ABSTRACT
Posttranscriptional processing of RNA molecules is a common strategy to enlarge the structural and functional repertoire of RNomes observed in all 3 domains of life. Fragmentation of RNA molecules of basically all functional classes has been reported to yield smaller non-protein coding RNAs (ncRNAs) that typically possess different roles compared with their parental transcripts. Here we show that a valine tRNA-derived fragment (Val-tRF) that is produced under certain stress conditions in the halophilic archaeon Haloferax volcanii is capable of binding to the small ribosomal subunit. As a consequence of Val-tRF binding mRNA is displaced from the initiation complex which results in global translation attenuation in vivo and in vitro. The fact that the archaeal Val-tRF also inhibits eukaryal as well as bacterial protein biosynthesis implies a functionally conserved mode of action. While tRFs and tRNA halves have been amply identified in recent RNA-seq project, Val-tRF described herein represents one of the first functionally characterized tRNA processing products to date.
Introduction
Over the last few decades various non-protein-coding RNA (ncRNA) classes have been identified as pivotal players in regulating gene expression (reviewed in [ref. Citation1,2]). Recent deep sequencing studies have uncovered thousands of ncRNA candidates in all 3 domains of life thus indicating a so far buried layer of transcriptome complexity.Citation1,3 Thereby it is intriguing that ncRNA transcripts with well assigned or proposed functions, such as small nucleolar RNAs or vault RNAs, can serve as precursors for downstream cleavage events generating yet another functional class of RNA pieces such as micro RNAs (miRNAs).Citation4,5 Subsequent transcriptome-wide studies showed that post-transcriptional cleavage events of RNAs with assigned cellular functions into smaller fragments fulfilling other, mostly regulatory, roles is rather the rule than the exception (reviewed in [ref. Citation6]). Posttranscriptional fragmentation to generate smaller functional RNAs involves all sorts of transcripts including protein coding (e.g. [ref. Citation7]) as well as ncRNAs (e.g., [ref. Citation8-10]) and is thus potentiating the regulatory and functional repertoire of cellular RNomes significantly.Citation6 One of the most amply reported substrates for such RNA fragmentation events in recent years involves tRNAs (reviewed in [ref. Citation11-15]).
tRNA molecules have been shown to be cut in the anticodon loop thus giving rise to tRNA halves. Fragmentation can also occur elsewhere in the molecule therefore generating shorter pieces typically referred to as tRNA-derived fragments (tRFs) with a size between ∼18–26 nucleotides. tRNA-derived fragments (tRFs) were first observed already several decades ago in human tumor tissueCitation16 and E. coli cells,Citation17 and have subsequently been reported in model organisms from all 3 domains of life. Nevertheless, tRFs and tRNA halve molecules were initially regarded as either by-products of tRNA biogenesis or random degradation intermediates. Only recently more dedicated research uncovered that tRNA pieces can fulfill regulatory roles in cell metabolism and thus some represent functional ncRNA molecules. Typically tRNA fragmentation is not a global intracellular phenomenon but affects only particular tRNA isoacceptors and has often been shown to be stress dependent or cell-type specific (reviewed in [ref. Citation11,12]). While some of the endonucleases involved in tRNA halves production in eukaryal species have been identified, such as angiogenin, Rny1 or Dicer, tRF biogenesis in prokaryal organisms is largely unknown. It has been discussed that post-transcriptional tRNA modifications can guide or expel processing RNases during tRNA fragmentation.Citation12,18 Also the functional repertoire of tRFs and tRNA halves is huge and seems to vary between cell types and incubation conditions. For example, recently 2 studies in human breast cancer cells reported that tRNA-derived fragments harbor tumor- and metastasis-suppressive activityCitation19 while Honda et al. revealed tRNA halves to have proliferation effects on estrogen receptor-positive breast cancer cells.Citation20 In mouse sperm tRNA halves have been identified to represent a paternal epigenetic factor that may contribute to intergenerational inheritance.Citation21 Even though our understanding of the tRF and tRNA halves biology is still in its infancy, the emerging picture form a plethora of recent studies is that tRNA pieces represent biologically relevant entities rather than mere degradation products of full-length tRNAs.
Even though recent transcriptome-wide RNA-seq studies led to the discovery of tRNA pieces in many different model organisms, functional or mechanistic insight into their physiological roles remained sparse with only few exceptions (e.g., [ref. Citation10,19-23]). Here we report on the functional characterization of a tRF deriving from Val-tRNA (Val-tRF) of the halophilic archaeon Haloferax volcanii. In a recent publication we could show that this 26 residue long 5′ tRF is produced under alkaline stress and is capable of binding to the ribosome in vitro and in vivo.Citation24 Here we provide functional evidence that this ribosome-associated small ncRNA (rancRNA; [ref. Citation25]) attenuates global protein biosynthesis by competing with mRNA binding to the small ribosomal subunit and thus affecting translation initiation.
Results
The inhibitory potential of Val-tRF is functionally conserved in all 3 domains of life
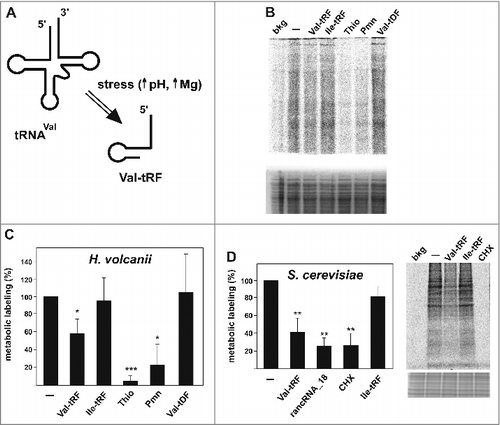
In a recent genomic screen designed at identifying small ribosome-associated ncRNAs (rancRNAs) in H. volcanii, we detected several tRFs, especially originating from the 5′ part of a subset of tRNAs (5′ tRFs), to co-purify with ribosomal particles in a stress-dependent manner.Citation24 A 26 nucleotide long 5′ fragment deriving from valine tRNA showed high abundance during alkaline stress and inhibited H. volcanii-based in vitro translation and in vitro peptide bond formation assays upon binding to the small ribosomal subunit.Citation24 To study the role of this Val-tRF in a more physiological setting we established a metabolic labeling approach to study protein biosynthesis in vivo. To this end we introduced the synthetic Val-tRF into H. volcanii cells using a polyethylene glycol-mediated transformation procedureCitation26 and subsequently assessed the amounts of newly synthesized proteins by means of in vivo 35S-methionine incorporation. Indeed after introduction of chemically synthesized Val-tRF markedly reduced levels of newly synthesized proteins were observed (). Inhibition efficiency was in a similar range compare with well-known ribosome-targeted antibiotics such as puromycin or thiostrepton. Importantly, introduction of an Ile-tRF, an analogous tRF deriving from a comparable region of isoleucine tRNA, served as specificity control. Significantly, Ile-tRF, a fragment that was not present in our rancRNA screen,Citation24 did not inhibit metabolic labeling. Similarly, a DNA-variant of the Val-tRF (Val-tDF) had no effect on the in vivo protein production in H. volcanii cells ( and Fig. S1). Since both tRNAs and ribosomes are universally conserved components of the translational machinery, we tested if the Val-tRF-mediated translation repression is functionally conserved in other species as well. In order to elucidate this we tested the H. volcanii Val-tRF in eukaryotic and bacterial systems, namely in S. cerevisiae and E. coli, respectively. To study the in vivo effect of the Val-tRF in yeast we performed an analogous metabolic labeling approach using S. cerevisiae spheroplasts.Citation27 Introduction of the Val-tRF into the yeast spheroplasts by electroporation resulted in a significant reduction of protein biosynthesis (). The Val-tRF-mediated reduction in the yeast metabolic labeling assay was comparable to the effects of a previously characterized yeast rancRNA (rancRNA_18) which is an mRNA exon-derived ncRNA shown to target and regulate yeast ribosomes.Citation7 The translation inhibitor cycloheximide served as protein synthesis inhibition control (). Importantly, also in this setting Ile-tRF was not able to inhibit protein biosynthesis in yeast cells in a comparable manner. To corroborate these findings, in vitro translation reactions were performed using yeast cell extracts or a commercially available E. coli in vitro translation system as well as a peptidyl transferase assay based on purified E. coli ribosomes. Addition of the H. volcanii Val-tRF, but not the Ile-tRF, reproducibly inhibited in vitro protein biosynthesis in both the eukaryal and bacterial systems (Fig. S2A, B, C). Additionally, during the peptidyl transferase assay the Val-tRF was able to decrease the extent of peptide bond formation by about 40% (Fig. S2D). Cumulatively, these data suggest that the archaeal Val-tRF is capable of also inhibiting bacterial and eukaryotic translation systems. This implies that the mode of action, and thus likely also the ribosomal target site, is conserved in all 3 domains of life.
