ABSTRACT
Seven-deazapurine modifications were thought to be highly specific of tRNAs, but have now been discovered in DNA of phages and of phylogenetically diverse bacteria, illustrating the plasticity of these modification pathways. The intermediate 7-cyano-7-deazaguanine (preQ0) is a shared precursor in the pathways leading to the insetion of 7-deazapurine derivatives in both tRNA and DNA. It is also used as an intermediate in the synthesis of secondary metabolites such as toyocamacin. The presence of 7-deazapurine in DNA is proposed to be a protection mechanism against endonucleases. This makes preQ0 a metabolite of underappreaciated but central importance.
KEYWORDS:
Introduction
DNA and RNA, the two cellular information polymers are made of very similar building blocks: nucleotides composed of a nitrogenous base, a five carbon sugar and a phosphate group. The seemingly small differences between the sugars (ribose in RNA and 2′-deoxyribose in DNA) and one of the four bases (thymine (T) in DNA and uracil (U) in RNA) do however dramatically change the properties of the two molecules. DNA is more stable as a double stranded helix, and has been recruited as the core matrix of genetic information. RNA is generally single stranded and more flexible, harboring different shapes and functions in the cell. Historically, the fields of DNA and RNA research had been kept quite separate with the two communities rarely interacting, but this has recently changed with 1) the popularity of the RNA world hypothesis, and the realization that DNA is a modified form of RNA; 2) the growing interest in RNA modifications that has revealed how similar many of the RNA and DNA modification machineries are (see reference [1] for a detailed discussion).
While the first nucleoside modifications were found in genomic DNA, with the discovery of 5-methyl-deoxycytosine (m5dC or 5mC) in Mycobacteria in 1925,Citation1 they are more diverse and chemically complex in RNA. Around twenty modifications have been found to date in DNA.Citation2-3 Most of the known genomic DNA modifications are methylations, although more complex modifications do exist, particularly in phages.Citation2-4 In contrast, more than one hundred modifications have been found in RNA,Citation3 and some of these modifications are very complex, e.g., wybutosine which requires five enzymes for its synthesis.Citation5 Modifications are mainly found in stable RNAs such as transfer-RNAs (tRNAs) and ribosomal-RNA (rRNAs),Citation3 and a few are also found in messenger-RNA (mRNAs).Citation6-7 The molecular and biochemical characterization of RNA modifying enzymes has revealed multiple cases of broad specificity for various RNA substrates. For example, some pseudouridine synthases have been shown to modify both mRNA and tRNA substrates,Citation8 whereas methylases of the RlmD family methylate rRNAs in some organisms and tRNAs in others.Citation9
More recently, examples of crosstalks have been observed between the RNA and DNA modification machineries. It is the case in the eukaryotic DNMT family of methyltransferases that is subdivided in three groups: DNMT1, 2 and 3. DNMT1 and DNMT3 methylate the carbon 5 of cytosines in DNA CpG sequences, playing key roles in epigenetic regulation.Citation10 DNMT2, despite its high sequence similarity to DNMT1 and 3, modifies specific tRNAs, e.g., tRNAAspGUC, at the position C38.Citation11 Crosstalks also exist in the APOBEC family of proteins that deaminate cytosines into uridines. Although the first proteins discovered in the APOBEC family were mRNA modifying proteins, almost all members of the APOBEC family also modify single stranded DNA.Citation12 Similarly, some members of the ADAT family, first discovered as a family of deaminases responsible for conversion of adenosine to inosine in tRNAs, deaminate cytosine into uridine in DNA.Citation13 Finally, the AlkB proteins are a family of oxidative dealkylases that are broadly distributed among sequenced organisms. The founding members of the AlkB family were initially identified as proteins involved in the repair of damaged DNA and RNA molecules (see ref [14] for review). It was later shown that members of this family, such as the mammalian ALKBH8, are responsible for the hydroxylation of mcm5U in tRNAs.Citation15 In addition, members of the FTO subgroup were found to demethylate mRNAs, adding regulatory functions to the AlkB family.Citation16
In retrospect, these examples of crosstalk are not surprising, as deamination and methylation reactions were already known for both RNA and DNA. The novelty mainly derives from the switch in specificity within an enzyme superfamily from a RNA to a DNA substrate. However, the recent discovery that DNA contains 7-deazaguanine derivatives,Citation17 modifications that were believed to exist exclusively in tRNA, is a novel and unexpected example of crosstalk between the RNA and DNA modification machineries, which is the focus of this review.
Queuosine and archaeosine function in tRNA
Queuosine (Q) and archaeosine (G+) are long-known 7-deazaguanine derivatives that were thought to exist exclusively in tRNA molecules. Q is found in many eukaryotes and bacteria at the wobble position of tRNAs harboring G34U35N36 anticodon sequences. These tRNAs are responsible for the insertion of Asn, Asp, His and Tyr amino acids in proteins.Citation18 A role for Q in decoding efficiency was demonstrated over 30 years agoCitation19 and recent work by the Farabaugh laboratory has also shown that Q affects decoding accuracy in both directions depending on the codon.Citation20-21 However, Q is not required for growth under most tested conditions and its physiological importance remains elusive, particularly as it was repeatedly lost in the course of evolution.Citation22 In bacteria, the absence of Q does not seem to be critical in exponential growth but affects growth under stress conditions in a diverse set of organismsCitation23-26 and is required for the synthesis of a virulence factor in Shigella.Citation25 In mammals, the queuine base is acquired from the diet or microfloraCitation27 and the absence of both Q and tyrosine is lethal.Citation28-29 A recent study in flies led to a model in which Q plays a key role in avoiding miscoding.Citation30 Q is also a positive determinant for the activity of the tRNA methyltransferase DNMT2.Citation31
G+ is found at position 15 of most archaeal tRNAs that harbor a G at that positionCitation32 and was recently found at position 13 in Thermoplasma acidophilum tRNAs.Citation33 Theoretical studies predict that G+15 plays a structural role, stabilizing the L-shaped tRNA structure through strengthening the G15-C48 interaction.Citation34 G+ is not essential in Haloferax volcaniiCitation35 nor in Thermococcus kodakarensis,Citation33 but its absence leads to a cold sensitivity phenotype in extreme halophiles.Citation35
Both the Q and G+ modifications require complex pathways for their synthesis and are derived from the 7-cyano-7-deazaguanine intermediate preQ0.
