ABSTRACT
Hammerhead ribozymes represent the most common of the 9 natural classes of self-cleaving RNAs. The hammerhead catalytic core includes 11 highly-conserved nucleotides located largely within the unpaired regions of a junction formed by stems I, II and III. The vast majority of previously reported examples carry an additional pseudoknot or other tertiary interactions between nucleotides that precede stem I and nucleotides in the loop of stem II. These extra contacts are critical for high-speed RNA catalysis. Herein, we report the discovery of ∼150,000 additional variant hammerhead representatives that exhibit diminished stem III substructures. These variants are frequently associated with Penelope-like retrotransposons, which are a type of mobile genetic element. Kinetic analyses indicate that these RNAs form dimers to cleave RNA.
Introduction
Of the 15 known classes of natural ribozymes, 9 catalyze RNA self-cleavage by a phosphoester transfer reaction.Citation1-4 This reaction involves the nucleophilic attack by a 2′-oxygen atom of ribose on the adjacent phosphorus center, which leads to departure of the neighboring 5′-oxygen atom and splitting of the RNA phosphodiester backbone.Citation1,5 Hammerhead ribozymes were the first of these 9 classes to be discovered,Citation6-8 which is not surprising given that members of this ribozyme class are the most common of all natural self-cleaving RNAs. Indeed, many thousands of hammerhead ribozyme representatives have been discovered in recent years among species from all 3 domains of life,Citation9-13 whereas some classes such as the hairpinCitation14 and the Neurospora VSCitation15 ribozymes appear to be exceedingly rare.
The catalytic core of hammerhead ribozymes is formed by 11 highly-conserved nucleotides encompassed by a 3-stem junction ().Citation8,10,16 Importantly, the vast majority of hammerhead ribozymes reported previously also include accessory structural elementsCitation10,17-19 that enhance the global fold of the ribozyme and promote high-speed catalysis.Citation20 Specifically, the first high-speed hammerhead constructs were predicted to form tertiary contacts that helped stabilize the parallel positioning of stems I and II in the active conformation of the ribozyme.Citation17 Bioinformatics analyses have since revealed that it is common for a pseudoknot to be formed between nucleotides preceding stem I and nucleotides in the loop of stem II.Citation10 Some hammerhead ribozymes with these natural accessory structures exhibit greater than 100 min−1 rate constants,Citation21 reflecting a rate enhancement of over a billion fold from the uncatalyzed value of ∼10−7 min−1 under cell-like conditions.Citation22 By contrast, hammerhead ribozyme constructs usually exhibit rate constants of ∼1 min−1 or lower when they have been trimmed to leave just the core 3-stem junction.
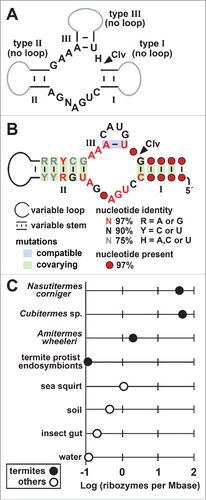
Figure 1. Identification of short stem III (SSIII) hammerhead ribozyme variants. (A) Conserved sequence and secondary structure model of the catalytic core of hammerhead ribozymes. The consensus sequence and structure model was used as the basis for the RNAMotif search, wherein variations were tolerated. N represents any nucleotide and H represents any nucleotide except G, while I, II and III identify 3 essential stems. Gray lines represent optional hairpin loops whose absence define the 3 major types as described previously.Citation10 The cleavage site (Clv) is designated with an arrowhead. (B) Conserved sequence and secondary structure model for hammerhead variants identified in our study, wherein stem III is formed by a single base-pair. (C) Distribution of identified variants in metagenomic data sets. Plotted are the number of predicted hammerhead ribozyme variants per megabase of DNA sequence searched from the sources indicated. Only those with a frequency above 0.1 are depicted (Table S1). These groups of DNA sequences are named after the host organism (N. corniger, Cubitermes sp, A. wheeleri, protists living in termites) or are comprised of metagenome sequences taken from sea squirt species Ecteinascidia turbinata and Lissoclinum patella (sea squirt), a variety of soil samples originating from forest or arctic peat environments (soil), different insect species such as Cardinium hertigii and Anoplophora glabripennis (insect gut), and freshwater samples or groundwater samples from a well contaminated with coal-tar waste (water). Ribozymes identified in moths (Agrotis sp.) and wasps (Encarsia pergandiella) are not shown because their frequency is below 0.1. Other details are as reported elsewhere (Table S1, Supplementary File 2).
Despite the reduced activity of hammerhead constructs retaining just the catalytic core, there have been examples of similarly small hammerhead ribozymes found in nature. For example, some of the first hammerhead ribozymes to be discovered carry greatly diminished stem III substructures,Citation23,24 appear to lack the largest and most prominent accessory features, and can be aided in their function by small peripheral regions.Citation25,26 Some of these RNAs appear to cleave RNA as obligate dimers, wherein each individual hammerhead domain overcomes the lack of a strong stem III by forming this essential substructure in collaboration with a partner.Citation23 More recently, many additional examples of short hammerhead representatives have been reported,Citation27 suggesting that these RNAs overcome the absence of these structural features that are critical for the high-speed function of other hammerhead ribozymes.
