ABSTRACT
Circular RNAs (circRNAs) have been identified in diverse eukaryotic species and are characterized by RNA backsplicing events. Current available methods for circRNA identification are able to determine the start and end locations of circRNAs in the genome but not their full-length sequences. In this study, we developed a method to assemble the full-length sequences of circRNAs using the backsplicing RNA-Seq reads and their corresponding paired-end reads. By applying the method to an rRNA-depleted/RNase R-treated RNA-Seq dataset, we for the first time identified full-length sequences of nearly 3,000 circRNAs in rice. We further showed that alternative circularization of circRNA is a common feature in rice and, surprisingly, found that the junction sites of a large number of rice circRNAs are flanked by diverse non-GT/AG splicing signals while most human exonic circRNAs are flanked by canonical GT/AG splicing signals. Our study provides a method for genome-wide identification of full-length circRNAs and expands our understanding of splicing signals of circRNAs.
Introduction
Circular RNAs (circRNAs) are generated by a non-linear backsplicing event between a downstream splice donor and an upstream splice acceptor. Although several examples of circRNAs have been known for several decades, the phenomenon of widespread of circRNAs in eukaryotes has only recently been revealed.Citation1-4 In humans and animals, extensive attentions have been given to circRNAs, which enhanced our understanding of biogenesis and functions of circRNAs.Citation5-8 CircRNAs can arise from exons, introns and intergenic regions. The junction sites of the circRNAs identified among various eukaryotic species are mostly flanked by canonical splice sites, i.e. a 5′ donor site of GT and a 3′ acceptor site of AG.Citation6 It was reported that, in humans, some exonic circRNAs are flanked by paired ALU elements and long introns.Citation9 Further works showed that biogenesis of some animal circRNAs is mediated by complementary sequences in their flanking introns.Citation10-13 Competitive generation of circular and linear transcripts from the same pre-transcript has been observed and the splicing factor MBNL1 has been found to be involved in circRNA biogenesis.Citation14 The RNA binding protein Quaking regulates the formation of circRNA.Citation15 Another work in yeast uncovered a mechanism that may account for circularization of RNA in genes lacking noticeable flanking intronic secondary structure, showing that circRNA can be generated through an exon-containing lariat precursor.Citation16 For functions of circRNAs, the most prominent examples are human CDR1as (CDR1 antisense) and mouse Sry, which were experimentally validated to function as miRNA sponges.Citation17,18 Human CDR1as contains dozens of miR-7 binding sites and its expression in zebrafish impaired midbrain development, a phenotype similar to that observed in zebrafish with a knocking-down level of miR-7.Citation18 Sry circRNA was found to function as a decoy of miR-138.Citation17 A recent work further showed that human circHIPK3 serves as a sponge for multiple miRNAs.Citation19 Another possible function of circRNAs is to regulate expression of their parental genes.Citation20,21 For example, exon-intron-derived circRNAs have been shown to promote transcription of their parental genes via interaction with U1 snRNA based on complementary base pairing.Citation20
In contrast to human/animal circRNAs, little attention has been given to circRNAs in plants. We have previously performed genome-wide identification of circRNAs in rice and Arabidopsis, and found that some features, such as conservation and harboring long flanking introns, are common in plant and animal circRNAs.Citation22 We also found some distinct features of plant circRNAs, such as much less repetitive elements and complementary sequences in their flanking introns. These results have been confirmed by a recent study in rice, which further showed that overexpression of a circRNA reduced the expression level of its parental gene, suggesting that plant circRNA might act as a negative regulator of its parental gene.Citation23
Several bioinformatics tools, such as find_circ, circRNA_finder, CIRCexplorer and CIRI, have been developed for circRNA identification.Citation5,24 After removing RNA-Seq reads that aligned contiguously and full length to the target genome, find_circ aligns 20-nt fragments (anchors) extracted from both ends of the unmapped reads to the genome sequence to find backsplicing events defined by alignment of the anchors in a reverse orientation.Citation18 CIRCexplorer determines the precise positions of downstream donor and upstream acceptor splice sites by aligning splice junction reads to annotated genes.Citation13 circRNA_finder uses the STAR read aligner to identify chimeric junction reads.Citation25 CIRI is an algorithm based on detection of paired chiastic clipping signal in the SAM alignment in combination with systematic filtrations to remove false positives.Citation26 Other algorithms used in identification of backsplicing transcripts include MapSplice, TopHat-Fusion and segemehl.Citation27-29 More recently, a statistical approach has also been used in detection of backsplicing events and identification of circRNAs.Citation30 It was reported that the loop portion of lariats is resistant to RNase R and that branch point reads could be detected in RNase R-treated RNA-Seq data. Most methods could not distinguish lariats and circRNAs, however, lariats usually contain a significant portion of intronic sequence.
