock out mice show defects in memory consolidation associated with spatial learning and disruption of CPEB3 function after consolidation of memory leads to defect in long-term maintenance of memory. Consistently disruption of CPEB3 in adult hippocampus leads to defects in synaptic plasticity as well. The function of CPEB3 in memory and synaptic plasticity is mediated by the N-terminal prion like domain that is responsible for activity-induced aggregation of CPEB3.Citation158 CPEB3 aggregation is regulated by sumoylation. Under resting conditions, the CPEB3 stays a soluble monomer and is sumoylated. Upon activity in neuron, desumoylation leads to enhanced aggregate formation and there occurs learning induced decrease in sumoylation of CPEB3.Citation159
Do self-perpetuating assemblies of CPEB1/Orb2/CPEB3 conformers contribute to LTM?
The supportive evidence detailed above can be summarized as follows. 1. The proteins themselves are required for long-term memory formation or its neurophysiological correlates like LTF. 2. In yeast cells, cultured cells and neurons, the proteins are capable of forming aggregates or multimers that show differing subsets of properties expected of amyloids. 3. Mutations that block aggregate formation in reduced test systems, also impair persistence of long-term memory formation of LTM.
Yet these fall short of proving the core elements of the prion-memory hypothesis. The oligomerization of CPEB/Orb2 in response to training that results in LTM has not been shown in neurons or synapses that encode memory in vivo. We do not know whether activity-induced CPEB/Orb2 aggregates are prion-like and capable of self-propagation. It has not been shown in-vivo if the multimers formed in response to activity are capable of recruiting synaptic mRNAs into these complexes and if these multimers are translation hotspots in the neurons and whether such recruitment and sustained complex activity is necessary for the persistence of LTM.
Of several alternative models that could potentially account for many of the key experimental observations, we suggest one as being particularly important to address experimentally. N-terminal domains of CPEB/Orb2 could mediate transient oligomerization that is necessary for activity-induced translation of synaptic mRNAs. This could be accomplished by de-repression, e,g. by disengaging from granules containing target mRNAs and/or by promoting translation e.g. providing, through oligomerization, a multivalent binding site for core translational activators. Variants of this latter model, even if proven to be incorrect for Orb2/CPEB, may yet provide a more general framework for understanding how prion-like domains in other RNA-binding proteins could contribute to neuronal translational control and long-term plasticity.
Prion like domains in neuronal RNP mediated translation
Prion-like domains are strikingly enriched in RNA-binding proteins, suggesting that they provide an activity particularly relevant to the biology of RNA regulation.Citation121,160 Whole genome screens of yeast, humans and Aplysia using different prediction algorithms for identifying prion domains show enrichment of proteins with RNA binding motifs.Citation161 RNA binding components of RNPs possess Intrinsically Disordered Regions (IDRs) that are characterized by low amino acid complexity (LC), absence of hydrophobic residues and enrichment of charged residues. In macromolecular assemblies, IDRs often interact weakly and transiently with other binding partners through short linear motifs composed of 2-8 amino acids. ID domains are capable of adopting conformations that allow weak and potentially multivalent protein-protein interactions.Citation5,162,163 Co-operative binding between LC domains can however transform them to stronger and stable interactions.Citation122 PrDs of prions that exhibit self assembly and aggregation are found to exist in disordered state and are also predicted to be IDRs/LC domains.Citation164-166
The biophysical nature and structural details of interaction between these domains in vivo remains to be clearly identified. Strikingly although only 1% of human genome contain RNA recognition motif (RRM), 11% of human prion domain containing proteins possess RRM, indicating their enrichment in the subset. Notable among these are TDP-43, FUS, TAF15 and hnRNP isoforms A2B1 and A1 that are implicated in the pathology of amyotrophic lateral sclerosis (ALS) and Fronto Temporal Degeneration (FTD). Mutations in the LC domains of these RNP components show genetic or pathologic association with neurodegenerative disorders, and hyper aggregation induced inclusion bodies comprising of these RNP components are also found associated with disease pathology, both suggestive of the role of LC domains in driving amyloid like assemblies.Citation161,167,168
RNA granules in cells
RNP particles are cytoplasmic foci formed by interaction of RNA and RNA interacting proteins and are globally involved in various cellular functions. RNPs enable membrane-free compartmentalization of cytoplasm and function as cytoplasmic microdomains wherein RNA protein interactions are enriched by aggregation of proteins and sequestered RNAs. RNA granules therefore function both in RNA localization as well as in local translation, enabling spatiotemporal regulation of protein synthesis.Citation71,169,170
Much of what is known about RNA granule functions comes from studies of P-bodies (PBs) and stress granules (SGs) in eukaryotic cells that contain non-translating mRNAs, together with various translational regulators. PBs contain RNA-degradative enzymes and translational repressors whereas SGs contain translational initiation factors stalled at specific stage of translational initiation.Citation171-173 Phosphorylation, methylation, acetylation, ubiquitinylation and O-Glc-NAc modifications are all reported to have effect on PB or SG formation.Citation169,174
Efficient and rapid translation of mRNAs in response to neuronal activity requires mRNA to be conveniently localized at synaptic regions in the resting state. They also need to be transported in a translationally repressed state to enable protein synthesis in accordance with neuronal acivity. Neuronal granules are a potentially diverse collection of RNP assemblies that occur in neurons associated with mRNA transport, localization, and local translation.Citation42,67,175 Neuronal granules are compositionally related to P bodies and stress granules.Citation176 The dynamic movement of RNA granules to dendrites were first observed in neurons using localization with fluorescent RNA dye SYTO14. These granules were found to contain translational machinery indicating that neuronal RNA granules were competent of regulating translation.Citation42 Later microtubule dependent bidirectional movement of RNA binding proteins like Staufen and ZBP1 protein and dynamic trafficking of RNA granules containing both ZBP1 and its target β-actin mRNA were reported.Citation64,177 A seminal study by Kanai et al showed that kinesin heavy chain (KIF5b) interact with large number of RNA binding proteins and major dendritically localized mRNAs like CaMKII and Arc.Citation67 The composition of these granules suggested that apart from RNA transport, neuronal RNPs are capable of regulating diverse aspects of RNA metabolism. Depolarization induced enhanced transport of RNPs to dendrites suggest that neuronal RNP transport is regulated by synaptic input.Citation178 RNA granules biochemically isolated from rat brain identified many RNA binding proteins like Staufen, SYNCRIP and dead box helicases to be prominent RNP components along with translational machinery and cytoskeletal proteins.Citation179 Fragile-X Mental Retardation Protein (FMRP) is another well established RNP component implicated in neuronal translational regulation and activity induced RNP transport. FMRP interaction with miRNA machinery is closely associated with its role in regulating the translation of mRNAs housed in RNPs.Citation180,181 In cultured hippocampal neurons, neuronal activation results in disassembly of specific neuronal RNA granules concurrently with increased local translation of RNAs localized to these granules. Stimulation releases free mRNA as well as ribosomes indicative of derepression.Citation4,70,182 These and several other arguments indicate that neural activity triggers signaling pathways that first cause granule disassembly and associated mRNA release. This enables mRNA translation and local protein synthesis required for synaptic plasticity underlying memory. Consistently several neuronal RNA-granule components such as Ataxin2, Staufen and FMRP have been shown to be required for long-term memory formation as well.Citation85,86,183-185
Prion-like domains may regulate RNA granule assembly
There is a growing chain of evidence to suggest that prion-like domains on RBPs have an important role in assembly and disassembly of RNA granules Prion like domains of, TIA-1 and TIA-R proteins are required for mammalian stress-granule assembly.Citation186 Similarly, Q/N domains of the Lsm4 protein that is required for P body formation in yeast and GW182 that is required for P body assembly in human and Drosophila cells mediate RNA granule aggregation.Citation186-189
