ABSTRACT
Metazoan replication-dependent (RD) histone genes encode the only known cellular mRNAs that are not polyadenylated. These mRNAs end instead in a conserved stem-loop, which is formed by an endonucleolytic cleavage of the pre-mRNA. The genes for all 5 histone proteins are clustered in all metazoans and coordinately regulated with high levels of expression during S phase. Production of histone mRNAs occurs in a nuclear body called the Histone Locus Body (HLB), a subdomain of the nucleus defined by a concentration of factors necessary for histone gene transcription and pre-mRNA processing. These factors include the scaffolding protein NPAT, essential for histone gene transcription, and FLASH and U7 snRNP, both essential for histone pre-mRNA processing. Histone gene expression is activated by Cyclin E/Cdk2-mediated phosphorylation of NPAT at the G1-S transition. The concentration of factors within the HLB couples transcription with pre-mRNA processing, enhancing the efficiency of histone mRNA biosynthesis.
Proper utilization of genetic information in eukaryotes requires extensive three-dimensional organization of the genome and its associated proteins within the nucleus. The basic unit of genome organization is the nucleosome, an octamer of two molecules of each of the four core histone proteins (H2A, H2B, H3 and H4) that packages ∼147 base pairs of DNA. In mammalian cells, approximately 4×108 molcules of each core histone must be synthesized during S phase of the cell cycle to package the newly replicated DNA. Defects in this process result in genome instability that can contribute to human pathologies like cancer. Histone protein synthesis results from a large increase in production at the beginning of S phase of equal amounts of mRNAs encoding the four core nucleosomal histones, as well as histone H1. This highly coordinated gene expression event is performed by a set of transcription and pre-mRNA processing factors unique to replication dependent (RD) histone mRNA biosynthesis that assemble into a nuclear body tightly associated with RD histone genes called the histone locus body (HLB). HLB formation involves a combination of ordered and stochastic assembly steps, and cell cycle regulated changes in the HLB occur to activate histone gene expression during S phase. Here we describe the history of how the HLB was discovered followed by a discussion of HLB assembly mechanism and how the HLB functions in RD histone mRNA biosynthesis.
Discovery of the Histone Locus Body: a nuclear body dedicated to replication dependent histone mRNA biosynthesis
Metazoan histone genes have unique properties
RD histone genes differ from all other genes in animal cells in several important ways. First, animal genomes contain multiple copies of each of the 5 different RD histone genes. There are over 100 gene copies in Drosophila melanogaster,Citation1,2 400 in sea urchins,Citation3,4 and 10–20 in mice and humans.Citation5 Second, RD histone genes have remained tightly physically clustered throughout evolution, unlike other genes (e.g. E2F targets) that are coordinately regulated during the cell cycle. The tight clustering of genes encoding different proteins suggests that there has been selective pressure during evolution to maintain a local environment in the nucleus (i.e. the HLB) that is optimal for histone mRNA biosynthesis. Third, RD histone genes encode the only eukaryotic cellular mRNAs that are not polyadenylated, ending instead in a conserved stem-loop sequence.Citation6,7 Hence they require a set of factors dedicated to histone mRNA biosynthesis. Because RD histone genes do not contain introns, only a single pre-mRNA processing step, an endonucleolytic cleavage that produces the 3′ end, is needed to form mature histone mRNA (). This processing reaction requires two cis elements, a stem-loop that binds stem-loop binding protein (SLBP) and a purine-rich sequence just downstream of the cleavage site termed the Histone Downstream Element (HDE)Citation8 that binds the 5′ end of U7 snRNA via base pairing.Citation9,10 U7 snRNA is part of the U7 snRNP, which assembles the processing machinery within the HLB. The only known functions of SLBP and U7 snRNP are in histone mRNA metabolism.
Figure 1. Processing of histone pre-mRNA. (A) The histone pre-mRNA processing reaction. As the 3′ end of the histone mRNA is transcribed, SLBP binds to the stem-loop and the U7 snRNP binds to the HDE through base-pairing between the 5′ end of U7 snRNA and the Histone Downstream Element. This binding is stabilized by interaction of U7 snRNP with SLBP. The U7 snRNP consists of the U7 snRNA and 7 Sm proteins including Lsm10 and Lsm11, which replace SmD1 and SmD2 of the spliceosomal snRNPs. FLASH binds to Lsm11, and the FLASH/Lsm11 complex recruits the histone cleavage complex (HCC; blue) containing the Symplekin/CstF64 heterodimer and the CPSF73/CPSF100 heterodimer (dark blue). Cleavage of pre-mRNA is catalyzed by CPSF73. Other CPSF components are bound to the HCC but they may be sub-stoichiometric and not essential for processing (light blue). (B) U7 snRNP and FLASH are constitutive components of the HLB. In G1-phase they are either present separately or bound together in an inactive form. Processing is activated only during S phase after recruitment of the HCC by FLASH and U7 snRNP.
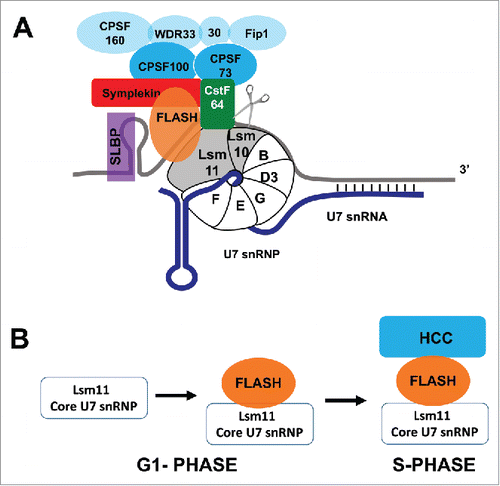
Histone mRNA biosynthetic factors are concentrated in a unique nuclear body
Following the identification of U7 snRNA by the Birnstiel, Schumperli, and Steitz laboratories as a processing factor for sea urchin and mouse histone mRNA,Citation9-12 Joe Gall's laboratory found that U7 snRNA is concentrated near the histone genes in amphibian oocytes.Citation13,14 These bodies were termed “C snurposomes” and are distinct from particles containing the snRNPs required for mRNA splicing. In Xenopus oocytes C snurposomes contain over 95% of the U7 snRNA in the oocyte and lack spliceosomal snRNAs.Citation15 The role of C snurposomes in oocytes is not clear. They might participate in the synthesis of the large store of histone mRNA produced earlier in oogenesis and/or serve as a stockpile of factors to allow histone mRNA synthesis in embryos at the mid-blastula transition. Subsequently, the Matera laboratory identified U7 snRNA-containing nuclear bodies in mammalian cells that were only found at the RD histone gene loci.Citation16 Many of these bodies also contained Coilin, which had been defined as a marker of the Cajal body.Citation17 Thus, the interpretation of these initial data was that there is a special Cajal body associated with RD histone genes.
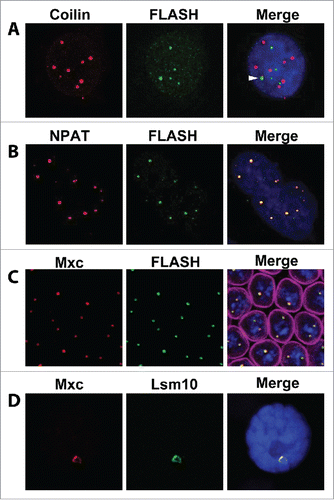
The explicit recognition that the nuclear body associated with RD histone genes was distinct from the Cajal body and other nuclear bodies came from a 2006 study by the Gall laboratory showing that U7 snRNP localized to a specific nuclear body at the Drosophila RD histone genes.Citation18 The core of snRNP particles is composed of an snRNA bound to a ring of seven highly conserved Sm proteins.Citation19 The core U7 snRNP particle differs from other snRNPs in the composition of the Sm ring. The U7 snRNP consists of the small (55–70 nt) U7 snRNA bound by a ring of 7 Sm proteins, five of which are also found in the Sm ring that binds to the spliceosomal snRNAs. The other two, Lsm10 and Lsm11, replace SmD1 and SmD2 and are found only in U7 snRNP.Citation20,21 Liu et al. expressed transgenic YFP-Lsm11 and found that it concentrated in a histone gene-associated nuclear body that also contained U7 snRNA and Lsm1018. They named this structure the “histone locus body” and showed it was distinct from the Drosophila Cajal body, which they detected by co-localized U85 scaRNA, U2 snRNA, or YFP-tagged SMN protein, and subsequently with anti-Coilin antibodies after they identified Drosophila Coilin.Citation22 Shortly thereafter, Cajal bodies and HLBs were shown to be distinct structures also in mammalian cells.Citation23,24 Although Coilin is sometimes observed at HLBs in both Drosophila and mammalian cells,Citation16,25-27 HLBs that do not contain Coilin are frequently observed. Moreover, genetic experiments show that HLB assembly occurs normally in the absence of Coilin.Citation22,28 Conversely, disruption of the HLB does not eliminate formation of Coilin-marked Cajal bodies, although it can alter their appearance, perhaps because of effects on cell cycle progression.Citation28-30 Thus, the cytological and genetic evidence indicates that HLBs and Cajal bodies are distinct nuclear structures (), with Cajal bodies involved in snRNP metabolism and HLBs in histone mRNA metabolism.