Basically, these bioinformatics tools first find fusion junction sites and then use different filtration strategies to identify the corresponding circRNAs. RNA-Seq reads supporting backsplicing events are crucial for circRNA identification. The aforementioned methods can identify the start and end locations of circRNAs in a genome based on the position of the backsplicing site, but cannot determine the full-length sequences of circRNAs, which can be obtained by PCR-based experimental method but usually only for a limited number of circRNAs.Citation31,32 Considering the importance of full-length sequences for functional and evolutionary analyses of circRNAs, it is necessary to find a way for large-scale identification of full-length sequences of circRNAs. In this study, we developed such a bioinformatics pipeline, which first identifies backsplicing junctions and then assembles the full-length sequences of circRNAs using the paired-end reads aligned to the backsplicing junctions. As a proof of concept, we identified approximately 3,000 full-length circRNAs using this approach in rice and found that non-GT/AG splicing signals are common in rice circRNAs.
Results
Assembly of full-length sequences of circular RNAs
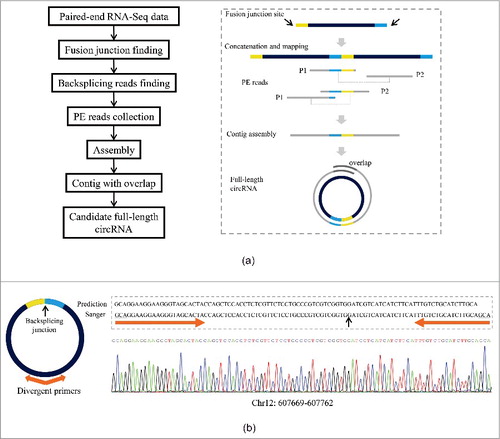
A method called circseq_cup was developed in this study to identify circRNAs and to assemble full-length circRNA sequences based on paired-end (PE) RNA-Seq data (; details see Methods). Briefly, circRNA-enriched clean RNA-Seq reads were aligned to genome sequence and fusion junction sites were identified using TopHat-Fusion.Citation29 The genomic sequence (referred to ‘n’) between 2 fusion junction sites was extracted and 2 such genomic sequences were concatenated to form a new sequence (referred to ‘2n’). The unmapped reads collected during alignment using TopHat were aligned to the ‘2n’ sequences and those covering the middle joint of the ‘2n’ sequences () were considered as backsplicing reads. The candidate backsplicing reads and their corresponding PE ones were then collected. As backsplicing reads are crucial for the identification of circRNAs, we thus only used the backsplicing reads satisfying the criteria detailed in the Methods section in contig assembly to make sure the accuracy of the assembled circRNAs. Additionally, considering the difficulty in distinguishing linear and circular transcripts, we only collected the backsplicing reads and their corresponding PE reads that were fully aligned to the ‘2n’ sequences to make sure that all collected reads are from candidate circular transcripts. The collected reads were then assembled for each candidate circRNA using Cap3.Citation33 A contig with overlap at its both ends was considered as a circRNA. The contig sequence (excluding the overlap at one of its ends) was designated the full-length circRNA sequence (). The tool circseq_cup we developed can also use the mapping and fusion junction detection results generated by other aligners, including STAR-FusionCitation34 and segemehl,Citation27 as the input of Cap3 assembly to suit different requirements for the sensitivity and accuracy of circRNA identification.
Figure 1. Identification of circRNAs with full-length sequences. (a) Workflow for computational identification of circRNAs with full-length sequences based on paired-end RNA-Seq data. Left panel, a pipeline for assembly of full-length circRNA sequence implemented in circseq_cup. Right panel, a model illustrating the assembly method by circseq_cup. (b) Validation of full-length circRNA sequences by divergent RT-PCR. Left panel, a diagram showing a circRNA and the positions of divergent primers used in amplification of circRNA. Forward and reverse primers were designed close to each other or even overlapping at their 5′ ends to make sure that the PCR product was or nearly was the full size of circRNA. Right panel, an example showing the full-length sequence of a circRNA (Chr12: 607,669–607,762) confirmed by Sanger sequencing. In this case, the forward and reverse primers overlapped 3-nt at their 5′ ends.
Full-length sequences of circRNAs in rice
To evaluate the method, we generated PE RNA-Seq reads from 3 libraries (replicates) using total RNA isolated from rice roots and treated to deplete rRNA and linear RNA molecules (see Methods and Supplementary Table S1). Two RNA-Seq libraries had an insertion size of approximately 300 bp, while the third library had an insertion size of ∼400 bp. Following the steps detailed in Methods and the last section, we identified 1,046, 1,368 and 1,225 circular full-length transcripts in the 3 libraries, with a total of 3,011 non-redundant ones. This result could indicate a poor repeatability of circRNA identification in different RNA-Seq libraries with a decent sequencing depth. However, when we considered 2 or more circRNA isoforms from the same circRNA locus as a single circRNA, we found that over 60% circRNAs were reproducibly detected in at least 2 libraries (Supplementary Table S2). circRNA isoforms refer to any overlapping circRNAs generated from the same circRNA locus (see details in the next section). Analysis of the number of PE reads supporting the detected circRNAs showed that most circRNAs (∼70%) have only ≤5 PE reads in each of the 3 libraries, demonstrating that most identified circRNAs are lowly expressed in rice (Fig. S1), which in part could address the inconsistency of circRNA isoforms identified in different libraries that were generated from the same tissue. RNA-Seq data with a much higher depth are preferred for robust and repeatable circRNA identification in rice, especially for consistent identification of all circRNA isoforms.
