ABSTRACT
Recently, a role for the anticodon wobble uridine modification 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U) has been revealed in the suppression of translational +1 frameshifts in Saccharomyces cerevisiae. Loss of either the mcm5U or s2U parts of the modification elevated +1 frameshift rates and results obtained with reporters involving a tRNALysUUU dependent frameshift site suggested these effects are caused by reduced ribosomal A-site binding of the hypomodified tRNA. Combined loss of mcm5U and s2U leads to increased ribosome pausing at tRNALysUUU dependent codons and synergistic growth defects but effects on +1 frameshift rates remained undefined to this end. We show in here that simultaneous removal of mcm5U and s2U results in synergistically increased +1 frameshift rates that are suppressible by extra copies of tRNALysUUU. Thus, two distinct chemical modifications of the same wobble base independently contribute to reading frame maintenance, loss of which may cause or contribute to observed growth defects. Since the thiolation pathway is sensitive to moderately elevated temperatures in yeast, we observe a heat-induced increase of +1 frameshift rates in wild type cells that depends on the sulfur transfer protein Urm1. Furthermore, we find that temperature-induced frameshifting is kept in check by the dehydration of N6-threonylcarbamoyladenosine (t6A) to its cyclic derivative (ct6A) at the anticodon adjacent position 37. Since loss of ct6A in elp3 or urm1 mutant cells is detrimental for temperature stress resistance we assume that conversion of t6A to ct6A serves to limit deleterious effects on translational fidelity caused by hypomodified states of wobble uridine bases.
Introduction
tRNA molecules from all three domains of life undergo extensive post-transcriptional modification, some of which are capable of modulating certain tRNA functions during the translation process.Citation1 One of these is the conserved wobble base modification 5-methoxy-carbonyl-methyl-2-thio-uridine (mcm5s2U), which is found in the anticodons of tRNAGlnUUG, tRNAGluUUC and tRNALysUUU.Citation2-4 Three distinct enzyme sets are required to convert the unmodified wobble uridine to mcm5s2U.Citation5 The six subunit Elongator complex (Elp1-Elp6) along with accessory proteins is required for the addition of a chemical side chain (likely a 5-carboxymethyl-chain, cm5) that is subsequently converted to mcmCitation5 by the action of the Trm9-Trm112 methyltransferase complex.,Citation2,6–19 The thiolation (s2U) involves members of the URM1 pathway, a sulfur transfer system consisting of Nfs1, Tum1, Urm1, Uba4 and the thiolase complex Ncs2-Ncs6.Citation7,20–22 In this pathway, sulfur is mobilized from cysteine and transferred to the thiolase complex via the sulfur transfer protein Urm1, which also acts as an ubiquitin-like protein modifier.Citation23–26 In budding yeast, the modifications of the uracil base at C2 (s2U) and C5 (mcm5U) occur independently of each other and inactivation of either alone results in shared pleiotropic phenotypes including sensitivities to various stressors and growth defects at elevated temperature.Citation5 Simultaneous inactivation of both s2U and mcm5U modifications results in synergistic growth defects that are partly suppressible by elevated copy numbers of the three tRNAs normally carrying mcm5s2U.Citation27,28 Suppression efficiency is maximal when copy numbers of all three tRNAs are jointly elevated but detectable phenotypic rescue became already apparent when tRNALysUUU was individually overexpressed, suggesting a major part of the growth defects does result from tRNALysUUU malfunction in the absence of the wobble uridine modification.Citation27 Recently, the mcm5U and s2U parts of the mcm5s2U modification were further implicated in the maintenance of translational accuracy.Citation29 Absence of either increased ribosomal +1 frameshifting at tRNAGluUUC and tRNALysUUU dependent test codons in dual luciferase based reporter constructs. To clarify whether induced frameshifting in the absence of mcm5U is due to weakened A-site or P-site binding, a series of modified Ty1 frameshift sites fused to a lacZ reporter gene were employed.Citation29 