Figure 2. Visualization of the HLB by immunofluorescence. (A) HeLa cell stained with antibodies recognizing Coilin (red) to detect Cajal bodies or FLASH (green) to detect HLBs. Note in this cell line these two bodies are distinct, although occasionally Coilin can be detected in the HLB (arrowhead). The number of HLBs in cultured cells depends on the ploidy. (B) HeLa cell stained with antibodies recognizing NPAT (red) or FLASH (green). Large HLBs correspond to the Hist1 cluster and smaller HLBs to the Hist2 cluster. Panels A and B were reproduced with permission from reference 42. (C) A Drosophila syncytial embryo stained with antibodies recognizing Mxc (red) and FLASH (green) and lamin (magenta; merged image). The chromosomes either are paired and nuclei contain one large HLB, or are separated and nuclei contain 2 smaller HLBs. (D) A Drosophila salivary gland cell stained with antibodies recognizing Mxc (red) and Lsm10 (green). Note that the HLB forms on the histone locus on polytene chromosomes. In all merge panels DNA is stained with DAPI (blue). © Elsevier. Reproduced by permission of Elsevier. Permission to reuse must be obtained from the rightsholder.
Another seminal observation that ultimately helped define the HLB was the discovery by the Harlow laboratory of NPAT as a Cyclin E/Cdk2 substrate that promoted S-phase entry.Citation31 Work from the Harlow and Harper laboratories showed that NPAT concentrated in foci that localized to histone genes and was necessary for histone gene expression.Citation26,27 These and subsequent studies demonstrated that NPAT is phosphorylated by Cyclin E/Cdk2 as cells approach entry into S-phase, resulting in activation of histone gene expression. The foci observed in this early work were clearly what we now know as HLBs, and NPAT together with U7 snRNP have become definitive markers for the HLB.
Our laboratories identified the product of the multi-sex combs gene (mxc) as the Drosophila ortholog of NPAT.Citation32 NPAT/Mxc is a scaffolding protein for the HLB, and may also directly participate in histone gene expression. The N-terminus of NPAT activates histone gene promoters 3–4-fold in reporter assays,Citation33 but mutations in NPAT that interfere with this activity do not affect the ability of full length NPAT to support histone gene expression, suggesting that the role of NPAT is more complex than simply participating in activation of transcription.Citation34 Consistent with its critical role in histone gene expression, NPAT is essential for cell viability and for both mouse and Drosophila development.Citation32,34-37 In pursuing the serendipitous observation of Brian Calvi and Allan Spradling that a monoclonal antibody raised against mitotic phosphoproteins from HeLa cells (MPM-2) detected nuclear foci in Drosophila ovarian follicle cells only when Cyclin E/Cdk2 was active,Citation38 we found that the MPM-2 foci contained Lsm11 and localized to the histone genes, indicating that the MPM-2 antibody recognized a component of the Drosophila HLB.Citation39 We subsequently determined that MPM-2 recognizes a Cyclin E-Cdk2 dependent phosphoepitope on Mxc (multi sex combs), identifying the product of the mxc locus as the Drosophila ortholog of human NPAT.Citation32 As both NPAT and Mxc are targets of Cyclin E/Cdk2, activation of Cyclin E is thus essential for initiation of both DNA replication and histone mRNA expression,Citation40 effectively coordinating these two events.
An expanding list of HLB components
Additional HLB components have been identified in a number of ways, including immunofluorescence with antibodies raised against newly identified proteins, genome-wide RNAi screens for factors required for histone pre-mRNA processing or HLB formation, and biochemical characterization of factors involved in histone gene transcription and pre-mRNA processing (). An example is FLASH, a large protein with low complexity regions that was identified by De Laurenzi and co-workers as concentrating in nuclear bodies containing NPAT and shown to be essential for cells to enter S-phase.Citation28,41 The Marzluff laboratory subsequently identified FLASH in a two-hybrid screen for proteins that interact with the N-terminus of Lsm11 and showed that it was essential for histone pre-mRNA processing.Citation42 A second example is muscle wasted (Mute), discovered by the Chia lab in a screen for factors required for muscle development and shown to concentrate in the HLB,Citation43 as does its mammalian ortholog YARP.Citation44 Mute may function as a negative regulator of histone gene expression.Citation43 Dominski and colleagues also showed that cleavage of histone pre-mRNA is catalyzed by CPSF-73, the same protein that catalyzes cleavage of pre-mRNAs encoding poly(A) mRNAs, demonstrating a role for polyadenylation factors in histone pre-mRNA processing,Citation45 including SymplekinCitation46 and CPSF100.Citation47,48 These discoveries opened up new avenues for understanding histone pre-mRNA processing and HLB composition and assembly.
Table 1. Proteins that can concentrate in the HLB and their role in histone gene expression.
As determined by immunofluorescence studies, HLB proteins can be grouped into at least two general categories: those that are present in the HLB throughout the cell cycle and that are not detected at other sites in the nucleus, and those that are recruited to the HLB during S phase when the RD histone genes are expressed. NPAT/Mxc, FLASH and U7 snRNP are factors in the first category that best define the HLB. The S phase category has been largely studied in Drosophila, where HLBs can be visualized in a developing organism and there is much tighter transcriptional regulation of histone genes than in mammals.Citation49 This category includes proteins required for general RNA synthesis such as TBP and RNA pol II,Citation25,49,50 the elongation factor Spt6,Citation32 transcription factors like Myc,Citation51 and factors required for histone pre-mRNA processing like Symplekin.Citation25,52 These proteins are also present in the nucleoplasm throughout the cell cycle because they also function in general mRNA biosynthesis (e.g., Symplekin) or in the expression of other genes (e.g., Myc). The recruitment of these factors to the HLB may be triggered or influenced by cell cycle regulated signals, including phosphorylation of NPAT/Mxc by Cyclin E/Cdk2. HLB components likely exchange continuously between the HLB and the nucleoplasm, as do components of the Cajal body, but the rates of exchange can vary for different components.Citation49,53,54 Finally, some HLB components are not detected in all HLBs (e.g., Coilin and the chaperone protein Cpn10) or are also recruited to other nuclear bodies (e.g., the Cajal Body in the case of Coilin).Citation18,55
Interestingly, several genes encoding Drosophila HLB components were originally discovered in genetic screens as mutations that affect organ development. The best examples are mute and mxc. The mxc locus was identified by hypomorphic mutations that cause homeotic transformation of adult male legs, and consequently mxc was originally classified as a polycomb group gene.Citation36 Only more than a decade later did we show it encodes the Drosophila ortholog of NPAT.Citation32 Mute was identified as a gene required for muscle development during embryogenesis,Citation43 and encodes a large protein with predicted intrinsically disordered (IDR) regions that localizes to the HLB. Winged eye (wge) was identified in a gain-of-function screen as a gene that causes eye to wing transformations when overexpressed.Citation56 wge encodes a bromo-adjacent homology (BAH) domain protein conserved in mammals that was recently shown to localize to the HLB in Drosophila.Citation57 Wge binds to Mute and the two appear to act together as negative regulators of histone gene expression. They can recruit co-repressors, but their specific mechanism of action is not known. Finally, abnormal oocyte (abo) was identified as a maternal effect mutation that reduces the viability of offspring. abo encodes an HLB protein that has no obvious functional domains, but is conserved in mammals. Abo appears to negatively regulate histone gene expression in the Drosophila female germ line.Citation58
An emerging theme from these observations is that alterations to histone gene expression via mutation of HLB components often result in widespread gene expression changes that affect the development of particular tissues or cell lineages. In the case of hypomorphic mutations in mxc, a reduction in replication dependent histone gene expression may result in a reduction of histone protein abundance, or an alteration of the proportion of canonical and variant histones, that attenuates the establishment of polycomb-mediated repressive chromatin environments controlling cell identity. Likewise, some transposons are de-repressed in wge mutants, perhaps because repressive heterochromatin is not properly established.Citation57 Another possibility is that some HLB components act directly to regulate gene expression at loci other than the RD-histone genes, consistent with the observation that Wge binds to multiple sites on salivary gland polytene chromosomes,Citation56 although Mute and Mxc are only found at the histone locus. It is important to also keep in mind that mutation of any factor that affects entry into or progression through S phase may result in alteration of histone mRNA levels, even though the factor may have no direct role in histone gene expression or histone mRNA metabolism.
HLB assembly
Work in a variety of experimental systems over the last 15 y has coalesced into three major concepts for how nuclear bodies may assemble, each of which likely applies to the HLB.Citation59 Importantly, the molecular mechanisms underlying these three concepts are not mutually exclusive. The first is that nuclear body components have an intrinsic ability for self-organization, resulting in complex macromolecular structures that can be visualized by light microscopy.Citation60 An important aspect of nuclear body assembly remains unclear: whether self-organization occurs stochastically or through an ordered, hierarchical process.Citation61 The second concept is that a specific “seeding” event initiates the self-organization process.Citation62,63 In several instances, most notably the nucleolus, the seeding event is transcription of a specific RNA (e.g., rRNA), although other types of seeding events are possible.Citation62,64-67 Identifying seeding events is key to understanding assembly of particular nuclear bodies during development and in specific cell types. The third concept postulates that nuclear body assembly results in a phase transition characterized by liquid-liquid demixing. This process results in a distinct liquid-like compartment within the nucleus with different physical properties than the surrounding nucleoplasm, and subsequent nuclear body function is mediated by this phase transition.Citation68 In this section we consider evidence suggesting that all three may contribute to the properties and assembly of the HLB.