Of the 3,011 circular full-length transcripts, 213 had their fusion joint sites (both the splicing donor and acceptor) from introns, of which 8 and 205 had the canonical GT/AG splicing signals and non-GT/AG splicing signals, respectively. The latter might be derived from lariats during splicing and were therefore excluded from further analyses.Citation9 Of the remaining 2,806 circRNAs (Supplementary Table S3), 1,846 (accounting for 65.8%) contain exons (exonic circRNAs) and 952 had their entire full-length sequences matching intergenic regions (intergenic circRNAs).
To confirm the circRNA identification results, we did RT-PCR with divergent primers (i.e., the forward primer being located downstream of the reverse primer when they are aligned to genomic sequence) followed by Sanger sequencing for 30 exonic circRNAs and 5 intergenic circRNAs (, Supplementary Table S4). The divergent primers were designed in a ‘back-to-back’ manner with the 5′ ends of the forward and reverse primers overlapping by less than 5-nt or separated by a distance of no more than 5-nt (). Of the 30 selected exonic circRNAs, 15 were validated successfully for their full-length sequences. Two of the 5 intergenic circRNAs were also validated successfully, including the longest (680 bases) circRNA (Chr4: 35,476,588–35,477,345) predicted by our method (Supplementary Table S4). Additionally, we randomly selected 5 circRNA loci generating multiple isoforms (details in the next section) for validation. At least one predicted circRNA isoform was validated successfully for each locus (Supplementary Table S4). These validation results demonstrated that our method is efficient in identification of full-length sequences of circRNAs.
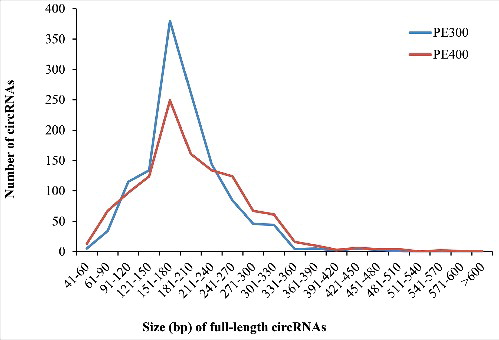
Our method used only RNA-Seq reads covering backsplicing sites and their corresponding PE reads in contig assembly, the sizes of the full-length circRNAs identified thus depend on the read length and the insertion sizes of RNA-Seq library. As expected, we identified more full-length circRNAs with a longer size (e.g. >240 bases) in the PE400 library than in the PE300 library (). The longest full-length circRNAs identified in the PE300 and PE400 libraries were 488 and 680 bases in length, respectively. The majority of full-length circRNAs identified in both RNA-Seq libraries had a length of 150–180 bases (). It seems to be that 150–180-nt circRNAs could be the dominant ones in rice. However, the number of shorter circRNAs could have been underestimated in our experiment, considering that the insertion sizes we used were 300 and 400 bases and that most small circRNAs (e.g., <100 bases) were not harvested for sequencing library construction.
Alternative circularization of circRNAs in rice
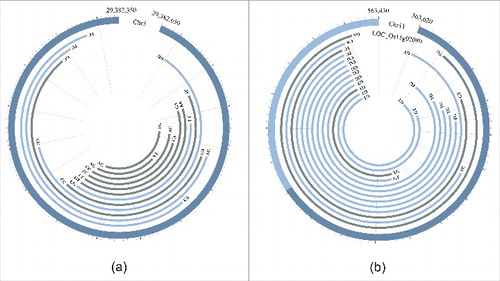
Previous result showed that alternative circularization of circRNA is a common feature in rice.Citation23 Any 2 partial overlapping circRNAs derived from the same locus were defined here as circRNA isoforms generated by alternative circularization. Among the 2,806 rice circRNAs (from 1,269 loci) presented in Supplementary Table S3, more than half (1,733, accounting for 61.8%) were generated from only 196 loci (15.4% of the total circRNA-generating loci; ). For 31 circRNA loci, over 10 isoforms were detected at each locus, of which 10 loci generated 16–30 circRNA isoforms, 7 loci produced 31–60 isoforms and 4 loci generated over 60 isoforms with one of them having 192 isoforms (). These results indicate that a large number of circRNA isoforms could be generated from the same locus by alternative circularization. Sequencing of RT-PCR products amplified from 5 loci by divergent primers was used to validate the predicted circRNA isoforms. For each locus, in addition to the validated circRNA isoform(s), new isoforms not predicted by our method were also found experimentally. For example, 192 isoforms with a length ranging from 67 to 311 bases were predicted by our method in a 314-nt intergenic region in chromosome 3 (the 2nd locus in Supplementary Table S4b). After sequencing 28 positive clones containing the expected insert, 12 circRNA isoforms were identified with some of them confirmed by multiple clones. Of the 12 circRNA isoforms, 4 were consistent with the prediction results and 8 were new circRNA isoforms that were not predicted by our method (). In another locus (306-nt in length; the 3rd locus in Supplementary Table S4b) corresponding to the 3′-UTR and an exon of the gene LOC_Os11g02080, which contains a hypoxia induced protein domain, 41 isoforms were predicted. After sequencing 27 positive clones containing the expected size of insert, we successfully validated 10 predicted isoforms and found 3 new isoforms that were not predicted. In this case, we found that one end of the experimentally identified 13 circRNA isoforms share the same or very close positions located at the 3′-UTR of LOC_Os11g02080 (). Furthermore, a circRNA locus (the 4th locus in Supplementary Table S4b) corresponding to the 3′-UTR and an exon of the gene LOC_Os12g02040 was predicted to generate 38 isoforms. The full-length sequences of 8 isoforms from this locus were validated successfully and 4 new isoforms that were not predicted were also found (Supplementary Table S4). Interestingly, the parental gene LOC_Os12g02040 of this circRNA locus is a homolog of LOC_Os11g02080, which was caused by large-scale duplication between chromosomes 11 and 12 (Plant genome duplication database).Citation35 These results confirmed that alternative circularization is quite common for circRNA loci in rice and that not all circRNA isoforms from the same locus could be bioinformatically predicted, probably due to their very low expression levels.