PreQ0 synthesis in bacteria and archaea
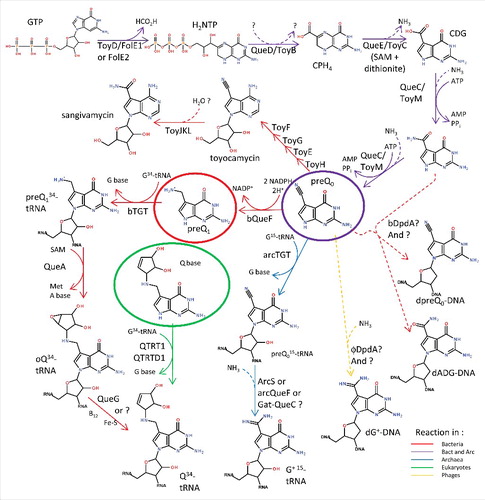
PreQ0 is synthesized de novo from GTP in many bacteria and archaea by a series of complex reactions catalyzed by four enzymesCitation36-37 (). The first step of the preQ0 pathway, the formation of dihydroneopterin triphosphate (H2NTP), is not a dedicated step but is shared with the tetrahydrofolate (THF) and biopterin (BH4) pathways. It is catalyzed by GTP cyclohydrolase I (EC 3.5.4.16).Citation38 Most organisms use a Zn2+-dependent GTP cyclohydrolase I (FolE), a member of the Tunnel-fold (T-fold) structural superfamily.Citation39 However, in a third of sequenced bacteria and in most archaea, FolE is replaced by FolE2, another T-fold superfamily member that utilizes other metals.Citation38,40 The first dedicated step of the preQ0 pathway is the conversion of H2NTP to 6-carboxy-5,6,7,8-tetrahydropterin (CPH4) by CPH4 synthase (EC 4.1.2.50, QueD).Citation41 QueD is a member the COG0720 family that also contains close homologs involved in THF and BH4 synthesis, revealing a catalytic promiscuityCitation42-43 that make these genes difficult to annotate by sequence similarity alone.Citation44 CPH4 is then converted to 5-carboxydeazaguanine (CDG) by CDG synthase (EC 4.3.99.3, QueE), a SAM dependent iron-sulfur cluster protein.Citation45 7-Cyano-7-deazaguanine synthase (EC 6.3.4.20, QueC), then catalyzes the formation of preQ0 from CDG through the recently discovered intermediate 7-amido-7-deazaguanine (ADG) in two ATP dependent reactions.Citation46
Figure 1. Deazaguanine derivative synthesis pathways. GTP is the preQ0 precursor in both bacteria and archaea (purple arrows). In most bacteria, four more enzymatic steps lead to the insertion of Q in tRNAs at position 34 (red arrows). In a few organisms, preQ0 can be transformed to secondary metabolites such as toyacamycin or sangivamycin antibiotics (red arrows, toy genes). In eukaryotes, queuine is salvaged (green circle) and directly transferred to tRNAs (green arrows). Bacteria salvage preQ1 (red circle), and both bacteria and archaea salvage preQ0 (purple circle). In archaea, preQ0 is transferred to position 15 of tRNA before being modified to G+ (blue arrows). PreQ0 and ADG have been found in bacterial DNA (dashed red arrows) and G+ in phage DNA (dashed yellow arrow). All dashed arrows represent uncharacterized reactions. All molecule abbreviations and protein names are described in the text.
Crystal structures have been determined for all preQ0 synthesis enzymes. FolE, FolE2 and QueD are members of the T-fold structural superfamily of multimeric pterin and purine binding proteins. T-fold enzymes are classified into two structural subfamilies: a unimodular subfamily, composed of proteins that are formed from subunits possessing a single T-fold domain, and a bimodular subfamily, composed of proteins formed from subunits possessing tandem T-fold domains. FolE and QueD are, respectively, homodecameric and homohexameric unimodeular T-fold enzymes.Citation43,47; whereas FolE2 is a homotetrameric bimodular enzymeCitation40 In all three proteins, the active sites lie at the interfaces between monomeric subunits. QueC exists as a homodimer, and its monomeric subunit is constituted of an N-terminal domain possessing the Rossman fold architecture characteristic of many nucleotide binding proteins, and a helical zinc-binding C-terminal domain, with the active site predicted to be at the interface between the two domains.Citation48 QueE is a homodimer built around a modified AdoMet radical fold.Citation49 Except for QueC, the structural details of substrate and cofactor recognition have been elucidated for all the dedicated preQ0 synthesis enzymes.Citation43,45,50-51
Q synthesis in bacteria
Three more steps are required to synthesize Q in bacteria (). The preQ0 precursor is first reduced to 7-aminomethyl-7-deazaguanine (preQ1) by the NADPH-dependent 7-cyano-7-deazaguanine reductase (EC 1.7.1.13) enzyme QueF.Citation52-53 QueF is closely related to FolE in primary structure, and the two families can be distinguished at the sequence level by a QueF-specific motif involved in NADPH binding and by the FolE-specific zinc binding residues.Citation52
QueF is also a T-fold enzyme and, like GTP cyclohydrolase I, is represented by the unimodular and bimodular T-fold subfamilies. In unimodular QueF, a homodecameric enzyme, catalysis occurs at the intersubunit interfaces; whereas in bimodular QueF, a homodimer, catalysis occurs at the intrasubunit interface between the two T-fold modules, not at the intersubunit interface as in the bimodular FolE2.Citation50,51
The entire pathway to preQ1 is independent of tRNA. tRNA comes into play when the bacterial tRNA-guanine transglycosylase (EC 2.4.2.29, bTGT) enzyme removes the G base at position 34 from the target tRNAs and replaces the base with preQ1 (). Extensive biochemical and structural studies on bTGT have shown that the enzyme is a homodimer that binds one tRNA molecule with its U33G34U35 sequence as the recognition element.Citation54-58 The monomer consists of an irregular (β/α)8 TIM barrel domain that harbors specific loop insertions that guarantee proper tRNA recognition, and a tightly attached C-terminal zinc-binding domain involved in dimerization (). A reaction mechanism has been proposed in which G34 of tRNA first binds in the purine recognition pocket, constituted of the residues D102, S103, D156, Q203 and G230 (Zymomonas mobilis TGT protein numbering, ). Nucleophilic attack by an active-site aspartate side chain (D280 in Z. mobilis TGT) on the ribosyl C1′ of G34 detaches the guanine base from the RNA, and results in formation of a covalent intermediate between TGT and RNA. A subsequent conformational change in the active site facilitates the release of the detached guanine base from the pocket and the binding of preQ1 in the same pocket with L231 and A232 as specificity determinants. Deprotonation of N9 of the incoming preQ1 by another active-site aspartate (D102) allows nucleophilic attack by N9 on ribose C1′ to form the preQ1-tRNA product.
Figure 2. Catalytic and substrate recognition residues in different TGT subgroups. Partial alignments of Zymomonas mobilis (Zm) bTGT (PDB 1WKF), Escherichia coli (Ec) bTGT (RefSeq AMK98755), Homo sapiens (Hm) QTRT1 (RefSeq AAH15350), Homo sapiens QTRTD1 (RefSeq EAW79613), Pyrococcus horikoshii (Ph) arcTGT (PDB 1IQ8), Salmonella enterica serovar Montevideo (Sm) bDpdA (Genbank EFY12575), and E. coli phage 9g pDpdA (RefSeq YP_009032326). The conserved residue for substrate recognition, zinc binding, and catalytic activity are shown in bold and numbered. The arcTGT exhibits three supplementary C-terminal domains (C1, C2 and PUA), represented in black on the archaea line.

Two subsequent reactions are required to synthesize the final Q product. First, S-adenosylmethionine tRNA ribosyltransferase-isomerase (EC 2.4.99.17, QueA) catalyzes the addition of an epoxycyclopentandiol ring to preQ1 to form the epoxyqueuosine (oQ).Citation59 QueA uses the ribose moiety from S-adenosyl-methionine (SAM), and transfers it to the ammonium moiety of preQ1. Finally, the epoxyqueuosine reductase (EC 1.17.99.6) reduces oQ into Q60. In E. coli, this reaction is catalyzed by QueG,Citation61 a B12-dependent enzyme with iron-sulfur clusters. Because QueG is missing in many organisms that harbor Q in tRNA (Zallot and de Crécy-Lagard, unpublished), it is expected that non-orthologous enzymes catalyzing the same reaction are yet to be discovered in these organisms.
