ABSTRACT
Eukaryotic cells produce a variety of non-coding RNAs (ncRNAs), many of which have been shown to play pivotal roles in biological processes such as differentiation, maintenance of pluripotency of stem cells, and cellular response to various stresses. Genome-wide analyses have revealed that many ncRNAs are transcribed around regulatory DNA elements located proximal or distal to gene promoters, but their biological functions are largely unknown. Recently, it has been demonstrated in yeast and mouse that ncRNA transcription around gene promoters and enhancers facilitates DNA binding of transcription factors to their target sites. These results suggest universal roles of promoter/enhancer-associated ncRNAs in the recruitment of transcription factors to their binding sites.
Abbreviations
| ChIP-seq | = | chromatin immunoprecipitation sequencing |
| CLIP-seq | = | crosslinking immunoprecipitation combined with deep sequencing |
| DRB | = | 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole |
| eRNA | = | enhancer RNA |
| ESR1 | = | estrogen receptor 1 |
| mlonRNA | = | metabolic stress-induced long non-coding RNA |
| ncRNA | = | non-coding RNA |
| RNAP | = | RNA polymerase |
| TF | = | transcription factor |
| TSS | = | transcription start site |
| VIM | = | vimentin |
| YY1 | = | Ying Yang 1 |
Introduction
Technological advancement over the past 2 decades has revealed that eukaryotic cells synthesize a plethora of RNAs that do not encode canonical proteins (non-coding RNAs, ncRNAs). Studies using DNA microarray or deep sequencing techniques have reported widespread transcription of ncRNAs in diverse organisms ranging from yeast to humans.Citation1,2 They are not merely nonfunctional “junk” transcripts, but a significant number of ncRNAs serve as functional molecules that play key roles in complex cellular events such as stress response and development.Citation3
Recent transcriptome analyses have also revealed that many ncRNAs are transcribed around cis-regulatory DNA elements involved in transcriptional control. For example, promoter regions (consisting of core promoter elements and proximal regulatory elements) of protein-coding genes not only produce mRNAs but also trigger ncRNA transcription near the mRNA transcription start sites (TSSs).Citation4 These promoter-associated ncRNAs can be transcribed in the unidirectional or divergent orientation with respect to the direction of mRNA transcription. ncRNAs also originate from enhancers, which can activate gene transcription at a distance in multicellular organisms.Citation5 These ncRNAs in particular are referred to as enhancer RNAs (eRNAs).
Considering that they are transcribed near regulatory units for transcription, it is plausible that those promoter/enhancer-associated ncRNAs play roles in gene regulation, and a growing body of evidence is supporting this notion. For instance, transcription of promoter-associated ncRNAs affects the expression of cognate genes through transcriptional interference or modulation of chromatin structure in promoter regions.Citation6,7 The eRNA transcription also regulates gene expression via several mechanisms, such as the stabilization of enhancer-promoter loops and the stimulation of transcription elongation at their target genes.Citation5,8,9 However, the function of the vast majority of ncRNAs transcribed from gene regulatory regions and their mechanisms of action are largely unknown.
Recently, it has been demonstrated that ncRNA expression around gene regulatory elements in fission yeast and mouse can enhance the loading of transcription factors (TFs) to their target sites.Citation10,11 Such a mechanism may be conserved among eukaryotes. This review introduces 2 such examples and discusses the mechanism by which ncRNA transcription around regulatory elements locally promotes TF binding.
Transcription of promoter-associated ncRNAs facilitates TF binding in fission yeast
In the fission yeast Schizosaccharomyces pombe, we previously identified ncRNAs that are transcribed by RNA polymerase II (RNAP II) from the promoter region of fbp1+ (fructose-1,6-bisphosphatase 1), encoding a key enzyme for gluconeogenesis.Citation12,13 These non-coding transcripts are referred to as “mlonRNAs” (metabolic stress-induced long non-coding RNAs).Citation14 When cells are grown in glucose-rich media, the longest version of mlonRNA (mlonRNA-a) is weakly transcribed across the TSS of fbp1+ mRNA (). In response to glucose depletion, the TSS of mlonRNA is shifted to the 3′ direction, leading to the production of shorter mlonRNA species (mlonRNA-b and -c). This cascade-like transcription of mlonRNAs triggers a 5′ to 3′ stepwise chromatin remodeling along the fbp1+ promoter, thereby activating the expression of fbp1+ mRNA.
