ABSTRACT
Nucleic acids, especially RNA, naturally contain a diversity of chemically modified nucleosides. To understand the biological role of these modified nucleosides, nucleic acid scientists need tools to specifically label, detect and enrich modified nucleic acids. These tools comprise a diverse set of chemical reagents which have been established in the early years of nucleic acid research. Recent developments in high-throughput sequencing and mass spectrometry utilize these chemical labeling strategies to efficiently detect and localize modifications in nucleic acids. As a consequence the transcriptome-wide distribution of modified nucleosides, especially 5-methylcytosine and pseudouridine, in all domains of life could be analyzed. With the help of these techniques and the gained knowledge, it becomes possible to understand the functions of modifications and even study their connections to human health and disease. Here, the differential chemical reactivity of modified nucleosides and their canonical counterpart is reviewed and discussed.
Introduction
In all domains of life, genetic information is stored by the sequence of the canonical nucleosides cytidine, guanosine, adenosine and thymidine in DNA and uridine in RNA, respectively. In addition, modified nucleosides in both DNA and RNA form a second layer of information, regulating major life processes like transcription and translation. To understand the biological role of these modified nucleosides, nucleic acid scientists need tools to specifically label, detect and enrich thus modified nucleic acids. These tools comprise a diverse set of chemical reagents which have been established in the early years of nucleic acid research. While their use has been nearly forgotten after the introduction of more sensitive techniques like mass spectrometry, some have been re-discovered in recent years. For example, the uridine isomer pseudouridine (Ψ, ) is labeled with carbodiimidesCitation1 () or new carbodiimide derivativesCitation2 to enable the modification's detection by either mass spectrometryCitation3 or sequencing.Citation4-6 The return of this reagent has led to the discovery of Ψ in many mRNAs which resulted in renewed interest to study not only the distribution and function of Ψ, but also other modifications in mRNA and non-coding RNAs. Another example is bisulfite, which is now commonly used in RNA and DNA sequencing for detection and localization of 5-methylcytosine,Citation7 the so-called 5th base of DNA and a common RNA modification.
Figure 1. Overview of modified nucleosides. The arrows indicate the target sites in the modified nucleosides responsible for their reactivity with the described reagents. Red arrows: covalent reaction, green arrows: converting reaction, blue arrows: physicochemical interaction.
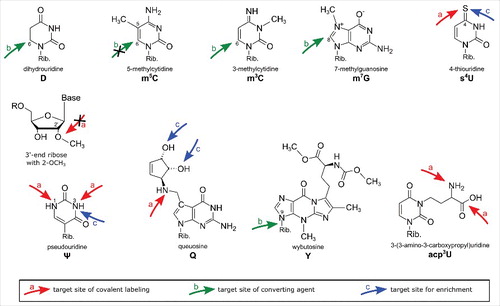
Table 1. Overview of all reagents used for covalent labeling and detection of modified nucleosides. Red arrows indicate the active groups responsible for the reaction with the target modification. The term “target modification” lists the modified nucleosides known to react with the reagent from the literature. “Potential side reactions” lists all nucleosides that are chemically able to react with the reagent but were not described in the literature.
Table 2. Overview of conversion products after bisulfite reaction of cytidine and its modified derivatives. Without pretreatment (2nd column from left) C, f5C and ca5C convert into uracil after bisulfite treatment. Catalytic oxidation of hm5C into f5C (3rd column from the left) leads to its conversion into uracil after bisulfite treatment while all cytidines are protected from conversion after enzymatic methylation (first column from the right). Thus f5C and ca5C become detectable.
Here, chemical reagents used for the detection of modified nucleosides will be classified into (a) reagents used to covalently label modified nucleosides, (b) reagents converting nucleosides or inducing strand breaks and (c) reagents for enrichment of modified nucleic acids. This review summarizes these reagents and nucleoside targets which lead to the differential reactivity of modified nucleosides and highlight recent breakthroughs achieved by the combination of chemical labeling with modern sequencing techniques. gives an overview of modified nucleosides detectable by chemical labeling approaches and the reactive groups of the modified nucleoside are indicated by arrows.
Covalent labeling of modified nucleosides
Chemical reagents, used for labeling of modified nucleotides, target certain functionalities in the modified residue. This target is, in the optimal case, only present in the modified nucleotide but not in the canonical, like the sulfur of 4-thiouridine (s4U), which allows a high selectivity of the reagent. Structures of the reagents with arrows indicating their reactive center can be found in . The exact mechanisms for all reagents have been summarized by the Motorin lab.Citation8
Thiols
In general, thiols are strong nucleophiles that attack all sorts of electrophiles like halomethylated fluorophores. So far an iodo-(9) and bromoacetamide.Citation10 have been used to label s2U and s4U (2-thio- and 4-thiouridine, respectively). In addition, a bromomethyl-coumarin.Citation11 was reported to react with s4U, however the selectivity towards other thiolated nucleosides like s2U or s2C (2-thiocytidine) was not studied yet.