Although conventional high-affinity interactions also contribute to RNA granule assembly,Citation173 recent evidence suggest that prion-like domains mediate weak interactions among RBPs that drive a “liquid-liquid phase transition” from a dispersed soluble state in the cytoplasm to a condensed state that resemble “liquid droplets..” Consistent with such a model, RNA granules are dynamic in that, like liquid droplets, their component proteins exchange and move freely within the granule and also exchange with the cytoplasmic pool. The importance of such liquid–liquid demixing phase transition is best demonstrated in C.elegans P-body assembly, where the granules show typical liquid-droplet characteristics like wetting of membranes, fusion and dripping during assembly.Citation5 Interaction with RNA targets can drive and enhance phase separation by altering the characteristics of liquid droplet assemblies of the regulatory proteins, as in the case of Whi3 protein.Citation190 Multivalent weak interactions among domains of protein assemblies are capable of promoting phase separation.Citation122,148,191 Similar multivalent interactions leading to phase separation are also implicated in the formation and of the post synaptic densitiy (PSD).Citation192
Several studies indicate that LC domains can drive assembly of RNP like aggregates. Candidate prion domains identified from yeast screens can act as independent domains to form cytoplasmic aggregates.Citation121 Two other studies have demonstrated how LC domains can drive phase separation. B-isox when added to cell lysates yield a precipitate that is enriched with RNA binding proteins most of which were found to possess LC domain. LC domains can be necessary and sufficient to phase separate and induce formation of hydrogels, in this cell free system. Invitro, these hydrogels are also capable of retaining other LC sequence containing proteins suggestive of prion-like propensities of LC domains. Also, disordered domains form different proteins co- phase separate to form liquid droplets in cytoplasm. Phosphorylation of the low-complexity domains render them unable to form hydrogel, suggestive of how such cellular assemblies may be similarly regulated.Citation7,8
The ability of the LC domains to polymerize and drive regulated phase separation is fundamental to the formation of RNP aggregates as well as RNA metabolism. Mutations in the LC domains of RNA binding proteins like hnRNPA2B1/A1 or FUS, TDP-43, TAF15 possibly affect the nucleation as well as polymerization of LC domains, leading to enhanced assembly of RNP like aggregates/stress granules and increased incorporation of these proteins to stress granules leading to disruption in the RNA homeostasis.Citation161,167,168,193
Structural basis for prion-like domain interactions in RNA granules
Electron microscopy and X-ray diffraction studies show that hydrogels contain morphologically uniform amyloid like fibers that possess characteristic cross β sheet structure.Citation8 However unlike them, LC induced fibers are very unstable and sensitive to low concentrations of SDS. This could indicate that amyloid like assemblies that are formed in physiological conditions are transient and reversible unlike irreversible pathological assemblies. More recent using NAI (N-acetylimidazole) based chemical footprinting assay provided further evidence that stacked β-sheet interactions, typical of amyloid type assembly, may be involved in RBP-assembly in vivo. These studies demonstrate that polymers formed by the prion-like domain containing protein hnRNPA1 in nuclei are required for interactions mediated by the same residues that are necessary for fiber assembly in hydrogels as well as in liquid droplets formed by LC domains of recombinant hnRNPA1.Citation194 However, recent studies of FUS and RNPA1 variants have argued that mutations that prevent amyloid-type associations in these proteins, do not interfere with their ability to associate with and function in RNA granules.Citation6,168 Thus, the current data are consistent with the idea that prion-like domains form transient oligomeric assemblies. These assemblies could arise either from weak multivalent interactions as proposed most clearly for polyglutamine domainsCitation148,195,196 or by reversible stacking of monomers into reversible oligomeric assemblies mediated by transient β-sheet interactions.Citation8
Reversible amyloid assembly mediated by prion domains?
Toxic and pathologic prions are characterized by formation of stable and irreversible amyloids. Thus, the concept of reversible amyloids appears almost oxymoronic. However other than described earlier many cellular proteins demonstrate template driven self-assembly leading to formation of transient and reversible macromolecular complexes. Such instances of functional “amyloid-like” assembly govern many cellular processes and occur in response to cellular cues and signals ensuring spatio temporal regulation.
The transcription factor GAF (GAGA factor) in Drosophila possesses a prion-forming PrD that function in nucleosome remodeling thereby linking transcriptional regulation to epigenetic control. Considering several yeast prions are also transcriptional factors that enable heritable phenotypic diversification in response to environmental signals, this indicates that prion mechanisms are widespread in effecting epigenetic inheritance.Citation197
Prion-like mechanisms are also found functional in immune system. In case of mammals, in response to viral infection pattern recognition receptors RIG-1 and NLRP-3 induce prion-like conformational switching and polymerization of their adaptors MAVS and ASC thereby enabling downstream signaling cascade resulting in cytokine secretion and cell death. An analogous phenomenon is observed in yeast cells, where NWD2 protein induces similar conversion of downstream adaptor protein HET-S/sCitation198. These fibers are self propagating and heritable but are notably different from conventional prions in that the fibers do not have the typical cross-β structures of amyloid fibers, but instead retain α helical conformation.Citation199 However, there is considerable conceptual similarity: indeed, it is now postulated that the persistence of prion characteristics of MAVS/ASC/HET-S/s polymers after infection provides the cell with molecular memory of the infection.
Concluding remarks
Does the mechanism by which CPEB regulates LTM involve the formation of self-sustaining amyloid assemblies? Is this mechanism unique for CPEB or is it shared by other proteins with similar molecular and biochemical attributes? Or are alternative mechanisms involved either for CPEB or for other prion-domain containing proteins also necessary for long-term memory? Why so many different prion-domain containing RBPs exist. Do multiple components promote different macromolecular assemblies with distinct characteristics and spatial distribution? These questions are still open. They may be addressed not only by detailed analysis of CPEB, but also by mechanistic analyses of similar proteins. Interesting candidates are Atx2 and FMRP, both of which are RNA-granule proteins, involved in neuronal translational control and preferentially required for long-term memory formation.Citation85,86,200-202 Algorithms that are designed to detect prion like Q/N domains identify both Atx2 and FMRP as prion like domain containing proteins.Citation160,166,184,203
We suggest alternative models for how these proteins could function, being aware not only that these models represent extreme and not mutually exclusive positions when intermediate and compromise positions are tenable, but also that different RBPs may differ in their mechanism of action.
CPEB and/or other RBPs undergo prion-like transformation into stable self-sustaining amyloids capable of sustained translational activation. The persistence of this biochemically altered state of CPEB underlies the persistence of memory. CPEB induces other proteins with LC domains to undergo such transition, thereby initiating a cascade of assemblies pertinent to memory formation. These possibilities need to be explored.
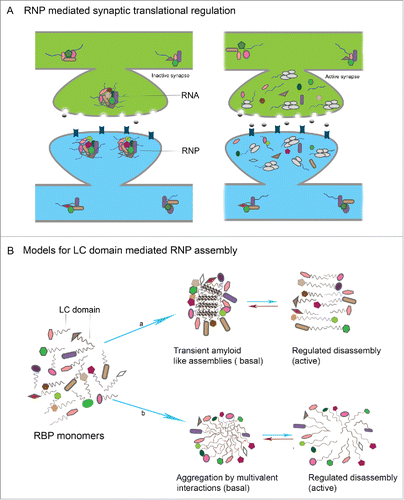
RBPs form prion-domain mediated assemblies wherein RNA is held in a translationally repressed state in basal conditions. These may function one of 2 ways. (a) Neuronal activity can lead to disassembly of the aggregates, leading to increased accessibility and translation of stored dormant mRNAs. Thus self assembly mediated by conformational switching and its coupling with neuronal activity in such proteins could provide a mechanism which enable synapse-specific translation that is crucial for induction of long-term memory (). (b) In a related schema, granule disassembly may be associated with the active change of RBPs into translationally competent complexes. For example, by freeing RBPs from granules, they could allow the formation of CPEB (or other RBP) oligomers that provide an oligomeric disordered protein interface with which that translational machinery associates to activate local translation.
The structural basis for prion-domain mediated interactions in vivo is particularly unclear, although there is consensus that these are regulated by intracellular signaling. Three possibilities exist: (a) liquid-like states driven by weak multivalent interactions among disordered protein domains; (b) stable amyloid fibers arising from β-sheet stacking of RBP monomers via the prion-like domain; (c) reversible oligomers arising from small scale β-sheet stacking among RBP monomers (). This last notion that conformations and residues involved in forming stable, self-sustaining amyloid fibers in various test systems, could in fact normally mediate small and reversible oligomeric assemblies in vivo is important for the interpretation and design of experiments to test the prion-hypothesis of memory.