G+ synthesis in archaea
ArcTGT is the archaeal bTGT homolog that inserts preQ0 at position 15 (or 13 in specific organisms) in the dihydrouridine arm (D-arm) of target tRNAs (). Like bTGT, arcTGT functions as a homodimer. The monomer is constituted of an N-terminal catalytic domain, and three arcTGT-specific C-terminal domains C1, C2 and C3/PUA (PseudoUridine synthase and Archaeosine transglycosylase domain). The catalytic domain is built around an (α/β)8 TIM barrel subdomain and a zinc-binding subdomain, making it very similar in structure to the bTGT monomer (). This catalytic domain also mediates the homodimerization of arcTGT through the zinc-binding sites, similar to bTGT homodimerization.Citation56,62 The C1 domain provides an additional homodimerization interface, while the C2 and C3/PUA domains provide a scaffold for tRNA binding. However, unlike the bTGT homodimer which binds one tRNA molecule, the arcTGT homodimer binds two tRNA molecules such that each tRNA interacts with both protein subunits. One subunit provides binding capacity through its C-terminal domains, while the other subunit performs the reaction.Citation63
ArcTGT distorts the tRNA structure from the canonical L-shaped form to the λ-form in which the D-arm is single stranded and protruded, allowing G15 to be accessible for transglycosidation. The enzyme presumably positions the exposed G15 in the active site by “counting” the nucleotides from G1 to G15 in the λ-form. Although the substrate binding pocket and catalytic residues are conserved between the bacterial and archaeal enzymes, the replacement of L231/A232 in bTGTs by V197/V198 in arcTGTs () allows for the change of substrate specificity from preQ1 to preQ056.
Once preQ0 has been inserted into the target archaeal tRNA, one last amidotransferase step is required to produce the G+ modification. Surprisingly, a great diversity of amidotranferase enzymes have been recruited to catalyze this reaction (). The first to be discovered was archaeosine synthase (EC 2.6.1.97, ArcS).Citation64 ArcS catalyzes the conversion of preQ0–tRNA to G+-tRNA in an ATP independent manner and may use glutamine, asparagine as well as free ammonia as ammonium donors in vitro.Citation64 ArcS is evolutionarily and structurally related to arcTGT. It has retained the three tRNA binding domains found in arcTGT but has acquired an insertion of four α-helices and 3 β-strands in its C1 domain that confer a Rossman fold architecture to the domain. This domain extension contains a conserved PCX3KPYX2SX2H motif proposed to be involved in catalytic activity. Like the TGT family, ArcS proteins form homodimers but are predicted to bind the L-form, not the λ-form tRNA.Citation64
Many archaea that synthesize G+ lack ArcS homologs and, in those organisms, two non-orthologous enzymes were found to be responsible for the formation of G+ 65. The first is QueF-Like (QueF-L) which converts preQ0 base of preQ0-modified tRNA to G+, unlike the bacterial preQ1 synthase QueF which acts on the free preQ0 base as substrate.Citation66 QueF-Like is also a homodecameric T-fold enzyme with a similar structure to that of QueF, except that its pentameric subunits are tightly wound, resulting in elimination of the QueF-specific NADPH binding site.Citation66 The second enzyme that replaces ArcS in some archaea is Gat-QueC, a homolog of QueC fused to a glutamine amidotransferase class-II domain (GATase). This enzyme is yet to be biochemically characterized, and it remains unclear whether it acts before or after integration of preQ0 in tRNA and, consequently, whether the corresponding arcTGT integrates the G+ base or the preQ0 base in the tRNAs of these organisms.
Q salvage in eukaryotes
Most eukaryotes harbor Q in their tRNAs but do not produce the modification de novo. Instead, they salvage the queuine base (q) from their diet or microflora (see reference [67] for recent review) using transporters that are yet to be discovered. Members of the DUF219 family have recently been implicated in q salvage in eukaryotes.Citation22 Structural modeling suggests a possible ribonucleotide hydrolase function for this family, but its exact role is yet to be determined.
The eukaryotic TGTs (eTGT) insert the q base directly in tRNAs (). eTGTs are heterodimers composed of a catalytic subunit (queuine tRNA-ribosyltransferase catalytic subunit 1 or QTRT1) and an accessory subunit that lacks the active site Asp residue (queuine tRNA-ribosyltransferase accessory subunit 2 or QTRT2, previously called QRTD1).Citation68 Key differences between the eukaryotic and prokaryotic TGTs are in the substrate binding pocket where Val233 is replaced by a glycine and Cys158 is replaced by a valine. These changes enlarge the binding pocket and allow the binding of a larger substrate (q versus preQ0 or preQ1).Citation69
Seven-deazapurines salvage in bacteria
bTGT is the signature gene of the Q pathway. All organisms that harbor a bTGT homolog can harbor Q in tRNA and no organism lacking bTGT has ever been found to contain Q in tRNA (de Crécy-Lagard, unpublished). However, many organisms with bTGT homologs lack preQ0 synthesis enzymes and/or QueF.Citation70 In these cases, the preQ0 or preQ1 precursor must be salvaged. preQ0 is certainly available in natural environments as an intermediate of the Q and G+ pathway as described above, but also as a secondary metabolite or secondary metabolite precursor.Citation71-72 It also may be recycled from tRNA degradation products, because the free G+ base spontaneously deamidates to preQ0.Citation73
Specific preQ1/preQ0 transporters of the ECF family (QueT and QrtT) have been predicted to be involved in 7-deazapurine salvageCitation74 but have never been experimentally validated as such, and their sparse distribution suggests that other transporter families exist. We recently reported that the COG1738/YhhQ family is a preQ0-specific transporter found in E. coli and is widespread in bacteria.Citation75 Of note, some bacteria such as Chlamydia trachomatis not only lack the precursor pathway but also QueA homologs. Queuine should therefore be salvaged in these bacteria. In summary, while the de novo Q pathway is well characterized, many open questions remain regarding the possibility of Q salvage in bacteria.
Discovery of dADG and dpreQ0 in bacterial DNA
Comparative genomic analyses led to the observation that two copies of the tgt gene are present in specific bacteria.Citation17 One copy encodes the canonical bTGT, while the other copy encodes a larger, more divergent protein that exhibited common structural features with both arcTGT and bTGT enzymes. Careful sequence analysis showed that the catalytic residues and substrate binding pocket of arcTGT (e.g., D95, D249, V217, and V218 in P. horikoshii arcTGT numbers) are conserved in this protein (), suggesting that it may bind preQ0. In addition, the tRNA-binding C3/PUA domain characteristic of arcTGT is missing from this protein, and the zinc binding motif is present in a supplementary domainCitation17 ().