Figure 1. Regulation of TF binding by promoter-associated ncRNAs in fission yeast. (A) Schematic diagram of the fbp1+ locus. Each number represents a location relative to the start site of the fbp1+ open reading frame. UAS1 and UAS2 are cis-acting elements involved in transcriptional activation of fbp1+.Citation16 (B) Models for how promoter-associated ncRNAs (including mlonRNAs) enhance Atf1 binding. (i) Groucho/Tup1-like corepressors Tup11 and Tup12 (represented by “Tup” in the figure for simplification) repress Atf1-DNA association, and this inhibition is locally attenuated by ncRNAs transcribed near Atf1-binding sites. (ii) ncRNAs can also facilitate Atf1 binding independently of Tup11/12.
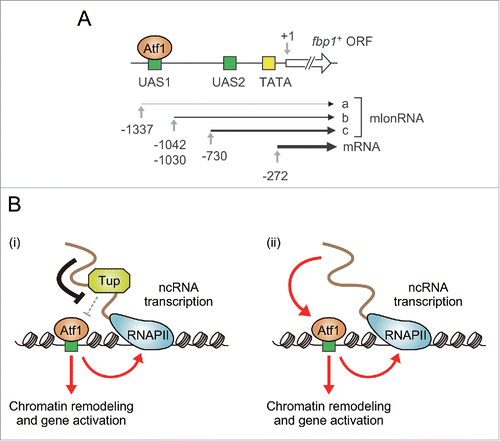
Recently, we have shown that mlonRNA transcription locally facilitates chromatin remodeling and fbp1+ expression by promoting the association of the TF Atf1 on the fbp1+ promoter.Citation10 Atf1 is a member of ATF/CREB family proteins and a master regulator of stress responses in S. pombe.Citation15 Atf1 is known to bind to a cAMP-responsive element (CRE)-like cis-acting element named UAS1 in the fbp1+ promoter region, located between the TSSs of mlonRNA-a and -b ().Citation12,16 We found that Atf1 binding is severely reduced when mlonRNA transcription is inhibited by transcription inhibitors or by a mutation in the mlonRNA promoter which is deficient for the cascade mlonRNA transcription.Citation10 It should also be noted that deletion of the Atf1-binding site in the fbp1+ promoter greatly decreased mlonRNA expression. These results suggest that mlonRNA transcription and Atf1 binding mutually promote each other to form a positive feedback loop, which is assumed to establish a sharp and robust induction of fbp1+ in response to glucose shortage.
To explore the generality of this mechanism, we performed genome-wide analysis of Atf1 binding using chromatin immunoprecipitation sequencing (ChIP-seq).Citation10 In the S. pombe cells cultured under low-glucose conditions, we identified 50 genomic regions at which Atf1 binding is markedly impaired in the presence of a transcription inhibitor. We referred to these sites as “transcription-enhanced sites.” Further comparison of the ChIP-seq data with the published RNA sequencing dataCitation17 revealed that many such transcription-enhanced sites express ncRNAs in response to glucose starvation. In addition, transcription of these ncRNAs occurs concomitantly with an enhanced binding of Atf1 to its target sites near the transcribed segments. These observations support that Atf1 binding is facilitated by nearby ncRNA expression at many of these transcription-enhanced sites.
How does ncRNA transcription promote Atf1 binding? It should be noted that Atf1-DNA association is blocked by Groucho/Tup1-like corepressors Tup11 and Tup12.Citation10,18,19 Moreover, fbp1+ mlonRNA transcription can locally attenuate corepressor functions of the Tup proteins, thereby facilitating Atf1 binding to the fbp1+ promoter.Citation10 Importantly, Tup11 and Tup12 were co-purified with mlonRNA and some other glucose starvation-induced ncRNAs transcribed near other transcription-enhanced sites, suggesting that Tup-ncRNA interaction may locally downregulate the Tup functions as transcriptional corepressors (). Such inhibition is assumed to occur only in cis around the site of ncRNA transcription, since ectopic expression of mlonRNAs could not result in any enhancement of Atf1 binding in a trans-acting manner. Another mutually non-exclusive mechanism is that the ncRNA transcription per se can facilitate the loading of Atf1 to the target sites (), possibly through the local alteration of chromatin structure.Citation10
Trapping of a TF by ncRNAs in gene regulatory elements in mice
Similar ncRNA-mediated TF recruitment has been described in embryonic stem cells by Sigova and colleagues.Citation11 In this study, the authors focused on the TF YY1 (Ying Yang 1), which is ubiquitously expressed in mammalian cells and plays roles in various biological processes such as development and cellular proliferation.Citation20 ChIP-seq and CLIP-seq (crosslinking immunoprecipitation combined with deep sequencing) analyses revealed that YY1 not only occupies enhancers and promoter-proximal elements but also interacts with RNAs transcribed from these loci.Citation11 In addition, the association of YY1 with chromatin was impaired upon treatment by the transcription inhibitor 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) and RNase. Furthermore, artificial tethering of RNA in the vicinity of YY1-binding sites increased the YY1 occupancy at the regions. These results suggest that ncRNAs transcribed from gene regulatory elements locally function in the form of nascent transcripts to trap YY1 on chromatin and assist its association with DNA.