5-methyl-ribopyrimidine
Deoxythymidine and 5-methyldeoxycytidine (m5dC or just mC) in DNA can be specifically labeled with OsO4 or osmate complexes, which react with the diastereotopic 5–6 double bond.Citation12 of the nucleobase. Only recently, this approach was presented in context of RNA where the osmium tetroxide-bipyridine complex reacted with both m5C and m5U on the nucleoside level. Interestingly, a change in diastereoselectivity leads to an almost complete loss of selectivity towards 5-methylcytosine (m5C) in a 5-mer ribonucleotide context, while 5-methyluridine (m5U) remained about 8 times more reactive than the canonical pyrimidines.Citation13
Carboxylic acids
Although common in other macromolecules, free carboxy groups are only found in modified nucleosides conjugated with amino acids like 3-(3-amino-3-carboxypropyl)uridine (acp3U), N6-threonylcarbamoyladenosine (t6A) or N6-methyl-N6-threonylcarbamoyladenosine (m6t6A). It was shown that these nucleosides react selectively with primary amines in e.g. aniline or ethylendiamine in soluble carbodiimide.Citation14 Many more carboxyl functionalized nucleosides (e.g. N6-glycinylcarbamoyladenosine (g6A), uridine 5-oxyacetic acid (cmo5U), 5-carboxymethylaminomethyluridine (cmnm5U), etc.) have been discovered, since this report was published, and it is very likely that these might also be labeled under the same conditions.
Aliphatic amines
Similar to carboxyl groups, aliphatic amines are only found in some modified nucleosides like acp3U, 5-methylaminomethyl-2-thiouridine (mnm5s2U) and queuosine (Q). Fluorescein-isothiocyanate (FITC) was reported to react with both Q15 and acp3U,Citation16 while the latter also reacts with N-hydroxysuccinimides.Citation10,17,18 The aliphatic amino group of an uncommon bacterial wobble modification mnm5s2U was found to react with a spin-labeled anhydrideCitation19 and in addition showed reactivity with a carbodiimide derivative.Citation10
Aromatic amines
shows the N-acylation of usually aromatic amines by the carbodiimde CMCT (N-cyclohexyl-N’-β-(4-methylmorpholinium)ethylcarbodiimide p-tosylate) which leads to products of guanosine, uridine, inosine (I), 2-methlythio-6-isopentenyladenosine (ms2i6A) and pseudouridine (Ψ). In a second step, the acyl-moieties can be removed from guanosine, uridine and inosineCitation20 residues by alkaline treatment, leaving only N3-acylated pseudouridinesCitation21 and an unknown derivative of 2-methylthio-N6-isopentenyladenosine (ms2i6A).Citation3 The acylation product is relatively bulky, which allows detection not only by mass spectrometry but additionally by reverse transcription (RT).Citation1,3 This reagent was recently used for sequencing which allowed a transcriptome wide mapping of Ψ in yeast,6 human cellsCitation4 and dyskeratosis congenital patient samples.Citation5 In addition, an azide modified CMC derivative was developed which allows enrichment of pseudouridylated RNA prior to sequencing by azide-alkyne cycloaddition and subsequent Biotin pulldown (encircled in ).Citation2
Figure 2. Reaction of carbodiimides with pseudouridine and uridine. Top: acylation of pseudouridine (Ψ) with the carbodiimide CMCT (full name in text). The carbodiimide group reacts with both N1 and N3, but after alkaline treatment the N1-CMCT is cleaved. The remaining CMC at N3 enables detection by mass spectrometry, reverse transcription, sequencing or even biotin pulldown in case of the CMC-azide (encircled, blue). Bottom panel: Like pseudouridine, uridine gets labeled at the N3 position, which is removed after alkaline treatment. The same was found to be true for guanosine and inosine. Insert: Structure of CMCT.
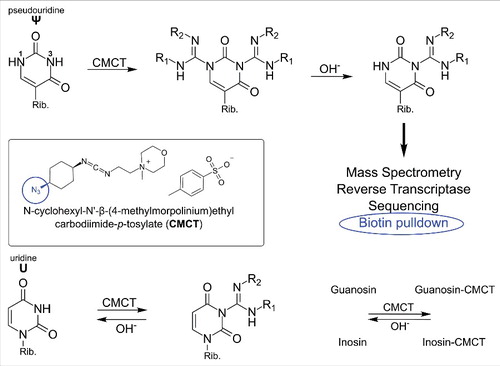
CMC labeling is a multi-step process with precisely defined conditions necessary to achieve the desired pseudouridine selectivity. Therefore special attention is required at all steps of the labeling procedure to avoid false positive results. In addition to pseudouridine, it was shown that carbodiimides react with aliphatic amines e.g. in mnm5s2UCitation10 and ms2i6A.