Figure 3. LC domain mediated RNP assembly in neuronal translational control (A) In the resting state, RNPs formed by interaction of individual RBPs and RNAs, sequester the RNA within RNPs in a translationally repressed state. Upon activity, the RNP disassembles leading to release of suppression resulting in translation of mRNAs with key synaptic functions (B) Models for LC mediated RNP assembly in neurons: in the resting state of synapses, individual RBPs interact by virtue of LC domains that are intrinsically disordered and form either a) transient and reversible amyloid like assemblies or b) reversible aggregates that are formed as result of weak multivalent interactions between LC domains.
Key to discriminating among these models will be both a wide range of experiments on different prion domain containing RBPs as well as the identification and analysis of mutations that clearly block one or other biochemical activity of each protein and the assessment of the effect of these mutations on definitive cell biological and behavioral assays for RNA granule assembly, disassembly and function as well as for long-term synaptic plasticity and long-term memory in vivo.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank K. VijayRaghavan, Baskar Bakthavachalu, Jens Hillebrand, Joern Heulsmeier and Sankar Narayanan for insights, helpful comments and discussions. We also thank David Glanzman and Kausik Si for conversations that have been constructive in drafting this review. Indulekha P.S was supported by research fellowship from CSIR-UGC and NCBS-TIFR core funding. Mani Ramaswami acknowledges Science Foundation of Ireland (SFI) for research support.
References
- Kandel ER. The molecular biology of memory storage: A dialog between genes and synapses. Bioscience Rep 2001; 21:565-611; PMID:16134023; http://dx.doi.org/10.1023/A:1014775008533
- Kandel ER, Dudai Y, Mayford MR. The molecular and systems biology of memory. Cell 2014; 157:163-86; PMID:24679534; http://dx.doi.org/10.1016/j.cell.2014.03.001
- Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling (vol 169, pg 871, 2005). J Cell Biol 2005; 170:847; PMID:15967811; http://dx.doi.org/10.1083/jcb.200502088
- Kiebler MA, Bassell GJ. Neuronal RNA granules: Movers and makers. Neuron 2006; 51:685-90; PMID:16982415; http://dx.doi.org/10.1016/j.neuron.2006.08.021
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Jülicher F, Hyman AA. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 2009; 324:1729-32; PMID:19460965; http://dx.doi.org/10.1126/science.1172046
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 2015; 163:123-33; PMID:26406374; http://dx.doi.org/10.1016/j.cell.2015.09.015
- Han TNW, Kato M, Xie SH, Wu LC, Mirzaei H, Pei JM, Chen M, Xie Y, Allen J, Xiao G, et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 2012; 149:768-79; PMID:22579282; http://dx.doi.org/10.1016/j.cell.2012.04.016
- Kato M, Han TNW, Xie SH, Shi K, Du XL, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753-67; PMID:22579281; http://dx.doi.org/10.1016/j.cell.2012.04.017
- Si K. Prions: what are they good for? Annu Rev Cell Dev Bi 2015; 31:149-69; PMID:26407211; http://dx.doi.org/10.1146/annurev-cellbio-100913-013409
- Si K, Kandel ER. The role of functional prion-like proteins in the persistence of memory. Cold Spring Harb Perspect Biol 2016; 8(4):a021774; PMID:27037416; http://dx.doi.org/10.1101/cshperspect.a021774
- Mcgaugh JL. Time-dependent processes in memory storage. Science 1966; 153:1351-&; PMID:5917768; http://dx.doi.org/10.1126/science.153.3742.1351
- Ebbinghaus H. Memory - a contribution to experimental psychology. Sci Am 1966; 214:144-&; PMID:25206041; http://dx.doi.org/10.1038/scientificamerican0666-144
- Tully T, Preat T, Boynton SC, Delvecchio M. Genetic dissection of consolidated memory in drosophila. Cell 1994; 79:35-47; PMID:7923375; http://dx.doi.org/10.1016/0092-8674(94)90398-0
- Bailey CH, Kandel ER, Si KS. The persistence of long-term memory: A molecular approach to self-sustaining changes in learning-induced synaptic growth. Neuron 2004; 44:49-57; PMID:15450159; http://dx.doi.org/10.1016/j.neuron.2004.09.017
- Kandel ER. Neuroscience - The molecular biology of memory storage: A dialogue between genes and synapses. Science 2001; 294:1030-8; PMID:11691980; http://dx.doi.org/10.1126/science.1067020
- Dudai Y. Neurogenetic dissection of learning and short-term-memory in drosophila. Annu Rev Neurosci 1988; 11:537-63; PMID:3129981; http://dx.doi.org/10.1146/annurev.ne.11.030188.002541
- Glanzman DL. Common mechanisms of synaptic plasticity minireview in vertebrates and invertebrates. Curr Biol 2010; 20:R31-R6; PMID:20152143; http://dx.doi.org/10.1016/j.cub.2009.10.023
- Malenka RC, Bear MF. LTP and LTD: An embarrassment of riches. Neuron 2004; 44:5-21; PMID:15450156; http://dx.doi.org/10.1016/j.neuron.2004.09.012
- Lynch MA. Long-term potentiation and memory. Physiol Rev 2004; 84:87-136; PMID:14715912; http://dx.doi.org/10.1152/physrev.00014.2003
- Hawkins RD, Kandel ER, Bailey CH. Molecular mechanisms of memory storage in Aplysia. Biol Bull-Us 2006; 210:174-91; PMID:16801493; http://dx.doi.org/10.2307/4134556
- Goelet P, Castellucci VF, Schacher S, Kandel ER. The long and the short of long-term-memory - a molecular framework. Nature 1986; 322:419-22; PMID:2874497; http://dx.doi.org/10.1038/322419a0
- Bailey CH, Bartsch D, Kandel ER. Toward a molecular definition of long-term memory storage. P Natl Acad Sci USA 1996; 93:13445-52; PMID:8942955; http://dx.doi.org/10.1073/pnas.93.24.13445
- Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature 1993; 361:31-9; PMID:8421494; http://dx.doi.org/10.1038/361031a0
- Mauelshagen J, Parker GR, Carew TJ. Dynamics of induction and expression of long-term synaptic facilitation in Aplysia. J Neurosci 1996; 16:7099-108; PMID:8929419
- Abrams TW, Castellucci VF, Camardo JS, Kandel ER, Lloyd PE. 2 endogenous neuropeptides modulate the gill and siphon withdrawal reflex in aplysia by presynaptic facilitation involving camp-dependent closure of a serotonin-sensitive potassium channel. P Natl Acad Sci-Biol 1984; 81:7956-60; PMID:6096869; http://dx.doi.org/10.1073/pnas.81.24.7956
- Brunelli M, Castellucci V, Kandel ER. Synaptic facilitation and behavioral sensitization in Aplysia: possible role of serotonin and cyclic AMP. Science 1976; 194:1178-81; PMID:186870; http://dx.doi.org/10.1126/science.186870
- Mauelshagen J, Sherff CM, Carew TJ. Differential induction of long-term synaptic facilitation by spaced and massed applications of serotonin at sensory neuron synapses of Aplysia californica. Learn Memory 1998; 5:246-56; PMID:10454368
- Frost WN, Castellucci VF, Hawkins RD, Kandel ER. Mono-synaptic connections made by the sensory neurons of the gill-withdrawal and siphon-withdrawal reflex in aplysia participate in the storage of long-term-memory for sensitization. P Natl Acad Sci USA 1985; 82:8266-9; PMID:16593630; http://dx.doi.org/10.1073/pnas.82.23.8266