Figure 3. The distribution of 2,806 circRNA isoforms identified by circseq_cup showing alternative circularization of circRNAs in rice.
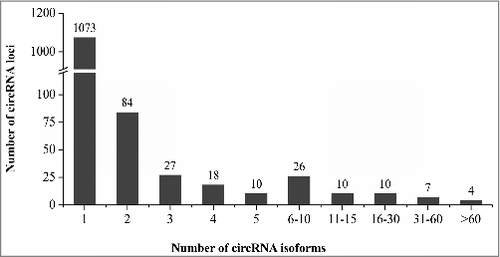
Figure 4. Two validated circRNA loci generating multiple isoforms with non-GT/AG splicing signals. (a) Validated full-length circRNA isoforms generated by alternative circularization at the circRNA locus from an intergenic region of the rice genome (Chr3: 29,382,350–29,382,650). Outer thick circle indicates genomic position. (b) Validated full-length circRNA isoforms generated by alternative circularization at the circRNA locus from the UTR and exonic region of the gene LOC_Os11g02080 (Chr11: 563,430–563,620). Outer thick circle indicates positions of UTR (light blue) and exon (dark blue). Each inner arc represents a validated circRNA isoform. The blue and gray arcs represent isoforms predicted by circseq_cup and identified only by amplification, respectively. Splicing signals were labeled at 2 ends of each isoform.
A large number of circRNAs with diverse non-GT/AG splicing signals in rice
The U2-dependent spliceosome is responsible for splicing of the vast majority of introns in both plants and animals, with GT and AG terminal dinucleotides at their 5′ and 3′ termini, respectively.Citation36 We analyzed the splice sites of 2,806 rice circRNAs identified in this study. The full-length sequences of some circRNAs could not be exactly mapped to a certain genome position showing several bases shuffling. In these cases, the exact backsplicing sites could not be determined. In view of this uncertainty, we first required the full-length circRNA sequences mapped to the rice genome satisfying both GT (AC) and AG (CT) splicing signal criteria. If the paired GT/AG splicing signal was not found, we then designated the annotated ones of the rice genome as the splice sites. If both the GT/AG splicing signal and the annotated splice sites were not found, the backsplicing position was randomly designated. Based on this mapping and annotation strategy, we surprisingly found that, of the 2,806 circRNAs, only a small portion (206 or 7.3%) were flanked by the canonical GT/AG (CT/AC) splicing signals (). A diverse set of non-GT/AG splicing signals were found for the 1,057 circRNAs with a determined mapping position (Supplementary Table S5). The top 5 non-GT/AG terminal dinucleotides pairs were GC/GG, CA/GC, GG/AG, GC/CG, and CT/CC. A certain splicing signal could not be assigned to the remaining 1,543 circRNAs due to their shuffling mapping on the rice genome, but no GT/AG was found among them.
Figure 5. Diverse splicing signals of full-length circRNAs in rice. (a) Splicing signals of circRNAs identified in our rRNA-depleted/RNase R-treated RNA-Seq dataset from rice roots. (b) Splicing signals of circRNAs identified in the public rRNA-depleted RNA-Seq data from rice roots under phosphate starvation stress. Splicing signal “GT/AG” also includes its complementary partner “AC/CT.” In the non-GT/AG group, the top 10 splicing signals were shown individually, and others represent the total number of the remaining non-GT/AG splicing signals of circRNAs with a determined mapping position.