Similar site reactivity (here I, m5C and m5U) was observed for Ψ labeling with the N-alkylating reagents acrylonitrile and methylvinylsulfone which are mostly applied in mass-spectrometric studiesCitation22,23 since the reverse transcriptase is not stopped by these labels. For mass spectrometry these reagents are greatly beneficial since Ψ as a uridine isomer has the same m/z as the canonical nucleoside. By labeling of Ψ it can be detected as the conjugate and the thereby increased m/z. The side reactivity of inosine with acrylonitrile has been exploited for inosine mapping in the human transcriptome.Citation24
In 1974, the fluorescent reagent 7-methoxy-4-bromomethylcoumarin was reported to react selectively with pseudouridine under specific reaction conditions.Citation25 The development of an azide functionalized coumarin derivative did not exert the same selectivity towards Ψ, but can be used to functionalize any native RNA or DNA instead.Citation26
Blocking target sites
All techniques mentioned above exploit the higher reactivity of functional groups introduced by the natural nucleoside modification. However, it is also possible that a modification e.g. a methyl group, blocks a functional group of a canonical nucleoside. N-methylisatoic anhydride (NMIA) is a reagent used for RNA structural analysis which reacts with free unrestrained ribose 2′OH.Citation27 In theory, methylation of the 2′ OH will omit the labeling with NMIA. Using this reagent, mapping of 2′O-methylated nucleosides should be possible as the label leads to a stop of reverse transcriptase.
Conversion of modified nucleosides
The second class of chemical reagents used for the detection of modified nucleosides comprises all reactions of modified nucleosides leading to their conversion or subsequent strand-breaks. In comparison to the first class of reagents, no bulky labels are introduced which allow direct detection of the modification by e.g. primer extension, sequencing or fluorescence. Therefore the techniques described in the following section are usually combined with primer extension assays or sequencing approaches.
Bisulfite conversion
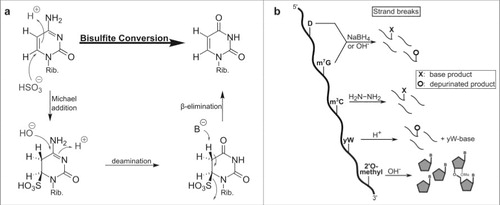
In both RNA and DNA, the C6 of cytosine reacts readily with bilsulfite which leads to its deamination into uracil (see ). In contrast, methylation of cytosine C5 increases the electron density in the pyrimidine ring, which makes it less prone to reaction with bisulfite.Citation28 The comparison of bisulfite and mock treated samples after high throughput sequencing is used to locate all 5-methylated cytosines since they still pair with guanosine, while the converted cytosines pair with adenosine. The technique of bisulfite sequencing was first developed for DNA and adjusted for RNA just a few years ago.Citation7 Since then it has been used for mapping of m5C in all domains of life and various RNA species.Citation29-32 A more detailed description of the sequencing method is reviewed by the Motorin lab in the same issue.
Figure 3. Conversion of different modified nucleobases and reagents. (a) Mechanism of cytosine to uracil conversion by bisulfite treatment. 5-methylated cytosine (m5C) does not react with bisulfite, due to its lower electrophilicity, and thus is not converted to uracil. (b) Strand break analysis reveals the presence of modified nucleosides after treatment with various reagents. These reactions with modified bases lead to e.g. depurination which results in strand breaks after anilin treatment. In contrast, 2′O-methylated ribose is protected from alkaline induced cleavage which allows detection of the methylated site.
Variations of bisulfite reaction
With the discovery of the oxidized derivatives of m5C in DNA and RNA, namely 5-hydroxymethylcytosine (hm5C), 5-formylcytosine (f5C) and 5-carboxycytosine (ca5C), the reactivity of bisulfite has been re-analyzed. It was found that both f5C and ca5C undergo the bisulfite induced conversion into uracil, while hm5C is sulfonated but not deaminated. This finding showed that regular bisulfite sequencing is not sufficient to distinguish m5C from hm5C or even detect f5C and ca5C. Therefore chemical and enzymatic pre-treatments have been developed. After catalytic oxidation to f5C,Citation33 hm5C converts into uracil after bisulfite treatment while m5C stays unaffected. By comparison of non-oxidized and oxidized samples, the localization of hm5C ca be assigned. Although this technique has been developed for DNA, it is likely that it can be applied to hm5C detection in RNA.
Similarly f5C can be reduced by NaBH4 into hm5C in DNACitation34 or RNACitation35,36 before bisulfite treatment. The comparison of reduced and untreated sample can be used to locate f5C in the nucleic acid of interest (see ).