- Montarolo PG, Goelet P, Castellucci VF, Morgan J, Kandel ER, Schacher S. A critical period for macromolecular-synthesis in long-term heterosynaptic facilitation in aplysia. Science 1986; 234:1249-54; PMID:3775383; http://dx.doi.org/10.1126/science.3775383
- Martin KC, Casadio A, Zhu H, Yaping E, Rose JC, Chen M, Bailey CH, Kandel ER. Synapse-specific, long-term facilitation of aplysia sensory to motor synapses: a function for local protein synthesis in memory storage. Cell 1997; 91:927-38; PMID:9428516; http://dx.doi.org/10.1016/S0092-8674(00)80484-5
- Bailey CH, Kandel ER. Structural-changes accompanying memory storage. Annu Rev Physiol 1993; 55:397-426; PMID:8466181; http://dx.doi.org/10.1146/annurev.ph.55.030193.002145
- Bacskai BJ, Hochner B, Mahautsmith M, Adams SR, Kaang BK, Kandel ER, Tsien RY. Spatially resolved dynamics of camp and protein kinase-a subunits in aplysia sensory neurons. Science 1993; 260:222-6; PMID:7682336; http://dx.doi.org/10.1126/science.7682336
- Martin KC, Michael D, Rose JC, Barad M, Casadio A, Zhu HX, Kandel ER. MAP kinase translocates into the nucleus of the presynaptic cell and is required for long-term facilitation in Aplysia. Neuron 1997; 18:899-912; PMID:9208858; http://dx.doi.org/10.1016/S0896-6273(00)80330-X
- Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell 1998; 95:211-23; PMID:9790528; http://dx.doi.org/10.1016/S0092-8674(00)81752-3
- Wang HD, Tiedge H. Translational control at the synapse. Neuroscientist 2004; 10:456-66; PMID:15487031; http://dx.doi.org/10.1177/1073858404265866
- Rao A, Steward O. Evidence that protein constituents of postsynaptic membrane specializations are locally synthesized - analysis of proteins synthesized within synaptosomes. J Neurosci 1991; 11:2881-95; PMID:1880554
- Steward O, Levy WB. Preferential localization of polyribosomes under the base of dendritic spines in granule cells of the dentate gyrus. J Neurosci 1982; 2:284-91; PMID:7062109
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell 2006; 127:49-58; PMID:17018276; http://dx.doi.org/10.1016/j.cell.2006.09.014
- Tiedge H, Brosius J. Translational machinery in dendrites of hippocampal neurons in culture. J Neurosci 1996; 16:7171-81; PMID:8929426
- Sutton MA, Schuman EM. Local translational control in dendrites and its role in long-term synaptic plasticity. J Neurobiol 2005; 64:116-31; PMID:15883999; http://dx.doi.org/10.1002/neu.20152
- Kuhl D, Skehel P. Dendritic localization of mRNAs. Curr Opin Neurobiol 1998; 8:600-6; PMID:9811623; http://dx.doi.org/10.1016/S0959-4388(98)80087-1
- Knowles RB, Sabry JH, Martone ME, Deerinck TJ, Ellisman MH, Bassell GJ, Kosik KS. Translocation of RNA granules in living neurons. J Neurosci 1996; 16:7812-20; PMID:8987809
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of alpha-CaMKII mRNA at synapses. Neuron 1998; 21:1129-39; PMID:9856468; http://dx.doi.org/10.1016/S0896-6273(00)80630-3
- Richter JD. CPEB: a life in translation. Trends Biochem Sci 2007; 32:279-85; PMID:17481902; http://dx.doi.org/10.1016/j.tibs.2007.04.004
- Ouyang Y, Rosenstein A, Kreiman G, Schuman EM, Kennedy MB. Tetanic stimulation leads to increased accumulation of Ca2+/calmodulin-dependent protein kinase II via dendritic protein synthesis in hippocampal neurons. J Neurosci 1999; 19:7823-33; PMID:10479685
- Wang DO, Kim SM, Zhao YL, Hwang H, Miura SK, Sossin WS, Martin KC. Synapse- and stimulus-specific local translation during long-term neuronal plasticity. Science 2009; 324:1536-40; PMID:19443737; http://dx.doi.org/10.1126/science.1173205
- Sherff CM, Carew TJ. Coincident induction of long-term facilitation in Aplysia: cooperativity between cell bodies and remote synapses. Science 1999; 285:1911-4; PMID:10489370; http://dx.doi.org/10.1126/science.285.5435.1911
- Sherff CM, Carew TJ. Coincident induction of long-term facilitation at sensory-motor synapses in Aplysia: presynaptic and postsynaptic factors. Neurobiol Learn Mem 2002; 78:498-507; PMID:12559830; http://dx.doi.org/10.1006/nlme.2002.4092
- Ashraf SI, McLoon AL, Sclarsic SM, Kunes S. Synaptic protein synthesis associated with memory is regulated by the RISC pathway in Drosophila. Cell 2006; 124:191-205; PMID:16413491; http://dx.doi.org/10.1016/j.cell.2005.12.017
- Bramham CR, Wells DG. Dendritic mRNA: transport, translation and function. Nat Rev Neurosci 2007; 8:776-89; PMID:17848965; http://dx.doi.org/10.1038/nrn2150
- Aakalu G, Smith WB, Nguyen N, Jiang CG, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron 2001; 30:489-502; PMID:11395009; http://dx.doi.org/10.1016/S0896-6273(01)00295-1
- Casadio A, Martin KC, Giustetto M, Zhu HX, Chen M, Bartsch D, Bailey CH, Kandel ER. A transient, neuron-wide form of CREB-mediated long-term facilitation can be stabilized at specific synapses by local protein synthesis. Cell 1999; 99:221-37; PMID:10535740; http://dx.doi.org/10.1016/S0092-8674(00)81653-0
- Cai D, Chen S, Glanzman DL. Postsynaptic regulation of long-term facilitation in Aplysia. Curr Biol 2008; 18:920-5; PMID:18571411; http://dx.doi.org/10.1016/j.cub.2008.05.038
- Glanzman DL. Common mechanisms of synaptic plasticity in vertebrates and invertebrates. Curr Biol 2010; 20:R31-6; PMID:20152143; http://dx.doi.org/10.1016/j.cub.2009.10.023
- Martin KC. Synaptic tagging during synapse-specific long-term facilitation of Aplysia sensory-motor neurons. Neurobiol Learn Mem 2002; 78:489-97; PMID:12559829; http://dx.doi.org/10.1006/nlme.2002.4088
- Sossin WS. Mechanisms for the generation of synapse specificity in long-term memory: the implications of a requirement for transcription. Trends Neurosci 1996; 19:215-8; PMID:8761954; http://dx.doi.org/10.1016/0166-2236(96)20016-5
- Frey U, Morris RGM. Weak before strong: dissociating synaptic tagging and plasticity-factor accounts of late-LTP. Neuropharmacology 1998; 37:545-52; PMID:9704995; http://dx.doi.org/10.1016/S0028-3908(98)00040-9
- Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature 1997; 385:533-6; PMID:9020359; http://dx.doi.org/10.1038/385533a0
- Redondo RL, Morris RG. Making memories last: the synaptic tagging and capture hypothesis. Nat Rev Neurosci 2011; 12:17-30; PMID:21170072; http://dx.doi.org/10.1038/nrn2963
- Alarcon JM, Barco A, Kandel ER. Capture of the late phase of long-term potentiation within and across the apical and basilar dendritic compartments of CA1 pyramidal neurons: synaptic tagging is compartment restricted. J Neurosci 2006; 26:256-64; PMID:16399695; http://dx.doi.org/10.1523/JNEUROSCI.3196-05.2006
- Kim S, Martin KC. Neuron-wide RNA transport combines with netrin-mediated local translation to spatially regulate the synaptic proteome. Elife 2015; 4; PMID:25569157; http://dx.doi.org/10.7554/eLife.04158
- Kiebler MA, Hemraj IA, Verkade P, Kohrmann M, Fortes P, Marion RM, Ortín J, Dotti CG. The mammalian Staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: Implications for its involvement in mRNA transport. J Neurosci 1999; 19:288-97; PMID:9870958
- Le Hir H, Moore MJ, Maquat LE. Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Gene Dev 2000; 14:1098-108; PMID:10809668