Figure 1. Val-tRF inhibits translation in vivo. (A) Under specific stress conditions, particularly under alkaline stress and elevated magnesium conditions (24), the valine tRNA(GAC) isoacceptor is processed to give rise to a 26-residue long 5′-tRF. (B) Metabolic labeling in H. volcanii spheroplasts in the absence (−) or presence of Val-tRF, Ile-tRF or an all DNA version of Val-tRF, named Val-tDF. The antibiotics thiostrepton (Thio) and puromycin (Pmn) served as translation inhibition controls. The background (bkg) represents radioactive bands measured in the absence of any incubation. A representative SDS page of the newly synthesized 35S-labled proteins is shown (upper gel). The coomassie stained gel part underneath the autoradiogram serves as loading control. (C) Quantification of the lane intensities obtained in the metabolic labeling experiments in H. volcanii are shown whereas the activity in the absence of any transformed synthetic RNA (−; mock) is set to 100%. The background values (see above) were subtracted from all other samples. The mean and standard deviations of 4 independent experiments are shown. (D) The H. volcanii Val-tRF also inhibits metabolic labeling in S. cerevisiae spheroplasts. The mean and standard deviations of 4 independent experiments are shown. Cycloheximide (CHX) and an already characterized yeast rancRNA (rancRNA_18) served as inhibition controls. A representative SDS PAGE of newly synthesized proteins after the 35S-methionine spike is shown. The coomassie stained gel part underneath the autoradiogram serves as loading control. In (C) and (D) significant differences relative to the mock control (−) were determined using the 2-tailed paired Student's t-test (***p < 0.001, **p < 0.01, *p < 0.05).
Val-tRF competes with efficient translation initiation
Protein biosynthesis proceeds in 4 phases, namely translation initiation, elongation, termination and recycling. In principle all of these 4 phases, which are characterized by complex interactions between the ribosome, tRNAs, mRNA, and additional protein factors could be affected by Val-tRF. It has been shown over the years that translation regulation mainly occurs on the level of initiation, since this is the most critical and rate-limiting step of protein biosynthesis.Citation28,29 Preferential binding of the Val-tRF to the small ribosomal subunit in vitro and in vivoCitation24 led to the hypothesis that this tRF might interfere with the formation of the translation initiation complex. In order to test this we performed toeprinting experiments using purified H. volcanii 70S (or 30S) particles, deacylated E. coli initiator tRNAfMet and an mRNA analog carrying a unique AUG codon.Citation30 The formation of the initiation-like complexes 30S/tRNAfMet/mRNA or 70S/tRNAfMet/mRNA was assessed by the reverse transcription inhibition-based toeprinting assay in the absence or presence of Val-tRF (). The primer extension stops at positions +15/+16 (whereas the A in the start codon is assigned to position +1) provides a measure for the formation of the initiation-like complex, since it is dependent on both 70S ribosomes (or 30S subunits, respectively) and initiator tRNA ( and Fig. S3A). Addition of Val-tRF interfered with this complex formation in a dose dependent manner. Importantly, under the highest tested concentration, the Ile-tRF in contrast had no effect on complex formation ( and Fig. S3B). Thus this assay indicates that the Val-tRF specifically interferes with the formation of translation initiation-like complexes.
Figure 2. Val-tRF interferes with the efficient establishment of a translation initiation-like complex as monitored by toeprinting. (A) Schematic representation of the toeprinting assay using H. volcanii 30S subunits (gray) or 70S ribosomes (not shown), initiator tRNAfMet and an mRNA. Reverse transcription (dotted arrow) of the [32P]-radiolabeled primer (solid arrow) is terminated in case of a stable 30S/tRNAfMet/mRNA complex formation. (B) The obtained cDNA products were separated on denaturing polyacrylamide gels and visualized by phosphorimaging. The toeprinting signal depends on the presence of tRNAfMet and H. volcanii 70S. U, A, C, G denote dideoxy sequencing lanes. The relevant mRNA sequence is shown left to the gel and the start codon (AUG) and the toeprinting sites are highlighted. (C) The toeprinting signals obtained in the presence of H. volcanii 70S ribosomes and tRNAfMet can be inhibited by increasing amounts of Val-tRF. The Ile-tRF at the highest tested concentration did not interfere with toeprinting and served as specificity control. A representative autoradiograph of a toeprinting gel is shown in the lower panel. Quantification of 4 independent toeprinting experiments is shown above the gel whereas the toeprinting signals in the absence of any added tRF was taken as 100%.