Enzymatic demethylation
An enzymatic approach is used for detection of f5C and ca5C in DNA by bisulfite sequencing. Here, the sample is methylated by a CpG methyltransferase M.Sssl in vitro. After this step all cytidines are methylated and no longer convert into uridine, while f5C and ca5C still undergo conversion after bisulfite treatment.Citation37
Using the bacterial enzyme AlkB, tRNA modifications like 1-methyladenosine, 3-methylcytidine or 1-methylguanosine are demethylated and become detectable by comparative sequencing.Citation38,39
Chemical conversion
For 1-methyladenosine (m1A), it has long been known, that dimroth rearrangement in alkaline conditions leads to formation of 6-methyladenosine (m6A).Citation40 This chemistry was recently rediscovered to identify the number of false-positive m1A signals during m1A-seq of eukaryotic mRNA.Citation41
RNA cleavage/depurination
While bisulfite is the most prominent nucleoside detection reagent, other chemicals were established to detect modifications in RNA. After disruption of the nucleobase's aromaticity by chemical reagents or after depurination, strand break analysis after aniline induced strand cleavage is a useful tool for the detection of modified nucleosides. Some modifications decrease the electron density in the nucleobase leaving the base more electrophilic and therefore prone to e.g. nucleophilic reactions or reduction (see ). 7-methylguanosine (m7G).Citation42 and dihydrouridine (D)Citation43 can be detected by reduction with NaBH4 or under mild alkaline conditions. In case of m7G these treatments lead to depurination, which makes the RNA prone to aniline induced strand breaks. The reaction of dihydrouridine leads to a ring opening of the nucleobase and subsequent strand breaks. Strand breaks are detectable by primer extension assays, where a radiolabeled primer is extended by a polymerase until it reaches the strand break and stops.Citation44 The length of the extended primer is compared on a gel to an alkaline RNA digest which assigns the location of the modification in the original sample.
Methylation of cytidine at position 3 decreases the electron density in the pyrimidine ring. Therefore, m3C reacts more readily with hydrazine than e.g. cytidine which leads to aniline induced strand breaks.Citation45
As mentioned above, alkaline treatment of RNA leads to strand breaks. Ribose 2′OH methylation is a common modification of RNA which prevents alkaline RNA cleavage which allows detection of 2′OH methylated nucleotides by primer extension. Only recently, the alkaline stability of 2′OH methylation was exploited by combining the treatment with high throughput sequencing.Citation46,47 The described method detects all ribosomal RNA methylations with high accuracy. Surprisingly, the alkaline treatment provided information on other modifications like pseudouridine or base methylated nucleosides as mentioned here.Citation48
The hypermodified nucleoside Wybutosin (yW) is prone to depurination after mild acidic treatmentCitation49 which makes it easily detectable by aniline induced strand-break analysis.
Detection and enrichment by physicochemical properties
Most modified nucleosides are low abundant compared with the canonical nucleosides, which makes it difficult to study their distribution and function among RNA species. To study these modifications it can be quite useful to enrich the modified RNA species before analysis. As described above, pseudouridinylated RNA can be enriched after covalent labeling with an azide functionalized CMCT derivative.Citation2 While this enrichment is based on covalent reaction with the modification, it is also possible by non-covalent interactions between the modification and a reagent. Affinity purification of e.g. thiolated RNA or cis-diols in RNA is commonly used and simply exploits the features introduced by a modification.
Mercury forms a coordinate covalent bond with sulfur as a ligand, retaining thiolated nucleic acids and reducing the migration of thus modified RNAs. Since 1977 organomercury functionalized agarose and 1978 mercury containing celluloseCitation50 has been used for the purification of synthetically thiolated RNACitation51 and later natural thiol and selenium containing RNAs.Citation52,53 Here the strong interaction of sulfur and selenium with mercury is used to retain the modified RNA on the agarose beads. After stringent washing and removal of non-modified RNA, the RNA is eluted by addition of organic sulfur compounds like dithiothreitol.
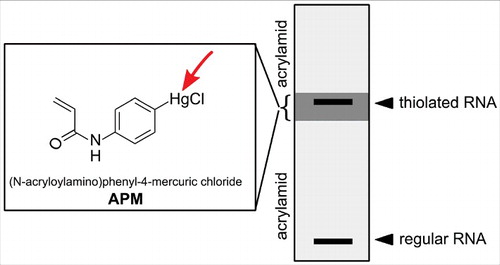
Later Igloi and co-workers developed an organomercury variant that could be co-polymerized into polyacrylamide gels, thus allowing separation of thiolated and non-thiolated RNA.Citation54 Furthermore, the authors showed that the degree of retardation depends on the type of RNA modification with 4-thiouridine being more retarded than 2-thiouridine or 5-methylaminomethyl-2-thiouridine (mnm5s2U). For purification of all thiol containing RNAs regardless of their affinity, a method involving 3-layered polyacrylamide gels in which only the middle layer contains a high amount of the organomercurial compound was developed (see ).Citation55 Organomercury gels are now commonly used to study the position 34 thiolated uridines in tRNAs.Citation56,57
Figure 4. Enrichment of thiolated RNA on a 3 layered acrylamide gel. The upper and lower gel layer allow normal migration of RNA, while the 2nd layer contains APM (structure shown in the box) which retains all thiolated RNA.