In the original Ty1 sequence, frameshifting is induced by a “hungry” AGG (Arg) codon following the CUU (Leu) codon.Citation30 The hungry Arg codon was replaced by the tRNALysUUU dependent AAA codon and strongly increased frameshift rates were recorded when the mcmCitation5 part of the mcm5s2U modification was lacking in tRNALysUUU of an Elongator (elp3) mutant.Citation29 Furthermore, elevated copy numbers of tRNALysUUU suppressed the increase in frameshifting in the elp3 mutant, suggesting that loss of mcm5U in tRNALysUUU induces +1 frameshifts mainly by reduced A-site binding of the hypomodified tRNA rather than causing effects by affecting efficiency of P-site binding.Citation29 This conclusion is further supported by in vitro assays demonstrating a reduced A-site binding capacity of tRNALysUUU in the absence of either s2U or mcm5U.Citation31
Recently, the combination of mcm5U and s2U modification defects was shown to induce an increased ribosomal stalling at mcm5s2U dependent AAA (Lys) and CAA (Gln) codons. This has a negative impact on co-translational folding of nascent polypeptide chains and triggers a widespread protein aggregation phenomenon.Citation28 Ribosomal stalling and protein aggregation were significantly suppressed by elevated levels of the three tRNAs normally carrying the mcm5s2U modification.Citation28 Since growth phenotypes and protein aggregation are suppressible by elevated tRNA copy numbers, it seems likely that phenotypes in the mcm5s2U minus mutant are caused by the protein aggregation effect.Citation28 However, in light of the contribution of mcm5U and s2U to the suppression of translational frameshifting,Citation29 it seemed possible that combined loss of mcm5U and s2U not only leads to increased protein aggregation but also impairs translational accuracy, which was investigated in this study. Further, wobble uridine thiolation was recently found in several independent studies to decrease in budding yeast under conditions of mild heat stress,Citation32–35 suggesting that elevated temperature may already cause measurable effects on +1 translational frameshifting. Since tRNALysUUU, additionally harbors a modified adenosine at position 37 (cyclic N6-threonyl-carbamoyl-adenosine, ct6A), which is already implicated in suppression of ribosomal frameshiftsCitation36,37 we monitored how this modification impacts on the translational effects of heat induced suppression of wobble uridine thiolation.
Results and discussion
Independent suppression of +1 frameshifts by s2U and mcm5U
To further check for a role of the mcm5U or s2U parts of the wobble uridine modification mcm5s2U in translational accuracy, we utilized +1 frameshift reporters containing a modified Ty1 frameshift site ().Citation29 Different yeast strains of the BY4741 background were transformed with an in-frame control construct (F12) and the test construct (W12). Both constructs contain the modified Ty1 frameshift site CTT-AAA-C decoded by tRNALeuUAG (CTT) and tRNALysUUU (AAA) in a HIS4A::lacZ reporter cassette (). F12 and W12 constructs differ in the position of the lacZ gene, which is in the 0 frame in F12 and in the +1 frame in W12. Previous work established that in the W303 strain background, loss of elp3, which removed the mcm5U side chain from tRNALysUUU decoding the AAA codon in the frameshift site, strongly induced normalized frameshift rates in comparison to the wild type.Citation28 We also detected a significant induction of +1 frameshift rates (4.5-fold) in the BY4741 elp3 mutant (Tab. 1; ). Deletion of the gene for the sulfur carrier Urm1, which is essential for wobble uridine thiolation, similarly increased frameshift rates (5.6-fold) (Tab. 1; ). To test for a potential contribution of the protein modifier function of Urm1, we compared frameshift rates between urm1 and ncs2. Loss of URM1 disables both protein modification and tRNA thiolation, whereas loss of the thiolase subunit Ncs2 disables tRNA thiolation alone.Citation7,22,23,25 We detected no significant difference between frameshift rates of both mutants (Tab. 1), suggesting the effects of URM1 deletion on translational fidelity exclusively result from loss of s2U formation. Since double deletion of elp3 and