Possible mechanisms for ncRNA-based enhancement of TF recruitment
The number of reports describing varied molecular functions of ncRNAs characterized so far has been steadily increasing, and the functions include RNA sponges, cis-acting tethers, and scaffolds to recruit chromatin modulators.Citation3,21 In light with previous research, we propose several possible molecular mechanisms for the ncRNA-based enhancement of TF recruitment (see ).
Figure 2. Possible models for how TF binding is driven by on-site transcription of ncRNAs. (A) Nascent ncRNAs trap TFs at their target DNA regions. (B) ncRNAs recruit proteins that assist TF binding (e.g., histone modifiers and chromatin remodelers that create open chromatin structure). (C) ncRNAs attenuate functions of proteins that play inhibitory roles for TF binding (e.g., corepressors that establish chromatin states refractory to TF binding). (D) ncRNA transcription leads to the formation of R-loops that facilitate TF binding. (E) Transcription-coupled chromatin reorganization promotes TF binding independently of RNA products.
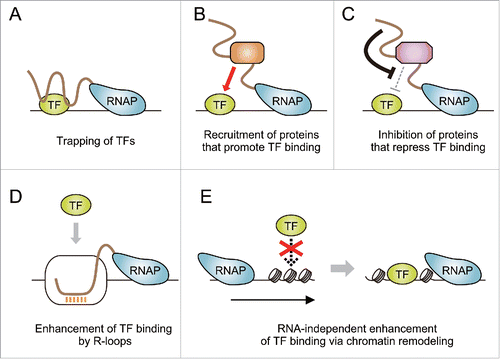
First, as seen in the case of mouse YY1, TFs can be trapped by nascent ncRNAs synthesized in the vicinity of their target sites, and this TF trapping enables efficient binding of the TFs to the regions (). It has been demonstrated that some TFs can bind both DNA and RNA.Citation22 Such dual binding capacity likely enables the TF trapping mechanism. It should be noted that Atf1 can physically interact with RNA as well as DNA.Citation23 Thus, nascent ncRNAs likely tether Atf1 to nearby target sitesCitation10 at least in some transcription-enhanced loci. Second, it is possible that promoter/enhancer-associated ncRNAs locally stimulate TF binding by modulating the action of proteins that promote TF binding (). A number of ncRNAs are known to interact with histone modifiers and chromatin remodelers.Citation3 It is therefore likely that promoter/enhancer-associated ncRNAs help specific and local entry of these chromatin modifiers to establish high competency for subsequent TF recruitment. The third possibility is that these ncRNAs attenuate the functions of proteins that inhibit TF binding (), such as corepressors, as was suggested in the case of fbp1+ mlonRNA.Citation10 Fourth, ncRNA transcription enables efficient TF binding through the formation of R-loops (RNA:DNA hybrids), which are often observed at promoters and enhancers ().Citation24,25 In humans, head-to-head antisense transcription leads to R-loop formation at the promoter of the vimentin (VIM) gene, and this structure indeed facilitates the binding of NF-κB to the promoter.Citation26 Lastly, promoter/enhancer-associated ncRNA transcription itself may enhance TF association, because the transcriptional machinery can affect the spatial distribution of nucleosomes ().Citation27,28 Such transcription-coupled chromatin reorganization may locally facilitate TF recruitment.