A non-mercury containing approach was presented for the enrichment of 4-thiouridine modified RNAs using activated disulfide reagents, which allow reductive release after enrichment.Citation58 Here, methylthiosulfonate-activated biotin (MTS-biotin) was used to efficiently react with s4U (after biosynthetic introduction into miRNA in HEK cells) and enrich the modified RNA.
The ribose of free nucleosides has vicinal OH groups on the 2′ and 3′ position which form a complex with boronic acids. RNAs with a non-phosphorylated 3′ end can be retained by boronic acid. tRNAs of Bacteria and Eukarya contain queosine (Q), a hypermodified 7-deazaguanosine nucleoside with a cis diol on an additional cyclopenten. Queosine containing RNA is more strongly retained on boronate polyacrylamide gels compared with unmodified RNA,Citation59 which was recently used to study the degree of Q scavenging and incorporation into RNA as an effect of nutrient availability.Citation60 This study revealed a strikingly direct mechanism by which recoding of entire genomes results from changes in utilization of the nutrient queosine. The second exception is capped RNA, where the 5′ cap-structure is connected with its 5′OH to the 5′ end of the RNA chain, leaving the 3′ and 2′-OH free. In 2015, the bacterial NAD cap was discovered, which was the first report of bacterial epitranscriptomics.Citation61 The connection of NAD to the RNA via a pyrophosphate bridge leaves a free 3′OH on the cap structure next to the 2′OH which interacts with boronate. Only recently, this technique (APB copolymerization with acrylamide) was utilized to purify bacterial cofactor-capped RNAs, e.g. NAD-RNA and study the demodification kinetics by NudC and which could potentially be used to study bacterial cofactor modification kinetics in general.Citation62
In this review we have shown small chemical reagents for RNA modification labeling, chemicals for conversion and enrichment of modified RNA. Modern techniques like RNA sequencing and mass spectrometry utilize the presented strategies to deepen our understanding of nucleic acid modifications.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
S.K. acknowledges funding by the Fonds der chemischen Industrie and the DFG (SPP1784).
References
- Bakin A, Ofengand J. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 1993; 32(37):9754-62. Epub 1993/09/21; PMID:8373778; http://dx.doi.org/10.1021/bi00088a030
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 2015; 11(8):592-7; PMID:26075521; http://dx.doi.org/10.1038/nchembio.1836
- Durairaj A, Limbach PA. Improving CMC-derivatization of pseudouridine in RNA for mass spectrometric detection. Analytica Chimica Acta 2008; 612(2):173-81; PMID:18358863; http://dx.doi.org/10.1016/j.aca.2008.02.026
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014; 515(7525):143-6; PMID:25192136; http://dx.doi.org/10.1038/nature13802
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, Leon-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014; 159(1):148-62; PMID:25219674; http://dx.doi.org/10.1016/j.cell.2014.08.028
- Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 2014; 9(10):e110799; http://dx.doi.org/10.1371/journal.pone.0110799
- Schaefer M, Pollex T, Hanna K, Lyko F. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res 2009; 37(2):e12. Epub 2008/12/09; PMID:19059995; http://dx.doi.org/10.1093/nar/gkn954
- Behm-Ansmant I, Helm M, Motorin Y. Use of specific chemical reagents for detection of modified nucleotides in RNA. J Nucleic Acids 2011; 2011:408053; PMID:21716696; http://dx.doi.org/10.4061/2011/408053
- Watson BS, Hazlett TL, Eccleston JF, Davis C, Jameson DM, Johnson AE. Macromolecular arrangement in the aminoacyl-tRNA.elongation factor Tu.GTP ternary complex. A fluorescence energy transfer study. Biochemistry 1995; 34(24):7904-12. Epub 1995/06/20; PMID:7794902; http://dx.doi.org/10.1021/bi00024a015