urm1, which concomitantly removes both mcm5U and s2U from wobble uridines is permitted in BY4741 (but not in the W303 strain used in the previous study)Citation27,38 we also analyzed frameshift rates in the double mutant and found a synergistic ∼16-fold increase compared with the wild type. Frameshift rates in the double mutant were significantly increased (p < 0.05) in comparison to either single mutant (Tab. 1; ), indicating the sulfur at position 2 and the mcm side chain at position 5 of the wobble base (U34) do contribute to translational accuracy independently of each other. To test whether loss of s2U and combined loss of mcm5U and s2U increase frameshift rates through an A site effect (), we analyzed whether the increase in the frameshift rates is suppressible by an extra tRNALysUUU gene copy, tK(UUU), as was previously described for rescue of elp3 phenotypes by higher-than-normal levels of tRNALysUUU.Citation29 Indeed, the extra tK(UUU) copy present in the reporter constructs significantly reduced frameshift levels in urm1 and elp3 urm1 (Table. 1; ) mutants, consistent with the interpretation that a delayed A-site entry of hypomodified tRNALysUUU induces a reading frame shift of the tRNALeuUAG occupying the ribosomal P-site (). Since combination of mcm5U and s2U modification defects has deleterious effects on growth and stress resistance and these effects are suppressible by elevated levels of those three tRNAs that normally harbor the modification,Citation27 we conclude that the observed strong increase in frameshift rates may contribute to growth defects, in addition to the described effects on protein homeostasis.Citation28 Complete loss of another anticodon loop modification (ct6A) similarly causes growth defects, stress sensitivity, protein aggregation and ribosomal frameshift induction, which may be due to changes in the efficiency of codon occupancy.Citation39 Hence it seems likely that tRNA modifications other than mcm5s2U independently add to the efficiency of A-site binding and thereby explain synergistic phenotypes observed upon combined removal of different modifications from bases in tRNA anticodon loops.Citation40
Figure 1. Anticodon loop modifications of tRNALysUUU and translational +1 frameshift models. (A) Schematic representation of tRNALysUUU anticodon loop with ct6A (6) and mcm5s2U (*) modifications. Gene-products relevant for synthesis of the modifications and addressed in this study are indicated. (B) Schematic representation of the modified Ty1 frameshift construct sensitive to tRNALysUUU modification defects.Citation28 Shown is the W12 construct where lacZ is in the +1 frame with respect to HIS4A.Citation28 The shift site as well as amino acids specified and +1 and 0 frame UGA stop codons are indicated. (C) With fully modified tRNALysUUU frameshift rates are low and termination at the UGA downstream of the Lys codon occurs efficiently. (D) Frameshift induction by tRNALysUUU modification defects via an A-site effect. Hypomodified tRNALysUUU (absence of * and 6, stippled) inefficiently binds to the AAA codon while peptidyl-tRNALeuUAG occupies the P-site. This pausing allows the peptidy-tRNALeuUAG to shift to the +1 frame and the appearance of an AAC codon in the A site (read by tRNAAsnGUU). This effect would be suppressible by extra copy of tRNALysUUU.Citation28 (E) Frameshift induction by tRNALysUUU modification defects via a P-site effect. Hyopmodified tRNALysUUU (absence of * and 6) binds to the AAA codon and translocation occurs. Once present in the P-site as peptidyl tRNALysUUU, binding is weakened (loosing P-site grip) and +1 frameshift occurs. This effect would not be suppressible by extra copy of tRNALysUUU.
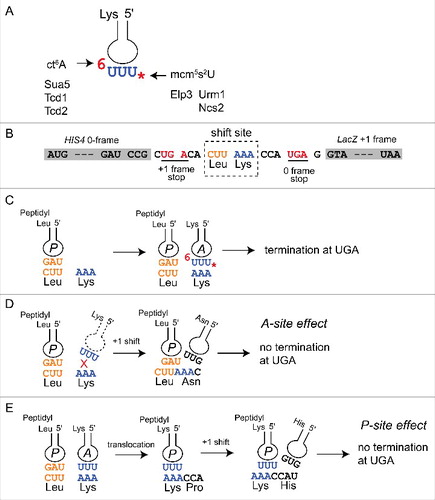
Figure 2. Induction of +1 translational frameshift rates relative to the wild type. Fold induction rates were calculated based on normalized frameshift values [%] shown in and significance of differences determined using two-tail t-test. n.s.: not significantly different. Heat indicates moderate thermal stress condition during growth (37°C) and + tK(UUU) refers to frameshift levels measured with the WA-2 and FA-1 constructsCitation28 that carry an additional copy of the tK(UUU) gene.