- Kohrmann M, Luo M, Kaether C, DesGroseillers L, Dotti CG, Kiebler MA. Microtubule-dependent recruitment of Staufen-Green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol Biol Cell 1999; 10:2945-53; PMID:10473638; http://dx.doi.org/10.1091/mbc.10.9.2945
- Puthanveettil SV. RNA transport and long-term memory storage. Rna Biol 2013; 10:1765-70; PMID:24356491; http://dx.doi.org/10.4161/rna.27391
- Wells DG. RNA-binding proteins: A lesson in repression. J Neurosci 2006; 26:7135-8; PMID:16822967; http://dx.doi.org/10.1523/JNEUROSCI.1795-06.2006
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron 2004; Aug 19;43(4):513-25; PMID: 15312650; http://dx.doi.org/10.1016/j.neuron.2004.07.022
- Buxbaum AR, Wu B, Singer RH. Single beta-actin mRNA detection in neurons reveals a mechanism for regulating its translatability. Science 2014; 343:419-22; PMID:24458642; http://dx.doi.org/10.1126/science.1242939
- Iacoangeli A, Tiedge H. Translational control at the synapse: role of RNA regulators. Trends Biochem Sci 2013; 38:47-55; PMID:23218750; http://dx.doi.org/10.1016/j.tibs.2012.11.001
- Krichevsky AM, Kosik KS. Neuronal RNA granules: A link between RNA localization and stimulation-dependent translation. Neuron 2001; 32:683-96; PMID:11719208; http://dx.doi.org/10.1016/S0896-6273(01)00508-6
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell 2009; 136:719-30; PMID:19239891; http://dx.doi.org/10.1016/j.cell.2009.01.044
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 2009; 136:731-45; PMID:19239892; http://dx.doi.org/10.1016/j.cell.2009.01.042
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Bio 2004; 5:827-35; PMID:17913617; http://dx.doi.org/10.1038/nrm1488
- Lisman J. The CaM kinase II hypothesis for the storage of synaptic memory. Trends Neurosci 1994; 17:406-12; PMID:7530878; http://dx.doi.org/10.1016/0166-2236(94)90014-0
- Richter JD, Klann E. Making synaptic plasticity and memory last: mechanisms of translational regulation. Gene Dev 2009; 23:1-11; PMID:19136621; http://dx.doi.org/10.1101/gad.1735809
- Stratton M, Lee IH, Bhattacharyya M, Christensen SM, Chao LH, Schulman H, Groves JT, Kuriyan J. Activation-triggered subunit exchange between CaMKII holoenzymes facilitates the spread of kinase activity (vol 3, e01610, 2014). Elife 2014; 1;3:e01610; PMID: 24473075; http://dx.doi.org/10.7554/eLife.01610
- Bhattacharyya M, Stratton MM, Going CC, McSpadden ED, Huang Y, Susa AC, Elleman A, Cao YM, Pappireddi N, Burkhardt P, et al. Molecular mechanism of activation-triggered subunit exchange in Ca2+/ calmodulin-dependent protein kinase II. Elife 2016; Mar 7;5.pii:e13405; PMID: 26949248; http://dx.doi.org/10.7554/eLife.13405.
- Lisman J, Yasuda R, Raghavachari S. Mechanisms of CaMKII action in long-term potentiation. Nat Rev Neurosci 2012; 13:169-82; PMID:22334212; http://dx.doi.org/10.1038/nrn3192
- Mori Y, Imaizumi K, Katayama T, Yoneda T, Tohyama M. Two cis-acting elements in the 3′ untranslated region of alpha-CaMKII regulate its dendritic targeting. Nat Neurosci 2000; 3:1079-84; PMID:11036263; http://dx.doi.org/10.1038/80591
- Mayford M, Baranes D, Podsypanina K, Kandel ER. The 3′-untranslated region of CaMKII alpha is a cis-acting signal for the localization and translation of mRNA in dendrites. P Natl Acad Sci USA 1996; 93:13250-5; PMID:8917577; http://dx.doi.org/10.1073/pnas.93.23.13250
- Miller S, Yasuda M, Coats JK, Jones Y, Martone ME, Mayford M. Disruption of dendritic translation of CaMKIIalpha impairs stabilization of synaptic plasticity and memory consolidation. Neuron 2002; 36:507-19; PMID:12408852; http://dx.doi.org/10.1016/S0896-6273(02)00978-9
- Lee SJR, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature 2009; 458:299-U58; PMID:19295602; http://dx.doi.org/10.1038/nature07842
- Lisman J, Schulman H, Cline H. The molecular basis of CaMKII function in synaptic and behavioural memory. Nat Rev Neurosci 2002; 3:175-90; PMID:11994750; http://dx.doi.org/10.1038/nrn753
- Buard I, Coultrap SJ, Freund RK, Lee YS, Dell'Acqua ML, Silva AJ, Bayer KU. CaMKII “Autonomy” is required for initiating but not for maintaining neuronal long-term information storage. J Neurosci 2010; 30:8214-20; PMID:20554872; http://dx.doi.org/10.1523/JNEUROSCI.1469-10.2010
- McCann C, Holohan EE, Das S, Dervan A, Larkin A, Lee JA, Rodrigues V, Parker R, Ramaswami M. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. P Natl Acad Sci USA 2011; 108:E655-E62; PMID:21795609; http://dx.doi.org/10.1073/pnas.1107198108
- Sudhakaran IP, Hillebrand J, Dervan A, Das S, Holohan EE, Hulsmeier J, Sarov M, Parker R, VijayRaghavan K, Ramaswami M. FMRP and Ataxin-2 function together in long-term olfactory habituation and neuronal translational control. P Natl Acad Sci USA 2014; 111:E99-E108; PMID:24344294; http://dx.doi.org/10.1073/pnas.1309543111
- Kosik KS. The neuronal microRNA system. Nat Rev Neurosci 2006; 7:911-20; PMID:17115073; http://dx.doi.org/10.1038/nrn2037
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development (vol 439, pg 283, 2006). Nature 2006; 441:902-; PMID:16421561; http://dx.doi.org/10.1038/nature04909
- Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Gene Dev 2002; 16:2497-508; PMID:12368261; http://dx.doi.org/10.1101/gad.1022002
- Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol 2008; 15:346-53; PMID:18345015; http://dx.doi.org/10.1038/nsmb.1405
- Banerjee S, Neveu P, Kosik KS. A coordinated local translational control point at the synapse involving relief from silencing and MOV10 degradation. Neuron 2009; 64:871-84; PMID:20064393; http://dx.doi.org/10.1016/j.neuron.2009.11.023
- Dudai Y. Molecular bases of long-term memories: a question of persistence. Curr Opin Neurobiol 2002; 12:211-6; PMID:12015239; http://dx.doi.org/10.1016/S0959-4388(02)00305-7
- Crick F. Neurobiology - memory and molecular turnover. Nature 1984; 312:101-; PMID:6504122; http://dx.doi.org/10.1038/312101a0
- Meagher RB. Memory and molecular turnover,' 30 years after inception. Epigenet Chromatin 2014; Dec 9;7(1):37 7; PMID: 25525471; http://dx.doi.org/10.1186/1756-8935-7-37
- Lisman JE. A mechanism for memory storage insensitive to molecular turnover - a bistable autophosphorylating kinase. P Natl Acad Sci USA 1985; 82:3055-7; PMID:2986148; http://dx.doi.org/10.1073/pnas.82.9.3055
- Pastalkova E, Serrano P, Pinkhasova D, Wallace E, Fenton AA, Sacktor TC. Storage of spatial information by the maintenance mechanism of LTP. Science 2006; 313:1141-4; PMID:16931766; http://dx.doi.org/10.1126/science.1128657
- Ling DSF, Benardo LS, Serrano PA, Blace N, Kelly MT, Crary JF, Sacktor TC. Protein kinase M zeta is necessary and sufficient for LTP maintenance. Nat Neurosci 2002; 5:295-6; PMID:11914719; http://dx.doi.org/10.1038/nn829
- Serrano P, Yao YD, Sacktor TC. Persistent phosphorylation by protein kinase M zeta maintains late-phase long-term potentiation. J Neurosci 2005; 25:1979-84; PMID:15728837; http://dx.doi.org/10.1523/JNEUROSCI.5132-04.2005