- Caron M, Dugas H. Specific spin-labeling of transfer ribonucleic acid molecules. Nucleic Acids Res 1976; 3(1):19-34. Epub 1976/01/01; PMID:175353; http://dx.doi.org/10.1093/nar/3.1.19
- Yang CH, Soll D. Covalent attachment of a fluorescent group to 4-thiouridine in transfer RNA. J Biochem 1973; 73(6):1243-7. Epub 1973/06/01; PMID:4579541
- Umemoto T, Okamoto A. Synthesis and characterization of the 5-methyl-2′-deoxycytidine glycol-dioxoosmium-bipyridine ternary complex in DNA. Org Biomol Chem 2008; 6(2):269-71; PMID:18174995; http://dx.doi.org/10.1039/B716400A
- Tserovski L, Helm M. Diastereoselectivity of 5-Methyluridine Osmylation Is Inverted inside an RNA Chain. Bioconjug Chem 2016; 27(9):2188-97; PMID:27540864; http://dx.doi.org/10.1021/acs.bioconjchem.6b00403
- Gornicki P, Judek M, Wolanski A, Krzyzosiak WJ. Hypermodified nucleoside carboxyl group as a target site for specific tRNA modification. Eur J Biochem 1986; 155(2):371-5; PMID:3956493; http://dx.doi.org/10.1111/j.1432-1033.1986.tb09500.x
- Pingoud A, Kownatzki R, Maass G. Fluoresceinylthiocarbamyl-tRNATyr: a useful derivative of tRNATyr (E.coli) for physicochemical studies. Nucleic Acids Res 1977; 4(2):327-38. Epub 1977/02/01; PMID:14327; http://dx.doi.org/10.1093/nar/4.2.327
- Plumbridge JA, Baumert HG, Ehrenberg M, Rigler R. Characterisation of a new, fully active fluorescent derivative of E. coli tRNA Phe. Nucleic Acids Res 1980; 8(4):827-43; PMID:6776491
- Schiller PW, Schechter AN. Covalent attachment of fluorescent probes to the X-base of Escherichia coli phenylalanine transfer ribonucleic acid. Nucleic Acids Res 1977; 4(7):2161-7; PMID:333386; http://dx.doi.org/10.1093/nar/4.7.2161
- Friedman S, Li HJ, Nakanishi K, Van Lear G. 3-(3-amino-3-carboxy-n-propyl)uridine. The structure of the nucleoside in Escherichia coli transfer ribonucleic acid that reacts with phenoxyacetoxysuccinimide. Biochemistry 1974; 13(14):2932-7; PMID:4601538; http://dx.doi.org/10.1021/bi00711a024
- McIntosh AR, Caron M, Dugas H. A specific spin labeling of the anticodon of E. coli tRNA-Glu. Biochem Biophys Res Commun 1973; 55(4):1356-63; PMID:4358937; http://dx.doi.org/10.1016/S0006-291X(73)80043-9
- Ho NW, Gilham PT. Reaction of pseudouridine and inosine with N-cyclohexyl-N';-beta-(4-methylmorpholinium)ethylcarbodiimide. Biochemistry 1971; 10(20):3651-7; PMID:4328867; http://dx.doi.org/10.1021/bi00796a002
- Naylor R, Ho NW, Gilham PT. Selective chemical modifications of uridine and pseudouridine in polynucleotides and their effect on the specificities of ribonuclease and phosphodiesterases. J Am Chem Soc 1965; 87(18):4209-10; PMID:4284810; http://dx.doi.org/10.1021/ja01096a050
- Mengel-Jorgensen J, Kirpekar F. Detection of pseudouridine and other modifications in tRNA by cyanoethylation and MALDI mass spectrometry. Nucleic Acids Res 2002; 30(23):e135. Epub 2002/12/06; PMID:12466567; http://dx.doi.org/10.1093/nar/gnf135
- Emmerechts G, Herdewijn P, Rozenski J. Pseudouridine detection improvement by derivatization with methyl vinyl sulfone and capillary HPLC-mass spectrometry. J Chromatography B, Analytical technologies in the biomedical and life sciences 2005; 825(2):233-8. Epub 2005/07/21; PMID:16029966; http://dx.doi.org/10.1016/j.jchromb.2005.06.041
- Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat Chem Biol 2010; 6(10):733-40; PMID:20835228; http://dx.doi.org/10.1038/nchembio.434
- Yang C, Soll D. Covalent attachment of fluorescent groups to transfer ribonucleic acid. Reactions with 4-bromomethyl-7-methoxy-2-oxo-2H-benzopyran. Biochemistry 1974; 13(17):3615-21. Epub 1974/08/13; PMID:4367729; http://dx.doi.org/10.1021/bi00714a033
- Kellner S, Seidu-Larry S, Burhenne J, Motorin Y, Helm M. A multifunctional bioconjugate module for versatile photoaffinity labeling and click chemistry of RNA. Nucleic Acids Res 2011; 39(16):7348-60; PMID:21646334; http://dx.doi.org/10.1093/nar/gkr449
- Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat Protoc 2006; 1(3):1610-6; PMID:17406453; http://dx.doi.org/10.1038/nprot.2006.249