![Figure 2. Val-tRF interferes with the efficient establishment of a translation initiation-like complex as monitored by toeprinting. (A) Schematic representation of the toeprinting assay using H. volcanii 30S subunits (gray) or 70S ribosomes (not shown), initiator tRNAfMet and an mRNA. Reverse transcription (dotted arrow) of the [32P]-radiolabeled primer (solid arrow) is terminated in case of a stable 30S/tRNAfMet/mRNA complex formation. (B) The obtained cDNA products were separated on denaturing polyacrylamide gels and visualized by phosphorimaging. The toeprinting signal depends on the presence of tRNAfMet and H. volcanii 70S. U, A, C, G denote dideoxy sequencing lanes. The relevant mRNA sequence is shown left to the gel and the start codon (AUG) and the toeprinting sites are highlighted. (C) The toeprinting signals obtained in the presence of H. volcanii 70S ribosomes and tRNAfMet can be inhibited by increasing amounts of Val-tRF. The Ile-tRF at the highest tested concentration did not interfere with toeprinting and served as specificity control. A representative autoradiograph of a toeprinting gel is shown in the lower panel. Quantification of 4 independent toeprinting experiments is shown above the gel whereas the toeprinting signals in the absence of any added tRF was taken as 100%.](/cms/asset/e6154fe2-3a21-47cc-834e-1151b8d849cd/krnb_a_1257470_f0002_b.gif)
Val-tRF interferes with mRNA binding to the small ribosomal subunit
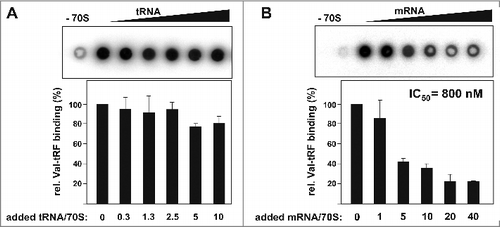
The most likely explanation for blocking the formation of the initiation complex in the above setting is that the Val-tRF either interferes with P-site tRNA or mRNA binding to the ribosome. Intuitively, the first alternative seems to be more plausible since tRFs arise from mature and thus fully functional tRNA molecules.Citation11 Even though secondary structure predictions of Val-tRF did not suggest any stable structure resembling a tRNA anticodon-stem loop,Citation24 we experimentally tested binding competition between ribosome bound tRNAs and Val-tRF. Therefore 5′-[32P]-labeled Val-tRF was incubated with purified H. volcanii 70S ribosomes in the presence of increasing amounts of yeast tRNA and binding efficiencies were analyzed via nitrocellulose filter binding. Even at a massive molar excess of tRNA over ribosomes no significant reduction in Val-tRF binding was observed (). Under the applied binding conditions (no mRNA and high tRNA excess) one P-site located tRNA molecule per ribosome is expected.Citation31 Indeed under these conditions we observed tRNA binding saturation to H. volcanii ribosome at one molecule per 70S at the highest tRNA concentration used (). However, when the ribosome binding experiments were repeated in the presence of increasing amounts of in vitro transcribed mRNA markedly reduced and dose-dependent Val-tRF binding became apparent (). The half maximum inhibitory mRNA concentration (IC50) was determined to be 800 nM. Importantly, this effect was apparently not dependent on the mRNA sequence since a very similar binding competition was evident when the homopolymeric mRNA-analog poly(U) was used (Fig. S4A). These data therefore indicate that Val-tRF does not inhibit ribosome-mRNA association by interfering with the Shine-Dalgarno/anti-Shine-Dalgarno interaction, a principle often exploited by other prokaryal small ncRNAs regulators (reviewed in [ref. Citation32]). This interpretation is in line with the fact that H. volcanii does actually not use the Shine-Dalgarno mechanism for translation initiation.Citation33
Figure 3. tRNA and mRNA binding competition with Val-tRF. (A) Radiolabeled Val-tRF was bound to H. volcanii 70S ribosomes in the absence or presence of increasing amounts of unlabeled yeast tRNA. The mean and the standard deviations of 3 independent binding competition experiments are shown underneath the dot blot. Signals measured in the absence of ribosomal particles (−70S) were subtracted from all experimental points. The added molar excess of tRNA over ribosomes is indicated below. (B) Val-tRF binding was monitored as a function of increased mRNA concentration. The mean and the standard deviations of 2 independent binding competition experiments are shown. The added molar excess of mRNA coding for r-protein L12 over ribosomes is shown. The half maximum inhibitory concentration (IC50) is given.
Val-tRF cross-links to nucleotides in close proximity to the 30S mRNA channel
Since the inhibitory effect of the Val-tRF on translation was observed not only in H. volcanii but also in eukaryal and bacterial systems, we assumed that a functionally conserved region of the small ribosomal subunit is targeted. Saturation binding experiments with radiolabeled Val-tRF and purified 30S or 70S H. volcanii particles revealed a single binding site in the small ribosomal subunit (Fig. S4B). Employing Cy5 end-labeled Val-tRF in solution binding experiments using microscale thermophoresis allowed us to determine a Kd of 773 ± 17 nM. The binding mode of Val-tRF to the 30S subunit does not entirely depend on simple sequence complementarity to 16S rRNA since the all-DNA variant Val-tDF did not bind significantly to 30S subunits and was also inactive in attenuating protein synthesis and peptide bond formation ( and Fig. S2D and S5A).
To gain a more comprehensive insight into the Val-tRF binding site on the ribosome and thus about the functional mechanism of tRF-mediated translation regulation, crosslinking experiments utilizing 4-thio uracil containing Val-tRF was performed. A synthetic version of the 5′-[32P]-end labeled Val-tRF RNA carrying 3 photo-reactive 4-thio uracil bases (named Val-tRF(4SU)) was bound to H. volcanii 70S ribosomes and cross-linked at 366 nm. Subsequently free Val-tRF(4SU) was removed via gel filtration and rRNAs as well as the ribosomal proteins from the cross-linked 70S complex were extracted and analyzed by scintillation counting. It turned out that 99% of the radiolabel was found in the rRNA rather than in the r-protein fraction thus suggesting the Val-tRF 30S binding site to be composed of rRNA. Running the cross-linked rRNAs on a denaturing gel revealed a preferentially cross-linking and therefore 32P-labeling of the 16S rRNA (Fig. S5B). This is consistent with our previously published data showing that Val-tRF predominantly binds to and acts on the small ribosomal subunit.Citation24 To narrow down the 16S rRNA cross-linking site, cDNA oligo-driven RNase H cleavage assays were performed using anti-sense strands distributed all over the 16S rRNA. By tracing the 32P label on the two 16S rRNA cleavage products on denaturing gels (data not shown) allowed us to narrow down the cross-linking position(s) between nucleotides 829 and 1473 in the 16S rRNA 3′ region (E. coli rRNA nomenclature is used here and throughout). To identify the Val-tRF(4SU) cross-linking sites on the nucleotide level, primer extension analyses on the 3′ region of 16S rRNA were performed. Specific primer extension stops occurring only after irradiation at 366 nm in the presence of Val-tRF(4SU) were reproducibly identified at three 16S rRNA regions, namely at nucleotides A968/A969 and A978/U979 close to helix 31, as well as at C1403/C1404 in the heart of the decoding center (). Consistent with the data shown in , , an mRNA analog was able to compete with cross-linking of Val-tRF(4SU) to these 16S rRNA sites, while Ile-tRF had no effect ( and Fig. S6). To substantiate the functional relevance of the cross-linking sites, additional competition experiments were performed. Since all 3 cross-linking regions are located close to functional centers of the small ribosomal subunit that are also targeted by antibiotics, we repeated the Val-tRF crosslinking in the presence of kanamycin, neomycin, spectinomycin or tetracyclin. It turned out that the latter 2 antibiotics, which have been shown to bind in close proximity to the crosslinking sites surrounding helix 31 ([ref. Citation34]), efficiently inhibited Val-tRF(4SU) crosslinking (). The aminoglycosides kanamycin and neomycin that bind to helix 44 in the decoding center had, however, no effect on cross-linking efficiencies. Highlighting the 3 crosslinking regions in the context of the available bacterial 30S crystal structure showed that they all surround the mRNA channel of the small ribosomal subunit (). Especially the 2 cross-linked regions C1403/C1404 in helix 44 and A968/A969 in the loop of helix 31 are of special interest since they are located in immediate proximity to the mRNA path. Thus these data further corroborate the observation that Val-tRF is competing with mRNAs for ribosome binding.