Self-organization of the HLB
Self-organization refers to the ability of many molecules of one or more proteins to spontaneously assemble into large networks or polymers. In the case of nuclear bodies like the HLB that are composed of several different types of factors, there is likely a dense network of weak interactions among individual components that are necessary for assembling and maintaining the body. Scaffold protein oligomerization is well established in the nuclear body literature, and the N-terminal self-interaction domain of Coilin provides a good example of a scaffold for Cajal Bodies.Citation69 Drosophila Mxc and mammalian NPAT contain a conserved N-terminal LisH domain and an adjacent SIF (self-interaction facilitator) domain that in Drosophila mediate self-interaction among Mxc molecules.Citation70 The LisH and SIF domains of Mxc do not mediate simple dimerization, but instead provide multivalent binding capability that could help oligomerize Mxc into a large molecular network that provides a scaffold for the recruitment of other HLB components (). Importantly, the oligomerization mediated by the LisH and SIF domains is required for Mxc function in vivo, as substitution mutations of 3 amino acids in either domain render Mxc completely unable to support HLB assembly and are lethal.Citation70 The LisH of NPAT domain is necessary for NPAT to stimulate histone gene expression.Citation33 However, an ectopically expressed NPAT lacking the LisH domain can localize to the HLB, perhaps because other domains can interact with endogenous, wild type NPAT at the HLB.Citation33
Figure 3. Schematic of Drosophila HLB assembly. The Drosophila replication dependent histone locus is organized as a tandem array of ∼100 copies of a 5 kb sequence containing one copy of each of the five RD histone genes. A sequence between the H3/H4 genes recruits Mxc and FLASH. Subsequent recruitment of other HLB components including U7 snRNP and Mute requires transcription initiation and results in maturation of a complete HLB. The result is a molecular environment that organizes the histone gene array and its accompanying trans-acting factors for efficient histone mRNA transcription and pre-mRNA processing.
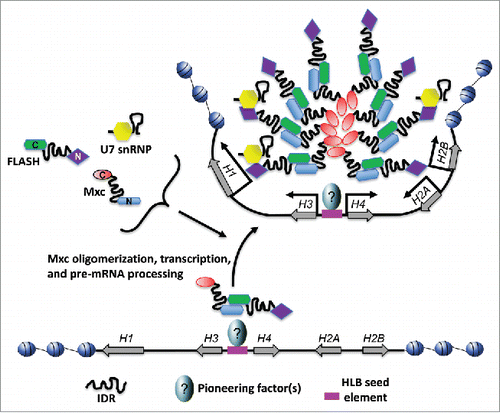
Multiple components can potentially self-organize to form nuclear bodies via two different mechanisms. The first is a stochastic recruitment mechanism in which factors are not required to assemble in any particular order, resulting in many different potential pathways to nuclear body assembly. This process contrasts with a hierarchical or ordered mechanism in which nuclear body assembly follows an invariant order of events (i.e., A recruits B, followed by recruitment of C, etc.). Interestingly, the HLB exhibits properties of both mechanisms. For instance, the Dundr laboratory has demonstrated that tethering a single HLB component or histone pre-mRNA to a specific locus in mammalian cells can trigger recruitment of other HLB components.Citation64,71 Moreover, several different HLB components are capable of nucleating HLB-like structures when tethered, supporting a model for stochastic assembly. In contrast, we have shown using genetic manipulation in Drosophila that some HLB factors (e.g., Mxc) are necessary for the recruitment of others (e.g., Mute, U7 snRNP), but not vice versa, consistent with an ordered assembly process.Citation32 In addition, we have microscopically observed the ordered appearance of components upon initial HLB assembly in the syncytial cycles of early Drosophila embryogenesis.Citation32,39 However, these two ideas need not be mutually exclusive, and the HLB could form by initial recruitment of certain scaffolding factors such as Mxc and FLASH followed by stochastic recruitment of additional factors like U7 snRNP and Mute ().
A feature of some HLB proteins is that the domains required for recruitment are separable from the domains required for biochemical function in histone mRNA biosynthesis. For instance, an N-terminal fragment of Mxc containing the LisH and SIF domains that is unable to support HLB formation or transcription on its own is nonetheless able to concentrate in the HLB in the presence of wild type Mxc, likely both by self-associating and interacting with endogenous full length Mxc within the HLB.Citation70 Similarly, a direct interaction between the NH2 terminus of Lsm11 (in the U7 snRNP) and the NH2 terminus of FLASH is necessary for pre-mRNA processing, but not for either U7 snRNP or FLASH to concentrate in the HLB.Citation72,73 In addition, the sequence in histone pre-mRNA that binds U7 snRNP is not required for U7 snRNP recruitment to the Drosophila HLB.Citation74 Instead, we found that the C terminus of FLASH, likely together with Mxc, is necessary for recruitment of U7 snRNP to the HLB.Citation72 Thus, there are interactions among HLB components involved in assembling the HLB that are distinct from the interactions required for histone mRNA biosynthesis. Such modularity could explain how a protein like FLASH participates directly and separately in HLB formation and pre-mRNA processing.
A specific DNA sequence is required for seeding Drosophila HLB assembly
What triggers self-organization of the HLB? In the context of combining ordered and stochastic models of assembly, a “seed and grow” mechanism best describes HLB assembly.Citation62 In this model a specific nucleation event results in an increase in local concentration of a specific factor(s) (“seed”) that provides a platform for recruitment of more factors via the properties of oligomerization and self-organization (“and grow”). But what provides the seed? Since HLBs are tightly associated with histone gene clusters, an obvious answer could be histone encoding DNA or RNA. Indeed, we discovered a specific sequence in the Drosophila RD histone gene array that is necessary and sufficient to nucleate HLB assembly.Citation74 The Drosophila RD histone locus is composed of ∼100 copies of a tandemly arrayed 5kb sequence containing one copy of H1 and one copy of divergently transcribed H3/H4 and H2A/H2B gene pairsCitation1(). A ∼300 bp sequence between the H3/H4 coding sequences and containing the H3/H4 promoter(s) nucleates HLB assembly, but the corresponding sequence containing the H2A/H2B promoter(s) does not.Citation74 In addition to driving HLB assembly, the H3/H4 promoter sequence is required for efficient transcription from the H2A/H2B gene pair, even though the H2A/H2B pair contains intact promoters.Citation74 Together these results suggest that the H3/H4 promoter is required for HLB assembly, which in turn is critical for histone gene expression.
Blocking transcription of the Drosophila H3/H4 genes by mutating their TATA boxes results in the recruitment of Mxc and FLASH but not Mute and U7 snRNP, suggesting that transcription is essential for assembly of the complete HLB.Citation74 At first glance, this result appears consistent with the observation that histone RNA can recruit HLB components in a tethering assay.Citation64 In addition, mature HLBs appear for the first time in early Drosophila development near the time of activation of zygotic histone gene expression.Citation32,39 However, a transgene containing a 300 nt fragment with the H3/H4 promoter is capable of ectopically recruiting the core HLB components, including Mxc, FLASH, Mute and U7 snRNP in the absence of any histone coding or 3′ end processing sequences.Citation74 Furthermore, in Drosophila tissues HLBs are present in G1 and G2 phase cells that do not express histone mRNA, as well as in cells that have exited the cell cycle.Citation18,32,39 Similarly, NPAT, FLASH and U7 snRNP continue to be present in mammalian tissues that do not express histone mRNA.Citation75 Thus, we favor a model in which transcription from the H3/H4 promoter facilitates HLB assembly rather than the actual histone mRNA sequence. In addition, the pre-mRNA processing signals are not required to recruit FLASH or U7 snRNP to the HLB. The initial recruitment of Mxc and FLASH followed by transcription dependent recruitment of Mute and U7 snRNP in Drosophila is also consistent with an ordered assembly model. As noted above, however, ordered HLB assembly steps are not mutually exclusive with stochastic self-organization. A dramatic example of this point is that FLASH and Mxc can temporarily organize into foci that resemble HLBs in embryos where the RD histone genes have been deleted, demonstrating the self-organizational properties of these molecules.Citation39,74
How do HLB components initially find the histone locus? Since NPAT/Mxc does not bind DNA specifically, a likely possibility is recruitment to histone genes by interaction with a DNA binding protein. Another possibility is that a specific chromatin structure could be established on histone genes in both humans and flies that promotes recruitment of HLB components, including NPAT/MxcCitation76,77. Conserved sequences in the Drosophila H3/H4 promoter that are not present in the H2A/H2B promoter might serve to bind a specific factor or establish a specific chromatin structure.Citation74 The assembly of the HLB has only been studied in detail in Drosophila, and mammalian histone genes are organized very differently than in flies. Humans contain a major histone gene cluster on chromosome 6 with 55 genes spread over several megabases as well as an unlinked, smaller, and more compact histone gene cluster on chromosome 1 with 10 genes in about 100 kB.Citation5 This general organization is conserved in all mammals and contrasts with the 500 Drosophila genes in 500 kB of DNA. The much greater distance between histone genes in mammals and different organization (e.g., there are no H3/H4 pairs) makes it less likely that there is a single DNA sequence that seeds the mammalian HLB. Thus, the mechanism for recruiting NPAT to the histone locus is likely to be different in mammals than in flies. However, the histone genes on each chromosome organize into a single HLB, as human cells in S phase have two large and two small HLBs that assemble at the large and small histone gene clusters, respectively.Citation26,27 Human NPAT can bind directly to HiNF-P, a sequence specific DNA binding protein that is required for histone H4 transcription.Citation78,79 Deletion of HinfP from mouse embryos abolishes histone H4 mRNA expression but does not abolish formation of the HLB as detected by staining for NPAT and FLASH.Citation80 Similarly, depletion of HiNF-P from cultured cells results in a dispersion of HLBs into smaller bodies, although they still form.Citation81,82 These data indicate that HiNF-P is not required for mammalian HLB formation but may promote complete HLB assembly via its role in H4 transcription.