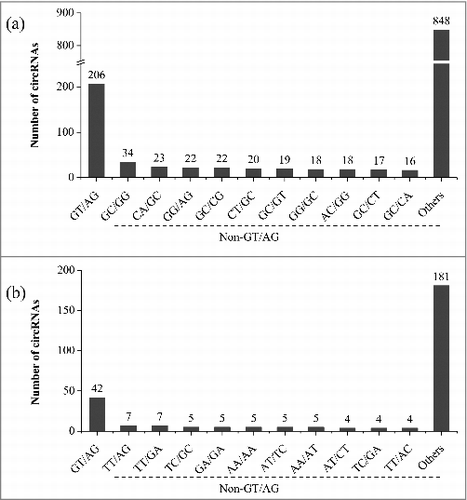
Existence of the non-GT/AG splicing signals was confirmed by our experimental validation of circRNAs. From the aforementioned 5 circRNA loci with multiple isoforms, a total of 44 circRNA isoforms were identified. We found that all of them were flanked by non-GT/AG splicing signals and that only a few of them could be uniquely mapped (Supplementary Table S4). For example, 3 of the 12 circRNA isoforms identified from the intergenic circRNA locus in chromosome 3, which had a unique mapping position in the rice genome, contained the splicing signals of CC/AC, GT/AA or TT/GA (). The remaining 9 isoforms could be aligned to a different position with several bases of shifting at the same locus. A definite splicing signal could not be assigned and no GT/AG was found for these circRNAs. Similarly, the GT/AG pair was not found in all of the 13 identified isoforms from LOC_Os11g02080 () and in all of the 12 identified isoforms from LOC_Os12g02040 (Supplementary Table S4). Additionally, we applied our method to another publicly available RNA-Seq data set (GenBank accession number SRR1005311) generated from rRNA depleted RNA from rice roots under phosphate starvation stress conditionsCitation37 and also found a large number of circRNAs with non-GT/AG splicing signals. In this analysis, a total of 943 circRNAs with full-length sequences were identified and only 42 contained the canonical GT/AG splicing signals (). These results demonstrated that diverse non-GT/AG splicing signals are common for rice circRNAs.
Full-length circRNAs in humans
To further evaluate our method in identification of human full-length circRNAs, we applied circseq_cup to 2 publicly available human RNA-Seq datasets (GenBank accession numbers SRR444974 and SRR445016) enriched for circRNAs.Citation9 After excluding possible lariats that had their fusion joint sites from introns and were flanked by non-GT/AG splicing signals, a total of 3,371 and 9,538 full-length circRNAs were identified in the 2 libraries, respectively (Supplementary Table S6). Analysis of the splice signals of these human circRNAs showed that the majority (88.3% and 85.0%, respectively) of exonic circRNAs have the canonical GT/AG splicing signals and only a small portion are flanked by the non-GT/AG splicing signals (Supplementary Table S6), which is in contrast to rice circRNAs (). For intergenic circRNAs in humans, we also found a large number of them containing non-GT/AG splicing signals (61.2% and 94.1% in the 2 examined RNA-Seq data sets).
Discussion
Circular RNAs are a new type of non-coding RNA that have been identified in a growing number of eukaryotes. Apart from the breakthrough in deep-sequencing technology, development of bioinformatical algorithms for genome-wide identification of circRNAs has played a critical role in discovery and studies of circRNAs. Genome-wide identification of circRNAs is an initial and crucial step toward understanding biogenesis, function and evolution of circRNAs. Based on the presence of backsplicing RNA-Seq reads, current available methods for circRNA identification are able to define the start and end locations of circRNA in a reference genome, but none of them provide solution for determination of the full-length sequences of circRNAs probably due to the difficulty in distinguishing linear and circular transcripts from the same locus. In this study, we developed a bioinformatics pipeline for identification of circRNA with full-length sequences, which are important for their functional characterization and evolutionary analyses ().
Our approach first used publicly available tools, such as TopHat-Fusion, STAR-Fusion and segemehl,Citation27,29,Citation34 to identify fusion joints, and then identified backsplicing reads. Based on backsplicing reads and their corresponding paired-end RNA-Seq reads we assembled the full-length sequences of circRNAs. We used a set of stringent criteria to define the PE reads used in assembly to guarantee that all the reads used are from circular transcripts rather than linear ones. Because of this, our approach does not have advantages regarding the sensitivity of circRNA identification compared to other current available methods. In addition, the length of circRNAs defined using our approach depends on the read length and the insertion sizes of the sequencing libraries. Nevertheless, our method has the following advantages. First, for the first time full-length sequences of circRNAs can be genome-widely predicted using the paired-end RNA sequencing strategy. Second, apart from the presence of backsplicing reads, a circRNA is determined by the acquisition of full-length sequences, which increases the accuracy of the identified circRNAs. For example, it could eliminate other types of non-co-linear events, such as trans-splicing and genetic rearrangements, for which we could find reads with a chiastic order but could not assemble successfully the full-length sequences. Third, our method has a good ability to identify circRNAs with multiple isoforms that shuffle only several bases at a certain circRNA locus (). Finally, the first step of our pipeline, i.e. identification of candidate backsplicing sites, can be done by adopting different fusion junction finding algorithms. Because of variable sensitivity of different algorithms, it is possible to identify more candidate full length circRNAs by pooling all backsplicing sites identified using different algorithms as the input in contig assembly.