Physical clustering analyses strongly linked this tgt paralog to DNA metabolism genes, leading to the prediction that this enzyme might be involved in inserting preQ0 or a derivative in DNA.Citation17,76 This prediction was tested by analyzing genomic DNA, extracted from different bacteria that contained copies of the tgt paralog, by mass-spectrometry, specifically searching for the deoxyribose forms of the Q pathway intermediates. Deoxy-preQ0 (dpreQ0) was detected as predicted but seemed to be a minor product. The main deazapurine modification found in the DNA of these organisms was dADG, the deoxyribose form of the preQ0 precursor ADG. The discovery of these modifications led to renaming these tgt paralogs as dpdA, for DeoxyPurine in DNA.Citation17
Discovery of dG+ in bacteriophage 9g DNA
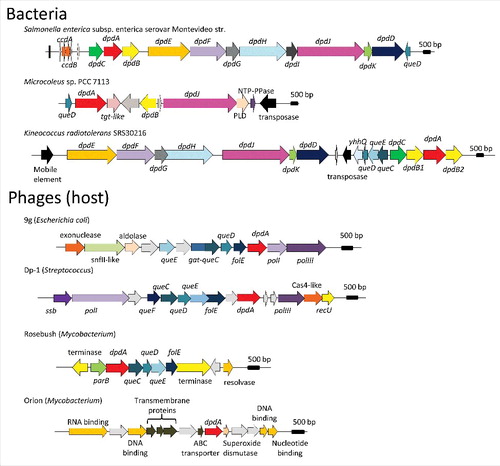
Many bacteriophages (or phages) encode TGT-like proteinsCitation17 (). Early studies of the Streptococcus Dp-1 phage,Citation77 and the Mycobacterium phage RosebushCitation78 had led to the proposal that these genes are involved in the synthesis of Q in tRNA, interfering with or enhancing the translation of the host or phage proteins. Another possible function was proposed by the Letarov group, who suggested that Q could be incorporated in the DNA of Escherichia coli phage 9g, causing its resistance to cleavage by several endonucleases.Citation79 Consistent with this proposal is the observation that most phage TGT-like proteins are closer in sequence to DpdA than to the canonical TGT enzymes. Although the catalytic core and substrate recognition residues of arcTGT are conserved in this protein (), two out of the four residues constituting the zinc binding sites are not conserved ().
Figure 3. Genome neighborhoods of dpdA in bacteria and phages. The dpd and the queuosine related genes are defined in the text, general family memberships were noted for the other genes. The host of each phage is indicated between parenthesis aside of each phage name. Accession numbers for the corresponding DpdA proteins are: WP_001542917.1 for Salmonella enterica subsp. enterica serovar Montevideo str., WP_015211588 for Microcoleus sp PCC 7113, ABS05840 for Kineococcus radiotolerans SRS30216, YP_009032326 for Escherichia coli phage 9g, YP_004306895 for Streptococcus phage Dp-1, NP_817763 for Mycobacterium phage Rosebush, and YP_655116 for Mycobacterium phage Orion.
Physical clustering analysis of the 9g phage genome revealed that the dpdA gene clustered with a gat-queC homolog, suggesting that G+ might be inserted in this phage. This prediction was validated by mass-spectrometry and it was shown that 25% of the dG residues were modified to dG+ in the 9g DNA, a first occurrence of the G+ base outside the archaeal kingdom.Citation17
The Dpd cluster and horizontal gene transfer
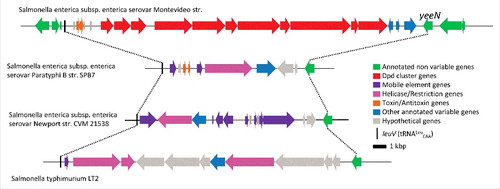
Similarly to tgt being the signature gene for Q in tRNA, dpdA is the signature gene for the presence of deazapurine derivatives in DNA. dpdA is part of a genomic island of about 20 kb in size (). Synteny analyses of closely related strains as well as co-distribution analysis of cluster genes with dpdA helped define the island boundaries.Citation17 Deletion of the whole dpd cluster in Salmonella enterica serovar Montevideo was shown to abolish the insertion of ADG and preQ0 in DNA.Citation17 In the S. Montevideo genomes, the cluster is inserted at the leuX locus, an hypervariable region that is a known landing site for pathogenicity virulence and defense genesCitation80 (). 70% of the available S. Montevideo sequences in the SEED database (SEED viewer version 2.081) contain the dpd cluster. This data, in combination with the phylogenetic analysis of the DpdA family that carries many deviations from the species treeCitation17 and the wide and sporadic distribution of the dpd cluster around the bacterial tree,Citation17 makes a strong case for the great mobility of this cluster and its propagation by horizontal gene transfer.
Figure 4. Schematic representation of the variable region between leuV and yeeN genes in different Salmonella genomes. The genes are colored by functional groups as indicated on the figure.
Three genes, dpdB, dpdC and dpdD, are nearly always associated with dpdA in the bacterial dpd clusters; while the dpdEFJK genes, all related to DNA metabolism, have a more variable distributionCitation17 (). One or more preQ0 synthesis genes are also present in some clusters ().
Based on the sequence similarity with TGT, DpdA is the most promising candidate for the enzyme that inserts preQ0 and/or ADG in DNA. DpdB is found in 92% of the genomes that harbor dpdA. Sequence similarity analyses showed that DpdB is a member of the ParB nuclease superfamily,Citation76 more specifically related to the DndB subgroup of proteins.Citation17 DndB is part of the dnd cluster that introduces a phosphorothioate modification in the DNA backbone.Citation82 DndB is not part of the core modification pathway but is a DNA binding protein with a regulatory role,Citation83 suggesting that DpdB might bind DNA and could be involved in target recognition. dpdC was found in 88% and DpdD in 90% of the genome containing dpdA. Both encoded proteins were similar to proteins of unknown function, DUF328 for DpdC and DUF3225 for a small portion of dpdD,Citation17 making any functional prediction difficult at this stage.
Function of deazapurines in DNA
Up to 25% of the G bases in 9g phage DNA are converted to G+.Citation17 The modification event in the bacterial genomes is more rare because only 0.01% of the G bases were observed to be modified to ADG/preQ0 in the DNA analyzed. 9g DNA is also resistant to cleavage by a variety of endonucleases,Citation79 hence dG+ might have an anti-restriction function allowing the phage to escape host restriction. S. Montevideo DNA on the other hand is not resistant to cleavage by restriction enzymes. However, at least one gene in the dpd cluster is involved in cutting DNA that lacks the deazapurine modification based on transformation efficiency assays,Citation17 suggesting that the cluster carries a restriction/modification system that protects the bacteria from foreign DNA. The identity of the restriction enzyme remains an open question. Candidates includes genes like dpdD or dpdC that are of unknown function; or some of the other Dpd proteins, like DpdE that is part of the SNF-II superfamily that contains endonucleases from type I and III modification/restriction systems.Citation84
Predicted diversity of deazapurine modification of phages
G+ is present in E. coli phage 9g DNA, and the presence of very similar gene clusters, including the gat-queC gene, suggests that G+ incorporation should also be the case for E. coli phages JenK1, and JenP1/2.Citation85 Analysis of the genomic context of DpdA-like encoding genes in all sequenced phages suggests that other deazapurine derivatives might be found in phages. Some phages, e.g., Mycobacteriophage Rosebush, encode all enzymes of the preQ0 biosynthesis pathway (from FolE to QueC, ), and are predicted to insert preQ0 or ADG in their DNA. Other phages like Streptococcus phage Dp-1 also encode QueF, and could therefore insert preQ1. Finally, some phages like Mycobacteriophage Orion only harbor a dpdA-like gene raising the possibility of salvage of Q precursors from the host.