- Hayatsu H. Discovery of bisulfite-mediated cytosine conversion to uracil, the key reaction for DNA methylation analysis–a personal account. Proc Japan Academy Series B, Physical and Biol Sci 2008; 84(8):321-30. Epub 2008/10/23; PMID:18941305; http://dx.doi.org/10.2183/pjab.84.321
- Squires JE, Patel HR, Nousch M, Sibbritt T, Humphreys DT, Parker BJ, Suter CM, Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res 2012; 40(11):5023-33; PMID:22344696; http://dx.doi.org/10.1093/nar/gks144
- Edelheit S, Schwartz S, Mumbach MR, Wurtzel O, Sorek R. Transcriptome-wide mapping of 5-methylcytidine RNA modifications in bacteria, archaea, and yeast reveals m5C within archaeal mRNAs. PLoS Genet 2013; 9(6):e1003602; PMID:23825970; http://dx.doi.org/10.1371/journal.pgen.1003602
- Hussain S, Sajini AA, Blanco S, Dietmann S, Lombard P, Sugimoto Y, Paramor M, Gleeson JG, Odom DT, Ule J, et al. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep 2013; 4(2):255-61; PMID:23871666; http://dx.doi.org/10.1016/j.celrep.2013.06.029
- Burgess AL, David R, Searle IR. Conservation of tRNA and rRNA 5-methylcytosine in the kingdom Plantae. BMC Plant Biol 2015; 15:199; PMID:26268215; http://dx.doi.org/10.1186/s12870-015-0580-8
- Fukuzawa S, Takahashi S, Tachibana K, Tajima S, Suetake I. Simple and accurate single base resolution analysis of 5-hydroxymethylcytosine by catalytic oxidative bisulfite sequencing using micelle incarcerated oxidants. Bioorg Med Chem 2016; 24(18):4254-62; PMID:27460669; http://dx.doi.org/10.1016/j.bmc.2016.07.016
- Song CX, Szulwach KE, Dai Q, Fu Y, Mao SQ, Lin L, Street C, Li Y, Poidevin M, Wu H, et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell 2013; 153(3):678-91; PMID:23602153; http://dx.doi.org/10.1016/j.cell.2013.04.001
- Van Haute L, Dietmann S, Kremer L, Hussain S, Pearce SF, Powell CA, Rorbach J, Lantaff R, Blanco S, Sauer S, et al. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat Commun 2016; 7:12039; PMID:27356879; http://dx.doi.org/10.1038/ncomms12039
- Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S, Rehling P, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNAMet to expand codon recognition in mitochondrial translation. EMBO J 2016; 35(19):2104-19; PMID:27497299; http://dx.doi.org/10.15252/embj.201694885
- Neri F, Incarnato D, Krepelova A, Parlato C, Oliviero S. Methylation-assisted bisulfite sequencing to simultaneously map 5fC and 5caC on a genome-wide scale for DNA demethylation analysis. Nat Protoc 2016; 11(7):1191-205; PMID:27281647; http://dx.doi.org/10.1038/nprot.2016.063
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 2015; 12(9):879-84; PMID:26237225; http://dx.doi.org/10.1038/nmeth.3508
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. Efficient and quantitative high-throughput TRNA sequencing. Nat Methods 2015; 12(9):835-7; PMID:26214130; http://dx.doi.org/10.1038/nmeth.3478
- Macon JB, Wolfenden R. 1-Methyladenosine. Dimroth rearrangement and reversible reduction. Biochemistry 1968; 7(10):3453-8; PMID:5681457; http://dx.doi.org/10.1021/bi00850a021
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016; 530(7591):441-6; PMID:26863196; http://dx.doi.org/10.1038/nature16998
- Wintermeyer W, Zachau HG. A specific chemical chain scission of tRNA at 7-methylguanosine. FEBS Lett 1970; 11(3):160-4; PMID:11945476; http://dx.doi.org/10.1016/0014-5793(70)80518-X
- Igo-Kemenes T, Zachau HG. On the specificity of the reduction of transfer ribonucleic acids with sodium borohydride. Eur J Biochem 1969; 10(3):549-56; PMID:4899928; http://dx.doi.org/10.1111/j.1432-1033.1969.tb00723.x
- Peattie DA. Direct chemical method for sequencing RNA. Proc Natl Acad Sci U S A 1979; 76(4):1760-4; PMID:377283; http://dx.doi.org/10.1073/pnas.76.4.1760
- Duh JL, Punzalan RR, Pless RC. Reactivity of pyrimidine nucleosides under the conditions of the pyrimidine sequencing reactions of Maxam and Gilbert. Anal Biochem 1988; 168(2):231-8; PMID:3364724; http://dx.doi.org/10.1016/0003-2697(88)90312-0