![Figure 2. Induction of +1 translational frameshift rates relative to the wild type. Fold induction rates were calculated based on normalized frameshift values [%] shown in Table 1 and significance of differences determined using two-tail t-test. n.s.: not significantly different. Heat indicates moderate thermal stress condition during growth (37°C) and + tK(UUU) refers to frameshift levels measured with the WA-2 and FA-1 constructsCitation28 that carry an additional copy of the tK(UUU) gene.](/cms/asset/c658530b-7f29-4d63-8e5b-92da53b12fe1/krnb_a_1267098_f0002_b.gif)
Elevated temperature impacts on translational accuracy via the Urm1 pathway
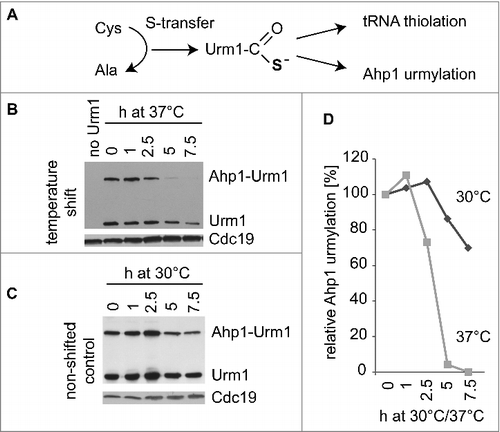
Recent work revealed an unanticipated sensitivity of several of the thiolation pathway components to elevated temperature.Citation32–35 This in turn leads to significantly reduced wobble uridine thiolation levels which are already detectable in BY4741 wild type cells after modest increase of temperatures to 33°C.Citation33 Hence, it can be expected that mild heat stress increases translational +1 frameshift levels due to its destabilizing effects on the tRNA thiolation pathway. To test this assumption, we grew wild type cells harboring the W12 or F12 constructs at mild heat stress conditions (37°C), measured +1 frameshift rates and compared them to rates observed under non-stress condition. We found that such elevated growth temperature increases +1 frameshift levels ∼5-fold (Tab 1; ), a value similar to that seen in urm1 cells grown under non-stressed conditions. Furthermore, extra copies of tRNALysUUU significantly reduce the frameshift inducing effect of temperature stress (Tab. 1; ). Since both elevated temperature and loss of URM1 have comparable effects on +1 frameshift levels and similarly respond to increased tRNALysUUU doses, it seemed likely that the temperature effect is largely due to inactivation of the Urm1 dependent tRNA thiolation pathway. We took advantage of the fact that urm1 mutants are hardly growth affected under such mild heat stress conditions 2527 and tested whether temperature effects on +1 frameshift levels depend on a functional Urm1 pathway. urm1 mutants harboring the W12 and F12 construct were grown at 37°C, +1 frameshift rates were measured and compared with levels measured under non-stress conditions. As shown in and , there is no significant difference in +1 frameshift levels between urm1 cells grown at 30°C or at 37°C. These results suggest that moderate heat stress causes a significant increase in translational inaccuracy via the destabilization of the Urm1 pathway components. To test whether moderate heat stress (37°C) indeed reduces sulfur flow through the Urm1 pathway, we monitored the relative efficiency of Ahp1 urmlyation, a process that is dependent on the thiocarboxylate formation at the Urm1 C-terminus. We have recently shown that more harsh heat stress (39°C) reduces both sulfur dependent Ahp1 urmylation and Urm1 stability without affecting Ahp1 levels.Citation26 Thus, in order to check whether temperature shift to 37°C already affects both the tRNA thiolation and urmylation branches of the Urm1 pathway, we analyzed levels of Ahp1-Urm1 conjugates and free Urm1 after temperature shift from 30°C to 37°C by Western analysis (). We detected a clear reduction of Ahp1 urmylation already after 2.5h at 37°C and a reduction of detectable free Urm1 after 7.5 h. This result suggests that mild heat stress which increases +1 frameshift rates (Tab. 1, ) has a direct effect on the Urm1 pathway, which might result in part from a destabilization of the sulfur carrier and a reduction of sulfur flow to Urm1. The latter notion is supported by a more rapid decline of Ahp1-Urm1 conjugates compared with Urm1 itself () and a previously reported instability of Tum1 at 37°C, which operates upstream of Urm1 in the sulfur transfer pathway.Citation34 Since neither wild type nor the urm1 mutant show a significant growth defect under mild heat stress conditions that induce +1 frameshift rates via downregulation of the Urm1 pathway, we assume that cells are well capable of handling the consequences of such disturbance of the translation machinery. Most likely, this occurs by proteasomal degradation of non-functional polypeptides generated due to translational frameshift events. In support, proteins induced in urm1 and elp3 cells are strongly enriched in proteasomal subunits and regulators,Citation31 suggesting an increased need for protein degradation and turnover in the absence of s2U modification. We show here that upon combination of mcm5U and s2U defects, frameshift rates synergistically increase to much higher levels compared with those seen in heat stressed cells; at the same time, a general growth defect becomes apparent,Citation27 suggesting that growth defects may correlate with an overflow of the cell's capacity to handle non-functional polypeptides. In support of this notion, it was demonstrated that cells lacking mcm5U and s2U display a severely reduced ability to respond to acute proteotoxic stress,Citation28 a condition that puts an additional burden on the cellular resources for handling damaged proteins.
Table 1. Normalized frameshift rates in different yeast strains and growth conditions.