As an extension of our ongoing effortsCitation2,28,29 to discover novel noncoding RNAs, we have identified ∼150,000 examples of hammerhead ribozymes with a short (1 bp) stem III (hereafter called SSIII), representing ∼30,000 unique sequence variants. Numerous examples were identified among metagenomic or environmental DNA sequences (e.g. soil or termite gut). However, the examples primarily appear to be derived from eukaryotic genomes and some hammerhead ribozyme representatives even appear to be of fungal origin. As reported previously,Citation27 examples of these SSIII hammerhead variants are frequently associated with Penelope-like elements (PLEs),Citation30 which are a type of eukaryotic retrotransposon. Furthermore, we provide additional in vitro evidence that these RNAs cleave predominantly as dimers.
Results
Using a previously described computational pipeline,Citation29,31 we searched through fully sequenced genomes (RefSeq version 63)Citation32 and several environmental sequence data sets to find new structured RNAs. This search led to the identification of novel examples of SSIII hammerhead ribozyme variants in environmental sequences, and the rediscovery of examples recently identified in metazoans.Citation27 We then created a sequence alignment with these variants to guide the design of an RNAMotifCitation33 descriptor that retains the well-established characteristics of the hammerhead ribozyme core nucleotides and secondary structures (, Supplementary File 1). RNAMotif searches resulted in the discovery of approximately 30,000 unique hammerhead variants that retain key features of the catalytic core, but form an unusually short stem III consisting of a single base-pair ().
We speculated that single base-pair stem III structures would not be thermodynamically stable under cell-like conditions, as has previously been demonstrated for other hammerhead ribozymes with unusually short stem III structures.Citation23,24 However, this apparent defect in the secondary-structure model for the variant hammerhead RNAs presumably is not deleterious to their biological roles, given how many examples exist in biology. The majority of the variant ribozymes (about 99% of those we identified) were found in metagenome sequences from termite gut. Curiously, as described in greater detail below, these hammerhead ribozymes appear to be encoded by the termites, namely Nasutitermes corniger, Cubitermes sp. and Amitermes wheeleri, rather than their bacterial microbiomes (, Table S1). An additional possible source for these ribozymes, if transcribed, is the termite protist endosymbiont community. Other variants were found in metagenomic sequences taken from the sea squirt species Ecteinascidia turbinata and Lissoclinum patella, a variety of soil samples originating from forest or arctic peat environments, freshwater or groundwater samples from a well contaminated with coal-tar waste, and other insect species (Cardinium hertigii, Anoplophora glabripennis) (). Also presented (Supplementary File 2) is a complete list of searched metagenomes that contain SSIII hammerhead ribozyme variants, as well as calculated frequencies of the number of ribozymes per megabase (Mbase) of sequence.
The origin and biological utility of noncoding RNAs can sometimes be inferred by examining their genetic contexts. Unfortunately, most metagenomic DNA sequencing data is highly fragmented due to the short sequence reads that are then computationally assembled into longer segments called contigs. Even the generation of large numbers of contigs frequently provides only sparse coverage of the genomes present in the samples examined. As a result, many of the SSIII hammerhead ribozymes identified in our study cannot yet be placed into fully-sequenced genomes, and some are present in contigs that are too short to provide information on adjacent genes. However, on long contigs sometimes adjacent genes and others that are in close proximity to hammerhead variants can be identified.
From these gene associations, 3 tentative conclusions can be made. First, by using protein BLAST,Citation34 we determined that these associated genes frequently coded for retrotransposon-related proteins, such as reverse transcriptases (RTs). Since most metagenomic contigs are too small to observe genes, we restricted our analysis of gene associations to the 111 metagenomic contigs that were at least 4000 bp. We found 10 such contigs with predicted RT genes within 4000 bp of an SSIII hammerhead. There were no other gene classes that were more numerous than RT genes within this range. The distance between the ribozyme and the RT gene ranged from 43 to 4272 bp, wherein 7 of 10 examples were larger than 1000 bp.
Retrotransposons, such as Penelope-like elements, are mobile genetic elements that insert themselves into host DNA using an RNA intermediate.Citation35 These selfish nucleic acids usually encode proteins such as RTs or endonucleases (ENs) to facilitate insertion into the host genome. Our findings are analogous to those reported recentlyCitation27 for similar hammerhead ribozymes found in metazoan genomes, which are most commonly associated with Penelope-like elements (PLEs).Citation30 PLEs consist of direct, long terminal repeats (LTRs) that flank coding regions for RT and EN proteins. Phylogenetic reconstruction analysis indicates that RTs found in PLEs are most closely related to telomerase reverse transcriptases (TERTs).Citation36 When we compared amino acid sequences of RTs located near SSIII hammerheads found in our study to various known RTs, we observed that the RTs identified in our searches indeed are closely related to known PLE-RTs and TERTs (Fig. S1).
Second, sequence analyses strongly indicate that the hammerhead variants are not present in the species comprising the bacterial communities, but instead originate either from the eukaryotic organisms that host these microbiomes or from the protists that are also present. To assess their origin, we compared the RTs in the same contig as a hammerhead ribozyme to available genomes, using protein BLAST. Generally, we expect that the best (highest scoring) BLAST match of a sequence is a rough prediction of the species from which the sequence originates. We found that the best matches of the RTs were not to bacteria, but eukaryotes. For example, even though there was no genomic sequence available for termites at the time of our analyses, the RTs found near most hammerhead variants from their metagenomes are most closely related to RTs from psyllid genomes (Table S2). Psyllids are insects that are distantly related to termites.