- Sacktor TC. How does PKM zeta maintain long-term memory? Nat Rev Neurosci 2011; 12:9-15; PMID:21119699; http://dx.doi.org/10.1038/nrn2949
- Volk LJ, Bachman JL, Johnson R, Yu YL, Huganir RL. PKM-zeta is not required for hippocampal synaptic plasticity, learning and memory. Nature 2013; 493:420-U169; PMID:23283174; http://dx.doi.org/10.1038/nature11802
- Kwapis JL, Jarome TJ, Lonergan ME, Helmstetter FJ. Protein kinase Mzeta maintains fear memory in the amygdala but not in the hippocampus. Behav Neurosci 2009; 123:844-50; PMID:19634944; http://dx.doi.org/10.1037/a0016343
- Tsokas P, Hsieh C, Yao Y, Lesburgueres E, Wallace EJC, Tcherepanov A, Jothianandan D, Hartley BR, Pan L, Rivard B, et al. Compensation for PKM zeta in long-term potentiation and spatial long-term memory in mutant mice. Elife 2016; May 17;5. pii: e14846; PMID:27187150; http://dx.doi.org/10.7554/eLife.14846
- Tompa P, Friedrich P. Prion proteins as memory molecules: An hypothesis. Neuroscience 1998; 86:1037-43; PMID:9697111; http://dx.doi.org/10.1016/S0306-4522(98)00148-1
- Prusiner SB. Novel proteinaceous infectious particles cause scrapie. Science 1982; 216:136-44; PMID:6801762; http://dx.doi.org/10.1126/science.6801762
- Cohen FE, Prusiner SB. Pathologic conformations of prion proteins. Annu Rev Biochem 1998; 67:793-+; PMID:9759504; http://dx.doi.org/10.1146/annurev.biochem.67.1.793
- Prusiner SB, Mckinley MP, Bowman KA, Bolton DC, Bendheim PE, Groth DF, Glenner GG. Scrapie prions aggregate to form amyloid-like birefringent rods. Cell 1983; 35:349-58; PMID:6418385; http://dx.doi.org/10.1016/0092-8674(83)90168-X
- Prusiner SB. Molecular-structure, biology, and genetics of prions. Adv Virus Res 1988; 35:83-136; PMID:2906782; http://dx.doi.org/10.1016/S0065-3527(08)60709-5
- Prusiner SB. Molecular-biology of prion diseases. Science 1991; 252:1515-22; PMID:1675487; http://dx.doi.org/10.1126/science.1675487
- Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struc Biol 1998; 8:101-6; PMID:9519302; http://dx.doi.org/10.1016/S0959-440X(98)80016-X
- Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. P Natl Acad Sci USA 1993; 90:10962-6; PMID:7902575; http://dx.doi.org/10.1073/pnas.90.23.10962
- Horwich AL, Weissman JS. Deadly conformations - Protein misfolding in prion disease. Cell 1997; 89:499-510; PMID:9160742; http://dx.doi.org/10.1016/S0092-8674(00)80232-9
- Prusiner SB. Biology and genetics of prions causing neurodegeneration. Annu Rev Genet 2013; 47:601-23; PMID:24274755; http://dx.doi.org/10.1146/annurev-genet-110711-155524
- Gomes MPB, Cordeiro Y, Silva JL. The peculiar interaction between mammalian prion protein and RNA. Prion 2008; 2:64-6; PMID:19098437; http://dx.doi.org/10.4161/pri.2.2.6988
- True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 2000; 407:477-83; PMID:11028992; http://dx.doi.org/10.1038/35035005
- Wickner RB. [Ure3] as an altered Ure2 protein - evidence for a prion analog in saccharomyces-cerevisiae. Science 1994; 264:566-9; PMID:7909170; http://dx.doi.org/10.1126/science.7909170
- Patino MM, Liu JJ, Glover JR, Lindquist S. Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science 1996; 273:622-6; PMID:8662547; http://dx.doi.org/10.1126/science.273.5275.622
- Cox BS, Tuite MF, McLaughlin CS. The psi factor of yeast: a problem in inheritance. Yeast 1988; 4:159-78; PMID:3059716; http://dx.doi.org/10.1002/yea.320040302
- Cox BS. A cytoplasmic suppressor of super-suppressor in yeast. Heredity 1965; 20:505-+; PMID:26645632; http://dx.doi.org/10.1038/hdy.1965.65
- Shorter J, Lindquist S. Prions as adaptive conduits of memory and inheritance. Nat Rev Genet 2005; 6:435-50; PMID:15931169; http://dx.doi.org/10.1038/nrg1616
- Tuite MF. Yeast prions and their prion-forming domain. Cell 2000; 100:289-92; PMID:10676809; http://dx.doi.org/10.1016/S0092-8674(00)80663-7
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell 2009; 137:146-58; PMID:19345193; http://dx.doi.org/10.1016/j.cell.2009.02.044
- Malinovska L, Kroschwald S, Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Bba-Proteins Proteom 2013; 1834:918-31; PMID:23328411; http://dx.doi.org/10.1016/j.bbapap.2013.01.003
- Liu JJ, Lindquist S. Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature 1999; 400:573-6; PMID:10448860; http://dx.doi.org/10.1038/22919
- Liu JJ, Sondheimer N, Lindquist SL. Changes in the middle region of Sup35 profoundly alter the nature of epigenetic inheritance for the yeast prion [PSI+]. P Natl Acad Sci USA 2002; 99:16446-53; PMID:12461168; http://dx.doi.org/10.1073/pnas.252652099
- de la Paz ML, Serrano L. Sequence determinants of amyloid fibril formation. P Natl Acad Sci USA 2004; 101:87-92; PMID:14691246; http://dx.doi.org/10.1073/pnas.2634884100
- Ross ED, Minton A, Wickner RB. Prion domains: sequences, structures and interactions. Nat Cell Biol 2005; 7:1039-44; PMID:16385730; http://dx.doi.org/10.1038/ncb1105-1039
- Toombs JA, Petri M, Paul KR, Kan GY, Ben-Hur A, Ross ED. De novo design of synthetic prion domains. P Natl Acad Sci USA 2012; 109:6519-24; PMID:22474356; http://dx.doi.org/10.1073/pnas.1119366109
- Suzuki G, Tanaka M. Expanding the yeast prion world Active prion conversion of non-glutamine/asparagine-rich Mod5 for cell survival. Prion 2013; 7:109-13; PMID:23117914; http://dx.doi.org/10.4161/pri.22685
- Huang YS, Carson JH, Barbarese E, Richter JD. Facilitation of dendritic mRNA transport by CPEB. Gene Dev 2003; 17:638-53; PMID:12629046; http://dx.doi.org/10.1101/gad.1053003
- Hake LE, Richter JD. Cpeb is a specificity factor that mediates cytoplasmic polyadenylation during xenopus oocyte maturation. Cell 1994; 79:617-27; PMID:7954828; http://dx.doi.org/10.1016/0092-8674(94)90547-9
- Kim JH, Richter JD. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol Cell 2006; 24:173-83; PMID:17052452; http://dx.doi.org/10.1016/j.molcel.2006.08.016
- Jung MY, Lorenz L, Richter JD. Translational control by neuroguidin, a eukaryotic initiation factor 4E and CPEB binding protein. Mol Cell Biol 2006; 26:4277-87; PMID:16705177; http://dx.doi.org/10.1128/MCB.02470-05
- Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 2005; 433:477-80; PMID:15690031; http://dx.doi.org/10.1038/nature03205
- Si K, Lindquist S, Kandel ER. A neuronal isoform of the Aplysia CPEB has prion-like properties. Cell 2003; 115:879-91; PMID:14697205; http://dx.doi.org/10.1016/S0092-8674(03)01020-1
- Hervas R, Li L, Majumdar A, Fernandez-Ramirez Mdel C, Unruh JR, Slaughter BD, Galera-Prat A, Santana E, Suzuki M, Nagai Y, et al. Molecular basis of Orb2 amyloidogenesis and blockade of memory consolidation. Plos Biol 2016; 14:e1002361; PMID:26812143; http://dx.doi.org/10.1371/journal.pbio.1002361
- Stephan JS, Fioriti L, Lamba N, Colnaghi L, Karl K, Derkatch IL, Kandel ER. The CPEB3 protein is a functional prion that interacts with the actin cytoskeleton. Cell Rep 2015; 11:1772-85; PMID:26074072; http://dx.doi.org/10.1016/j.celrep.2015.04.060