Figure 3. Effect of temperature shift on Ahp1 urmylation and Urm1 levels. (A) Sulfur transfer from cysteine to Urm1 results in formation of Urm1 thiocarboxylate, which is required for covalent attachment of Urm1 to Ahp1 (urmylation) and tRNA thiolation. (B) Temperature shift assay. urm1 mutants expressing TAP-tagged Urm1Citation25 were cultivated at 30°C in YNB media to OD600nm = 1 after which one half of the culture was shifted to 37°C. At indicated time points, total protein extracts were prepared and subjected to anti-TAP Western analysis to detect free Urm1 as well as the slower migrating Ahp1-Urm1 conjugate. Previous studies with ahp1mutants confirmed the the slower migrating band to result from Ahp1 urmylation by the tagged Urm1 variant.Citation25,26 To verify equal loading, anti-Cdc19 antibodies were utilized. (C) As in (B) but with the culture part that remained at 30°C. (D) Signal intensities of slower migrating band in (B) and (C) were quantified and relative levels of Ahp1 urmylation at 30°C or 37°C were calculated by dividing signal intensity at time points 2.5h, 5h or 7.5h by the one of timepoint 0.
Role of the anticodon adjacent ct6A modification in the prevention of heat induced +1 frameshifts
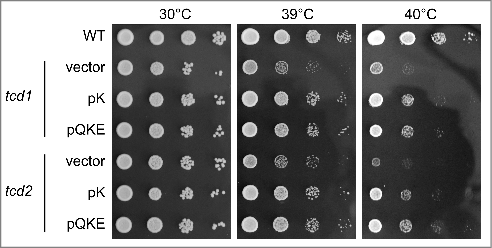
Since the frameshift events recorded in this study are caused by a wobble uridine modification defect in tRNALysUUU, we investigated the relevance of cyclic N6-threonyl-carbamoyl-adenosine which is found in the anticodon loop of this tRNA at position 37 (modomics.genesilico.pl). Complete loss of this modification is known to induce translational frameshift events in yeast,Citation36 however, the role of the dehydration of t6A to its cyclic form ct6A in yeast is unknown.Citation37 Conversion of t6A to ct6A is abolished in tcd1 and tcd2 mutants and both show negative genetic interaction with Urm1 pathway and Elongator genes.Citation37,40,41 In addition, combination of tcd1 and Elongator mutations was shown to increase the formation of protein aggregates, which is likely due to increased ribosomal pausing.Citation40 Hence, ct6A might play a functionally overlapping role with mcm5U and s2U in the suppression of translational frameshifts by independently contributing to the efficiency of A-site binding. To investigate this possibility, we introduced the W12 and F12 frameshift reporter constructs into a tcd1 mutant strain and measured frameshift rates at non-stress and mild heat stress conditions (37°C). While loss of the modification has no effect on frameshift levels in non-stressed cells, there is a significant increase in +1 frameshift events at 37°C when ct6A is absent compared with wild type cells at the same condition (Tab. 1; ). At further elevated temperatures, both tcd1 and tcd2 single mutants display a growth defect that can be rescued specifically by overexpression of tRNALysUUU (), suggesting that in the absence of ct6A (presence of t6A) and elevated temperature, malfunction of tRNALysUUU is responsible for the observed growth defect. Among the three tRNAs that are subject to heat induced shut-off of wobble uridine thiolation, tRNALysUUU is the single species that also carries ct6A (modomics.genesilico.pl). Since elevated temperature induces a loss of s2U, it appears likely that temperature sensitivity of tcd1 or tcd2 cells occurs due to further increased translational frameshifting by tRNALysUUU when both ct6A and s2U are lacking. To verify this assumption, we measured frameshift rates in tcd1 urm1 double mutants at non-stress conditions. Indeed, frameshift levels are elevated to similar levels in tcd1 single mutants grown under heat stress and tcd1 urm1 double mutants grown under non-stress conditions (Tab. 1). We conclude that the dehydration of t6A to ct6A counteracts the frameshift inducing effects of mild temperature stress in budding yeast. Since ct6A is the natural configuration of t6A in several, but not all cell types and temperature effects on the thiolation pathway occur only in some yeast species,Citation31 it would be interesting to see if this form of t6A has co-evolved with a temperature responsive thiolation pathway to limit its negative influence on translational accuracy. The finding that loss of ct6A does not influence +1 frameshift rates when tRNA thiolation is unaffected but increases rates of urm1 mutants or wild type cells grown under mild heat stress identifies ct6A as a modification that becomes crucial for translational efficiency particularly when s2U is missing or downregulated.