The canonical GT/AG splicing signals were usually used as a major filter parameter in current circRNA identification methods. There are 2 types of spliceosomes in plants and animals, i.e., the canonical U2-dependent and the non-canonical U12-dependent spliceosomes. It has been reported that the U12-type introns accounts for only ∼0.3% of the introns in humanCitation38 and ∼0.15% of introns in Arabidopsis thaliana.Citation39 Compared to the U2-type introns that possess the canonical GT and AG terminal dinucleotides, the U12-type introns possess very divergent terminal dinucleotides.Citation36 Szabo et al (2015) first found a small portion of U12-dependent circRNAs in humans.Citation30 Besides backsplicing reads, our method predicts circRNAs mainly depending on full-length sequence acquisition rather than filtration by the GT/AG splicing signals. To investigate the splicing signals involved in formation of circRNAs, we first excluded potential circular transcripts that were derived from introns and had a non-GT/AG splicing signals, as these circular transcripts might be lariats produced by a standard splicing event, which converts an intron into a lariat that is then removed by the splicing machinery and finally degraded.Citation16,40 Among the 2,806 rice exonic and intergenic circRNAs as well as intronic circRNAs with the canonical GT/AG splicing signals, we surprisingly found that only a very small portion (7.3%) of these circRNAs are flanked by the GT/AG splicing signals. The vast majority of them contain very diverse non-GT/AG splicing signals (Supplementary Table S5). This phenomenon was confirmed by our experimental validation (Supplementary Table S4). In contrast, the majority (88.3% and 85.0%, respectively) of human exonic circRNAs contain the canonical GT/AG splicing signals, although a small portion with non-GT/AG splicing signals could be found in our 2 examined RNA-Seq data from human fibroblasts (Supplementary Table S6). At this stage, we do not know whether circular splicing in rice circRNAs is mainly derived from the minor U12 spliceosome dependent splicing or caused by other mechanisms. But our results suggest that it should be tentative for using the GT/AG splicing signals as a major filtration in circRNA identification, particularly in rice, as this will significantly underestimate the number of authentic circRNAs.
Materials and methods
Plant materials
Rice (Oryza sativa cv Nipponbare) seedlings growing in the hydroponic solution were used in RNA extraction and RNA-Seq library preparation. Rice seeds were first pre-germinated in tap water for 2 d before being transferred into the hydroponic solution. Hydroponic experiments were performed under a day/night temperature of 30/22°C and a 12-h photoperiod. The composition of the hydroponic solution was the same as that described by Yoshida et al. (1976).Citation41 During the experimental time, the hydroponic solution was renewed every 3 d. After growing for 5 weeks, roots of rice seedlings were harvested directly into liquid nitrogen and stored at −80°C until use. Three biological experiments were performed.
Circular RNAs enriched RNA-Seq in rice
Total RNA was extracted from rice roots using RNeasy Mini Kit (QIAGEN) and treated by DNase I in order to remove residual genomic DNA. For preparation of RNA-Seq libraries enriched for circRNAs, total RNA was first treated with the Ribo-Zero rRNA Removal Kit (Epicentre) to remove rRNA according to the manufacturer's instructions, and then treated by RNase R (Epicentre) to deplete linear RNA. RNase R treatment was done by incubating 1 µg of rRNA-depleted total RNA with 5 U RNase R in 1× RNase R buffer at 37°C for 30 min. Three RNA sequencing libraries (PE300_r1, PE300_r2 and PE400) with approximately 300 or 400 bp insertion size for Illumina Hiseq 3000 platform were constructed according to the manufacturer's instructions (Illumina). Over 10 Gb of paired-end (PE, 2×150 bp) sequence data were generated for each library (Supplementary Table S1).
We used FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to check the quality of the sequencing reads and the fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) to remove the adaptor and low quality reads. RNA-Seq data have been deposited in GenBank under the accession number SRX1688668.
Development of the bioinformatics method for assembly of full-length sequences of circRNAs
To obtain full-length sequences of circRNAs, we developed a bioinformatics pipeline called circseq_cup, which is available at http://ibi.zju.edu.cn/bioinplant/. The first step of the pipeline is to align RNA-Seq reads and to identify candidate fusion junction sites using publicly available tools, such as TopHat-Fusion, STAR-Fusion and segemehl.Citation27,29,Citation34 In this study, TopHat-Fusion was used to identify candidate fusion junction sites with the same parameters used in CIRCexplorer.Citation13,29 Distance between 2 fusion junction sites was set to be ≤5 kb and ≤50 kb for rice and humans, respectively.
After candidate fusion junction sites were determined, the genomic sequence between the fusion junction sites was extracted. Two such genomic sequences were concatenated to form a single sequence fragment so the junction between these 2 genomic sequences represents a backsplicing site, to which RNA-Seq reads from the corresponding circRNA can be contiguously aligned. The concatenated sequences were then used as the reference to align unmapped reads collected in the first round of alignment using TopHat with the default parameters to confirm backsplicing sites. The reads mapped to the reference and satisfied the following criteria were considered as backsplicing reads and were collected for contig assembly. Firstly, both reads (P1 and P2 in ) of the same pair should be mapped to the reference. Secondly, at least one of the PE reads should cover the joint of the 2 concatenated sequences by ≥20 -nt with at least 10-nt mapped to each side of the joint in the reference. Thirdly, the 20-nt sequences from the PE read (i.e. backsplicing read) aligned to the junction should be fully matched to the corresponding 20-nt sequences flanking the joint in the reference. It aims to exclude the reads that were mapped to the flanking sequence of the joint in the reference with mismatches introduced by the parameter settings used in TopHat.