Figure 4. Val-tRF photo-crosslinking to the 30S ribosomal subunit. (A) The sites of crosslinking at 366 nm of Val-tRF carrying 4-thio uracil, Val-tRF(4SU), to the 970 and 980 regions of the 3′-domain of 16S rRNA of H. volcanii 30S subunits was monitored by primer extension analysis. Experiments in the presence of Val-tRF(4SU) without irradiation or in the presence of unmodified Val-tRF with irradiation served as controls. U, A, C, G denote dideoxy sequencing reactions. The relevant rRNA sequence is shown left to the gel and the crosslinked nucleotides are indicated by asterisks. Note that the reverse transcriptase always stops one nucleotide 3′ of the actual crosslinking site. (B) Sites of Val-tRF(4SU) crosslinking to positions 1403/1404 of 16S rRNA helix 44 in the decoding center were identified by primer extension analysis. U, A, C denote dideoxy sequencing reactions. The relevant rRNA sequence is shown left to the gel and the crosslinked nucleotides are indicated by asterisks. (C) The Val-tRF(4SU)-dependent crosslinks can be competed by the addition of unmodified Val-tRF or mRNA, but not by Ile-tRF. Note that all lanes shown originate from the same gel. Below the autoradiogram image the quantification of the crosslinked bands compare with control bands is given. The intensities of the primer extension stops in the reaction containing Val-tRF(4SU) in the absence of any competitor was taken as 1.0. ‘n.d.’ denotes an experiment not performed in this particular crosslinking experiment shown (refer to Fig. S6 for more data). (D) Val-tRF(4SU) crosslinking in the presence of the antibiotics kanamycin (Kan), neomycin (Neo), spectinomycin (Spt), or tetracycline (Tet).
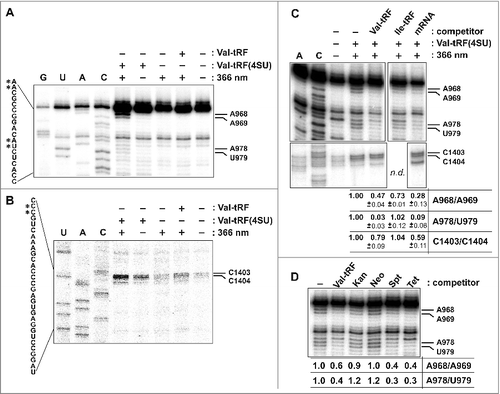
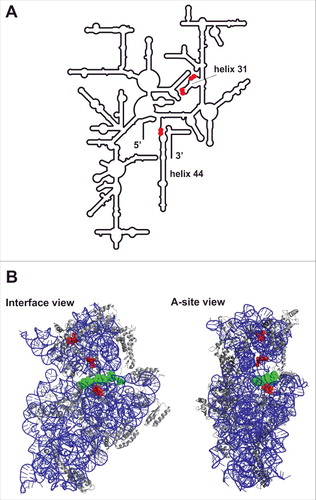
Figure 5. Val-tRF crosslinks to sites in close proximity to the 30S mRNA channel. (A) Sites of Val-tRF(4SU) crosslinking in proximity to helix 31 and helix 44 are indicated by red dots in the schematic representation of the E. coli 16S rRNA secondary structure model. (B) Interface view (left) and A-site view (right) of the small ribosomal subunit from T. thermophilus (pdb file 2J00; Citation46). The Val-tRF crosslinking sites are depicted in red and a 30S-bound mRNA piece is shown in green. The 16S rRNA is in blue and the ribosomal proteins are depicted in gray.
Discussion
Here we report on the biochemical characterization of a 5′ tRNA fragment (tRF) deriving from the Val-tRNA(GAC) isoacceptor of H. volcanii regarding its function and cellular target. We provide evidence that this stress-dependent Val-tRF binds to the small ribosomal subunit in immediate vicinity of the mRNA channel and dims protein biosynthesis on a global level (). Since tRNAs are key molecules of protein biosynthesis, inhibition of translation as a consequence of full-length tRNA destruction seems to be a plausible scenario. However, this picture appears to be too simplistic, since it has been shown in several studies that the steady-state levels of genuine tRNAs do not change significantly upon tRNA fragmentation.Citation20,35-37 It follows that the mechanism by which tRNA pieces negatively affect protein biosynthesis is more intricate.Citation22,24,38 In fact several lines of experimental evidence indicate that H. volcanii Val-tRF competes with mRNA binding rather than with tRNA binding () and thus affects translation initiation. However we note that also peptide bond formation is inhibited (Fig. S2D and [ref. Citation24]), therefore a role of Val-tRF during the elongation phase of protein production cannot be excluded. The mechanism by which Val-tRF exerts its biological role seems to be highly conserved since the archaeal tRF is capable of attenuating protein biosynthesis in vivo and in vitro in a eukaryal (S. cerevisiae) and bacterial (E. coli) system as well ( and Fig. S2). While we do not have evidence that a similar tRNA fragment actually is produced in yeast or E. coli, the mere fact that the Val-tRF from the halophilic archaeon H. volcanii can function in species from other domains argues for a fundamental principle of translation regulation. In many prokaryal species translation regulation mediated by small ncRNAs (sRNAs) involves the ribosome binding site on the mRNAs including the Shine-Dalgarno sequence.Citation39 Thus it appears reasonable to assume that Val-tRF interferes with the Shine-Dalgarno/anti-Shine-Dalgarno interaction during translation initiation. However is has been demonstrated that the archaeon H. volcanii does not utilize the Shine-Dalgarno system to start protein biosynthesis.Citation33 This probably explains why Val-tRF was such a competent in vivo inhibitor of protein synthesis in the eukaryal model S. cerevisiae (). The main advantage of RNA regulators compare with protein-mediated processes is their immediate availability upon rapidly changing environmental conditions. tRFs are in that respect ideally suited to function as stress regulators since the mother molecule, the genuine tRNA, is already available and only a single phosphodiester cut, by a yet unknown processing regime, is sufficient to generate the regulatory ncRNA species. While the exact processing mechanism is so far unclear, we could show that a protein enzyme with RNAse activity is likely responsible for Val-tRF biogenesis (Fig. S7). H. volcanii Val-tRF is produced primarily upon alkaline stress and quickly attenuates global protein biosynthesis by targeting the small ribosomal subunit.Citation24 This allows the cells to reduce their metabolic activities under these harsh conditions, which is also reflected by a severely retarded growth phenotype.