Figure 4. Heat induced growth defects in tcd1 and tcd2 single mutants are suppressible by elevated levels of tRNALysUUU. tcd1 and tcd2 mutants were transformed with either pRS425 (vector), pK (multi copy tRNALysUUU) or pQKE (multi copy tRNALysUUU, tRNAGlnUUG and tRNAGluUUC) and serial dilutions of cultures transferred to YPD plates that were incubated at the indicated temperatures for two days.
Methods
Strains, plasmids and growth assay
Yeast strains used in this study are listed in . Constructs containing the HIS4A::lacZ reporter cassette and modified Ty1 frameshift sites are described inCitation29 and are based on pMB38–9merWT or pMB38–9merFF, respectively.Citation30 Construct F12 contains the frameshift site CUU-AAA-C and the lacZ gene in reading frame 0 (in frame control). Construct W12 contains the same frameshift site and the lacZ gene in frame +1 with respect to HIS4A. Suppression analysis of frameshift rates involved constructs WA-2 and FA-1, which are similar to W12 and F12 but carry an additional insert with the tK(UUU) gene.Citation29 Plasmids were transformed into yeast strains using the lithium acetate methodCitation42 and transformants selected on yeast nitrogen based media (Foremedium, Norfolk, UK) lacking uracil. Selection plates were incubated at 30°C until colonies were formed. Independent colonies from the selection plates were used to inoculate cultures for β galactosidase assays. Growth phenotypes of tcd1 and tcd2 single mutants at different temperatures and suppression by tRNA overexpression were determined by serial dilution and spot assay as described.Citation27 Overexpression plasmids for tRNALysUUU (pK) and for combined overexpression of tRNALysUUU, tRNAGlnUUG and tRNAGluUUCCitation10,43 were used along with pRS425Citation44 as the empty vector control. Assays were performed on solid yeast peptone dextrose medium (2% glucose, 1% yeast extract, 2% peptone) and plasmid carrying cells were precultured on yeast nitrogen based minimal medium (Foremedium, Norfolk, UK) lacking leucine.
Table 2. Strains used in this study.
Determination of frameshift levels
Freshly transformed cells were grown at 30°C or 37°C in liquid yeast nitrogen based media containing 2% glucose and lacking uracil to OD600 nm of 2–3 and harvested by centrifugation. Cells were washed with water and resuspended in Z-buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 50 mM 2-mercaptoethanol, pH 7). Dilutions of this suspension were used to determine cell density by measuring the absorbance at 600 nm. 500 µl aliquots of undiluted suspension were removed to new tubes and 2 drops of 0.01% SDS solution and chloroform added followed by vortex-aided mixing of samples for 30 sec each. Samples were then incubated at 37°C for 5 min and 100 µl of 4 mg ml−1 ortho-nitrophenyl-β-galactoside dissolved in Z-buffer was added. Upon color development during incubation at 37°C reactions were stopped by the addition of 250µl 1M Na2CO3 and centrifuged at 10.000 g for 5 min. Upper phases were transferred to optical cuvettes and absorbance at 420nm determined using a photometer. β galactosidase activity units were calculated using absorbance data at 600 and 420nm as well as reaction times by employing Miller's formula.Citation44 Frameshift rates [%] were calculated by dividing the β galactosidase activity measured with the W12 (or WA-2) construct by the one measured with the F12 (or FA-1) construct. For each strain at least three independent cultures were run with both the test constructs and the in-frame control constructs and measured. Normalized frameshift rates of the biological replicates were averaged and standard deviations calculated. Statistical significance was analyzed using two-tail t-test.
Ahp1 urmylation assay
TAP-tagged Urm1 was expressed in BY4741 urm1 mutantsCitation26 and cells were grown to OD600nm = 1 after which the culture was split. One half was further incubated at 30°, whereas the other half was shifted to 37°C incubation temperature. At time points 0, 1h, 2.5h 5h and 7.5h, aliquots were removed, total protein extracts generated and Ahp1 urmylation as well as free Urm1 levels determined as described previously.Citation25,26 In parallel, equal loading was verified by anti Cdc19 Western analysis as described.Citation25,26
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Anders S. Byström for providing plasmids. We gratefully acknowledge support by the DFG (Deutsche Forschungsgemeinschaft) to RS (SCHA750/15) and funding by DFG Priority Program SPP1784 ‘Chemical Biology of Native Nucleic Acid Modifications' to RS (SCHA750/20) and RK (KL2937/1).