Generality and biological consequences of TF binding enhanced by ncRNA transcription
The studies by our group and Sigova et al.Citation10,11 have suggested that ncRNA transcription near gene regulatory elements locally promotes TF recruitment to their target sites in diverse organisms. These observations also suggest that promoter/enhancer-associated ncRNAs function by common mechanisms to regulate gene expression. It has been reported that the expression of ncRNAs often occurs around cis-acting regulatory DNA elements prior to the induction of cognate genes, especially during a variety of cellular responses.Citation17,29 These observations lead us to speculate that the ncRNA transcription near these loci may regulate the local recruitment of TFs to their target sites in response to certain stimuli. It should be noted that transcriptional inhibition does not always affect binding of TFs: among others, estrogen receptor 1 (ESR1) binding to enhancers apparently is eRNA transcription-independent.Citation30
What is the biological significance of the role of ncRNA transcription in regulation of TF binding? As mentioned above, the fission yeast Atf1 binding and mlonRNA transcription mutually promote each other, establishing a local positive feedback.Citation10 This local regulation enables the topical activation of a self-reinforcing loop, leading to a switch-like robust activation of the downstream gene. The ncRNA-enhanced recruitment of mouse YY1 to DNA may also exhibit a similar feedback mechanism.Citation11 Another advantage of the ncRNA-based mechanism may be gene-specific activation in response to numerous types of cellular signals. In this model, TFs may be directed to limited binding sites by nearby ncRNA transcription induced by specific stimuli. Such a system only requires specific ncRNAs, reducing the number of specific TFs that would be necessary to trigger specific responses or developmental stages. By employing such ncRNA-based transcription control, eukaryotic cells may achieve higher complexity with a more simple system containing fewer TFs.
Concluding remarks
Higher eukaryotic cells are known to express numerous promoter/enhancer-associated ncRNAs. Recent studies suggest that ncRNA transcription plays an evolutionarily conserved key role in the locus-specific recruitment of TFs in response to various stimuli. Further investigation of ncRNA-dependent TF recruitment in various processes such as stress response, cellular differentiation, and occurrence of diseases are expected to shed light on context-specific molecular mechanisms that govern cellular adaptation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
We are grateful for Josephine Galipon for her comments on this review.
Funding
This work was supported by grants from the Japan Society for the Promotion of Science (JSPS) to K.O. (23114003, 21241046, and 26291018) and Grant-in-Aid for JSPS Fellows (3J08245) to N.T. The work was also supported by Platform for Dynamic Approaches to Living System from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al. Landscape of transcription in human cells. Nature 2012; 489:101-8; PMID:22955620; http://dx.doi.org/10.1038/nature11233
- Rhind N, Chen Z, Yassour M, Thompson DA, Haas BJ, Habib N, Wapinski I, Roy S, Lin MF, Heiman DI, et al. Comparative functional genomics of the fission yeasts. Science 2011; 332:930-6; PMID:21511999; http://dx.doi.org/10.1126/science.1203357
- Bonasio R, Shiekhattar R. Regulation of transcription by long noncoding RNAs. Annu Rev Genet 2014; 48:433-55; PMID:25251851; http://dx.doi.org/10.1146/annurev-genet-120213-092323
- Wei W, Pelechano V, Järvelin AI, Steinmetz LM. Functional consequences of bidirectional promoters. Trends Genet 2011; 27:267-76; PMID:21601935; http://dx.doi.org/10.1016/j.tig.2011.04.002
- Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat Rev Genet 2016; 17:207-23; PMID:26948815; http://dx.doi.org/10.1038/nrg.2016.4
- Bumgarner SL, Dowell RD, Grisafi P, Gifford DK, Fink GR. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc Natl Acad Sci U S A 2009; 106:18321-6; PMID:19805129; http://dx.doi.org/10.1073/pnas.0909641106
- Hainer SJ, Pruneski JA, Mitchell RD, Monteverde RM, Martens JA. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev 2011; 25:29-40; PMID:21156811; http://dx.doi.org/10.1101/gad.1975011