Conclusion
It has become quite clear recently that 7-cyano-7-deazaguanine, or preQ0, is a key metabolite that is recruited in different pathways and is used in a variety of ways. PreQ0 can be the final product, as recently described in the Streptomyces,Citation71 but its most frequent fate (in around 70% of sequenced bacteria and nearly all archaea) is to be inserted in tRNA where it is further modified to Q or G+ (). The preQ0 base is also used as precursor of secondary metabolites, such as toyocamycin and sangivamycinCitation86 in several Actinomycetes,Citation72 or can be inserted in DNA in organisms that harbor the dnd genomic island. It is still not clear yet if ADG is incorporated directly in DNA or if it is synthesized after preQ0 incorporation.
Some organisms will use the preQ0 precursor in multiple pathways like S. Montevideo that harbors both Q in tRNA and ADG in DNA, hence the preQ0 synthesis cluster is sometimes duplicated (). Finding the preQ0 transporter gene yhhQ imbedded in certain dpd clusters also suggests that salvaged preQ0 could be inserted in DNA.
The insertion of preQ0 in a nucleic acid polymer required the presence of a transglycosylase of the TGT family that must exchange the guanine base with the 7-deazaguanine derivative. The plasticity of the TGT family was well established as small variations in sequences allowed the different orthologs to favor preQ0, preQ1 or q substrates. It had also been shown that the bTGT could modify mRNA in vitroCitation87 and artificially modify DNA when thymines were replaced by deoxyuracils.Citation88 The discovery of the role of the TGT paralog DndA in modifying DNA has shown that this can happen in vivo and opens many questions on the identity of potential sequence specific endonuclease inhibited by the insertion of dADG, on the role of deazapurine in protecting DNA from restriction, and on how the normal replication/transcription machineries can deal with these modifications.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dirk Iwata-Reuyl for sharing unpublished results and Rémi Zallot for editing the manuscript.
Funding
This work was supported by the National Institutes of Health (grant number R01 GM70641 to V.d.C.-L.)
References
- Forterre P, Grosjean H. The interplay between RNA and DNA modifications. Back to the RNA world. In DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution, H Grosjean, Ed. Landes Bioscience, 2009; pp 259-274.
- Weigele P, Raleigh EA. Biosynthesis and function of modified bases in Bacteria and their viruses. Chemical Rev 2016; 116(20):12655-87; PMID:27319741; http://dx.doi.org/10.1021/acs.chemrev.6b00114
- Grosjean H. Nucleic Acids are not boring long polymers of only four types of nucleotides. In DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution, H Grosjean, Ed. Landes Bioscience, 2009; pp 1-18.
- Warren RA. Modified bases in bacteriophage DNAs. Ann Rev Microbiol 1980; 34:137-58; PMID:7002022; http://dx.doi.org/10.1146/annurev.mi.34.100180.001033
- Perche-Letuvee P, Molle T, Forouhar F, Mulliez E, Atta M. Wybutosine biosynthesis: structural and mechanistic overview. RNA Biol 2014; 11:1508-18; PMID:25629788; http://dx.doi.org/10.4161/15476286.2014.992271
- Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: Form, distribution, and function. Science 2016; 352:1408-1412; PMID:27313037; http://dx.doi.org/10.1126/science.aad8711
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. The dynamic N1-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016; 530:441-446; PMID:26863196; http://dx.doi.org/10.1038/nature16998
- Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS ONE 2014; 9:e110799; PMID:25353621; http://dx.doi.org/10.1371/journal.pone.0110799
- Auxilien S, Rasmussen A, Rose S, Brochier-Armanet C, Husson C, Fourmy D, Grosjean H, Douthwaite S. Specificity shifts in the rRNA and tRNA nucleotide targets of archaeal and bacterial m5U methyltransferases. RNA 2011; 17:45-53; PMID:21051506; http://dx.doi.org/10.1261/rna.2323411
- Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem 2011; 12:206-22; PMID:21243710; http://dx.doi.org/10.1002/cbic.201000195
- Jeltsch A, Ehrenhofer-Murray A, Jurkowski TP, Lyko F, Reuter G, Ankri S, Nellen W, Schaefer M, Helm M. Mechanism and biological role of Dnmt2 in nucleic acid methylation. RNA biology 2016; 1-16; PMID:27232191; http://dx.doi.org/10.1080/15476286.2016.1191737
- Salter JD, Bennett RP, Smith HC. The APOBEC protein family: united by structure, divergent in function. Trends Biochem Sci 2016; 41:578-594; PMID:27283515; http://dx.doi.org/10.1016/j.tibs.2016.05.001
- Rubio MAT, Pastar I, Gaston KW, Ragone FL, Janzen CJ, Cross GAM, Papavasiliou FN, Alfonzo JD. An adenosine-to-inosine tRNA-editing enzyme that can perform C-to-U deamination of DNA. Proc Natl Acad Sci USA 2007; 104:7821-7826; PMID:17483465; http://dx.doi.org/10.1073/pnas.0702394104
- Fedeles BI, Singh V, Delaney JC, Li D, Essigmann JM. The AlkB family of Fe(II)/α-ketoglutarate-dependent dioxygenases: repairing nucleic acid alkylation damage and beyond. J Biol Chem 2015; 290:20734-20742; PMID:26152727; http://dx.doi.org/10.1074/jbc.R115.656462
- van den Born E, Vagbo CB, Songe-Moller L, Leihne V, Lien GF, Leszczynska G, Malkiewicz A, Krokan HE, Kirpekar F, Klungland A, et al. ALKBH8-mediated formation of a novel diastereomeric pair of wobble nucleosides in mammalian tRNA. Nat Commun 2011; 2:172; PMID:21285950; http://dx.doi.org/10.1038/ncomms1173
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011; 7:885-7; PMID:22002720; http://dx.doi.org/10.1038/nchembio.687
- Thiaville JJ, Kellner SM, Yuan Y, Hutinet G, Thiaville PC, Jumpathong W, Mohapatra S, Brochier-Armanet C, Letarov AV, Hillebrand R, et al. Novel genomic island modifies DNA with 7-deazaguanine derivatives. Proc Natl Acad Sci USA 2016; 113:E1452-E1459; PMID:26929322; http://dx.doi.org/10.1073/pnas.1518570113
- Okada N, Noguchi S, Kasai H, Shindo-Okada N, Ohgi T, Goto T, Nishimura S. Novel mechanism of post-transcriptional modification of tRNA. Insertion of bases of Q precursors into tRNA by a specific tRNA transglycosylase reaction. J Biol Chem 1979; 254:3067-73; PMID:372186
- Meier F, Suter B, Grosjean H, Keith G, Kubli E. Queuosine modification of the wobble Base in tRNAHis influences ‘in vivo’ decoding properties. EMBO J 1985; 4:823-827; PMID:2988936
- Manickam N, Nag N, Abbasi A, Patel K, Farabaugh PJ. Studies of translational misreading in vivo show that the ribosome very efficiently discriminates against most potential errors. RNA 2014; 20:9-15; PMID:24249223; http://dx.doi.org/10.1261/rna.039792.113