Likewise, hammerhead variants are also present in the metagenomic datasets derived from other insect guts (Supplementary File 2). Again, the genes most closely related to those associated with hammerheads are of insect origin. For example, the gut metagenome of Nasutitermis corniger contained an RT whose best match was to an RT from the insect Diaphorina citri. Both N. corniger and D. citri are classified under the taxon Neoptera in Genbank, but immediately under Neoptera, N. corniger falls under Orthopteroidea, while D. citri is in Paraneoptera.Citation37 Since no Orthopteroidea genomes are available in the BLAST database, it is not surprising that the best match would be to another insect that fits into Neoptera but not Orthopteroidea. Thus, this BLAST result suggests that the metagenomics sequences are derived from the N. corniger host. By applying this reasoning to other host-associated metagenomic data sets, we concluded that hammerhead ribozyme variants with truncated stem III substructures are present in multiple termite species (e.g., Nasutitermes corniger, Cubitermes sp, Amitermes wheeleri), and also in moths (Agrotis sp.) and wasps (Encarsia pergandiella) (, Tables S1, S2).
Despite extensive searches, we did not find any of these novel hammerhead ribozyme variants in confirmed bacterial sequences. This finding, and the observation that metagenomic hammerhead variants are probably eukaryotic in origin, could mean that these variants are either extremely rare in bacteria, or do not exist at all in this domain of life. This might not be surprising as PLEs, which are highly associated with the SSIII hammerhead ribozyme variant, have not been reported in bacteria to date.Citation27
Third, we observed rare cases, from an environmental sample collected in a well contaminated with coal-tar waste,Citation38 in which RT sequences near SSIII hammerhead variants exhibit high similarity to RTs found in fungal genomes (Table S3). Previously, only type I hammerhead ribozymes with typically-sized stem III substructures have been identified in fungal PLEs.Citation9,10,24 Penelope-like elements previously have been identified from the genomes of fungi, as well as many animals, protists, plants.Citation39 The possible association of hammerhead variants with fungal PLEs suggests that the involvement of SSIII hammerheads might be exceedingly widespread and that similar mechanisms might be used by PLEs across different divisions of the eukaryotic domain of life.
To evaluate the performance characteristics of a SSIII hammerhead ribozyme, we chose a representative that retains all the typical features of the core of this ribozyme class, that was identified on a very long contig, and that has no close hammerhead ribozyme neighbors. The ribozyme, derived from a termite gut metagenome dataset and called env1 (), was predicted to be likely to form a homodimer as explained below. Short flanking regions comprised of the natural sequences were included both upstream and downstream of the wild-type (WT) ribozyme to retain possible tertiary RNA interaction sites typical of larger hammerhead ribozymes.Citation17,19
Figure 2. Sequence, predicted secondary structures, and functional characteristics of env1, a representative SSIII hammerhead. (A) Sequence and secondary structure prediction for a single WT env1 RNA (top) and 2 env1 RNAs forming a dimer (bottom). Constructs carrying mutations at specific sites (boxed) are designated M1 through M5. Note that M2 and M3 are insertions at the location indicated by the line. Encircled numbers indicate the length of added native nucleotides surrounding the conserved ribozyme core. 20 nucleotides were present on the 3′ terminus of the WT, M1, M2 and M3 constructs, whereas 30 nucleotides were present on constructs M4 and M5. Arrowhead designates the site of cleavage, and nucleotide numbering is relative to the in vitro transcription start site. (B) Co-transcriptional cleavage of WT env1 and its mutants was monitored by denaturing polyacrylamide gel electrophoresis (PAGE) of internally 32P-labeled RNA (α-32P-GTP) (see Materials and Methods for details). Full-length precursor (Pre) RNAs in nucleotides are 82 (WT and M1), 84 (M2), 86 (M3) and 92 (M4 and M5). Ribozyme cleavage product bands in nucleotides are 57 and 24 (WT and M1), 59 and 24 (M2), 61 and 24 (M3), and 57 and 35 (M4 and M5) as annotated with numbered arrowheads.
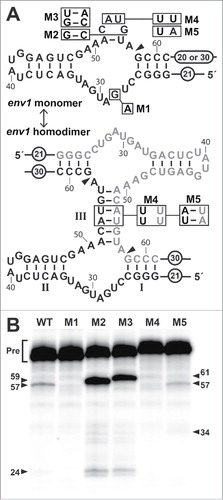
The RNA cleavage activities for the WT env1 ribozyme and for mutant constructs M1 through M5 were assessed by visualizing cleavage products during RNA preparation by in vitro transcription. The WT env1 construct cleaves very poorly when synthesized in a 2-hour transcription reaction (), whereas the same construct carrying a single G28A mutation (M1) in the conserved core completely eliminates activity as expected.Citation40 The poor performance of the WT construct suggests that the RNA does not efficiently adopt a catalytically active state as it exits RNA polymerase. This is unlike hammerhead ribozymes with stable stem structuresCitation41 or other ribozymesCitation2,42 that can undergo rapid self-cleavage when allowed to react during in vitro transcription. Therefore, some other folding step or other factor appears to be required for efficient self-processing of this SSIII hammerhead ribozyme.
An obvious structural deficiency in the WT env1 construct is its potential to form only a single base-pair in stem III in its unimolecular state, assuming that at least 4 nucleotides are required in the hairpin loop. To assess the importance of a stable stem III sub-structure, we examined the co-transcriptional ribozyme activities for env1-based constructs that carry 2 (M2) or 3 (M3) base-pairs. For these constructs, the formation of a thermodynamically stable stem III by a single molecule is likely, even though we cannot completely rule out that M2 and M3 also could form and cleave as dimers. As expected, these longer constructs exhibit more robust RNA cleavage, which supports the hypothesis that WT env1 on its own cannot form an efficient ribozyme structure.