References
- El Yacoubi B, Bailly M, de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 2012; 46:69-95; PMID:22905870; https://doi.org/10.1146/annurev-genet-110711-155641
- Huang B, Johansson MJ, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005; 11:424-36; PMID:15769872; https://doi.org/10.1261/rna.7247705
- Johansson MJ, Esberg A, Huang B, Björk GR, Byström AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 2008; 28:3301-12; PMID:18332122; https://doi.org/10.1128/MCB.01542-07
- Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, Jäger G, Gong Z, Byström AS, Schaffrath R, Breunig KD. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol 2010; 75:1082-94; PMID:20398216; https://doi.org/10.1111/j.1365-2958.2010.07163.x
- Karlsborn T, Tükenmez H, Mahmud AK, Xu F, Xu H, Byström AS. Elongator, a conserved complex required for wobble uridine modifications in eukaryotes. RNA Biol 2014; 11:1519-28; PMID:25607684; https://doi.org/10.4161/15476286.2014.992276
- Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol 2003; 23:9283-9292; PMID:14645538; https://doi.org/10.1128/MCB.23.24.9283-9292.2003
- Huang B, Lu J, Byström AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 2008; 14:2183-94; PMID:18755837; https://doi.org/10.1261/rna.1184108
- Jablonowski D, Butler AR, Fichtner L, Gardiner D, Schaffrath R, Stark MJ. Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics 2001; 159:1479-89; PMID:11779790
- Jablonowski D, Fichtner L, Stark MJR, Schaffrath R. The yeast Elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol Biol Cell 2004; 15:1459-69; PMID:14718557; https://doi.org/10.1091/mbc.E03-10-0750
- Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol 2006; 59:677-88; PMID:16390459; https://doi.org/10.1111/j.1365-2958.2005.04972.x
- Jablonowski D, Täubert JE, Bär C, Stark MJR, Schaffrath R. Distinct subsets of Sit4 holo-phosphatases are required for inhibition of yeast growth by rapamycin and zymocin. Eukaryot Cell 2009; 8:1637-47; PMID:19749176; https://doi.org/10.1128/EC.00205-09
- Fichtner L, Frohloff F, Bürkner K, Larsen M, Breunig KD, Schaffrath R. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol Microbiol 2002; 43:783-91; PMID:11929532; https://doi.org/10.1046/j.1365-2958.2002.02794.x
- Fichtner L, Jablonowski D, Schierhorn A, Kitamoto HK, Stark MJR, Schaffrath, R. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol 2003; 49:1297-307; PMID:12940988; https://doi.org/10.1046/j.1365-2958.2003.03632.x
- Bär C, Zabel R, Liu S, Stark MJ, Schaffrath R. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol Microbiol 2008; 69:1221-33; PMID:18627462; https://doi.org/10.1111/j.1365-2958.2008.06350.x
- Studte P, Zink S, Jablonowski D, Bär C, von der Haar T, Tuite MF, Schaffrath R. tRNA and protein methylase complexes mediate zymocin toxicity in yeast. Mol Microbiol 2008; 69:1266-77; PMID:18657261; https://doi.org/10.1111/j.1365-2958.2008.06358.x
- Zabel R, Bär C, Mehlgarten C, Schaffrath R. Yeast alpha-tubulin suppressor Ats1/Kti13 relates to the Elongator complex and interacts with Elongator partner protein Kti11. Mol Microbiol 2008; 69:175-87; PMID:18466297; https://doi.org/10.1111/j.1365-2958.2008.06273.x
- Mehlgarten C, Jablonowski D, Breunig KD, Stark MJ, Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbiol 2009; 73:869-81; PMID:19656297; https://doi.org/10.1111/j.1365-2958.2009.06811.x
- Mazauric MH, Dirick L, Purushothaman SK, Björk GR, Lapeyre B. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J Biol Chem 2010; 285:18505-15; PMID:20400505; https://doi.org/10.1074/jbc.M110.113100
- Abdel-Fattah W, Jablonowski D, Di Santo R, Scheidt V, Hammermeister A, ten Have SM, Thüring KL, Helm M, Schaffrath R, Stark MJR. Phosphorylation of Elp1 by Hrr25 is required for Elongator-dependent tRNA modification in yeast. PLoS Genet 2015; 11:e1004931; PMID:25569479; https://doi.org/10.1371/journal.pgen.1004931
- Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem 2008; 283:27469-76; PMID:18664566; https://doi.org/10.1074/jbc.M804043200
- Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Research 2009; 37:1335-52; PMID:19151091; https://doi.org/10.1093/nar/gkn1023
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold, R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 2009; 458:228-32; PMID:19145231; https://doi.org/10.1038/nature07643