The investigated Val-tRF is a bona fide ribosome-associated-ncRNA (rancRNA) since it targets and affects the functional competence of the small ribosomal subunit. Our data demonstrate that Val-tRF is capable of binding to the 30S subunit in the absence of additional protein cofactors with a Kd in the nM range, an affinity comparable to ribosome targeting antibiotics such as tetracyclin (Kd ∼1 µM; [ref. Citation40]) or neomycin (Kd ∼0.8 µM; [ref. Citation41]). Various rancRNAs have been identified in different organisms in several research groups and represent a so far poorly studied class of regulatory ncRNAs (reviewed in [ref. Citation25]). Val-tRF (this study) and the mRNA-derived rancRNA_18 in yeastCitation7 are among the best characterized rancRNAs so far and the picture that emerges is that rancRNAs are involved in the first wave of stress response. While Val-tRF in H. volcanii is produced and thus functional upon alkaline stress, rancRNA_18 is required under hyperosmotic growth conditions in S. cerevisiae. From an energetic point of view targeting the translation machinery directly for regulatory purposes is reasonable since it allows a swift and global effect on cell metabolism which is crucial for survival under challenging environmental conditions. The Val-tRF characterized in this study therefore fulfills all criteria to be considered as new entry in the ever-growing list of regulatory ncRNA.
Materials and methods
Strains and growth conditions
Haloferax volcanii strain H26 was grown aerobically at 42°C in complex medium (2.9 M NaCl, 150 mM MgSO4 × 7H2O, 60 mM KCl, 4 mM CaCl2, 50 mM Tris/HCl (pH 7.2), 0.45% (w/v) tryptone, 0.275% (w/v) yeast extract). S. cerevisiae strain BY4742 was grown in Sc-Leu medium (yeast synthetic complete drop out medium –Leu; Sigma Aldrich) at 30°C.
Metabolic labeling in H. volcanii and S. cerevisiae
For metabolic labeling in H. volcanii the protocol for spheroplast preparation and nucleic acid transformation has been adapted from previous publications.Citation26,42 H. volcanii cells were grown in 10 ml rich medium into the exponential phase to an OD600 of 0.6, pelleted during 10 min centrifugation at 4,400 × g at room temperature and resuspended in 2 ml buffered spheroplasting solution containing 1 M NaCl, 27 mM KCl, 50 mM Tris/HCl (pH 8.5) and 15% (w/v) sucrose. Cells were pelleted again and resuspended in 600 µl buffered spheroplasting solution. Cells were then divided into 3 equal aliquots in 2 ml Eppendorf tubes, 20 µl of 0.5 M EDTA (pH 8.0) was added and the tubes were inverted 3 times and incubated for 10 min at room temperature. 180 µl H. volcanii spheroplasts were then added to 30 µl of a premixed solution containing 15 µl unbuffered spheroplasting solution (1 M NaCl, 27 mM KCl, 15% (w/v) sucrose), 5 µl 0.5 M EDTA (pH 8.0), 1 µl 35S-methionine (1,000 Ci/mmol, 10 mCi/ml), RNA (200 pmol) or antibiotic (f.c. 0.6 mM thiostrepton or puromycin) and incubated for 5 min at room temperature. Transformation was then initiated by adding 250 µl 60% (v/v) polyethylene glycol 600 in unbuffered spheroplasting solution and samples were mixed by carefully inverting the tube and incubated at room temperature for 30 min. The samples were then diluted with 1 ml of spheroplast dilution solution (15% sucrose (w/v), 3.2 M NaCl, 109 mM MgSO4 × 7H2O, 112 mM MgCl2 × 6H2O, 71 mM KCl, 4 mM CaCl2, 15 mM Tris/HCl (pH 7.2)), and pelleted for 10 min at 3,300 × g at room temperature. Subsequently 500 µl regeneration solution containing 0.45% (w/v) tryptone, 0.275% (w/v) yeast extract, 0.1% (w/v) casamino acids, 15% (w/v) sucrose, 2.5 M NaCl, 85 mM MgSO4 × 7H2O, 88 mM MgCl2 × 6H2O, 56 mM KCl, 4 mM CaCl2, and 12 mM Tris/HCl (pH 7.2) was carefully added to the pellet (without resuspending the pellet). Spheroplasts were then incubated for 20 min at 42°C and subsequently resuspended. The extent of labeled proteins was monitored after pelleting the spheroplasts followed by lysing with the addition of 50 µl H2O and incubation at 95°C for 2 min. The lysate was passed over an illustra Microspin G25-Sepadex column (GE Healthcare) and the proteins were then separated by 11% Tricine-SDS-PAGE. The newly synthesized proteins were monitored by phosphorimaging (FLA-3000) and quantified with the densiometric program Aida Image Analyzer. The following synthetic oligonucleotides were introduced with this approach into H. volcanii spheroplasts: Val(GAC)-tRF: 5′-GGGUUGGUGGUCUAGUCUGGUUAUGA-3′, Ile-tRF: 5′- GGGCCAAUAGCUCAGUCAGGUUGAGC-3′, Val-tDF: 5′-GGGTTGGTGGTCTAGTCTGGTTATGA-3′.
Metabolic labeling in S. cerevisiae was performed as described.Citation7 Briefly, yeast spheroplasts were mixed with 100 pmol of synthetic RNA (rancRNA_18: 5′-AGGAAAAGGUGAAAAGAA-3′ or tRFs) and introduced into the cell via electroporation. After electroporation 1 ml YPD/1 M sorbitol and 1 μl 35S-methionine (1,000 Ci/mmol, 10 mCi/ml) were added and the reactions were incubated for 1 hour at 30°C. The extent of labeled proteins was monitored by TCA precipitation and subsequent liquid scintillation counting or by running the labeled proteins on an SDS polyacrylamide gel followed by quantification by phosphorimaging.Citation7
Peptidyl transferase assay
The assay was performed in 25 μl of appropriate reaction buffer containing 10 pmol 70S ribosomes, 0.8 pmol N-acetyl-[3H]Phe-tRNA (15,000 cpm/pmol), puromycin (f.c. 1 mM) and, as indicated, 100 pmol of the synthetic tRF or tDF (see above). For H. volcanii ribosomes a high salt buffer composed of 2 M KCl, 30 mM Tris/HCl (pH 7.6), 0.4 M NH4Cl, 60 mM Mg(OAc)2, 7 mM β-mercaptoethanol, 2 mM spermidine, 0.05 mM spermine was used. The transpeptidation reaction was initiated by the addition of 12.5 μl cold methanol (f.c. 33%) and incubated for 3.5 h on ice. For E. coli ribosomes binding buffer (f.c. 20 mM Hepes/KOH pH 7.6, 6 mM Mg(OAc)2, 150 mM NH4Cl, 4 mM β-mercaptoethanol, 2 mM spermidine, 0.05 mM spermine) was used. The reaction was started by the addition of puromycin and incubated for 30 min at 37°C. Reaction products were extracted into ethyl acetate and quantified by liquid scintillation counting as described previously.Citation24
In vitro translation
Yeast S30 extract preparation and subsequent in vitro translation was performed as described in detail previously.Citation7 In vitro translation reactions in E. coli (New England BioLabs) were performed according to the manufacturer's instructions.