Our bioinformatics data revealed that among variant ribozymes with stem III elements consisting of a single base-pair and a tetraloop, 89% have nucleotide sequences in this region that are perfectly palindromic. Encountering a 4-nucleotide palindromic arrangement by chance would only account for 14% of the sequences (Table S4, see also Materials and Methods). This palindromic feature is similar to that observed for other hammerhead ribozymes described previously.Citation9,10,23,27,43,44 Some of these previous reports also provided evidence that the RNAs exploited this palindromic sequence to form reactive hammerhead dimers that together form a stable stem III sub-structure.Citation10,23,27 If a similar dimer arrangement is exploited by env1 RNAs, they would form a 6-base-pair stem III (). We established the importance of dimer formation by testing env1 constructs that carry nucleotide changes that are predicted to either disrupt (M4) or restore (M5) stem III-mediated bimolecular complex formation. The assay results for M4 and M5 () are consistent with the hypothesis that the palindromic sequence, and therefore dimer formation, is necessary for env1 RNAs to generate any cleavage products.
To further assess the possibility that WT env1 and other SSIII hammerhead constructs might function efficiently as dimers in vitro, we examined the effects of ribozyme concentration on the rate constant for RNA cleavage (, Fig. S2). The env1 construct lacking any flanking sequences (called env1c, Fig. S2A) was compared with 2 ribozyme constructs based on previously reportedCitation23 SSIII hammerhead ribozymes called newt and newt-like (Fig. S2B, C). The rate constants exhibited at the highest RNA concentration (8 µM) by these ribozymes range from ∼0.0024 min−1 for the env1c construct to ∼0.033 min−1 for the newt construct (Fig. S2D-F). These values are extremely poor in comparison to most other hammerhead ribozyme constructs with typical-size stem III sub-structures, whose rate constants range from 1 min−1 to up to 900 min−1.Citation21,45 However, the rate constants for all 3 SSIII hammerhead constructs increase when the RNA concentration is increased. The simplest explanation for this finding is that increasing RNA concentrations progressively facilitate dimer formation, and that dimers are necessary for ribozyme function.Citation23 Since the rate constants measured in our in vitro assays are exceedingly poor, these RNAs might only achieve biologically relevant rate constants when efficiently forming dimers in their host cells. Another possibility to enable these RNAs to cleave at biologically relevant speeds is by forming complexes with auxiliary RNA-binding proteins as previously suggested.Citation46,47
Table 1. Observed rate constants (kobs) of newt and the newt-like hammerhead ribozyme variant previously describedCitation23 and the example env1 identified in our search (env1c). All reactions contained 0.08 µM 32P-labeled RNA and were supplemented with respective unlabeled RNA to achieve final concentrations listed in the center column. Newt-like designates a construct that is identical to the newt-like construct previously investigated.Citation23 Fig. S2B and C highlight the differences to the natural newt genomic sequence. No observed rate constant was determined for construct env1c at 0.08 µM RNA because of expected extremely slow speed of ribozyme self-cleavage.
Given that the env1 construct is derived from a possible natural RNA transcript that lacks a close hammerhead ribozyme neighbor, it seems likely that multiple transcript copies might be required to form homodimers. Unfortunately we cannot easily anticipate the nature of this larger assembly, and as a result we sought a more predictable system to evaluate the importance of dimer formation. We therefore chose to examine 2 newly discovered SSIII hammerhead ribozymes that naturally occur nearby to each other. An RNA construct, called env2 (), was created based on a natural arrangement of 2 hammerhead ribozymes wherein both can only form a single base-pair in stem III unless they collaborate as a dimer. When env2 folds to form 2 separate hammerhead monomers (), the 2 independently-folded ribozyme domains are separated by a 44-nucleotide linker. In the dimer configuration (), each ribozyme core is a combination of the first and second hammerhead sequences, and the 44-nucleotide linker forms the large hairpin loop of a stem I sub-structure ().
Figure 3. A tandem hammerhead cleaves as an obligate dimer. (A) Sequence and secondary structures of the WT tandem hammerhead construct env2 depicted as independently folded (top) and dimer (bottom) structures. Hammerhead depiction is rotated 90° from previous figures. Individual ribozyme sequences are shown in black (first ribozyme) and gray (second ribozyme). Mutations to active-site nucleotides in constructs M6, M7 and M8 are boxed. Two guanine nucleotides on the 5′ end were added for efficient transcription (see Materials and Methods). Other annotations are as described for . (B) Ribozyme cleavage reactions (Rxn) were conducted (+) at 37°C for 2 h using 50 mM Tris-HCl (pH 8.0 at 23°C), 0.5 mM EDTA, 10 mM MgCl2 and internally labeled RNA. The no reaction lane (−) yields only the full length RNA precursor (Pre). The expected 3 nt RNA cleavage fragment has been run off the gel.