Assembly of the collected backsplicing PE reads for each individual possible circRNA was performed using Cap3 with the default parameters except the settings of -o 20, -s 300 and -j 40.Citation33 An assembled contig possessing ≥5-nt overlap (the same sequence) at its 2 ends was considered as the candidate full-length sequence of circRNA. One mismatch was allowed if the overlap was ≥10-nt. If the length of a contig was less than 100 bases, the overlap was required to be ≥20-nt. The contigs were then filtered using the following criteria to exclude those 1) that do not contain backsplicing sites; 2) that contain a sequence that is repeated (allowing one mismatch) at the 20-nt upstream of both the start and the end of backsplicing sites; 3) that contain a sequence that is repeated (allowing one mismatch) at the 20-nt downstream of both the end and the start of backsplicing sites; 4) that are supported by less than 2 pairs of PE reads. Additionally, we compared the remaining contigs with a set of reference sequences, including a pool of all exons, every linear transcript and the genomic sequence between backsplicing sites, to remove the contigs that were not consistent with the reference and possessed <10-nt overlap. Finally, the remaining contigs were considered as full-length sequences of circRNAs.
Annotation of circRNAs
When the full-length sequence of a circRNA could not be uniquely mapped to the reference genome with shuffling several bases at a certain locus, we first mapped the full-length sequences of circRNAs to satisfy the canonical GT/AG (CT/AC) splicing signals and then the annotated splicing sites of the reference genome. The circRNAs with their full-length sequences not overlapping with any gene were defined as intergenic circRNAs. The circRNAs with their full-length sequences containing exons were defined as exonic circRNAs. The rice genome annotation information was obtained from RGAP7 (http://rice.plantbiology.msu.edu/). The human genome sequence and annotation information were obtained from Illumina iGenomes (hg19; http://support.illumina.com/sequencing/sequencing_software/igenome.html). Any two circRNAs from a circRNA locus with overlapping positions were defined as circRNA isoforms that were derived from alternative circularization. A circRNA locus may contain multiple isoforms (≥2 ), of which one should overlap with at least another one from the same locus.
Validation of full-length sequences of rice circRNAs
Total RNA was extracted from the same tissues as those used in RNA-Seq and then treated by RNase R. First-strand cDNA was synthesized from ∼1 μg of RNase R-treated RNA with random hexamer using the First Strand cDNA Synthesis kit (TaKaRa, Dalian, China). Genomic DNA was extracted using the conventional CTAB method. Divergent primers were designed for Polymerase Chain Reaction (PCR) with cDNA or genomic DNA as templates (Supplementary Table S4). The forward and reverse divergent primers were designed close to each other or even overlapping a few base pairs to make sure that the PCR product amplified was or nearly was the full size of the target circRNA. The expected results for a circular RNA would be positive and negative amplification for cDNA and genomic DNA, respectively. PCR conditions were as follows: an initial 3 min step at 94°C followed by 40 cycles of 45 s denaturing at 94°C, 35 s annealing at the appropriate annealing temperature (depending on the primer set used) and 30 s extension at 72°C. The final step was conducted at 72°C for 10 min. PCR products were separated using agarose gel and bands were individually excised, purified and directly Sanger-sequenced from both ends using forward or reverse primer. For the bands weakly amplified, small PCR products or smearing bands, they were introduced into pMD18-T vector (TaKaRa) and transformed into Escherichia coli strain DH5α. Clones were then sequenced with vector primers.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Data.zip
Download Zip (203.3 KB)Acknowledgment
We would like to thank Jiangsu Collaborative Innovation Center for Modern Crop Production (JCIC-MCP) for research support.
Funding
This work was supported by the National Basic Research Program of China (2015CB150200), the National Science Foundation of China (91435111, 31521064), and Shanghai Municipal Commission of Agriculture (2014-7-1-4).