In vitro binding studies
Binding studies of Val-tRF to ribosomal particles were performed using a dot blot-filtering deviceCitation24 or microscale thermophoresis. For the filter binding assay 5 pmol of H. volcanii 70S or 30S subunits were incubated with varying amounts of 5′-[32P]-end-labeled synthetic tRF in 25 μl 1x TMN buffer (f.c. 20 mM Tris acetate (pH 7.6), 100 mM NaOAc and 5 mM Mg(OAc)2) or 1x Hfx binding buffer (f.c. 20 mM Hepes/KOH pH 7.6, 150 mM Mg(OAc)2, 150 mM NH4Cl, 4 mM β-mercaptoethanol, 2 mM spermidine, 0.05 mM spermine), respectively. After 30 min of incubation at 42°C the reactions were filtered through a nitrocellulose membrane (0.45 μM diameter) using a vacuum device, followed by 2 washing steps with ice cold buffer. For binding competition assays 5 pmol of H. volcanii 70S ribosomes were incubated in a total volume of 25 μl with 4 pmol 5′-[32P]-labeled synthetic Val-tRF and indicated amounts (5 to 400 pmol) of unlabeled competitor RNA (tRFs, yeast bulk tRNA, poly(U), or in vitro transcribed mRNA coding for r-protein L12Citation43 in 1x TMN or 1x Hfx binding buffer (for tRNA competition). Reactions were assembled at room temperature and incubated at 42°C for 15 min before filtration through the nitrocellulose membrane. After two washing steps with the respective ice-cold buffer, membranes were exposed to phosphor-imaging screens. The signals were monitored and quantified as described above. Microscale thermophoresis measurements were performed on a Monolith NT.115 apparatus (NanoTemper Technologies, Munich, Germany). Therefore constant concentrations of a 5′-Cy5-end-labeled synthetic Val-tRF (10 nM) were assembled in a series of 16 1:1 dilution steps of H. volcanii 70S ribosomal particles dissolved in 1 × TMN (starting with 25 μM) in a final reaction volume of 20 μl. Reactions were soaked in standard capillaries and the thermophoretic movement was monitored with a laser (on for 30 s and off for 5 s; at a laser voltage of 20%). The Kd was calculated from 3 independent measurements with the according NanoTemper Software.
Toeprinting analysis
Toeprinting analysis was performed as described elsewhereCitation44 with some minor changes. For primer extension, 6.2 pmol of [32P]-labeled primer 5'- CGTTAATCTGTGATG-3' were annealed to 25 pmol in vitro transcribed and purified mRNA carrying a short open reading frame coding for MFKRSIYV in a reaction volume of 10 μl (20 mM Hepes/KOH pH 7.6, 150 mM NH4Cl, 4 mM β-mercaptoethanol, 2 mM spermidine, 0.05 mM spermine). 15 pmol H. volcanii 70S ribosomes (or H. volcanii 30S subunits) were incubated for 15 min at 40°C in a total volume of 7 µl in toeprinting buffer (29 mM Hepes/KOH pH 7.5, 29 mM NH4Cl, 29 mM Mg(OAc)2, 1.7 mM β-mercaptoethanol, 0.29 M KCl) in the presence of various amounts of synthetic Val-tRF RNA (20, 50, or 100 pmol) or Ile-tRF RNA (100 pmol). Then 2 µl of the above mentioned mRNA/primer mix was added as well as 1 µl of E. coli tRNAfMet (200 pmol) and the sample was incubated at 40°C for 15 min. Subsequently for primer extension 2.5 µl RT Mix (20 mM Tris/HCl pH 7.4, 20 mM NH4Cl, 100 mM MgCl2, 14.5 mM dNTPs, 1 U/µl AMV reverse transcriptase) was added and the sample was incubated for 15 min at 40°C. The reaction was terminated by the addition of 5 volumes 9:1 EtOH/0.3 M NaOAc (pH 5.5). Precipitated and pelleted cDNA was dissolved in formamide dye and separated on 7.5% polyacrylamide gels containing 7 M urea. Gels were vacuum-dried at 70°C for 2 h and exposed to phosphor-imaging screens. The signals were monitored and quantified as already described above.
Cross-linking studies
For the cross-linking experiments, a synthetic Val-tRF RNA carrying 3 photo-reactive 4-thiouridine residues at positions +5, +13, and +22 was used (Val-tRF(S4U)). Depending on the downstream analysis, the Val-tRF(S4U) was either radioactively 5′ end labeled or used as unlabeled RNA. 5 pmol of H. volcanii ribosomal particles (70S, 50S, 30S) were incubated with 10 - 50 pmol of Val-tRF(S4U) or competitor (tRFs, poly(U) or antibiotics) in a final volume of 25 μl 1x TMN for 15 min at 37°C or 42°C. Cross-linking was either performed during this incubation step or afterwards during an additional incubation for 15 min on ice. For cross-linking, reaction tubes were opened and a hand-held UV lamp was placed in a distance of 3.5 cm. Cross-linking was performed at 366 nm for 15 min. For subsequent primer extension analyses, 125 μl TE/SDS buffer (10 mM Tris/HCl pH 7.4, 100 mM NaCl, 5 mM EDTA, 0.5% (w/v) SDS) was added and phenol/chloroform extraction was performed. To determine whether rRNA or ribosomal proteins were crosslinked under these conditions, 32P 5′ end labeled Val-tRF(4SU) was crosslinked to H. volcanii 70S. Subsequently non-reactive Val-tRF(4SU) was removed via gel filtration using Illustra MicroSpin S-300 HR columns (GE Healthcare) according to the manufacturer's instructions. Subsequently a phenol/chloroform extraction was performed and the amount of crosslinked 32P-Val-tRF(4SU) in the aqueous as well as the organic phases was quantified by liquid scintillation counting.