During mitosis the HLB is disassembled, but a small amount of Mxc as well as FLASH can be detected on condensed chromosomes throughout mitosis in Drosophila.Citation32 By contrast, NPAT has not been observed associated with mitotic chromosomes in mammalian cells.Citation26,27,83 The mechanism of HLB disassembly during mitosis is not known, but perhaps involves changes in phosphorylation of HLB components by mitotic CDKs. In flies, a small pool of NPAT/Mxc may remain associated with mitotic chromosomes in order to “bookmark” the histone genes and rapidly nucleate full HLB reassembly at the beginning of interphase. This argument has also been made for TBP, which in S2 cells is tightly associated with the Drosophila core histone genes even after RNAi depletion of Mxc.Citation49 Note that a novel form of TFIID with a limited number of additional components beside TBP localizes to the H2a/H2b and H3/H4 genes,Citation49 and a TBP paralog, TRF2, is present in the HLB at the histone H1 genes.Citation50 The concept of bookmarking specific loci also holds true for nucleoli, where UBF “bookmarks” rDNA after mitosis and transcription of rDNA is necessary for full nucleolar formation.Citation66,84 Thus the HLB and the nucleolus and potentially other nuclear bodies that associate with specific loci may have a component that serves a bookmarking role during mitosis.
Does phase separation help drive HLB assembly?
Phase separation has recently emerged as a possible common principle in the assembly of both nuclear and cytoplasmic bodies.Citation68,85-87 In this process the physical chemical properties of body components promote a phase transition when the concentration of the component(s) exceeds their solubility limit, such as when molecules precipitate out of solution and form a crystal. The phase transition need not result in a solid structure: Many bodies display liquid like properties, similar to a lipid droplet within an aqueous environment. A spherical shape and fusing of body “droplets” are evidence for liquid like behavior in the context of phase separation. This model is consistent with the rapid exchange of nuclear body components with the nucleoplasm.
Although there is no direct evidence that HLBs form as a consequence of phase separation, several observations suggest that this process may be applicable to HLBs. In diploid cells of early Drosophila embryos, we typically observe either one or two HLBs in interphase that appear spherical by confocal microscopy, and this number correlates well with the frequency of homolog pairing ().Citation39 Thus, two HLBs, one on each homolog, might fuse into one HLB when the histone gene arrays on homologous chromosomes pair with each other, consistent with the HLB having liquid like properties. Human cells have four spherical HLBs because there are two histone gene clusters and the homologous chromosomes do not pair.Citation26,27 Intrinsically disordered regions (IDRs) are a common feature of proteins that undergo phase transition and form bodies.Citation85 IDRs do not fold into well-defined protein domains and are often composed of regions of low sequence complexity.Citation88,89 Both Mxc and NPAT as well as FLASH and Mute are large proteins with regions of low complexity, such as the large polyserine tract in the middle of the Mxc protein. Multivalent interactions driven by IDRs can reduce solubility and facilitate body formation, with the strong interactions governing polymerization and the weak interactions governing solubility.Citation86 Interestingly, conversion over time of phase separated droplets composed of IDRs and RNAs from a liquid like to a more fiber like state has been demonstrated in vitro suggesting that IDRs may contribute to the “maturing” of the HLB in response to transcriptional activation as described above.Citation90,91 Post-translational modifications may also modulate phase separation associated with gene expression.Citation92 Poly ADP ribose promotes phase separation,Citation93 and poly (ADP-ribose) polymerase has been detected in Drosophila HLBs.Citation94 Finally, given that IDRs might be promiscuous, some proteins might concentrate in the HLB but not have an essential biological or biochemical function in histone mRNA biosynthesis. Such promiscuity may explain the observation that Coilin, which contains regions of low sequence complexity that are likely IDRs,Citation95 sometimes appears within HLBs but is not required for HLB assembly or for histone mRNA biosynthesis.Citation22,25 Although these ideas are rather speculative, we suggest that phase separation may act together with self-organization and molecular seeding to promote HLB formation ().
HLB function
A prevailing model for nuclear body function postulates that nuclear bodies promote reactions by increasing the local concentration of necessary factors and/or promoting macromolecular assembly to allow efficient coupling of multiple reactions.Citation96 Indeed, recent data demonstrate that Cajal bodies enhance the assembly of mature snRNPs both in mammalian cells and early zebrafish embryos.Citation97,98 In this section we will summarize recent experiments that address how the HLB contributes to histone mRNA biosynthesis, with an emphasis on the role of FLASH and U7 snRNP in the pre-mRNA processing reaction.
Concentrating factors in the HLB promotes histone pre-mRNA processing
Since HLBs are tightly associated with RD histone genes, an important question is whether HLBs are actually sites of histone mRNA biosynthesis, or whether they might simply provide a stockpile of factors close to the histone genes. In large amphibian oocytes some C snurposomes (HLBs) are not associated with histone genes.Citation25 By contrast, in cultured mammalian cells and in Drosophila tissues the HLB is invariably found at the histone locus throughout the cell cycle.Citation18,41 More importantly, in Drosophila cells HLB components co-localize with nascent RD histone transcripts, as detected by in situ hybridization,Citation49 and RNA polymerase II becomes highly enriched in HLBs during S phase when histone mRNA production is high.Citation25,49 These data suggest that HLBs are the sites of histone mRNA biosynthesis in flies and mammals, although direct evidence for this situation requires co-localizing the HLB and the histone DNA by high resolution microscopy.
An ideal way to address whether concentrating factors within the HLB promotes reaction efficiency is to prevent a wild type, fully active protein involved in some aspect of histone mRNA biosynthesis from concentrating in the HLB without disrupting the integrity of the HLB. Taking advantage of our knowledge of the molecular interactions of HLB components,Citation44,70,72,73,99-101 we have performed genetic studies in Drosophila using mutant proteins that are either processing defective or unable to localize to the HLB. Unlike other metazoan genes, where transcription can continue well 3′ of the polyadenylation site, transcription termination is tightly coupled to histone mRNA processing in both Drosophila and mammals.Citation72,102,103 Drosophila RD histone pre-mRNA processing is normally very efficient, meaning that in wild type cells and animals only stem-loop RD histone mRNAs can be detected. RNA pol II pauses just 3′ of the HDE,Citation104 and likely terminates immediately after processing since there are no detectable transcripts downstream of the HDE.Citation72 Failure to process histone mRNA in Drosophila results in continued transcription past the HDE and polyadenylation at cryptic polyadenylation sites 3′ of the HDE in each histone gene.Citation105 Sufficient polyadenylated histone mRNA is produced in the absence of processing factors (e.g., U7 snRNP) to allow much of development to occur in Drosophila.Citation106,107 We can detect both poly A+ histone RNA and nascent read-through unprocessed RNAs by nuclease protection and in situ hybridization assays, allowing us to quantitatively determine the efficiency of histone pre-mRNA processing in various tissues.Citation72,108
Concentration of U7 snRNP and FLASH in the HLB appears to be particularly important for histone mRNA biosynthesis. There is a very small amount of U7 snRNA in the cell (about 500 copies cf. with ∼500,000 for U1 snRNA in mammals) and very small amounts of FLASH in Drosophila embryos and larvae.Citation72 FLASH is recruited to the HLB by an interaction between the C-terminus of FLASH and the C-terminus of NPAT/MxcCitation29,44,72,73. A hypomorphic allele of mxc (mxcG46) that encodes a truncated form of Mxc lacking the last 195 amino acids results in failure of full length, wild type FLASH protein and U7 snRNP to concentrate in the HLB, although an HLB containing the truncated Mxc protein forms.Citation72 Interestingly, in this genotype histone gene transcription is unaffected but histone pre-mRNA processing is inefficient, resulting in small amounts of both nascent, read-through and polyadenylated histone transcripts.Citation72 This result demonstrates that concentrating FLASH and/or U7 snRNP in the HLB is necessary for efficient coupling of histone mRNA processing and transcription termination.
A number of results suggest that the loss of coupling between processing and transcription termination may also occur in mammalian cells. Based on Pol II Chip assays, mammalian histone genes differ from other genes in that pol II terminates shortly after the 3′ end of histone mRNA.Citation72,102,103 Knockdown of a number of factors involved in either the early steps of elongation of RNA (e.g., NELF,Citation109 Cdk9110), factors that interact with FLASH (e.g., Ars2,Citation29,111), factors that catalyze histone H2b ubiquitinationCitation110 and Y3** RNACitation112 all result in expression of a small amount of polyadenylated histone mRNA, about 2–4% of the total histone mRNA, without affecting cell growth. This phenotype is similar to the molecular phenotype observed in Drosophila when FLASH or U7 snRNP is mislocalized,Citation72 and could arise from uncoupling processing and transcription termination. Disrupting the interaction between Ars2 and FLASH interferes with cell cycle progression, and depletion of Ars2 reduces the number of HLBs.Citation29 Knockdown of Y3** RNA results in perturbation of the size of the HLBs suggesting that it interacts directly with HLB components. We find that there is no Ars2 or Y3**RNA in our purified in vitro processing complexes (Dominski and Marzluff, unpublished observations), consistent with these factors affecting HLB function, and Y3** RNA has been suggested to play a role in recruiting CPSF to the HLB.Citation112
Functional changes in the HLB during S phase
As cells enter S-phase, there are at least two major events that together result in a massive increase in histone mRNA levels: initiation of histone gene transcription in response to activation of Cyclin E/Cdk2 in late G1 and activation of histone pre-mRNA processing. Phosphorylation of NPAT by Cyclin E/Cdk2 is essential for histone gene transcription,Citation26,27,34 and may also result in the recruitment of pre-mRNA processing factors to the HLB like Symplekin,Citation52 thereby activating histone pre-mRNA processing. Changes to the HLB during S phase can be visualized cytologically. For instance, HLBs in both Drosophila and human cells can be specifically detected during S phase with antibodies that recognize phosphorylated NPAT/MxcCitation26,27,32,39. In human cells the HLBs at the major cluster on chromosome 6 are readily visualized throughout the cell cycle, but clearly increase in size as cells enter S phase. RNA pol II,Citation25,49,50 and other factors (e.g., Spt6Citation32 and MycCitation51) required for transcription of the histone genes appear in the HLB during S phase, although TATA binding protein (TBP) is present constitutively in the HLB.Citation49 There is also recruitment of sequence specific DNA binding proteins: e.g., OCA-S/Pdm-1 for core histone expression,113114 and HiNF-P for human His4 expression,Citation78 although it is not known if they are only in the HLB in S phase.