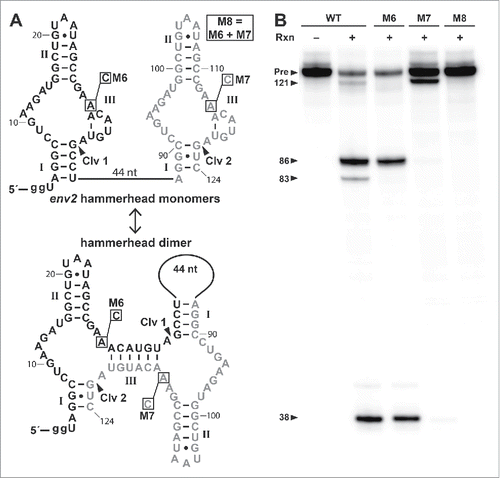
The WT env2 RNA was found to undergo efficient RNA cleavage at both cleavage sites 1 and 2 (Clv 1 and Clv 2) when incubated with Mg2+ (). Importantly, mutant M6, which carries an A28C mutation that inactivates the first hammerhead catalytic core, undergoes efficient cleavage only at Clv 1 and not at Clv 2. If the hammerheads functioned as independently folded ribozymes, then the M6 mutation should have abolished cleavage at Clv 1, but permitted processing of Clv 2 by the second hammerhead domain. Therefore, the 2 hammerheads must form chimeric ribozyme cores wherein the cleavage site of the first ribozyme is presented for cleavage by the core of the second ribozyme, and vice versa. This conclusion is further supported by the cleavage pattern observed for env2 mutant M7, which carries an A111C mutation that likewise inactivates the second hammerhead. M7 undergoes cleavage only at Clv 2, indicating that the cleavage site for the second ribozyme is presented to the first ribozyme core. Finally, construct M8, which combines the M6 and M7 mutations into a single RNA, fails to undergo cleavage at either site, as expected.
The more robust folding and function of the env2 construct compared to env1 compelled us to determine if SSIII hammerhead ribozymes are often found naturally in close proximity to each other. We estimated the frequency of nearby hammerhead arrangements by finding large contigs that contain a hammerhead surrounded by at least 1 kb of sequence both upstream and downstream, and then determined how many of these ribozymes have another ribozyme close by (Table S5). A total of 668 ribozymes were identified, of which 370 carried a second hammerhead within 500 nucleotides (Fig. S3). These adjacent hammerhead arrangements are similar to those that have been described in the literature previouslyCitation23,47 However, 247 of these 370 ribozymes associate with nearby ribozymes on the opposite strand. Our analysis of stem III sequences of nearby ribozymes located either on the same or opposite DNA strands revealed that, in many cases, stem III sequences from the different ribozymes would permit the folding of an extended, more stable stem III in a complex formed by neighboring ribozymes. However, hammerheads on opposite strands could also be explained by their association to PLEs and the PLE architecture and propagation mechanism.Citation27,30
Curiously, tandem hammerheads on the same strand are only rarely located close to each other, which might otherwise be an easy strategy to promote dimer formation. The majority are separated by more than 450 nucleotides, and these might need assistance (such as the involvement of long-distance base-pairing or protein factors) to form dimers between such distal partners. If tandem ribozymes form dimers within the same transcript, we would expect that closer proximity between the 2 hammerheads would be favored. Perhaps for some of the distally located ribozymes, obligate dimer formation occurs between 2 different transcripts of the same RNA sequence, or there are other mechanisms to ensure that a single SSIII hammerhead ribozyme finds a partner to promote catalysis. Regardless, in instances when an assembled contig is long enough for meaningful analysis, a SSIII hammerhead ribozyme is found nearby another about 55% of the time.
In some instances we even found multimeric ribozyme arrangements that include more than 2 in close proximity on the same or opposite strands. Among the many identified multimeric examples were some with an extensive number of hammerhead ribozyme variants on one strand. In one example (contig JGI20163J15578_10119844/1-1635) up to 20 nearly identical sequences were found in series. These arrangements might indicate the involvement of repetitive hammerhead ribozymes in a new biological role. However, these examples should be investigated with caution for the following reasons. Often, nucleotide differences within the otherwise highly conserved regions of the ribozyme prevented automated detection of all multimeric sequences, which required manual identification steps to be used with those contigs that were already enriched for multiple hammerheads. It is likely that the examples carrying mutation in the core regions are functionally inactive, or might require the assistance of an adjacent ribozyme core to undergo RNA cleavage. Moreover, these contigs are derived from the assembly of environmental sequencing data that is highly complex, and it is possible that they result from assembly errors.Citation48 Nevertheless, many discovered examples with highly conserved ribozyme core nucleotides appear valid and the tandem construct env2 can be taken as proof that there are catalytically active examples.
Discussion
Our bioinformatics searches have uncovered ∼150,000 examples of hammerhead ribozymes with unusually short stem III substructures. This number of hits in our searches is vastly larger in comparison to the number generated in control searches, in which the highly conserved CUGANGA core sequence was scrambled (Table S4). Therefore, we have reason to believe that only few of the putative ribozymes are false positives. It is also possible that some of these examples are not actively transcribed, and we cannot rule out the possibility that some hits might represent hammerhead ribozyme variants that are losing catalytic function through natural evolutionary processes.
Regardless, these hammerhead ribozymes are highly abundant in some termite gut metagenomes (), more so than in previously studied metazoans.Citation27 This observation suggests that PLEs are especially prolific in these termite species and many of the contigs are derived from eukaryotic cells and not bacterial cells. It is also conceivable these results are caused by artifacts in DNA preparation, sequencing or metagenomic assembly. A definitive answer will likely require a genome sequence of these termites.
Previous studiesCitation23,24 had uncovered the existence of similar SSIII hammerhead ribozymes, and had provided evidence that these natural constructs exhibit improved function by forming dimers wherein a longer stem III is shared between the 2 ribozyme cores. Our data extend these findings to reveal the existence of numerous tandem arrangements of these SSIII hammerhead ribozymes, and our data confirm that a representative natural tandem arrangement requires the formation of the heterodimer structure to promote robust RNA cleavage activity ().