RNase H cleavage assays
Prior to RNase H cleavage, 25 pmol 5′ [32P]-end-labeled Val-tRF(4SU) was cross-linked to 2.5 pmol 30S subunits and 16S rRNA was phenol/chloroform extracted. RNase H cleavage experiments were performed in a final volume of 10 μl 1x TMN buffer. 0.5 μg extracted cross-linked 16S rRNA was denatured at 95°C for 2 min. 10 pmol of DNA oligonucleotide complementary to specific sites on the 16S rRNA (16S_454–473: 5′-TATTACCGCGGCGGCTGGCA-3′, 16S_624–644: 5′-ATTACGGGATTTCACTCCTAC-3′, 16S_740–759: 5′-ATCGTTTACAGCTAGGACTA-3′, 16S_829–848: 5′-ATCCTTGCGGACGTACTTCC-3′, 16S_942–964: 5′-TCATCAACCTGATCGTCATCACT-3′, 16S_977–995: 5′-TGCACCTCCTCTCTACAGC-3′, 16S_1109–1130: 5′-CCTTCCTCCGCTTTAGCAGCGG-3′, 16S_1215–1237: 5′-TTTAGGAGATTAGCGTTCTCTTT-3′, 16S_1330–1351: 5′-GGCGGTGTGTGCAAGGAGCAGG-3′, 16S_1451–1473: 5′-AGGAGGTGATCCAGCCGCAGATT-3′) were added and incubated at 70°C for 2 min and for 30 min at 37°C. 1 U of RNase H was added and cleavage was performed at 37°C for 30 min. RNase H cleavage was stopped by snap freezing of reactions. Cleavage products were analyzed on 5% polyacrylamide gels containing 7 M urea. After staining with ethidium bromide, gels were vacuum-dried at 70°C for 2 h and exposed to phosphor-imaging screens. The signals were monitored with a phosphor imager (FLA-3000; Fuji Photo Film).
Primer extension analysis
In order to detect the sites of Val-tRF(4SU) cross-link on the 16S rRNA by reverse transcription,Citation45 the DNA primer (16S_454–473: 5′-TATTACCGCGGCGGCTGGCA-3′, 16S_624–644: 5′-ATTACGGGATTTCACTCCTAC-3′, 16S_740–759: 5′-ATCGTTTACAGCTAGGACTA-3′, 16S_829–848: 5′-ATCCTTGCGGACGTACTTCC-3′, 16S_942–964: 5′-TCATCAACCTGATCGTCATCACT-3′, 16S_977–995: 5′-TGCACCTCCTCTCTACAGC-3′, 16S_1109–1130: 5′-CCTTCCTCCGCTTTAGCAGCGG-3′, 16S_1215–1237: 5′-TTTAGGAGATTAGCGTTCTCTTT-3′, 16S_1330–1351: 5′-GGCGGTGTGTGCAAGGAGCAGG-3′, 16S_1451–1473: 5′-AGGAGGTGATCCAGCCGCAGATT-3′) was radioactively labeled in a first step. For ten reactions 50 pmol of primer were incubated with 2 μl γ-[32P]-ATP and 10 U T4 PNK in a final volume of 20 μl 1x PK buffer (70 mM Tris/HCl pH 7.5, 5 mM EDTA, 10 mM MgCl2) at 37°C for 20 min. Subsequently the reaction was stopped by heating at 95°C for 2 min. For annealing, 2 μl 16S rRNA (100–200 ng/μl) were mixed with 2 μl primer (corresponds to 5 pmol) and 4 μl 2.5x HY buffer (62.5 mM Tris/HCl pH 8.4, 75 mM KCl). After an initial denaturation at 95°C for 2 min, the annealing was performed at 42°C for 30 min. Samples were briefly spun down and 10 μl extension mixture, containing 1 μl dNTPs (1.25 mM each) and 0.4 U AMV reverse transcriptase in 1x EX buffer (0.1 M Tris/HCl pH 8.4, 10 mM MgCl2, 10 mM DTT), were added. For sequencing reactions, ddNTPs (f.c. 0.2 mM) were additionally added to the reaction. After incubation at 42°C for 45 min, the reactions were stopped by adding 2 volumes stop solution (4 M NH4Ac, 20 mM EDTA). Subsequently, nucleic acids were ethanol (120 μl) precipitated at −20°C for at least 20 min in the presence of 1 μl GlycoBlue (Ambion). Subsequently, samples were pelleted at 18,000 × g for 20 min, washed with 70% EtOH and resuspended in 12 μl loading buffer (60% (v/v) formamide, 0.1% (w/v) bromphenol blue, 0.1% (w/v) xylene cyanol). The cDNA products were run in 1x TBE at 40 W on denaturing 6 or 8% polyacrylamide gels containing 7 M urea, respectively. Gels were vacuum-dried at 70°C for 1 h and exposed to phosphor-imaging screens. The signals were monitored with a phosphor imager.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Data.docx
Download MS Word (22.8 MB)Acknowledgments
We thank Miriam Koch for comments on the manuscript. Our thanks are extended to Marek Zywicki for bioinformatics support, Andreas Pircher for experimental assistance and Anita Marchfelder for discussions and advice.
Funding
The work was partly supported by the NCCR ‘RNA & Disease’ funded by the Swiss National Science Foundation and by the Swiss National Science Foundation grant [31003A_166527 to N.P.]. J.R. is a recipient of a postdoctoral fellowship from the Deutsche Forschungsgemeinschaft DFG [RE 4041/1–1].
References
- Huttenhofer A, Schattner P, Polacek N. Non-coding RNAs: hope or hype? Trends Genet 2005; 21:289-97; PMID:15851066; https://doi.org/10.1016/j.tig.2005.03.007
- Morris KV, Mattick JS. The rise of regulatory RNA. Nature Rev 2014; 15:423-37; PMID:24776770; https://doi.org/10.1038/nrg3722
- Mattick JS, Makunin IV. Non-coding RNA. Human Mol Genet 2006; 15:R17-29; PMID:16651366; https://doi.org/10.1093/hmg/ddl046
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell 2008; 32:519-28; PMID:19026782; https://doi.org/10.1016/j.molcel.2008.10.017
- Persson H, Kvist A, Vallon-Christersson J, Medstrand P, Borg A, Rovira C. The non-coding RNA of the multidrug resistance-linked vault particle encodes multiple regulatory small RNAs. Nat Cell Biol 2009; 11:1268-71; PMID:19749744; https://doi.org/10.1038/ncb1972
- Tuck AC, Tollervey D. RNA in pieces. Trends Genet 2011; 27:422-32; PMID:21741109; https://doi.org/10.1016/j.tig.2011.06.001
- Pircher A, Bakowska-Zywicka K, Schneider L, Zywicki M, Polacek N. An mRNA-derived noncoding RNA targets and regulates the ribosome. Mol Cell 2014; 54:147-55; PMID:24685157; https://doi.org/10.1016/j.molcel.2014.02.024
- Luidalepp H, Berger S, Joss O, Tenson T, Polacek N. Ribosome shut-down by 16S rRNA fragmentation in stationary-phase Escherichia coli. J Mol Biol 2016; 428:2237-47; PMID:27067112; https://doi.org/10.1016/j.jmb.2016.01.033
- Vesper O, Amitai S, Belitsky M, Byrgazov K, Kaberdina AC, Engelberg-Kulka H, Moll I. Selective translation of leaderless mRNAs by specialized ribosomes generated by MazF in Escherichia coli. Cell 2011; 147:147-57; PMID:21944167; https://doi.org/10.1016/j.cell.2011.07.047
- Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 2012; 40:6787-99; PMID:22492706; https://doi.org/10.1093/nar/gks307
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 2013; 10:1798-806; PMID:24351723; https://doi.org/10.4161/rna.27177
- Kirchner S, Ignatova Z. Emerging roles of tRNA in adaptive translation, signalling dynamics and disease. Nature Rev 2015; 16:98-112; PMID:5534324; https://doi.org/10.1038/nrg3861