As cells enter S phase, a major change likely occurs in the U7 snRNP within the HLB, activating it for histone pre-mRNA processing. FLASH and Lsm11 interact to form a protein surface that recruits a set of proteins termed the histone cleavage complex (HCC).Citation100,101 The HCC includes the endonuclease core of the CPSF polyadenylation complex (CPSF73 and CPSF100) plus the protein Symplekin, which binds CstF64. The Symplekin/CstF64 interaction is likely specific for histone pre-mRNA processing, since CstF64 forms a complex with CstF77 and CstF50 that functions in polyadenylation using the same region it uses to interact with Symplekin.Citation115,116 U7 snRNP isolated from nuclear extracts contains not only the core Sm proteins, and FLASH, but also the HCC components, suggesting that the U7 snRNP is already associated with the HCC when it binds to the histone pre-RNA. Since U7 snRNP is present in very small amounts in mammalian or Drosophila cells, only a small fraction (<1%) of the total CPSF73, CPSF100, Symplekin and CstF64 are present in the HLB even in S phase.Citation100,101 We have found that Symplekin is concentrated in the HLB primarily in S phase,Citation52 while U7 snRNP is there constitutively. This result suggests that the active form of U7 snRNP, containing the HCC, may be assembled in the HLB during S phase as part of the mechanism of activating histone pre-mRNA processing ().
The recruitment of the HCC to the HLB suggests a regulatory step may drive the assembly of the active U7 snRNP and/or cause other important molecular changes within the HLB. Mammalian YARP, the Drosophila homolog of Mute, and FLASH each interact with the C-terminus of NPAT using a Myb-like domain, and this domain from either protein can localize a heterologous protein to the HLB.Citation44 The C-terminus of NPAT can bind either YARP or FLASH but not both at once. Detailed in vitro binding studies demonstrated that the C-terminal Myb-like domain of FLASH also binds to Lsm11 adjacent to the binding site on Lsm11 for the N-terminus of FLASH, and that this interaction prevents FLASH binding to NPAT.Citation99 Thus, it is tempting to speculate that reorganization of the Lsm11-FLASH-NPAT interactions in the HLB may be part of U7 snRNP activation and the onset of RD histone pre-mRNA processing during S phase. This idea also suggests that different pools of molecules may exist within the HLB. For example, some molecules of FLASH may interact with Mxc, and may be involved in scaffolding the HLB, while others could interact directly with U7 snRNP and not Mxc and participate in processing. The size of these pools could be dynamically regulated by Cyclin E/Cdk2 activity during S phase. The application of super high resolution microscopy might provide an avenue to begin exploring these issues.
Concluding remarks
Does HLB function matter for organism development or homeostasis? Null mutations of core HLB components (e.g., FLASH and NPAT/Mxc) in mammals and flies are lethal early in development,Citation35,36,117 likely in large part because little to no histone gene expression occurs. Because proteins like NPAT/Mxc and FLASH are necessary to scaffold HLB assembly and to stimulate histone mRNA biosynthesis, both of these functions will be lost in the absence of these proteins making it difficult to assign functional roles to HLB assembly per se. An important point to emphasize is that HLB assembly need not be absolutely essential, or may only be essential in specific cells when there is a particularly high demand for histone mRNA synthesis. For example, Coilin depletion is lethal in zebrafish due to the need for rapid assembly of snRNPs in embryogenesis,Citation97 whereas Coilin mutation and the loss of Cajal Bodies in flies is not lethal and doesn't impact scaRNA modification, which is thought to occur within Cajal bodies.Citation118 In Drosophila the maximal requirement for histone mRNA synthesis is likely to be during oogenesis when large amounts of maternal histone mRNA and protein are deposited into the egg. Several mutations that affect the HLB and histone pre-mRNA processing result in female sterility,Citation32,72,105,107 suggesting that the HLB may be particularly important during oogenesis, either in proper development of the eggCitation107 or in production of sufficient maternal histone mRNA for the embryo.Citation105,108
Interestingly, one well-studied organism, C. elegans, apparently lacks an HLB, although their histone mRNAs end in a stem loop and associate with SLBP.Citation119 In C. elegans the histone mRNA 3′ end is formed by cleavage of a longer polyadenylated precursor using small RNAs (analogous to RNAi).Citation120 Furthermore, the C. elegans histone genes are not tandemly arrayed,Citation121 but the 13–14 copies of each core histone gene are found in many small clusters spread over different chromosomes.Citation122 Moreover, homologs of U7 snRNP and FLASH are not identifiable in the C. elegans genome (Z. Dominski, personal communication). Perhaps these worms have lost the HLB because there is not a need to concentrate low abundance factors like U7 snRNP and FLASH for RD histone pre-mRNA processing due to the alternate cleavage mechanism.
Thus HLB assembly may provide a boost to efficiency of histone mRNA biosynthesis, rather than being absolutely essential. Such a boost in efficiency could provide an advantage that is selected during the evolution of particular animal lineages. This interesting question may be applicable to other nuclear bodies, and addressing it will require the further application of genetics, cell biology, and biochemistry that has been so important to our current understanding of HLB assembly and function.
Disclosure of potential conflicts of interest
The authors have no potential conflicts of interest.
Acknowledgments
We thank Dan McKay, Deirdre Tatomer, Leila Rieder, and Erica Larschan for critical reading of the manuscript, and Katy Curry for critical comments and assembling . HLB studies in the Duronio and Marzluff labs are funded by the NIH (GM058921).
References
- McKay DJ, Klusza S, Penke TJ, Meers MP, Curry KP, McDaniel SL, Malek PY, Cooper SW, Tatomer DC, Lieb JD, et al. Interrogating the function of metazoan histones using engineered gene clusters. Dev Cell 2015; 32:373-86; PMID:25669886; https://doi.org/10.1016/j.devcel.2014.12.025
- Lifton RP, Goldberg ML, Karp RW, Hogness DS. The organization of the histone genes in Drosophila melanogaster: functional and evolutionary implications. Cold Spring Harb Symp Quant Biol 1978; 42:1047-51; PMID:98262; https://doi.org/10.1101/SQB.1978.042.01.105
- Schaffner W, Gross K, Telford J, Birnstiel M. Molecular analysis of the histone gene cluster of psammechinus miliaris: II. The arrangement of the five histone-coding and spacer sequences. Cell 1976; 8:471-8; PMID:954100; https://doi.org/10.1016/0092-8674(76)90214-2
- Cohn RH, Lowry JC, Kedes LH. Histone genes of the sea urchin (S. purpuratus) cloned in E coli: order, polarity, and strandedness of the five histone-coding and spacer regions. Cell 1976; 9:147-61; PMID:788917; https://doi.org/10.1016/0092-8674(76)90060-X
- Marzluff WF, Gongidi P, Woods KR, Jin J, Maltais LJ. The human and mouse replication-dependent histone genes. Genomics 2002; 80:487-98; PMID:12408966; https://doi.org/10.1006/geno.2002.6850
- Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat Rev Genet 2008; 9:843-54; PMID:18927579; https://doi.org/10.1038/nrg2438
- Romeo V, Schumperli D. Cycling in the nucleus: regulation of RNA 3′ processing and nuclear organization of replication-dependent histone genes. Curr Opin Cell Biol 2016; 40:23-31; PMID:26895140; https://doi.org/10.1016/j.ceb.2016.01.015
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA: getting closer to the end. Gene 2007; 396:373-90; PMID:17531405; https://doi.org/10.1016/j.gene.2007.04.021
- Cotten M, Gick O, Vasserot A, Schaffner G, Birnstiel ML. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3′ RNA processing reaction. Embo J 1988; 7:801-8; PMID:3396543
- Soldati D, Schumperli D. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol Cell Biol 1988; 8:1518-24; PMID:3380087; https://doi.org/10.1128/MCB.8.4.1518
- Schaufele F, Gilmartin GM, Bannwarth W, Birnstiel ML. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature 1986; 323:777-81; PMID:3022153; https://doi.org/10.1038/323777a0