- Heinrich SU, Lindquist S. Protein-only mechanism induces self-perpetuating changes in the activity of neuronal Aplysia cytoplasmic polyadenylation element binding protein (CPEB). P Natl Acad Sci USA 2011; 108:2999-3004; PMID:21270333; http://dx.doi.org/10.1073/pnas.1019368108
- Si K, Giustetto M, Etkin A, Hsu R, Janisiewicz AM, Miniaci MC, Kim JH, Zhu H, Kandel ER. A neuronal isoform of CPEB regulates local protein synthesis and stabilizes synapse-specific long-term facilitation in Aplysia. Cell 2003; 115:893-904; PMID:14697206; http://dx.doi.org/10.1016/S0092-8674(03)01021-3
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Aplysia CPEB can form prion-like multimers in sensory neurons that contribute to long-term facilitation. Cell 2010; 140:421-U179; PMID:20144764; http://dx.doi.org/10.1016/j.cell.2010.01.008
- Miniaci MC, Kim JH, Puthanveettil SV, Si K, Zhu HX, Kandel ER, Bailey CH. Sustained CPEB-dependent local protein synthesis is required to stabilize synaptic growth for persistence of long-terra facilitation in Aplysia. Neuron 2008; 59:1024-36; PMID:18817739; http://dx.doi.org/10.1016/j.neuron.2008.07.036
- Chang JS, Tan LH, Schedl P. The Drosophila CPEB homolog, Orb, is required for Oskar protein expression in oocytes. Dev Biol 1999; 215:91-106; PMID:10525352; http://dx.doi.org/10.1006/dbio.1999.9444
- Chang JS, Tan LH, Wolf MR, Schedl P. Functioning of the Drosophila orb gene in gurken mRNA localization and translation. Development 2001; 128:3169-77; PMID:11688565
- McBride SMJ, Giuliani G, Choi C, Krause P, Correale D, Watson K, Baker G, Siwicki KK. Mushroom body ablation impairs short-term memory and long-term memory of courtship conditioning in Drosophila melanogaster. Neuron 1999; 24:967-77; PMID:10624959; http://dx.doi.org/10.1016/S0896-6273(00)81043-0
- Krashes MJ, Keene AC, Leung B, Armstrong JD, Waddell S. Sequential use of mushroom body neuron subsets during Drosophila odor memory processing. Neuron 2007; 53:103-15; PMID:17196534; http://dx.doi.org/10.1016/j.neuron.2006.11.021
- Heisenberg M, Borst A, Wagner S, Byers D. Drosophila mushroom body mutants are deficient in olfactory learning. J Neurogenet 1985; 2:1-30; PMID:4020527; http://dx.doi.org/10.3109/01677068509100140
- Keleman K, Kruttner S, Alenius M, Dickson BJ. Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat Neurosci 2007; 10:1587-93; PMID:17965711; http://dx.doi.org/10.1038/nn1996
- Kruttner S, Stepien B, Noordermeer JN, Mommaas MA, Mechtler K, Dickson BJ, Keleman K. Drosophila CPEB Orb2A mediates memory independent of Its RNA-binding domain. Neuron 2012; 76:383-95; PMID:23083740; http://dx.doi.org/10.1016/j.neuron.2012.08.028
- Majumdar A, Cesario WC, White-Grindley E, Jiang HQ, Ren FZ, Khan M, Li L, Choi EM, Kannan K, Guo F, et al. Critical role of amyloid-like oligomers of drosophila Orb2 in the persistence of memory. Cell 2012; 148:515-29; PMID:22284910; http://dx.doi.org/10.1016/j.cell.2012.01.004
- Mastushita-Sakai T, White-Grindley E, Samuelson J, Seidel C, Si K. Drosophila Orb2 targets genes involved in neuronal growth, synapse formation, and protein turnover. P Natl Acad Sci USA 2010; 107:11987-92; PMID:20547833; http://dx.doi.org/10.1073/pnas.1004433107
- White-Grindley E, Li LY, Khan RM, Ren FZ, Saraf A, Florens L, Si K. Contribution of Orb2A stability in regulated amyloid- like oligomerization of drosophila Orb2. Plos Biol 2014; 12Feb 11;12(2):e1001786; PMID: 24523662; http://dx.doi.org/10.1371/journal.pbio.1001786
- Khan MR, Li LY, Perez-Sanchez C, Saraf A, Florens L, Slaughter BD, Unruh JR, Si K. Amyloidogenic oligomerization transforms drosophila Orb2 from a translation repressor to an activator. Cell 2015; 163:1468-83; PMID:26638074; http://dx.doi.org/10.1016/j.cell.2015.11.020
- Kruttner S, Traunmuller L, Dag U, Jandrasits K, Stepien B, Iyer N, Fradkin LG, Noordermeer JN, Mensh BD, Keleman K. Synaptic Orb2A bridges memory acquisition and late memory consolidation in drosophila. Cell Rep 2015; 11:1953-65; PMID:26095367; http://dx.doi.org/10.1016/j.celrep.2015.05.037
- Alarcon JM, Hodgman R, Theis M, Huang YS, Kandel ER, Richter JD. Selective modulation of some forms of Schaffer collateral-CA1 synaptic plasticity in mice with a disruption of the CPEB-1 gene. Learn Memory 2004; 11:318-27; PMID:15169862; http://dx.doi.org/10.1101/lm.72704
- Theis M, Si K, Kandel ER. Two previously undescribed members of the mouse CPEB family of genes and their inducible expression in the principal cell layers of the hippocampus. P Natl Acad Sci USA 2003; 100:9602-7; PMID:12871996; http://dx.doi.org/10.1073/pnas.1133424100
- Berger-Sweeney J, Zearfoss NR, Richter JD. Reduced extinction of hippocampal-dependent memories in CPEB knockout mice. Learn Mem 2006; 13:4-7; PMID:16452649; http://dx.doi.org/10.1101/lm.73706
- Huang YS, Kan MC, Lin CL, Richter JD. CPEB3 and CPEB4 in neurons: analysis of RNA-binding specificity and translational control of AMPA receptor GluR2 mRNA. Embo J 2006; 25:4865-76; PMID:17024188; http://dx.doi.org/10.1038/sj.emboj.7601322
- Pavlopoulos E, Trifilieff P, Chevaleyre V, Fioriti L, Zairis S, Pagano A, Malleret G, Kandel ER. Neuralized1 activates CPEB3: a function for nonproteolytic ubiquitin in synaptic plasticity and memory storage. Cell 2011; 147:1369-83; PMID:22153079; http://dx.doi.org/10.1016/j.cell.2011.09.056
- Fioriti L, Myers C, Huang YY, Li X, Stephan JS, Trifilieff P, Colnaghi L, Kosmidis S, Drisaldi B, Pavlopoulos E, et al. The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron 2015; 86:1433-48; PMID:26074003; http://dx.doi.org/10.1016/j.neuron.2015.05.021
- Drisaldi B, Colnaghi L, Fioriti L, Rao N, Myers C, Snyder AM, Metzger DJ, Tarasoff J, Konstantinov E, Fraser PE, et al. SUMOylation is an inhibitory constraint that regulates the prion-like aggregation and activity of CPEB3. Cell Rep 2015; 11:1694-702; PMID:26074071; http://dx.doi.org/10.1016/j.celrep.2015.04.061
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol 2013; 201:361-72; PMID:23629963; http://dx.doi.org/10.1083/jcb.201302044
- King OD, Gitler AD, Shorter J. The tip of the iceberg: RNA-binding proteins with prion-like domains in neurodegenerative disease. Brain Res 2012; 1462:61-80; PMID:22445064; http://dx.doi.org/10.1016/j.brainres.2012.01.016
- Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell 2012; 149:1188-91; PMID:22682242; http://dx.doi.org/10.1016/j.cell.2012.05.022
- Lin Y, Protter DSW, Rosen MK, Parker R. Formation and maturation of phase-separated liquid droplets by RNA-binding proteins. Mol Cell 2015; 60:208-19; PMID:26412307; http://dx.doi.org/10.1016/j.molcel.2015.08.018
- Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. P Natl Acad Sci USA 2007; 104:2649-54; PMID:17299036; http://dx.doi.org/10.1073/pnas.0611503104
- Ngo S, Chiang V, Ho E, Le L, Guo ZF. Prion domain of yeast Ure2 protein adopts a completely disordered structure: a solid-support EPR study. Plos One 2012; 7(10):e47248; PMID: 23077577; http://dx.doi.org/10.1371/journal.pone.0047248
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. P Natl Acad Sci USA 2000; 97:11910-5; PMID:11050225; http://dx.doi.org/10.1073/pnas.97.22.11910