- Li W, Notani D, Ma Q, Tanasa B, Nunez E, Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al. Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 2013; 498:516-20; PMID:23728302; http://dx.doi.org/10.1038/nature12210
- Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell 2014; 56:29-42; PMID:25263592; http://dx.doi.org/10.1016/j.molcel.2014.08.023
- Takemata N, Oda A, Yamada T, Galipon J, Miyoshi T, Suzuki Y, Sugano S, Hoffman CS, Hirota K, Ohta K. Local potentiation of stress-responsive genes by upstream noncoding transcription. Nucleic Acids Res 2016; 44:5174-89; PMID:26945040; http://dx.doi.org/10.1093/nar/gkw142
- Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science 2015; 350:978-81; PMID:26516199; http://dx.doi.org/10.1126/science.aad3346
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 2008; 456:130-4; PMID:18820678; http://dx.doi.org/10.1038/nature07348
- Hoffman CS, Winston F. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev 1991; 5:561-71; PMID:1849107; http://dx.doi.org/10.1101/gad.5.4.561
- Galipon J, Miki A, Oda A, Inada T, Ohta K. Stress-induced lncRNAs evade nuclear degradation and enter the translational machinery. Genes Cells 2013; 18:353-68; PMID:23489294; http://dx.doi.org/10.1111/gtc.12042
- Chen D, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bähler J. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell 2003; 14:214-29; PMID:12529438; http://dx.doi.org/10.1091/mbc.E02-08-0499
- Neely LA, Hoffman CS. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Mol Cell Biol 2000; 20:6426-34; PMID:10938120; http://dx.doi.org/10.1128/MCB.20.17.6426-6434.2000
- Oda A, Takemata N, Hirata Y, Miyoshi T, Suzuki Y, Sugano S, Ohta K. Dynamic transition of transcription and chromatin landscape during fission yeast adaptation to glucose starvation. Genes Cells 2015; 20:392-407; PMID:25728061; http://dx.doi.org/10.1111/gtc.12229
- Mukai Y, Matsuo E, Roth SY, Harashima S. Conservation of histone binding and transcriptional repressor functions in a Schizosaccharomyces pombe Tup1p homolog. Mol Cell Biol 1999; 19:8461-8; PMID:10567571; http://dx.doi.org/10.1128/MCB.19.12.8461
- Asada R, Takemata N, Hoffman CS, Ohta K, Hirota K. Antagonistic controls of chromatin and mRNA start site selection by Tup family corepressors and the CCAAT-binding factor. Mol Cell Biol 2015; 35:847-55; PMID:25535331; http://dx.doi.org/10.1128/MCB.00924-14
- Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene 2006; 25:1125-42; PMID:16314846; http://dx.doi.org/10.1038/sj.onc.1209080
- Cech TR, Steitz JA. The noncoding RNA revolution - Trashing old rules to forge new ones. Cell 2014; 157:77-94; PMID:24679528; http://dx.doi.org/10.1016/j.cell.2014.03.008
- Cassiday LA. Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res 2002; 30:4118-26; PMID:12364590; http://dx.doi.org/10.1093/nar/gkf512
- Wahls WP. RNA associated with a heterodimeric protein that activates a meiotic homologous recombination hot spot: RL/RT/PCR strategy for cloning any unknown RNA or DNA. PCR Methods Appl 1994; 3:272-7; PMID:7518718; http://dx.doi.org/10.1101/gr.3.5.272
- Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. R-Loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell 2012; 45:814-25; PMID:22387027; http://dx.doi.org/10.1016/j.molcel.2012.01.017
- Pefanis E, Wang J, Rothschild G, Lim J, Kazadi D, Sun J, Federation A, Chao J, Elliott O, Liu ZP, et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015; 161:774-89; PMID:25957685; http://dx.doi.org/10.1016/j.cell.2015.04.034
- Boque-Sastre R, Soler M, Oliveira-Mateos C, Portela A, Moutinho C, Sayols S, Villanueva A, Esteller M, Guil S. Head-to-head antisense transcription and R-loop formation promotes transcriptional activation. Proc Natl Acad Sci 2015; 112:5785-90; PMID:25902512; http://dx.doi.org/10.1073/pnas.1421197112
- Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res 2010; 20:90-100; PMID:19846608; http://dx.doi.org/10.1101/gr.098509.109
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell 2010; 143:540-51; PMID:21074046; http://dx.doi.org/10.1016/j.cell.2010.10.004
- Arner E, Daub CO, Vitting-Seerup K, Andersson R, Lilje B, Drablos F, Lennartsson A, Ronnerblad M, Hrydziuszko O, Vitezic M, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 2015; 347:1010-4; PMID:25678556; http://dx.doi.org/10.1126/science.1259418
- Hah N, Murakami S, Nagari A, Danko CG, Lee Kraus W. Enhancer transcripts mark active estrogen receptor binding sites. Genome Res 2013; 23:1210-23; PMID:23636943; http://dx.doi.org/10.1101/gr.152306.112