Additionally, we found that the putative ribozymes in termite DNA are dominated by 4-nucleotide-palindromes plus the additional A-U base-pair that would otherwise form the predicted stem III substructure. By contrast, most SSIII hammerhead sequences derived from metazoans contain a 2 nucleotide palindrome in the loop of stem III.Citation24 Although palindromic sequences in the loop predominate, non-palindromic sequences do occur. These could be non-functional representatives, ribozymes that partner with others that contain non-palindromic but complementary loop III sequences, or perhaps the resulting mismatches in the shared stem III of dimers might be tolerated just like mismatches in other structured RNAs can sometimes be tolerated. In our example sequence env1, ribozyme self-cleavage was completely abolished when the palindromic loop of stem III was mutated and ribozyme activity was restored when the compensatory mutations restored palindromic character (). However, other examples have been previously described in which mismatches in the stem III substructure occur and are tolerated.Citation23
The biological utility of obligate hammerhead dimer formation remains uncertain. It is clear from our findings and others,Citation24 that numerous SSIII hammerhead ribozymes are associated with selfish genetic elements called PLEs.Citation27 Examples of PLEs are known to contain 2 Penelope long-terminal repeats (PLTRs), and it is common for SSIII hammerhead ribozymes to be encoded within these PLTRs. Therefore it is not surprising to often find these hammerhead ribozymes in tandem (within 3 kb) given the close proximity of PLTRs in the genomes that host PLEs. In cases where a second hammerhead cannot be found nearby, it is possible that the PLE has become truncated by mutation events or that the metagenomic contig is not long enough to contain the entire PLE including both PLTRs.
In instances when the sequenced contig containing the hammerhead ribozyme is long enough to identify associated genes, we commonly find RT genes are present. These RTs are phylogenetically most similar to those carried by PLEs, suggesting that the great number of SSIII hammerhead ribozymes is due to the spread and evolution of PLEs or of related elements. The architectures and replication mechanisms of these selfish genetic elements might naturally exploit obligate dimer-forming hammerhead ribozymes to control the timing of RNA processing during their lifecycles.
The relative abundance of self-cleaving RNA classes emphasizes their mysterious roles in biology. The continuing discovery of novel self-cleaving ribozyme motifsCitation2-4 as well as variants of long standing self-cleaving ribozymes such as the hammerhead class will give us greater opportunity to decipher their functions. These discoveries also highlight the fact that modern organisms continue to make extensive use of these catalytic RNAs.
Materials and methods
Computational methods
The final searches used in our analysis were performed on the bacterial and archaeal subsets of RefSeq version 63Citation29 and a collection of environmental nucleotide sequences that was previously used.Citation2 Searches used RNAmotifCitation30 with descriptors given in Supplementary File 1. Environmental data set metadata was taken from various sources, and most datasets relevant to the hammerhead ribozyme variants were taken from the IMG/M web site.Citation38 Drawings of consensus sequence and secondary structure predictions were performed using R2R.Citation49
Calculation of probability for random occurrence of 4-nucleotide palindrome
We calculated that, by random chance, ∼14% of 4-nucleotide sequences are expected to bind to themselves using Watson-Crick or G-U pairs. This calculation assumes that nucleotides are independently and uniformly distributed, i.e., that each nucleotide occurs with a 25% probability. The probability that a pair of nucleotides is Watson-Crick or G-U is 6/16, for the 6 favorable base pairs, out of the 16 possible combinations. Since there are 2 (assumed) independent base pairs in the 4 nucleotides, the probability is (6/16)2, or roughly 14%.
Phylogenetic analysis of known RTs and predicted RTs near hammerhead ribozyme variants
Known reverse transcriptase (RT) proteins,Citation33 and RT proteins in metagenome data sets nearby to variant hammerhead ribozymes were aligned to PfamCitation50 model PF00078 using the hmmalign program of HMMER3.Citation51 Amino acids outside of the PF00078 model were removed. Some sequences lacked many residues on their N- and C-termini in the HMM alignment, and in extreme cases sequences were removed. Columns were removed when 80% or more of the sequences contained a gap. We inferred a phylogenetic tree and branch confidence values using PhyMLCitation52 version 20110105 (command line: phyml -i alignment.phylip – –rand_start – –n_rand_rand_starts 10 -d aa -f e -t e -v e -a e -s SPR -o tlr – –no_memory_check -b -4). The phylogenetic tree in Fig. S1 was drawn using iTOL.Citation53
Construct design and synthesis using in vitro transcription
Core hammerhead ribozyme sequences were extended by natural sequences on their 5′ and 3′ ends to include possible tertiary RNA interaction sites. For efficient in vitro transcription a T7 RNA polymerase promoter sequence was added to the 5′ end as well as 2 guanosines where the natural sequence did not contain G-nucleotides. The example sequence env1 is found under nucleotide accession numberCitation37 JGI20163J15578_10008454 on the sense strand of nucleotides 1152–1191 and the tandem example env2 was found in JGI20172J14457_10225843 on the reverse strand of nucleotides 100-221. Double-stranded DNA templates were generated from oligonucleotides (Table S6) by overlap extension using Superscript II reverse transcriptase (Invitrogen) according to manufacturer's instructions. Transcriptions were performed for 2 h at 37°C in 80 mM HEPES-KOH (pH 7.5 at 23°C), 24 mM MgCl2, 2 mM spermidine, 40 mM DTT, 10 mM NTPs, 10µCi α-32P-GTP, T7 RNA polymerase (25 U/µl) and 1 µM dsDNA template. Transcriptions were stopped using 2x denaturing loading buffer (0.09 M Tris, 0.09 M borate, 10 mM EDTA pH 8.0, 8 M urea, 20% sucrose (w/v), 0.1% SDS (w/v), 0.05% bromophenol blue (w/v), 0.05% xylene cyanol (w/v)) and RNA products were separated by denaturing (8 M urea) 20% PAGE. RNA bands were visualized by autoradiography on a STORM imager (Molecular Dynamics).