- Mowry KL, Steitz JA. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 1987; 238:1682-7; PMID:2825355; https://doi.org/10.1126/science.2825355
- Wu CH, Murphy C, Gall JG. The Sm binding site targets U7 snRNA to coiled bodies (spheres) of amphibian oocytes. Rna 1996; 2:811-23; PMID:8752090
- Wu CH, Gall JG. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci U S A 1993; 90:6257-9; PMID:8327506; https://doi.org/10.1073/pnas.90.13.6257
- Abbott J, Marzluff WF, Gall JG. The stem-loop binding protein (SLBP1) is present in coiled bodies of the Xenopus germinal vesicle. Mol Biol Cell 1999; 10:487-99; PMID:9950690; https://doi.org/10.1091/mbc.10.2.487
- Frey MR, Matera AG. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc Natl Acad Sci U S A 1995; 92:5915-9; PMID:7597053; https://doi.org/10.1073/pnas.92.13.5915
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 2000; 16:273-300; PMID:11031238; https://doi.org/10.1146/annurev.cellbio.16.1.273
- Liu JL, Murphy C, Buszczak M, Clatterbuck S, Goodman R, Gall JG. The Drosophila melanogaster Cajal body. J Cell Biol 2006; 172:875-84; PMID:16533947; https://doi.org/10.1083/jcb.200511038
- Weichenrieder O. RNA binding by Hfq and ring-forming (L)Sm proteins: a trade-off between optimal sequence readout and RNA backbone conformation. RNA Biol 2014; 11:537-49; PMID:24828406; https://doi.org/10.4161/rna.29144
- Pillai RS, Grimmler M, Meister G, Will CL, Luhrmann R, Fischer U, Schümperli D. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev 2003; 17:2321-33; PMID:12975319; https://doi.org/10.1101/gad.274403
- Pillai RS, Will CL, Luhrmann R, Schumperli D, Muller B. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. Embo J 2001; 20:5470-9; PMID:11574479; https://doi.org/10.1093/emboj/20.19.5470
- Liu JL, Wu Z, Nizami Z, Deryusheva S, Rajendra TK, Beumer KJ, Gao H, Matera AG, Carroll D, Gall JG. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell 2009; 20:1661-70; PMID:19158395; https://doi.org/10.1091/mbc.E08-05-0525
- Bongiorno-Borbone L, De Cola A, Vernole P, Finos L, Barcaroli D, Knight RA, Melino G, De Laurenzi V. FLASH and NPAT positive but not Coilin positive Cajal Bodies correlate with cell ploidy. Cell Cycle 2008; 7:2357-67; PMID:18677100; https://doi.org/10.4161/cc.6344
- Alm-Kristiansen AH, Saether T, Matre V, Gilfillan S, Dahle O, Gabrielsen OS. FLASH acts as a co-activator of the transcription factor c-Myb and localizes to active RNA polymerase II foci. Oncogene 2008; 27:4644-56; PMID:18408764; https://doi.org/10.1038/onc.2008.105
- Nizami ZF, Deryusheva S, Gall JG. Cajal bodies and histone locus bodies in Drosophila and Xenopus. Cold Spring Harb Symp Quant Biol 2010; 75:313-20; PMID:21047905; https://doi.org/10.1101/sqb.2010.75.005
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle-regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in cajal bodies promotes histone gene transcription. Genes Dev 2000; 14:2298-313; PMID:10995387; https://doi.org/10.1101/gad.829500
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-Cdk2 to the regulation of replication-dependent histone gene transcription. Genes Dev 2000; 14:2283-97; PMID:10995386; https://doi.org/10.1101/gad.827700
- Barcaroli D, Dinsdale D, Neale MH, Bongiorno-Borbone L, Ranalli M, Munarriz E, Sayan AE, McWilliam JM, Smith TM, Fava E, et al. FLASH is an essential component of Cajal bodies. Proc Natl Acad Sci U S A 2006; 103:14802-7; PMID:17003126; https://doi.org/10.1073/pnas.0604225103
- Kiriyama M, Kobayashi Y, Saito M, Ishikawa F, Yonehara S. Interaction of FLASH with arsenite resistance protein 2 is involved in cell cycle progression at S phase. Mol Cell Biol 2009; 29:4729-41; PMID:19546234; https://doi.org/10.1128/MCB.00289-09
- Bongiorno-Borbone L, De Cola A, Barcaroli D, Knight RA, Di Ilio C, Melino G, De Laurenzi V. FLASH degradation in response to UV-C results in histone locus bodies disruption and cell-cycle arrest. Oncogene 2010; 29:802-10; PMID:19915611; https://doi.org/10.1038/onc.2009.388
- Zhao J, Dynlacht B, Imai T, Hori T, Harlow E. Expression of NPAT, a novel substrate of cyclin E-CDK2, promotes S- phase entry. Genes Dev 1998; 12:456-61; PMID:9472014; https://doi.org/10.1101/gad.12.4.456
- White AE, Burch BD, Yang XC, Gasdaska PY, Dominski Z, Marzluff WF, Duronio RJ. Drosophila histone locus bodies form by hierarchical recruitment of components. J Cell Biol 2011; 193:677-94; PMID:21576393; https://doi.org/10.1083/jcb.201012077
- Wei Y, Jin J, Harper JW. The Cyclin E/Cdk2 Substrate and Cajal Body Component p220(NPAT) Activates Histone Transcription through a Novel LisH-Like Domain. Mol Cell Biol 2003; 23:3669-80; PMID:12724424; https://doi.org/10.1128/MCB.23.10.3669-3680.2003
- Ye X, Wei Y, Nalepa G, Harper JW. The cyclin E/Cdk2 substrate p220(NPAT) is required for S-phase entry, histone gene expression, and Cajal body maintenance in human somatic cells. Mol Cell Biol 2003; 23:8586-600; PMID:14612403; https://doi.org/10.1128/MCB.23.23.8586-8600.2003
- Di Fruscio M, Weiher H, Vanderhyden BC, Imai T, Shiomi T, Hori TA, Jaenisch R, Gray DA. Proviral inactivation of the Npat gene of Mpv 20 mice results in early embryonic arrest. Mol Cell Biol 1997; 17:4080-6; PMID:9199343; https://doi.org/10.1128/MCB.17.7.4080
- Santamaria P, Randsholt NB. Characterization of a region of the X chromosome of Drosophila including multi sex combs (mxc), a Polycomb group gene which also functions as a tumour suppressor. Mol Gen Genet 1995; 246:282-90; PMID:7854313; https://doi.org/10.1007/BF00288600
- Saget O, Forquignon F, Santamaria P, Randsholt NB. Needs and targets for the multi sex combs gene product in Drosophila melanogaster. Genetics 1998; 149:1823-38; PMID:9691040
- Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes Dev 1998; 12:734-44; PMID:9499407; https://doi.org/10.1101/gad.12.5.734
- White AE, Leslie ME, Calvi BR, Marzluff WF, Duronio RJ. Developmental and cell cycle regulation of the Drosophila histone locus body. Mol Biol Cell 2007; 18:2491-502; PMID:17442888; https://doi.org/10.1091/mbc.E06-11-1033
- Lanzotti DJ, Kupsco JM, Marzluff WF, Duronio RJ. string(cdc25) and cyclin E are required for patterned histone expression at different stages of Drosophila embryonic development. Dev Biol 2004; 274:82-93; PMID:15355790; https://doi.org/10.1016/j.ydbio.2004.06.019
- Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De Laurenzi V. FLASH is required for histone transcription and S-phase progression. Proc Natl Acad Sci U S A 2006; 103:14808-12; PMID:17003125; https://doi.org/10.1073/pnas.0604227103
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell 2009; 36:267-78; PMID:19854135; https://doi.org/10.1016/j.molcel.2009.08.016
- Bulchand S, Menon SD, George SE, Chia W. Muscle wasted: a novel component of the Drosophila histone locus body required for muscle integrity. J Cell Sci 2010; 123:2697-707; PMID:20647374; https://doi.org/10.1242/jcs.063172
- Yang XC, Sabath I, Kunduru L, van Wijnen AJ, Marzluff WF, Dominski Z. A conserved interaction that is essential for the biogenesis of histone locus bodies. J Biol Chem 2014; 289:33767-82; PMID:25339177; https://doi.org/10.1074/jbc.M114.616466
- Dominski Z, Yang XC, Marzluff WF. The Polyadenylation Factor CPSF-73 Is Involved in Histone-Pre-mRNA Processing. Cell 2005; 123:37-48; PMID:16213211; https://doi.org/10.1016/j.cell.2005.08.002
- Kolev NG, Steitz JA. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev 2005; 19:2583-92; PMID:16230528; https://doi.org/10.1101/gad.1371105
- Wagner EJ, Burch BD, Godfrey AC, Salzler HR, Duronio RJ, Marzluff WF. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol Cell 2007; 28:692-9; PMID:18042462; https://doi.org/10.1016/j.molcel.2007.10.009
- Kolev NG, Yario TA, Benson E, Steitz JA. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep 2008; 9:1013-8; PMID:18688255; https://doi.org/10.1038/embor.2008.146
- Guglielmi B, La Rochelle N, Tjian R. Gene-specific transcriptional mechanisms at the histone gene cluster revealed by single-cell imaging. Mol Cell 2013; 51:480-92; PMID:23973376; https://doi.org/10.1016/j.molcel.2013.08.009
- Isogai Y, Keles S, Prestel M, Hochheimer A, Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes Dev 2007; 21:2936-49; PMID:17978101; https://doi.org/10.1101/gad.1608807