- Marchand V, Blanloeil-Oillo F, Helm M, Motorin Y. Illumina-based RiboMethSeq approach for mapping of 2′-O-Me residues in RNA. Nucleic Acids Res 2016; 44(16):e135; PMID:27302133; http://dx.doi.org/10.1093/nar/gkw547
- Krogh N, Jansson MD, Hafner SJ, Tehler D, Birkedal U, Christensen-Dalsgaard M, Lund AH, Nielsen H. Profiling of 2′-O-Me in human rRNA reveals a subset of fractionally modified positions and provides evidence for ribosome heterogeneity. Nucleic Acids Res 2016; 44(16):7884-95; PMID:27257078; http://dx.doi.org/10.1093/nar/gkw482
- Birkedal U, Christensen-Dalsgaard M, Krogh N, Sabarinathan R, Gorodkin J, Nielsen H. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew Chem Int Ed Engl 2015; 54(2):451-5; PMID:25417815; http://dx.doi.org/10.1002/anie.201408362
- Thiebe R, Zachau HG. A specific modification next to the anticodon of phenylalanine transfer ribonucleic acid. Eur J Biochem 1968; 5(4):546-55; PMID:5698615; http://dx.doi.org/10.1111/j.1432-1033.1968.tb00404.x
- Melvin WT, Milne HB, Slater AA, Allen HJ, Keir HM. Incorporation of 6-thioguanosine and 4-thiouridine into RNA. Application to isolation of newly synthesised RNA by affinity chromatography. Eur J Biochem 1978; 92(2):373-9; PMID:570106; http://dx.doi.org/10.1111/j.1432-1033.1978.tb12756.x
- Reeve AE, Smith MM, Pigiet V, Huang RC. Incorporation of purine nucleoside 5′-[gamma-S]triphosphates as affinity probes for initiation of RNA synthesis in vitro. Biochemistry 1977; 16(20):4464-70; PMID:334243; http://dx.doi.org/10.1021/bi00639a021
- Ching WM, Stadtman TC. Selenium-containing tRNAGlu from Clostridium sticklandii: correlation of aminoacylation with selenium content. Proc Natl Acad Sci U S A 1982; 79(2):374-7; PMID:6176991; http://dx.doi.org/10.1073/pnas.79.2.374
- Woodford TA, Schlegel R, Pardee AB. Selective isolation of newly synthesized mammalian mRNA after in vivo labeling with 4-thiouridine or 6-thioguanosine. Anal Biochem 1988; 171(1):166-72; PMID:3407913; http://dx.doi.org/10.1016/0003-2697(88)90138-8
- Igloi GL. Interaction of tRNAs and of phosphorothioate-substituted nucleic acids with an organomercurial. Probing the chemical environment of thiolated residues by affinity electrophoresis. Biochemistry 1988; 27(10):3842-9; PMID:3044450; http://dx.doi.org/10.1021/bi00410a048
- Biondi E, Burke DH. Separating and analyzing sulfur-containing RNAs with organomercury gels. Methods Mol Biol 2012; 883:111-20; PMID:22589128; http://dx.doi.org/10.1007/978-1-61779-839-9_8
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 2009; 458(7235):228-32; PMID:19145231; http://dx.doi.org/10.1038/nature07643
- Delaunay S, Rapino F, Tharun L, Zhou Z, Heukamp L, Termathe M, Shostak K, Klevernic I, Florin A, Desmecht H, et al. Elp3 links tRNA modification to IRES-dependent translation of LEF1 to sustain metastasis in breast cancer. J Exp Med 2016; 213(11):2503-2523; PMID:27811057; http://dx.doi.org/10.1084/jem.20160397
- Duffy EE, Rutenberg-Schoenberg M, Stark CD, Kitchen RR, Gerstein MB, Simon MD. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Mol Cell 2015; 59(5):858-66; PMID:26340425; http://dx.doi.org/10.1016/j.molcel.2015.07.023
- Igloi GL, Kossel H. Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronic acid. Nucleic Acids Res 1985; 13(19):6881-98; PMID:2414733; http://dx.doi.org/10.1093/nar/13.19.6881
- Zaborske JM, DuMont VL, Wallace EW, Pan T, Aquadro CF, Drummond DA. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol 2014; 12(12):e1002015; PMID:25489848; http://dx.doi.org/10.1371/journal.pbio.1002015
- Cahova H, Winz ML, Hofer K, Nubel G, Jaschke A. NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 2015; 519(7543):374-7; PMID:25533955; http://dx.doi.org/10.1038/nature14020
- Nubel G, Sorgenfrei FA, Jaschke A. Boronate affinity electrophoresis for the purification and analysis of cofactor-modified RNAs. Methods 2016; S1046-2023(16)30246-8; PMID:27645507; http://dx.doi.org/10.1016/j.ymeth.2016.09.008