- Megel C, Morelle G, Lalande S, Duchene AM, Small I, Marechal-Drouard L. Surveillance and cleavage of eukaryotic tRNAs. Int J Mol Sci 2015; 16:1873-93; PMID:25599528; https://doi.org/10.3390/ijms16011873
- Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett 2014; 588:4297-304; PMID:25220675; https://doi.org/10.1016/j.febslet.2014.09.001
- Keam SP, Hutvagner G. tRNA-Derived Fragments (tRFs): Emerging new roles for an ancient RNA in the regulation of gene expression. Life 2015; 5:1638-51; PMID:26703738; https://doi.org/10.3390/life5041638
- Borek E, Baliga BS, Gehrke CW, Kuo CW, Belman S, Troll W, Waalkes TP. High turnover rate of transfer RNA in tumor tissue. Cancer Res 1977; 37:3362-6; PMID:884680
- Levitz R, Chapman D, Amitsur M, Green R, Snyder L,Kaufmann G. The optional E. coli prr locus encodes a latent form of phage T4-induced anticodon nuclease. EMBO J 1990; 9:1383-9; PMID:1691706
- Durdevic Z, Schaefer M. tRNA modifications: necessary for correct tRNA-derived fragments during the recovery from stress? BioEssays 2013; 35:323-7; PMID:23315679; https://doi.org/10.1002/bies.201200158
- Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 2015; 161:790-802; PMID:25957686; https://doi.org/10.1016/j.cell.2015.02.053
- Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, Rigoutsos I, Kirino Y. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci USA 2015; 112:E3816-3825; PMID:26124144; https://doi.org/10.1073/pnas.1510077112
- Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Qian J, et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science 2016; 351:397-400; PMID:26721680; https://doi.org/10.1126/science.aad7977
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 2011; 43:613-23; PMID:21855800; https://doi.org/10.1016/j.molcel.2011.06.022
- Bakowska-Zywicka K, Kasprzyk M, Twardowski T. tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro. FEMS Yeast Res 2016; 16; PMID:27609601; https://doi.org/10.1093/femsyr/fow077
- Gebetsberger J, Zywicki M, Kunzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012; 2012:260909; PMID:23326205; https://doi.org/10.1155/2012/260909
- Pircher A, Gebetsberger J, Polacek N. Ribosome-associated ncRNAs: an emerging class of translation regulators. RNA Biol 2014; 11:1335-9; PMID:25692232; https://doi.org/10.1080/15476286.2014.996459
- Cline SW, Lam WL, Charlebois RL, Schalkwyk LC, Doolittle WF. Transformation methods for halophilic archaebacteria. Canadian J Microbiol 1989; 35:148-52; PMID:2497937; https://doi.org/10.1139/m89-022
- Russell PJ, Hambidge SJ, Kirkegaard K. Direct introduction and transient expression of capped and non-capped RNA in Saccharomyces cerevisiae. Nucleic Acids Res 1991; 19:4949-53; PMID:1656383; https://doi.org/10.1093/nar/19.18.4949
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 2004; 5:827-35; PMID:15459663; https://doi.org/10.1038/nrm1488
- Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochem 1990; 29:5881-9; PMID:2200518; https://doi.org/10.1021/bi00477a001
- Qin Y, Polacek N, Vesper O, Staub E, Einfeldt E, Wilson DN, Nierhaus KH. The highly conserved LepA is a ribosomal elongation factor that back-translocates the ribosome. Cell 2006; 127:721-33; PMID:17110332; https://doi.org/10.1016/j.cell.2006.09.037
- Rheinberger HJ, Sternbach H, Nierhaus KH. Three tRNA binding sites on Escherichia coli ribosomes. Proc Natl Acad Sci USA 1981; 78:5310-4; PMID:7029532; https://doi.org/10.1073/pnas.78.9.5310
- Desnoyers G, Bouchard MP, Masse E. New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet 2013; 29:92-8; PMID:23141721; https://doi.org/10.1016/j.tig.2012.10.004
- Kramer P, Gabel K, Pfeiffer F, Soppa J. Haloferax volcanii, a prokaryotic species that does not use the Shine Dalgarno mechanism for translation initiation at 5′-UTRs. PloS One 2014; 9:e94979; PMID:24733188; https://doi.org/10.1371/journal.pone.0094979
- Wilson DN. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat Rev Microbiol 2014; 12:35-48; PMID:24336183; https://doi.org/10.1038/nrmicro3155
- Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 2009; 23:2639-49; PMID:19933153; https://doi.org/10.1101/gad.1837609
- Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, Anderson P, Pan T, Hatzoglou M. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem 2012; 287:42708-25; PMID:23086926; https://doi.org/10.1074/jbc.M112.371799
- Thompson DM, Lu C, Green PJ, Parker R. tRNA cleavage is a conserved response to oxidative stress in eukaryotes. RNA 2008; 14:2095-103; PMID:18719243; https://doi.org/10.1261/rna.1232808
- Sobala A, Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol 2013; 10:553-63; PMID:23563448; https://doi.org/10.4161/rna.24285
- Svensson SL, Sharma CM. Small RNAs in bacterial virulence and communication. Microbiol Spect 2016; 4; PMID:27337442; https://doi.org/10.1128/microbiolspec.VMBF-0028-2015
- Buck MA, Cooperman BS. Single protein omission reconstitution studies of tetracycline binding to the 30S subunit of Escherichia coli ribosomes. Biochem 1990; 29:5374-9; PMID:2200507; https://doi.org/10.1021/bi00474a024
- Llano-Sotelo B, Hickerson RP, Lancaster L, Noller HF, Mankin AS. Fluorescently labeled ribosomes as a tool for analyzing antibiotic binding. RNA 2009; 15:1597-604; PMID:19553343; https://doi.org/10.1261/rna.1681609
- Allers T, Ngo HP, Mevarech M, Lloyd RG. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Applied Environ Microbiol 2004; 70:943-53; PMID:14766575; https://doi.org/10.1128/AEM.70.2.943-953.2004
- Erlacher MD, Chirkova A, Voegele P, Polacek N. Generation of chemically engineered ribosomes for atomic mutagenesis studies on protein biosynthesis. Nature Prot 2011; 6:580-92; PMID:21527916; https://doi.org/10.1038/nprot.2011.306
- Chirkova A, Erlacher MD, Clementi N, Zywicki M, Aigner M, Polacek N. The role of the universally conserved A2450-C2063 base pair in the ribosomal peptidyl transferase center. Nucleic Acids Res 2010; 38:4844-55; PMID:20375101; https://doi.org/10.1093/nar/gkq213
- Polacek N, Barta A. Metal ion probing of rRNAs: evidence for evolutionarily conserved divalent cation binding pockets. RNA 1998; 4:1282-94; PMID:9769102; https://doi.org/10.1017/S1355838298980347
- Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006; 313:1935-42; PMID:16959973PMID:; https://doi.org/10.1126/science.1131127