References
- Cocquerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155-60; PMID:7678559
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453-61; PMID:24811520; https://doi.org/10.1038/nbt.2890
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B. Scrambled exons. Cell 1991; 64:607-13; PMID:1991322; https://doi.org/10.1016/0092-8674(91)90244-S
- Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One 2014; 9:e90859; PMID:24609083; https://doi.org/10.1371/journal.pone.0090859
- Chen I, Chen CY, Chuang TJ. Biogenesis, identification, and function of exonic circular RNAs. RNA 2015; 6:563-79; PMID:26230526; https://doi.org/10.1002/wrna.1294
- Shen T, Han M, Wei G, Ni T. An intriguing RNA species-perspectives of circularized RNA. Protein & cell 2015; 6:871-80; PMID:26349458; https://doi.org/10.1007/s13238-015-0202-0
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014; 20:1829-42; PMID:25404635; https://doi.org/10.1261/rna.047126.114
- Glazar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA 2014; 20:1666-70; PMID:25234927; https://doi.org/10.1261/rna.043687.113
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu JZ, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:426-; PMID:23249747; https://doi.org/10.1261/rna.035667.112
- Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 2015; 10:170-7; PMID:25558066; https://doi.org/10.1016/j.celrep.2014.12.019
- Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28:2233-47; PMID:25281217; https://doi.org/10.1101/gad.251926.114
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015; 21:172-9; PMID:25449546; https://doi.org/10.1261/rna.048272.114
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell 2014; 159:134-47; PMID:25242744; https://doi.org/10.1016/j.cell.2014.09.001
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell 2014; 56:55-66; PMID:25242144; https://doi.org/10.1016/j.molcel.2014.08.019
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein Quaking regulates formation of circRNAs. Cell 2015; 160:1125-34; PMID:25768908; https://doi.org/10.1016/j.cell.2015.02.014
- Barrett SP, Wang PL, Salzman J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 2015; 4:e07540; PMID:26057830; https://doi.org/10.7554/eLife.07540
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; https://doi.org/10.1038/nature11993
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; https://doi.org/10.1038/nature11928
- Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 2016; 7:11215; PMID:27050392; https://doi.org/10.1038/ncomms11215
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 2015; 22:256-64; PMID:25664725; https://doi.org/10.1038/nsmb.2959
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular Intronic Long Noncoding RNAs. Mol Cell 2013; 51:792-806; PMID:24035497; https://doi.org/10.1016/j.molcel.2013.08.017
- Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol 2015; 208:88-95; PMID:26204923; https://doi.org/10.1111/nph.13585
- Lu T, Cui L, Zhou Y, Zhu C, Fan D, Gong H, Zhao Q, Zhou C, Zhao Y, Lu D, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA 2015; 21:2076-87; PMID:26464523; https://doi.org/10.1261/rna.052282.115
- Hansen TB, Veno MT, Damgaard CK, Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res 2016; 44:e58; PMID:26657634; https://doi.org/10.1093/nar/gkv1458
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014; 9:1966-80; PMID:25544350; https://doi.org/10.1016/j.celrep.2014.10.062
- Gao Y, Wang J, Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol 2015; 16:4; PMID:25583365; https://doi.org/10.1186/s13059-014-0571-3
- Hoffmann S, Otto C, Doose G, Tanzer A, Langenberger D, Christ S, Kunz M, Holdt LM, Teupser D, Hackermüller J, et al. A multi-split mapping algorithm for circular RNA, splicing, trans-splicing and fusion detection. Genome Biol 2014; 15:R34; PMID:24512684; https://doi.org/10.1186/gb-2014-15-2-r34
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, He X, Mieczkowski P, Grimm SA, Perou CM, et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 2010; 38:e178; PMID:20802226; https://doi.org/10.1093/nar/gkq622
- Kim D, Salzberg SL. TopHat-Fusion: an algorithm for discovery of novel fusion transcripts. Genome Biol 2011; 12:R72; PMID:21835007; https://doi.org/10.1186/gb-2011-12-8-r72
- Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 2015; 16:126; PMID:26076956; https://doi.org/10.1186/s13059-015-0690-5
- Fan X, Zhang X, Wu X, Guo H, Hu Y, Tang F, Huang Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol 2015; 16:148; PMID:26201400; https://doi.org/10.1186/s13059-015-0706-1
- You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 2015; 18:603-10; PMID:25714049; https://doi.org/10.1038/nn.3975
- Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res 1999; 9:868-77; PMID:10508846; https://doi.org/10.1101/gr.9.9.868
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15-21; PMID:23104886; https://doi.org/10.1093/bioinformatics/bts635
- Lee TH, Tang H, Wang X, Paterson AH. PGDD: a database of gene and genome duplication in plants. Nucleic Acids Res 2013; 41:D1152-8; PMID:23180799; https://doi.org/10.1093/nar/gks1104
- Szczesniak MW, Kabza M, Pokrzywa R, Gudys A, Makalowska I. ERISdb: a database of plant splice sites and splicing signals. Plant Cell Physiol 2013; 54:e10; PMID:23299413; https://doi.org/10.1093/pcp/pct001
- Secco D, Jabnoune M, Walker H, Shou HX, Wu P, Poirier Y, Whelan J. Spatio-temporal transcript profiling of rice roots and shoots in response to phosphate starvation and recovery. Plant Cell 2013; 25:4285-304; PMID:24249833; https://doi.org/10.1105/tpc.113.117325
- Levine A, Durbin R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res 2001; 29:4006-13; PMID:11574683; https://doi.org/10.1093/nar/29.1.300
- Zhu W, Brendel V. Identification, characterization and molecular phylogeny of U12-dependent introns in the Arabidopsis thaliana genome. Nucleic Acids Res 2003; 31:4561-72; PMID:12888517; https://doi.org/10.1093/nar/gkg492
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141-57; PMID:23249747; https://doi.org/10.1261/rna.035667.112
- Yoshida S, Forno D, Cock J, Gomez K. Laboratory Manual for Physiological Studies of Rice. 1976:Manila, The Philippines: The International Rice Research Institute. 3rd ed.