In vitro self-cleavage assays and determinations of observed rate constants
Full length hammerhead ribozyme bands were isolated from denaturing polyacrylamide gels by crushing gel slices, soaking them in crush-soak solution (200 mM NaCl, 10 mM Tris-HCl [pH 7.5 at 23°C], 1 mM EDTA pH 8.0) for 2 h at room temperature, passing eluate from gel pieces through a filtered Costar Spin-X centrifugation column (Corning) and precipitating the RNA with ethanol. Radiolabeled RNA concentrations were estimated using a scintillation counter (MicroBeta2, PerkinElmer). Time course experiments were performed for up to 2 h in 50 mM Tris-HCl, [pH 8.0 at 23°C], 10 mM MgCl2, 0.5 mM EDTA at 55°C containing 0.08 µM 32P-labeled RNA,Citation23 with samples being withdrawn at intermittent time intervals. These conditions were chosen to be able to compare our data to that previously obtained.Citation23 Tandem hammerhead ribozyme construct was incubated under the same buffer conditions at 37°C. Cleavage products were separated by PAGE and quantified using ImageQuant TL software (GE Healthcare Life Sciences). The observed rate constants (kobs) listed in were determined by plotting the natural logarithm of the fraction of substrate that remained uncleaved versus time, and establishing the negative slope of the resulting line. Example polyacrylamide gels and plots to obtain kobs are shown in Fig. S2.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Tables_and_Figures.docx
Download MS Word (496.4 KB)Acknowledgments
We thank the Breaker laboratory, especially Dr. Adam Roth, for helpful discussions and comments on this manuscript. C.E.L. was supported by the German Research Foundation (DFG grant LU1889/1-1) and NIH grant P01 GM022778 (awarded to R.R.B.). Research in the Breaker laboratory is also supported by the Howard Hughes Medical Institute and by Yale University. We also thank R. Bjornson for assisting our use of the Yale Life Sciences High Performance Computing Center (NIH grant RR19895-02).
References
- Jimenez RM, Polanco JA, Lupták A. Chemistry and biology of self-cleaving ribozymes. Trends Biochem Sci. 2015;40:648–61. doi:10.1016/j.tibs.2015.09.001
- Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lünse CE, Breaker RR. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol. 2015;11:606–10. doi:10.1038/nchembio.1846
- Li S, Lünse CE, Harris KA, Breaker RR. Biochemical analysis of hatchet self-cleaving ribozymes. RNA. 2015;21:1845–51. doi:10.1261/rna.052522.115
- Harris KA, Lünse CE, Li S, Brewer KI, Breaker RR. Biochemical analysis of pistol self-cleaving ribozymes. RNA. 2015;21:1852–8. doi:10.1261/rna.052514.115
- Emilsson GM, Nakamura S, Roth A, Breaker RR. Ribozyme speed limits. RNA. 2003;9:907–18. doi:10.1261/rna.5680603
- Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–40. doi:10.1093/nar/14.9.3627
- Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–80. doi:10.1126/science.231.4745.1577
- Forster AC, Symons RH. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987;49:211–20. doi:10.1016/0092-8674(87)90562-9
- de la Peña M, Garcìa-Robles I. Ubiquitous presence of the hammerhead ribozyme motif along the tree of life. RNA. 2010;16:1943–50. doi:10.1261/rna.2130310
- Perreault J, Weinberg Z, Roth A, Popescu O, Chartrand P, Ferbeyre G, Breaker RR. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput Biol. 2011;7:e1002031. doi:10.1371/journal.pcbi.1002031
- Jimenez RM, Delwart E, Lupták A. Structure-based search reveals hammerhead ribozymes in the human microbiome. J Biol Chem. 2011;286:7737–43. doi:10.1074/jbc.C110.209288
- Seehafer C, Kalweit A, Steger G, Gräf S, Hammann C. From alpaca to zebrafish: Hammerhead ribozymes wherever you look. RNA. 2011;17:21–6. doi:10.1261/rna.2429911
- Hamann C, Lupták A, Perreault J, de la Peña M. The ubiquitous hammerhead ribozyme. RNA. 2012;18:871–85. doi:10.1261/rna.031401.111
- Wilson TJ, Nahas M, Ha T, Lilley DMJ. Folding and catalysis of the hairpin ribozyme. Biochem Soc Trans. 2005;33:461–5. doi:10.1042/BST0330461
- Collins RA. The Neurospora varkud satellite ribozyme. Biochem Soc Trans. 2002;30:1122–6. doi:10.1042/bst0301122
- Hertel KJ, Pardi A, Uhlenbeck OC, Koizumi M, Ohtsuka E, Uesugi S, Cedergren R, Eckstein F, Gerlach WL, Hodgson R, et al. Numbering system for the hammerhead. Nucleic Acids Res. 1992;20:3252. doi:10.1093/nar/20.12.3252
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–12. doi:10.1038/nsb959
- Uhlenbeck OC. Less isn't always more. RNA. 2003;9:1415–7. doi:10.1261/rna.5155903
- de la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–70. doi:10.1093/emboj/cdg530
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–20. doi:10.1016/j.cell.2006.06.036