- Daneshvar K, Khan A, Goodliffe JM. Myc localizes to histone locus bodies during replication in Drosophila. PLoS One 2011; 6:e23928; PMID:21886841; https://doi.org/10.1371/journal.pone.0023928
- Tatomer DC, Rizzardi LF, Curry KP, Witkowski AM, Marzluff WF, Duronio RJ. Drosophila Symplekin localizes dynamically to the histone locus body and tricellular junctions. Nucleus 2014; 5:613-25; PMID:25493544; https://doi.org/10.4161/19491034.2014.990860
- Wagner EJ, Ospina JK, Hu Y, Dundr M, Matera AG, Marzluff WF. Conserved zinc fingers mediate multiple functions of ZFP100, a U7snRNP associated protein. RNA 2006; 12:1206-18; PMID:16714279; https://doi.org/10.1261/rna.2606
- Dundr M, Hebert MD, Karpova TS, Stanek D, Xu H, Shpargel KB, Meier UT, Neugebauer KM, Matera AG, Misteli T. In vivo kinetics of Cajal body components. J Cell Biol 2004; 164:831-42; PMID:15024031; https://doi.org/10.1083/jcb.200311121
- Ling Zheng L, Wang FY, Cong XX, Shen Y, Rao XS, Huang DS, Fan W, Yi P, Wang XB, Zheng L, et al. Interaction of heat shock protein Cpn10 with the Cyclin E/Cdk2 substrate Nuclear Protein Ataxia-Telangiectasia (NPAT) is involved in regulating histone transcription. J Biol Chem 2015; 290:29290-300; PMID:26429916; https://doi.org/10.1074/jbc.M115.659201
- Katsuyama T, Sugawara T, Tatsumi M, Oshima Y, Gehring WJ, Aigaki T, Kurata S. Involvement of winged eye encoding a chromatin-associated bromo-adjacent homology domain protein in disc specification. Proc Natl Acad Sci U S A 2005; 102:15918-23; PMID:16247005; https://doi.org/10.1073/pnas.0507945102
- Ozawa N, Furuhashi H, Masuko K, Numao E, Makino T, Yano T, Kurata S. Organ identity specification factor WGE localizes to the histone locus body and regulates histone expression to ensure genomic stability in Drosophila. Genes Cells 2016; 21:442-56; PMID:27145109; https://doi.org/10.1111/gtc.12354
- Berloco M, Fanti L, Breiling A, Orlando V, Pimpinelli S. The maternal effect gene, abnormal oocyte (abo), of Drosophila melanogaster encodes a specific negative regulator of histones. Proc Natl Acad Sci U S A 2001; 98:12126-31; PMID:11593026; https://doi.org/10.1073/pnas.211428798
- Machyna M, Heyn P, Neugebauer KM. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 2013; 4:17-34; PMID:23042601; https://doi.org/10.1002/wrna.1139
- Misteli T. The concept of self-organization in cellular architecture. J Cell Biol 2001; 155:181-5; PMID:11604416; https://doi.org/10.1083/jcb.200108110
- Matera AG, Izaguire-Sierra M, Praveen K, Rajendra TK. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell 2009; 17:639-47; PMID:19922869; https://doi.org/10.1016/j.devcel.2009.10.017
- Dundr M. Seed and grow: a two-step model for nuclear body biogenesis. J Cell Biol 2011; 193:605-6; PMID:21576389; https://doi.org/10.1083/jcb.201104087
- Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol 2010; 2:a000711; PMID:21068152; https://doi.org/10.1101/cshperspect.a000711
- Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol 2011; 13:167-73; PMID:21240286; https://doi.org/10.1038/ncb2157
- Mao YS, Sunwoo H, Zhang B, Spector DL. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol 2011; 13:95-101; PMID:21170033; https://doi.org/10.1038/ncb2140
- Grob A, Colleran C, McStay B. Construction of synthetic nucleoli in human cells reveals how a major functional nuclear domain is formed and propagated through cell division. Genes Dev 2014; 28:220-30; PMID:24449107; https://doi.org/10.1101/gad.234591.113
- Falahati H, Pelham-Webb B, Blythe S, Wieschaus E. Nucleation by rRNA Dictates the Precision of Nucleolus Assembly. Curr Biol 2016; 26:277-85; PMID:26776729; https://doi.org/10.1016/j.cub.2015.11.065
- Zhu L, Brangwynne CP. Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol 2015; 34:23-30; PMID:25942753; https://doi.org/10.1016/j.ceb.2015.04.003
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell 2000; 11:4159-71; PMID:11102515; https://doi.org/10.1091/mbc.11.12.4159
- Terzo EA, Lyons SM, Poulton JS, Temple BR, Marzluff WF, Duronio RJ. Distinct self-interaction domains promote Multi Sex Combs accumulation in and formation of the Drosophila histone locus body. Mol Biol Cell 2015; 26:1559-74; PMID:25694448; https://doi.org/10.1091/mbc.E14-10-1445
- Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science 2008; 322:1713-7; PMID:18948503; https://doi.org/10.1126/science.1165216
- Tatomer DC, Terzo E, Curry KP, Salzler H, Sabath I, Zapotoczny G, McKay DJ, Dominski Z, Marzluff WF, Duronio RJ. Concentrating pre-mRNA processing factors in the histone locus body facilitates efficient histone mRNA biogenesis. J Cell Biol 2016; 213:557-70; PMID:27241916; https://doi.org/10.1083/jcb.201504043
- Burch BD, Godfrey AC, Gasdaska PY, Salzler HR, Duronio RJ, Marzluff WF, Dominski Z. Interaction between FLASH and Lsm11 is essential for histone pre-mRNA processing in vivo in Drosophila. RNA 2011; 17:1132-47; PMID:21525146; https://doi.org/10.1261/rna.2566811
- Salzler HR, Tatomer DC, Malek PY, McDaniel SL, Orlando AN, Marzluff WF, Duronio RJ. A sequence in the Drosophila H3-H4 Promoter triggers histone locus body assembly and biosynthesis of replication-coupled histone mRNAs. Dev Cell 2013; 24:623-34; PMID:23537633; https://doi.org/10.1016/j.devcel.2013.02.014
- Lyons SM, Cunningham CH, Welch JD, Groh B, Guo AY, Wei B, Whitfield ML, Xiong Y, Marzluff WF. A subset of replication-dependent histone mRNAs are expressed as polyadenylated RNAs in terminally differentiated tissues. Nucleic Acids Res 2016; 44:9190-9205; PMID:27402160; https://doi.org/10.1093/nar/gkw620
- Ito S, Fujiyama-Nakamura S, Kimura S, Lim J, Kamoshida Y, Shiozaki-Sato Y, Sawatsubashi S, Suzuki E, Tanabe M, Ueda T, et al. Epigenetic silencing of core histone genes by HERS in Drosophila. Mol Cell 2012; 45:494-504; PMID:22365829; https://doi.org/10.1016/j.molcel.2011.12.029
- Mahajan K, Fang B, Koomen JM, Mahajan NP. H2B Tyr37 phosphorylation suppresses expression of replication-dependent core histone genes. Nat Struct Mol Biol 2012; 19:930-7; PMID:22885324; https://doi.org/10.1038/nsmb.2356
- Miele A, Braastad CD, Holmes WF, Mitra P, Medina R, Xie R, Zaidi SK, Ye X, Wei Y, Harper JW, et al. HiNF-P directly links the cyclin E/CDK2/p220NPAT pathway to histone H4 gene regulation at the G1/S phase cell cycle transition. Mol Cell Biol 2005; 25:6140-53; PMID:15988025; https://doi.org/10.1128/MCB.25.14.6140-6153.2005
- Mitra P, Xie RL, Medina R, Hovhannisyan H, Zaidi SK, Wei Y, Harper JW, Stein JL, van Wijnen AJ, Stein GS. Identification of HiNF-P, a key activator of cell cycle-controlled histone H4 genes at the onset of S phase. Mol Cell Biol 2003; 23:8110-23; PMID:14585971; https://doi.org/10.1128/MCB.23.22.8110-8123.2003
- Ghule PN, Xie RL, Colby JL, Rivera-Perez JA, Jones SN, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Maternal expression and early induction of histone gene transcription factor Hinfp sustains development in pre-implantation embryos. Dev Biol 2016; 419:311-320; PMID:27609454; https://doi.org/10.1016/j.ydbio.2016.09.003
- Ghule PN, Xie RL, Colby JL, Jones SN, Lian JB, Wijnen AJ, Stein JL, Stein GS. p53 checkpoint ablation exacerbates the phenotype of Hinfp dependent histone H4 deficiency. Cell Cycle 2015; 14:2501-8; PMID:26030398; https://doi.org/10.1080/15384101.2015.1049783
- Ghule PN, Xie RL, Medina R, Colby JL, Jones SN, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Fidelity of histone gene regulation is obligatory for genome replication and stability. Mol Cell Biol 2014; 34:2650-9; PMID:24797072; https://doi.org/10.1128/MCB.01567-13
- Ghule PN, Dominski Z, Yang XC, Marzluff WF, Becker KA, Harper JW, Lian JB, Stein JL, van Wijnen AJ, Stein GS. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc Natl Acad Sci U S A 2008; 105:16964-9; PMID:18957539; https://doi.org/10.1073/pnas.0809273105
- Grob A, McStay B. Construction of synthetic nucleoli and what it tells us about propagation of sub-nuclear domains through cell division. Cell Cycle 2014; 13:2501-8; PMID:25486191; https://doi.org/10.4161/15384101.2014.949124
- Bergeron-Sandoval LP, Safaee N, Michnick SW. Mechanisms and Consequences of Macromolecular Phase Separation. Cell 2016; 165:1067-79; PMID:27203111; https://doi.org/10.1016/j.cell.2016.05.026
- Banani SF, Rice AM, Peeples WB, Lin Y, Jain S, Parker R, Rosen MK. Compositional control of phase-separated cellular bodies. Cell 2016, 166:651-63; PMID:27374333; https://doi.org/10.1016/j.cell.2016.06.010
- Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J 2016; 35:1603-12; PMID:27357569; https://doi.org/10.15252/embj.201593517
- Toretsky JA, Wright PE. Assemblages: functional units formed by cellular phase separation. J Cell Biol 2014; 206:579-88; PMID:25179628; https://doi.org/10.1083/jcb.201404124
- Malinovska L, Kroschwald S, Alberti S. Protein disorder, prion propensities, and self-organizing macromolecular collectives. Biochim Biophys Acta 2013; 1834:918-31; PMID:23328411; https://doi.org/10.1016/j.bbapap.2013.01.003
- Lin Y, Protter DS, Rosen MK, Parker R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 2015; 60:208-19; PMID:26412307; https://doi.org/10.1016/j.molcel.2015.08.018
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 2015; 162:1066-77; PMID:26317470; https://doi.org/10.1016/j.cell.2015.07.047
- Kwon I, Kato M, Xiang S, Wu L, Theodoropoulos P, Mirzaei H, Han T, Xie S, Corden JL, McKnight SL. Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 2013; 155:1049-60; PMID:24267890; https://doi.org/10.1016/j.cell.2013.10.033
- Altmeyer M, Neelsen KJ, Teloni F, Pozdnyakova I, Pellegrino S, Grofte M, Rask MB, Streicher W, Jungmichel S, Nielsen ML, et al. Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat Commun 2015; 6:8088; PMID:26286827; https://doi.org/10.1038/ncomms9088
- Kotova E, Jarnik M, Tulin AV. Poly (ADP-ribose) polymerase 1 is required for protein localization to Cajal body. PLoS Genet 2009; 5:e1000387; PMID:19229318; https://doi.org/10.1371/journal.pgen.1000387
- Tucker KE, Massello LK, Gao L, Barber TJ, Hebert MD, Chan EK, Matera AG. Structure and characterization of the murine p80 coilin gene, Coil. J Struct Biol 2000; 129:269-77; PMID:10806077; https://doi.org/10.1006/jsbi.2000.4234
- Sawyer IA, Dundr M. Nuclear bodies: Built to boost. J Cell Biol 2016; 213:509-11; PMID:27241912; https://doi.org/10.1083/jcb.201605049
- Strzelecka M, Trowitzsch S, Weber G, Luhrmann R, Oates AC, Neugebauer KM. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol 2010; 17:403-9; PMID:20357773; https://doi.org/10.1038/nsmb.1783
- Novotny I, Blazikova M, Stanek D, Herman P, Malinsky J. In vivo kinetics of U4/U6.U5 tri-snRNP formation in Cajal bodies. Mol Biol Cell 2011; 22:513-23; PMID:21177826; https://doi.org/10.1091/mbc.E10-07-0560
- Skrajna A, Yang XC, Tarnowski K, Fituch K, Marzluff WF, Dominski Z, Dadlez M. Mapping the interaction network of key proteins involved in histone mRNA generation: a hydrogen/deuterium exchange study. J Mol Biol 2016; 428:1180-96; PMID:26860583; https://doi.org/10.1016/j.jmb.2016.01.031
- Yang XC, Sabath I, Debski J, Kaus-Drobek M, Dadlez M, Marzluff WF, Dominski Z. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3′-end processing. Mol Cell Biol 2013; 33:28-37; PMID:23071092; https://doi.org/10.1128/MCB.00653-12
- Sabath I, Skrajna A, Yang XC, Dadlez M, Marzluff WF, Dominski Z. 3′-End processing of histone pre-mRNAs in Drosophila: U7 snRNP is associated with FLASH and polyadenylation factors. RNA 2013; 19:1726-44; PMID:24145821; https://doi.org/10.1261/rna.040360.113
- Anamika K, Gyenis A, Poidevin L, Poch O, Tora L. RNA polymerase II pausing downstream of core histone genes is different from genes producing polyadenylated transcripts. PLoS One 2012; 7:e38769; PMID:22701709; https://doi.org/10.1371/journal.pone.0038769
- Cheng B, Li T, Rahl PB, Adamson TE, Loudas NB, Guo J, Varzavand K, Cooper JJ, Hu X, Gnatt A, et al. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol Cell 2012; 45:38-50; PMID:22244331; https://doi.org/10.1016/j.molcel.2011.10.022
- Adamson TE, Price DH. Cotranscriptional processing of Drosophila histone mRNAs. Mol Cell Biol 2003; 23:4046-55; PMID:12773550; https://doi.org/10.1128/MCB.23.12.4046-4055.2003
- Sullivan E, Santiago C, Parker ED, Dominski Z, Yang X, Lanzotti DJ, Ingledue TC, Marzluff WF, Duronio RJ. Drosophila stem loop binding protein coordinates accumulation of mature histone mRNA with cell cycle progression. Genes Dev 2001; 15:173-87; PMID:11157774; https://doi.org/10.1101/gad.862801
- Godfrey AC, White AE, Tatomer DC, Marzluff WF, Duronio RJ. The Drosophila U7 snRNP proteins Lsm10 and Lsm11 are required for histone pre-mRNA processing and play an essential role in development. RNA 2009; 15:1661-72; PMID:19620235; https://doi.org/10.1261/rna.1518009
- Godfrey AC, Kupsco JM, Burch BD, Zimmerman RM, Dominski Z, Marzluff WF, Duronio RJ. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA 2006; 12:396-409; PMID:16495235; https://doi.org/10.1261/rna.2270406
- Lanzotti DJ, Kaygun H, Yang X, Duronio RJ, Marzluff WF. Developmental control of histone mRNA and dSLBP synthesis during Drosophila embryogenesis and the role of dSLBP in histone mRNA 3′ end processing in vivo. Mol Cell Biol 2002; 22:2267-82; PMID:11884612; https://doi.org/10.1128/MCB.22.7.2267-2282.2002
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell 2007; 26:349-65; PMID:17499042; https://doi.org/10.1016/j.molcel.2007.04.011
- Pirngruber J, Shchebet A, Schreiber L, Shema E, Minsky N, Chapman RD, Eick D, Aylon Y, Oren M, Johnsen SA. CDK9 directs H2B monoubiquitination and controls replication-dependent histone mRNA 3′-end processing. EMBO Rep 2009; 10:894-900; PMID:19575011; https://doi.org/10.1038/embor.2009.108
- Gruber JJ, Olejniczak SH, Yong J, La Rocca G, Dreyfuss G, Thompson CB. Ars2 promotes proper replication-dependent histone mRNA 3′ end formation. Mol Cell 2012; 45:87-98; PMID:22244333; https://doi.org/10.1016/j.molcel.2011.12.020
- Kohn M, Ihling C, Sinz A, Krohn K, Huttelmaier S. The Y3** ncRNA promotes the 3′ end processing of histone mRNAs. Genes Dev 2015; 29:1998-2003; PMID:26443846; https://doi.org/10.1101/gad.266486.115
- Lee MC, Toh LL, Yaw LP, Luo Y. Drosophila octamer elements and Pdm-1 dictate the coordinated transcription of core histone genes. J Biol Chem 2010; 285:9041-53; PMID:20097756; https://doi.org/10.1074/jbc.M109.075358
- Zheng L, Roeder RG, Luo Y. S phase activation of the histone H2B promoter by OCA-S, a coactivator complex that contains GAPDH as a key component. Cell 2003; 114:255-66; PMID:12887926; https://doi.org/10.1016/S0092-8674(03)00552-X
- Ruepp MD, Schweingruber C, Kleinschmidt N, Schumperli D. Interactions of CstF-64, CstF-77, and symplekin: implications on localisation and function. Mol Biol Cell 2011; 22:91-104; PMID:21119002; https://doi.org/10.1091/mbc.E10-06-0543
- Takagaki Y, Manley JL. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol Cell Biol 2000; 20:1515-25; PMID:10669729; https://doi.org/10.1128/MCB.20.5.1515-1525.2000
- De Cola A, Bongiorno-Borbone L, Bianchi E, Barcaroli D, Carletti E, Knight RA, Di Ilio C, Melino G, Sette C, De Laurenzi V. FLASH is essential during early embryogenesis and cooperates with p73 to regulate histone gene transcription. Oncogene 2012; 31:573-82; PMID:21725362; https://doi.org/10.1038/onc.2011.274
- Deryusheva S, Gall JG. Small Cajal body-specific RNAs of Drosophila function in the absence of Cajal bodies. Mol Biol Cell 2009; 20:5250-9; PMID:19846657; https://doi.org/10.1091/mbc.E09-09-0777
- Kodama Y, Rothman JH, Sugimoto A, Yamamoto M. The stem-loop binding protein CDL-1 is required for chromosome condensation, progression of cell death and morphogenesis in Caenorhabditis elegans. Development 2002; 129:187-96; PMID:11782412
- Avgousti DC, Palani S, Sherman Y, Grishok A. CSR-1 RNAi pathway positively regulates histone expression in C. elegans. EMBO J 2012; 31:3821-32; PMID:22863779; https://doi.org/10.1038/emboj.2012.216
- Roberts SB, Sanicola M, Emmons SW, Childs G. Molecular characterization of the histone gene family of Caenorhabditis elegans. J Mol Biol 1987; 196:27-38; PMID:3656446; https://doi.org/10.1016/0022-2836(87)90508-0
- Keall R, Whitelaw S, Pettitt J, Muller B. Histone gene expression and histone mRNA 3′ end structure in Caenorhabditis elegans. BMC Mol Biol 2007; 8:51; PMID:17570845; https://doi.org/10.1186/1471-2199-8-51
- Strzelecka M, Oates AC, Neugebauer KM. Dynamic control of Cajal body number during zebrafish embryogenesis. Nucleus 2010; 1:96-108; PMID:21327108; https://doi.org/10.4161/nucl.1.1.10680
- Zhang J, Zhang F, Zheng X. Depletion of hCINAP by RNA interference causes defects in Cajal body formation, histone transcription, and cell viability. Cell Mol Life Sci 2010; 67:1907-18; PMID:20186459; https://doi.org/10.1007/s00018-010-0301-2