- Manickam N, Joshi K, Bhatt MJ, Farabaugh PJ. Effects of tRNA modification on translational accuracy depend on intrinsic codon-anticodon strength. Nucleic Acids Res 2016; 44:1871-81; PMID:26704976; http://dx.doi.org/10.1093/nar/gkv1506
- Zallot R, Brochier-Armanet C, Gaston KC, Forouhar F, Limbach PA, Hunt JF, De Crécy Lagard V. The plant, animal, and fungal micronutrient queuosine is salvaged by members of the DUF2419 protein family. ACS Chem Biol 2014; 9:1812-25; PMID:24911101; http://dx.doi.org/10.1021/cb500278k
- Noguchi S, Nishimura Y, Hirota Y, Nishimura S. Isolation and characterization of an Escherichia coli mutant lacking tRNA-guanine transglycosylase. Function and biosynthesis of queuosine in tRNA. J Biol Chem 1982; 257:6544-50; PMID:6804468
- Thibessard A, Borges F, Fernandez A, Gintz B, Decaris B, Leblond-Bourget N. Identification of Streptococcus thermophilus CNRZ368 genes involved in defense against superoxide stress. Appl EnvironMicrobiol 2004; 70:2220-9; PMID:15066816
- Durand JM, Dagberg B, Uhlin BE, Björk GR. Transfer RNA modification, temperature and DNA superhelicity have a common target in the regulatory network of the virulence of Shigella flexneri: the expression of the virF gene. Mol Microbiol 2000; 35:924-35; PMID:10692168; http://dx.doi.org/10.1046/j.1365-2958.2000.01767.x
- Marchetti M, Capela D, Poincloux R, Benmeradi N, Auriac MC, Le Ru A, Maridonneau-Parini I, Batut J, Masson-Boivin C. Queuosine biosynthesis is required for Sinorhizobium meliloti-induced cytoskeletal modifications on HeLa Cells and symbiosis with Medicago truncatula. PLoS ONE 2013; 8:e56043; PMID:23409119; http://dx.doi.org/10.1371/journal.pone.0056043
- Vinayak M, Pathak C. Queuosine modification of tRNA: its divergent role in cellular machinery. Bioscience Rep 2010; 30:135-48; PMID:19925456; http://dx.doi.org/10.1042/BSR20090057
- Marks T, Farkas WR. Effects of a diet deficient in tyrosine and queuine on germfree mice. Biochem Bioph Res Com 1997; 230:233-237; PMID:9016755; http://dx.doi.org/10.1006/bbrc.1996.5768
- Rakovich T, Boland C, Bernstein I, Chikwana VM, Iwata-Reuyl D, Kelly VP. Queuosine deficiency in eukaryotes compromises tyrosine production through increased tetrahydrobiopterin oxidation. J Biol Chem 2011; 286:19354-63; PMID:21487017; http://dx.doi.org/10.1074/jbc.M111.219576
- Zaborske JM, Bauer DuMont JL, Wallace EWJ, Pan T, Aquadro CF, Drummond DA. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein encoding across and animal genus. Plos Biol 2014; 12:e1002015; PMID:25489848; http://dx.doi.org/10.1371/journal.pbio.1002015
- Müller M, Hartmann M, Schuster I, Bender S, Thüring KL, Helm M, Katze JR, Nellen W, Lyko F, Ehrenhofer-Murray AE. Dynamic modulation of Dnmt2-dependent tRNA methylation by the micronutrient queuine. Nucleic Acids Res 2015; 43:10952-10962; PMID:26424849; http://dx.doi.org/10.1093/nar/gkv980
- Gregson JM, Crain PF, Edmonds CG, Gupta R, Hashizume T, Phillipson DW, McCloskey JA. Structure of Archaeal transfer RNA nucleoside G*-15 (2-Amino-4,7-dihydro-4-oxo-7-b-D-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximidamide (Archaeosine)). J Biol Chem 1993; 268:10076-10086; PMID:7683667
- Kawamura T, Hirata A, Ohno S, Nomura Y, Nagano T, Nameki N, Yokogawa T, Hori H. Multisite-specific archaeosine tRNA-guanine transglycosylase (ArcTGT) from Thermoplasma acidophilum, a thermo-acidophilic archaeon. Nucleic Acids Res 2016; 44:1894-1908; PMID:26721388; http://dx.doi.org/10.1093/nar/gkv1522
- Oliva R, Tramontano A, Cavallo L. Mg2+ binding and archaeosine modification stabilize the G15 C48 Levitt base pair in tRNAs. RNA 2007; 13:1427-1436; PMID:17652139; http://dx.doi.org/10.1261/rna.574407
- Blaby IK, Phillips G, Blaby-Haas CE, Gulig KS, El Yacoubi B, de Crécy-Lagard V. Towards a systems approach in the genetic analysis of Archaea: Accelerating mutant construction and phenotypic analysis in Haloferax volcanii. Archaea 2010; 2010; 426239; PMID:21234384; http://dx.doi.org/10.1155/2010/426239
- Reader JS, Metzgar D, Schimmel P, de Crécy-Lagard V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J Biol Chem 2004; 279:6280-5; PMID:14660578; http://dx.doi.org/10.1074/jbc.M310858200
- McCarty RM, Somogyi Ard, Lin G, Jacobsen NE, Bandarian V. The deazapurine biosynthetic pathway revealed: in vitro enzymatic synthesis of preQ0 from guanosine 5′-triphosphate in four steps. Biochemistry 2009; 48:3847-3852; PMID:19354300; http://dx.doi.org/10.1021/bi900400e
- Phillips G, El Yacoubi B, Lyons B, Alvarez S, Iwata-Reuyl D, de Crécy-Lagard V. Biosynthesis of 7-deazaguanosine-modified tRNA nucleosides: a new role for GTP Cyclohydrolase I. J Bacteriol 2008; 190:7876-7884; PMID:18931107; http://dx.doi.org/10.1128/JB.00874-08
- Colloc'h N, Poupon A, Mornon JP. Sequence and structural features of the T-fold, an original tunnelling building unit. Proteins 2000; 39:142-54; PMID:10737935; http://dx.doi.org/10.1002/(SICI)1097-0134(20000501)39:2%3c142::AID-PROT4%3e3.0.CO;2-X
- Sankaran B, Bonnett SA, Shah K, Gabriel S, Reddy R, Schimmel P, Rodionov DA, de Crécy-Lagard V, Helmann JD, Iwata-Reuyl D, et al. Zinc-independent folate biosynthesis: genetic, biochemical, and structural investigations reveal new metal dependence for GTP cyclohydrolase IB. J Bacteriol 2009; 191:6936-49; PMID:19767425; http://dx.doi.org/10.1128/JB.00287-09
- McCarty RM, Somogyi Ard, Bandarian V. Escherichia coli QueD is a 6-Carboxy-5,6,7,8-tetrahydropterin synthase. Biochemistry 2009; 48:2301-2303; PMID:19231875; http://dx.doi.org/10.1021/bi9001437
- Phillips G, Grochowski LL, Bonnett S, Xu H, Bailly M, Blaby-Haas C, El Yacoubi B, Iwata-Reuyl D, White RH, de Crécy-Lagard V. Functional promiscuity of the COG0720 family. ACS Chem Biol 2012; 7:197-209; PMID:21999246; http://dx.doi.org/10.1021/cb200329f
- Miles ZD, Roberts SA, McCarty RM, Bandarian V. Biochemical and structural studies of 6-Carboxy-5,6,7,8-tetrahydropterin synthase reveal the molecular basis of catalytic promiscuity within the Tunnel-fold superfamily. J Biol Chem 2014; 289:23641-23652; PMID:24990950; http://dx.doi.org/10.1074/jbc.M114.555680
- Zallot R, Harrison K, Kolaczkowski B, de Crécy-Lagard V. Functional annotations of paralogs: a blessing and a curse. Life 2016; 6:39; PMID:27618105; http://dx.doi.org/10.3390/life6030039
- Bandarian V, Drennan CL. Radical-mediated ring contraction in the biosynthesis of 7-deazapurines. Curr Opin Struc Biol 2015; 35:116-124; PMID:26643180; http://dx.doi.org/10.1016/j.sbi.2015.11.005