- Goehring AS, Rivers DM, Sprague GF Jr. Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol Biol Cell 2003; 14:4329-41; PMID:14551258; https://doi.org/10.1091/mbc.E03-02-0079
- Van der Veen AG, Schorpp K, Schlieker C, Buti L, Damon JR, Spooner E, Ploegh HL, Jentsch S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci USA 2011; 108:1763-70; PMID:21209336; https://doi.org/10.1073/pnas.1014402108
- Jüdes A, Ebert F, Bär C, Thüring KL, Harrer A, Klassen R, Helm M, Stark MJ, Schaffrath R. Urmylation and tRNA thiolation functions of ubiquitin-like Uba4·Urm1 systems are conserved from yeast to man. FEBS Lett 2015; 589:904-9; PMID:25747390; https://doi.org/10.1016/j.febslet.2015.02.024
- Jüdes A, Bruch A, Klassen R, Helm M, Schaffrath R. Sulfur transfer and activation by ubiquitin-like modifier system Uba4•Urm1 link protein urmylation and tRNA thiolation in yeast. Microb Cell 2016; 3:423-433; https://doi.org/10.15698/mic2016.11.539
- Klassen R, Grunewald P, Thüring KL, Eichler C, Helm M, Schaffrath R. Loss of anticodon wobble uridine modifications affects tRNA(Lys) function and protein levels in Saccharomyces cerevisiae. PLoS One 2015; 10:e0119261; PMID:25747122; https://doi.org/10.1371/journal.pone.0119261
- Nedialkova DD, Leidel SA. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015; 161:1606-18; PMID:26052047; https://doi.org/10.1016/j.cell.2015.05.022
- Tükenmez H, Xu H, Esberg A, Byström AS. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res 2015; 43:9489-99; PMID:26283182; https://doi.org/10.1093/nar/gkv832
- Belcourt MF, Farabaugh PJ. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell 1990; 62:339-52; PMID:2164889; https://doi.org/10.1016/0092-8674(90)90371-K
- Rezgui VA, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PG. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci U S A 2013; 110:12289-94; PMID:23836657; https://doi.org/10.1073/pnas.1300781110
- Alings F, Sarin LP, Fufezan C, Drexler HC. Leidel SA An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 2015; 21:202-12; PMID:25505025; https://doi.org/10.1261/rna.048199.114
- Han L, Kon Y, Phizicky EM. Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA 2015; 21:188-201; PMID:25505024; https://doi.org/10.1261/rna.048173.114
- Damon JR, Pincus D. Ploegh HL tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol Biol Cell 2015; 26:270-82; PMID:25392298; https://doi.org/10.1091/mbc.E14-06-1145
- Tyagi K, Pedrioli PG. Protein degradation and dynamic tRNA thiolation fine-tune translation at elevated temperatures. Nucleic Acids Res 2015; 43:4701-12; PMID:25870413; https://doi.org/10.1093/nar/gkv322
- El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crécy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J 2011; 30:882-93; PMID:21285948; https://doi.org/10.1038/emboj.2010.363
- Miyauchi K, Kimura S, Suzuki T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat Chem Biol 2013; 9:105-11; PMID:23242255; https://doi.org/10.1038/nchembio.1137
- Björk GR, Huang B, Persson OP, Byström AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 2007; 13:1245-55; PMID:17592039; https://doi.org/10.1261/rna.558707
- Thiaville PC, Legendre R, Rojas-Benítez D, Baudin-Baillieu A, Hatin I, Chalancon G, Glavic A, Namy O, de Crécy-Lagard V. Global translational impacts of the loss of the tRNA modification t6A in yeast. Microbial Cell 2015; 3:29-45; PMID:26798630; https://doi.org/10.15698/mic2016.01.473
- Klassen R, Ciftci A, Johanna Funk J, Bruch A, Butter F, Schaffrath R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res 2016; 44:10946-10959; PMID:27496282; https://doi.org/10.1093/nar/gkw705
- Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S, et al. The genetic landscape of a cell. Science 2010; 327:425-31; PMID:20093466; https://doi.org/10.1126/science.1180823
- Gietz D, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nature Protocols 2007; 2:31-4; PMID:17401334; https://doi.org/10.1038/nprot.2007.13
- Lu J, Huang B, Esberg A, Johansson MJ, Byström AS. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA 2005; 11:1648-54; PMID:16244131; https://doi.org/10.1261/rna.2172105
- Miller JH. Experiments in Molecular Genetics 1972; Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