- Ramaswami M, Taylor JP, Parker R. Altered ribostasis: RNA-protein granules in degenerative disorders. Cell 2013; 154:727-36; PMID:23953108; http://dx.doi.org/10.1016/j.cell.2013.07.038
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 2013; 495:467-+; PMID:23455423; http://dx.doi.org/10.1038/nature11922
- Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. Mol Cell 2014; 54:547-58; PMID:24856220; http://dx.doi.org/10.1016/j.molcel.2014.04.033
- Decker CJ, Parker R. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Csh Perspect Biol 2012; Sep 1;4(9):a012286; PMID: 22763747; http://dx.doi.org/10.1101/cshperspect.a012286
- Anderson P, Kedersha N. Stress granules. Curr Biol 2009; 19:R397-R8; PMID:19467203; http://dx.doi.org/10.1016/j.cub.2009.03.013
- Buchan JR, Muhlrad D, Parker R. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J Cell Biol 2008; 183:441-55; PMID:18981231; http://dx.doi.org/10.1083/jcb.200807043
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell 2009; 36:932-41; PMID:20064460; http://dx.doi.org/10.1016/j.molcel.2009.11.020
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol 2008; 10:1224-31; PMID:18794846; http://dx.doi.org/10.1038/ncb1783
- Fritzsche R, Karra D, Bennett KL, Ang FY, Heraud-Farlow JE, Tolino M, Doyle M, Bauer KE, Thomas S, Planyavsky M, et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep 2013; 5:1749-62; PMID:24360960; http://dx.doi.org/10.1016/j.celrep.2013.11.023
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron 2006; 52:997-1009; PMID:17178403; http://dx.doi.org/10.1016/j.neuron.2006.10.028
- Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron 2001; 31:261-75; PMID:11502257; http://dx.doi.org/10.1016/S0896-6273(01)00357-9
- Rook MS, Lu M, Kosik KS. CaMKII alpha 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J Neurosci 2000; 20:6385-93; PMID:10964944
- Elvira G, Wasiak S, Blandford V, Tong XK, Serrano A, Fan X, del Rayo Sánchez-Carbente M, Servant F, Bell AW, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteomics 2006; 5:635-51; PMID:16352523; http://dx.doi.org/10.1074/mcp.M500255-MCP200
- Antar LN, Afroz R, Dictenberg JB, Carroll RC, Bassell GJ. Metabotropic glutamate receptor activation regulates fragile x mental retardation protein and FMR1 mRNA localization differentially in dendrites and at synapses. J Neurosci 2004; 24:2648-55; PMID:15028757; http://dx.doi.org/10.1523/JNEUROSCI.0099-04.2004
- Zalfa F, Achsel T, Bagni C. mRNPs, polysomes or granules: FMRP in neuronal protein synthesis. Curr Opin Neurobiol 2006; 16:265-9; PMID:16707258; http://dx.doi.org/10.1016/j.conb.2006.05.010
- Zeitelhofer M, Karra D, Macchi P, Tolino M, Thomas S, Schwarz M, Kiebler M, Dahm R. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J Neurosci 2008; 28:7555-62; PMID:18650333; http://dx.doi.org/10.1523/JNEUROSCI.0104-08.2008
- Hillebrand J, Pan KY, Kokaram A, Barbee S, Parker R, Ramaswami M. The Me31B DEAD-box helicase localizes to postsynaptic foci and regulates expression of a CaMKII reporter mRNA in dendrites of Drosophila olfactory projection neurons. Front Neural Circuit 2010; Nov 3;4:121; PMID: 21267420; http://dx.doi.org/10.3389/fncir.2010.00121
- Banerjee P, Schoenfeld BP, Bell AJ, Choi CH, Bradley MP, Hinchey P, Kollaros M, Park JH, McBride SM, Dockendorff TC. Short- and long-term memory are modulated by multiple isoforms of the fragile X mental retardation protein. J Neurosci 2010; 30:6782-92; PMID:20463240; http://dx.doi.org/10.1523/JNEUROSCI.6369-09.2010
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol 2003; 13:286-96; PMID:12593794; http://dx.doi.org/10.1016/S0960-9822(03)00064-2
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 2004; 15:5383-98; PMID:15371533; http://dx.doi.org/10.1091/mbc.E04-08-0715
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol 2007; 179:437-49; PMID:17984320; http://dx.doi.org/10.1083/jcb.200704147
- Reijns MAM, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci 2008; 121:2463-72; PMID:18611963; http://dx.doi.org/10.1242/jcs.024976
- Kedersha N, Cho MR, Li W, Yacono PW, Chen S, Gilks N, Golan DE, Anderson P. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J Cell Biol 2000; 151:1257-68; PMID:11121440; http://dx.doi.org/10.1083/jcb.151.6.1257
- Zhang HY, Elbaum-Garfinkle S, Langdon EM, Taylor N, Occhipinti P, Bridges AA, Brangwynne CP, Gladfelter AS. RNA controls PolyQ protein phase transitions. Mol Cell 2015; 60:220-30; PMID:26474065; http://dx.doi.org/10.1016/j.molcel.2015.09.017
- Toretsky JA, Wright PE. Assemblages: Functional units formed by cellular phase separation. J Cell Biol 2014; 206:579-88; PMID:25179628; http://dx.doi.org/10.1083/jcb.201404124
- Zeng M, Shang Y, Araki Y, Guo T, Huganir RL, Zhang M. Phase transition in postsynaptic densities underlies formation of synaptic complexes and synaptic plasticity. Cell 2016; 166:1163-75 e12; PMID:27565345; http://dx.doi.org/10.1016/j.cell.2016.07.008
- Bowden HA, Dormann D. Altered mRNP granule dynamics in FTLD pathogenesis. J Neurochem 2016; 138 Suppl 1:112-33; PMID:26938019; http://dx.doi.org/10.1111/jnc.13601
- Xiang SH, Kato M, Wu LC, Lin Y, Ding M, Zhang YJ, Yu Y, McKnight SL. The LC domain of hnRNPA2 adopts similar conformations in hydrogel polymers, liquid-like droplets, and nuclei. Cell 2015; 163:829-39; PMID:26544936; http://dx.doi.org/10.1016/j.cell.2015.10.040
- Das RK, Ruff KM, Pappu RV. Relating sequence encoded information to form and function of intrinsically disordered proteins. Curr Opin Struc Biol 2015; 32:102-12; PMID:25863585; http://dx.doi.org/10.1016/j.sbi.2015.03.008
- Ruff KM, Khan SJ, Pappu RV. A coarse-grained model for polyglutamine aggregation modulated by amphipathic flanking sequences. Biophys J 2014; 107:1226-35; PMID:25185558; http://dx.doi.org/10.1016/j.bpj.2014.07.019
- Tariq M, Wegrzyn R, Anwar S, Bukau B, Paro R. Drosophila GAGA factor polyglutamine domains exhibit prion-like behavior. Bmc Genomics 2013; Jun 3;14:374; PMID:23324084; http://dx.doi.org/10.1186/1471-2164-14-374
- Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. P Natl Acad Sci USA 2002; 99:7402-7; PMID:12032295; http://dx.doi.org/10.1073/pnas.072199199
- Cai X, Chen JQ, Xu H, Liu SQ, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 2014; 156:1207-22; PMID:24630723; http://dx.doi.org/10.1016/j.cell.2014.01.063
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell 2007; 18:1385-96; PMID:17392519; http://dx.doi.org/10.1091/mbc.E06-12-1120
- Wang H, Dictenberg JB, Ku L, Li W, Bassell GJ, Feng Y. Dynamic association of the fragile X mental retardation protein as a messenger ribonucleoprotein between microtubules and polyribosomes. Mol Biol Cell 2008; 19:105-14; PMID:17978095; http://dx.doi.org/10.1091/mbc.E07-06-0583
- O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci 2002; 25:315-38; PMID:12052912; http://dx.doi.org/10.1146/annurev.neuro.25.112701.142909
- Harrison PM, Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol 2003; 4(6):R40; PMID: 12801414; http://dx.doi.org/10.1186/gb-2003-4-6-r40