- Nelp MT, Bandarian VA. single enzyme transforms a carboxylic acid into a nitrile through an amide Intermediate. Angew Chem Int Ed Engl 2015; 54:10627-10629; PMID:26228534; http://dx.doi.org/10.1002/anie.201504505
- Auerbach G, Herrmann A, Bracher A, Bader G, Gutlich M, Fischer M, Neukamm M, Garrido-Franco M, Richardson J, Nar H, et al. Zinc plays a key role in human and bacterial GTP cyclohydrolase I. Proc Natl Acad Sci USA 2000; 97:13567-72; PMID:11087827; http://dx.doi.org/10.1073/pnas.240463497
- Cicmil N, Huang RH. Crystal structure of QueC from Bacillus subtilis: an enzyme involved in preQ1 biosynthesis. Proteins 2008; 72:1084-8; PMID:18491386; http://dx.doi.org/10.1002/prot.22098
- Dowling DP, Bruender NA, Young AP, McCarty RM, Bandarian V, Drennan CL. Radical SAM enzyme QueE defines a new minimal core fold and metal-dependent mechanism. Nature Chem Biol 2014; 10:106-12; PMID:24362703; http://dx.doi.org/10.1038/nchembio.1426
- Kim Y, Zhou M, Moy S, Morales J, Cunningham MA, Joachimiak A. High-Resolution structure of the nitrile reductase QueF combined with molecular simulations provide insight into enzyme mechanism. J Mol Biol 2010; 404:127-137; PMID:20875425; http://dx.doi.org/10.1016/j.jmb.2010.09.042
- Chikwana VM, Stec B, Lee BWK, de Crécy-Lagard V, Iwata-Reuyl D, Swairjo MA. Structural basis of biological nitrile reduction. J Biol Chem 2012; 287:30560-30570; PMID:22787148; http://dx.doi.org/10.1074/jbc.M112.388538
- Van Lanen SG, Reader JS, Swairjo MA, de Crécy-Lagard V, Lee B, Iwata-Reuyl D. From cyclohydrolase to oxidoreductase: discovery of nitrile reductase activity in a common fold. Proc Natl Acad Sci USA 2005; 102:4264-9; PMID:15767583; http://dx.doi.org/10.1073/pnas.0408056102
- Lee BWK, Van Lanen SG, Iwata-Reuyl D. Mechanistic studies of Bacillus subtilis QueF, the nitrile oxidoreductase involved in Queuosine biosynthesis. Biochemistry 2007; 46:12844-12854; PMID:17929836; http://dx.doi.org/10.1021/bi701265r
- Romier C, Reuter K, Suck D, Ficner R. Mutagenesis and crystallographic studies of Zymomonas mobilis tRNA-guanine transglycosylase reveal aspartate 102 as the active site nucleophile. Biochemistry 1996; 35:15734-9; PMID:8961936; http://dx.doi.org/10.1021/bi962003n
- Ritschel T, Atmanene C, Reuter K, Van Dorsselaer A, Sanglier-Cianferani S, Klebe G. An integrative approach combining noncovalent mass spectrometry, enzyme kinetics and X-ray crystallography to decipher Tgt protein-protein and protein-RNA interaction. J Mol Biol 2009; 393:833-847; PMID:19627989; http://dx.doi.org/10.1016/j.jmb.2009.07.040
- Stengl B, Reuter K, Klebe G. Mechanism and substrate specificity of tRNA-Guanine transglycosylases (TGTs): tRNA-modifying enzymes from the three different kingdoms of life share a common catalytic mechanism. Chem Bio Chem 2005; 6:1926-1939; PMID:16206323; http://dx.doi.org/10.1002/cbic.200500063
- Biela I, Tidten-Luksch N, Immekus F, Glinca S, Nguyen TXP, Gerber H-D, Heine A, Klebe G, Reuter K. Investigation of specificity determinants in bacterial tRNA-guanine transglycosylase reveals queuine, the substrate of Its eucaryotic counterpart, as inhibitor. PLoS ONE 2013; 8:e64240; PMID:23704982; http://dx.doi.org/10.1371/journal.pone.0064240
- Romier C, Reuter K, Suck D, Ficner R. Crystal structure of tRNA-guanine transglycosylase: RNA modification by base exchange. Embo J 1996; 15:2850-2857; PMID:8654383
- Van Lanen SG, Iwata-Reuyl D. Kinetic Mechanism of the tRNA-Modifying Enzyme S-Adenosylmethionine:tRNA Ribosyltransferase-Isomerase (QueA). Biochemistry 2003; 42:5312-5320; PMID:12731872; http://dx.doi.org/10.1021/bi034197u
- Frey B, McCloskey JA, Kersten W, Kersten H. New function of vitamin B12: cobamide-dependent reduction of epoxyqueuosine to queuosine in tRNAs of Escherichia coli and Salmonella typhimurium. J Bacteriol 1988; 170:2078-2082; PMID:3129401; http://dx.doi.org/10.1128/jb.170.5.2078-2082.1988
- Miles ZD, McCarty RM, Molnar G, Bandarian V. Discovery of epoxyqueuosine (oQ) reductase reveals parallels between halorespiration and tRNA modification. Proc Natl Acad Sci USA 2011; 108:7368-7372; PMID:21502530; http://dx.doi.org/10.1073/pnas.1018636108
- Ishitani R, Nureki O, Fukai S, Kijimoto T, Nameki N, Watanabe M, Kondo H, Sekine M, Okada N, Nishimura S, et al. Crystal structure of archaeosine tRNA-guanine transglycosylase. J Mol Biol 2002; 318:665-77; PMID:12054814; http://dx.doi.org/10.1016/S0022-2836(02)00090-6
- Ishitani R, Nureki O, Nameki N, Okada N, Nishimura S, Yokoyama S. Alternative tertiary structure of tRNA for recognition by a posttranscriptional modification enzyme. Cell 2003; 113:383-94; PMID:12732145; http://dx.doi.org/10.1016/S0092-8674(03)00280-0
- Phillips G, Chikwana VM, Maxwell A, El-Yacoubi B, Swairjo MA, Iwata-Reuyl D, de Crécy-Lagard V. Discovery and characterization of an amidinotransferase involved in the modification of archaeal tRNA. J Biol Chem 2010; 285:12706-12713; PMID:20129918; http://dx.doi.org/10.1074/jbc.M110.102236
- Phillips G, Swairjo MA, Gaston KW, Bailly M, Limbach PA, Iwata-Reuyl D, de Crécy Lagard V. Diversity of archaeosine synthesis in crenarchaeota. ACS Chem Biol 2012; 7:300-5; PMID:22032275; http://dx.doi.org/10.1021/cb200361w
- Mei X, Alvarez J, Ramos AB, Samanta U, Iwata-Reuyl D, Swairjo MA. Crystal structure of the archaeosine synthase QueF-Like - insights into amidino transfer and tRNA Recognition by the tunnel fold. Proteins 2016; PMID:25884661; http://dx.doi.org/10.3390/nu7042897
- Fergus C, Barnes D, Alqasem M, Kelly V. The Queuine micronutrient: charting a course from microbe to man. Nutrients 2015; 7:2897; PMID:25884661; http://dx.doi.org/10.3390/nu7042897
- Boland C, Hayes P, Santa-Maria I, Nishimura S, Kelly VP. Queuosine formation in eukaryotic tRNA occurs via a mitochondria-localized heteromeric transglycosylase. J Biol Chem 2009; 284:18218-18227; PMID:19414587; http://dx.doi.org/10.1074/jbc.M109.002477
- Biela I, Tidten-Luksch N, Immekus F, Glinca S, Nguyen TX, Gerber HD, Heine A, Klebe G, Reuter K. Investigation of specificity determinants in bacterial tRNA-guanine transglycosylase reveals queuine, the substrate of its eucaryotic counterpart, as inhibitor. PLoS ONE 2013; 8:e64240; PMID:23704982; http://dx.doi.org/10.1371/journal.pone.0064240
- de Crécy-Lagard V, Olson G. RNA modification subsystems in the SEED database In DNA and RNA editing enzymes: comparative structure, mechanism, functions, cellular interactions and evolution, H Grosjean, Ed. Landes Bioscience: Austin, 2009; pp 624-228.