- Canny MD, Jucker FM, Kellogg E, Khvorova A, Jayasena SD, Pardi A. Fast cleavage kinetics of a natural hammerhead ribozyme. J Am Chem Soc. 2004;126:10848–9. doi:10.1021/ja046848v
- Li Y, Breaker RR. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J Am Chem Soc. 1999;121:5364–72. doi:10.1021/ja990592p
- Forster AC, Davies C, Sheldon CC, Jeffries AC, Symons RH. Self-cleaving viroid and newt RNAs may only be active as dimers. Nature. 1988;334:265–7; PMID:2456468; https://doi.org/10.1038/334265a0
- Epstein LM, Gall JG. Self-cleaving transcripts of satellite DNA from the newt. Cell. 1987;48:535–543
- Epstein LM, Pabón-Peña LM. Alternative modes of self-cleavage by newt satellite 2 transcripts. Nucleic Acids Res 1991; 19:1699–705. doi:10.1093/nar/19.7.1699
- Pabón-Peña LM, Zhang Y, Epstein LM. Newt satellite 2 transcripts self-cleave by using an extended hammerhead structure. Mol Cell Biol. 1991;11:6109–15. doi:10.1128/MCB.11.12.6109
- Cervera A, de la Peña M. Eukaryotic penelope-like retroelements encode hammerhead ribozyme motifs. Mol Biol Evol. 2014;31:2941–7. doi:10.1093/molbev/msu232
- Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–19. doi:10.1093/nar/gkm487
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:R31. doi:10.1186/gb-2010-11-3-r31
- Evgen'ev MB, Arkhipova IR. Penelope-like elements – a new class of retroelements: distribution, function and possible evolutionary significance. Cytogenet Genome Res. 2005;110:510–21. doi:10.1159/000084984
- Tseng HH, Weinberg Z, Gore J, Breaker RR, Ruzzo WL. Finding non-coding RNAs through genome-scale clustering. J Bioinform Comput Biol. 2009;7:373–88. doi:10.1142/S0219720009004126
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–45. doi:10.1093/nar/gkv1189
- Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–35. doi:10.1093/nar/29.22.4724
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Xhang Z, Miller W, Lipman DL. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402. doi:10.1093/nar/25.17.3389
- Craig NL, Craidie R, Gellert M, Lambowitz A., eds. Mobile DNA II. Washington DC, American Society for Microbiology Press 2002
- Arkhipova IR, Pyatkov KI, Meselson M, Evgen'ev MB. Retroelements containing introns in diverse invertebrate taxa. Nat Genet. 2003;33:123–4. doi:10.1038/ng1074
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res.. 2009;37(Database issue):D26–31. doi:10.1093/nar/gkn723
- Markowitz VM, Chen IM, Chu K, Szeto E, Palaniappan K, Pillay M, Ratner A, Huang J, Pagani I, Tringe S, et al. IMG/M 4 version of the integrated metagenome comparative analysis system. Nucleic Acids Res. 2014;42:D568–73. doi:10.1093/nar/gkt919
- Arkhipova IR. Distribution and phylogeny of Penelope-like elements in eukaryotes. Syst Biol. 2006;55:875–85. doi:10.1080/10635150601077683
- Ruffner DE, Stormo GD, Uhlenbeck OC. Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry. 1990;29:10695–702. doi:10.1021/bi00499a018
- Long DM, Uhlenbeck OC. Kinetic characterization of intramolecular and intermolecular Hammerhead RNAs with stem II deletions. Proc Natl Acad Sci USA. 1994;91:6977–81. doi:10.1073/pnas.91.15.6977
- Roth A, Weinberg Z, Chen AG, Kim PB, Ames TD, Breaker RR. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol. 2014;10:56–60. doi:10.1038/nchembio.1386
- Ferbeyre G, Smith JM, Cedergren R. Schistosome satellite DNA encodes active hammerhead ribozymes. Mol Cell Biol. 1998;18(7):3880–8. doi:10.1128/MCB.18.7.3880
- Rojas AA, Vazquez-Tello A, Ferbeyre G, Venanzetti F, Bachmann L, Paquin B, Sbordoni V, Cedergren R. Hammerhead-mediated processing of satellite pDo500 family transcripts from Dolichopoda cave crickets. Nucleic Acids Res.. 2000;28(20):4037–43. doi:10.1093/nar/28.20.4037
- Stage-Zimmermann TK, Uhlenbeck OC. Hammerhead ribozyme kinetics. RNA. 1998;4:875–89. doi:10.1017/S1355838298980876
- Luzi E, Eckstein F, Barsacchi G. The newt ribozyme is part of a riboprotein complex. Proc Natl Acad Sci U S A. 1997;94(18):9711–6. doi:10.1073/pnas.94.18.9711
- Denti MA, Martínez de Alba AE, Sägesser R, Tsagris M, Tabler M. A novel RNA-binding protein from Triturus carnifex identified by RNA-ligand screening with the newt hammerhead ribozyme. Nucleic Acids Res. 2000;28(5):1045–52. doi:10.1093/nar/28.5.1045
- Nagarajan N, Pop M. Sequence assembly demystified. Nat Rev Genet 2013;14:157–67. doi:10.1038/nrg3367
- Weinberg Z, Breaker RR. R2R-software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011;12:3. doi:10.1186/1471-2105-12-3
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–30. doi:10.1093/nar/gkt1223
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–11
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–52. doi:10.1080/10635150600755453
- Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39:W475–8. doi:10.1093/nar/gkr201