- Xu D, Ma M, Liu Y, Zhou T, Wang K, Deng Z, Hong K. PreQ0 base, an unusual metabolite with anti-cancer activity from Streptomyces qinglanensis 172205. Anticancer Agents Med Chem 2015; 15:285-90; PMID:25353335; http://dx.doi.org/10.2174/1871520614666141027144653
- McCarty RM, Bandarian V. Biosynthesis of pyrrolopyrimidines. Bioorg Chem 2012; 43:15-25; PMID:22382038; http://dx.doi.org/10.1016/j.bioorg.2012.01.001
- Watanabe M, Matsuo M, Tanaka S, Akimoto H, Asahi S, Nishimura S, Katze JR, Hashizume T, Crain PF, McCloskey JA, et al. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement on the tRNA polynucleotide chain. J Biol Chem 1997; 272:20146-51; PMID:9242689; http://dx.doi.org/10.1074/jbc.272.32.20146
- Rodionov DA, Hebbeln P, Eudes A, ter Beek J, Rodionova IA, Erkens GB, Slotboom DJ, Gelfand MS, Osterman AL, Hanson AD, et al. A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 2009; 191:42-51; PMID:18931129; http://dx.doi.org/10.1128/JB.01208-08
- Zallot R, De Crécy Lagard V. Comparative genomics of bacterial queuosine salvage identify COG1738 as a preQ0 transporter. Biomolecules 2017, submitted.
- Iyer LM, Zhang D, Maxwell Burroughs A, Aravind L. Computational identification of novel biochemical systems involved in oxidation, glycosylation and other complex modifications of bases in DNA. Nucleic Acids Res 2013; 41:7635-7655; PMID:23814188; http://dx.doi.org/10.1093/nar/gkt573
- Sabri M, Häuser R, Ouellette M, Liu J, Dehbi M, Moeck G, García E, Titz B, Uetz P, Moineau S. Genome annotation and intraviral interactome for the Streptococcus pneumoniae virulent Phage Dp-1. J Bacteriol 2011; 193:551-562; PMID:21097633; http://dx.doi.org/10.1128/JB.01117-10
- Pedulla ML, Ford ME, Houtz JM, Karthikeyan T, Wadsworth C, Lewis JA, Jacobs-Sera D, Falbo J, Gross J, Pannunzio NR, et al. Origins of highly mosaic Mycobacteriophage genomes. Cell 2003; 113:171-182; PMID:12705866; http://dx.doi.org/10.1016/S0092-8674(03)00233-2
- Kulikov E, Golomidova A, Letarova M, Kostryukova E, Zelenin A, Prokhorov N, Letarov A. Genomic sequencing and biological characteristics of a novel Escherichia coli bacteriophage 9g, a putative representative of a new Siphoviridae genus. Viruses 2014; 6:5077; PMID:25533657; http://dx.doi.org/10.3390/v6125077
- Moreno Switt AI, den Bakker HC, Cummings CA, Rodriguez-Rivera LD, Govoni G, Raneiri ML, Degoricija L, Brown S, Hoelzer K, Peters JE, et al. Identification and characterization of novel Salmonella mobile elements involved in the dissemination of genes linked to virulence and transmission. PLoS One 2012; 7:e41247; PMID:22911766; http://dx.doi.org/10.1371/journal.pone.0041247
- Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Research 2014; 42:D206-14; PMID:24293654; http://dx.doi.org/10.1093/nar/gkt1226
- Wang L, Chen S, Xu T, Taghizadeh K, Wishnok JS, Zhou X, You D, Deng Z, Dedon PC. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol 2007; 3:709-710; PMID:17934475; http://dx.doi.org/10.1038/nchembio.2007.39
- He W, Huang T, Tang Y, Liu Y, Wu X, Chen S, Chan W, Wang Y, Liu X, Chen S, et al. Regulation of DNA phosphorothioate modification in Salmonella enterica by DndB. Scientific Reports 2015; 5:12368; PMID:26190504; http://dx.doi.org/10.1038/srep12368
- Gorbalenya AE, Koonin EV. Endonuclease (R) subunits of type-I and type-III restriction-modification enzymes contain a helicase-like domain. FEBS Letters 1991; 291:277-81; PMID:1657645; http://dx.doi.org/10.1016/0014-5793(91)81301-N
- Carstens AB, Kot W, Hansen LH. Complete genome sequences of four novel Escherichia coli bacteriophages belonging to new phage groups. Genome Announc 2015; 3; PMID:26184932; http://dx.doi.org/10.1128/genomeA.00741-15
- McCarty RM, Bandarian V. Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem & Biol 2008; 15:790-8; http://dx.doi.org/10.1016/j.chembiol.2008.07.012
- Hurt JK, Olgen S, Garcia GA. Site-specific modification of Shigella flexneri virF mRNA by tRNA-guanine transglycosylase in vitro. Nucleic Acids Res 2007; 35:4905-13; PMID:17626052; http://dx.doi.org/10.1093/nar/gkm473
- Nonekowski ST, Kung FL, Garcia GA. The Escherichia coli tRNA-guanine transglycosylase can recognize and modify DNA. J Biol Chem 2002; 277:7178-82; PMID:11751936; http://dx.doi.org/10.1074/jbc.M111077200
