ABSTRACT
For a long time, eukaryotic stand-alone pseudouridine synthases (Pus enzymes) were neglected as non-essential enzymes adding seemingly simple modifications to tRNAs and small nuclear RNAs. Most studies were limited to the identification and initial characterization of the yeast Pus enzymes. However, recent transcriptome-wide mapping of pseudouridines in yeast and humans revealed pervasive modification of mRNAs and other non-coding RNAs by Pus enzymes which is dynamically regulated in response to cellular stress. Moreover, mutations in at least 2 genes encoding human Pus enzymes cause inherited diseases affecting muscle and brain function. Together, the recent findings suggest a broader-than-anticipated role of the Pus enzymes which are emerging as potential regulators of gene expression. In this review, we summarize the current knowledge on Pus enzymes, generate hypotheses regarding their cellular function and outline future areas of research of pseudouridine synthases.
Introduction
The cellular transcriptome contains far more than one hundred different modifications that play a variety of roles in the cell including the translation, localization and stabilization of RNA.Citation1 These post-transcriptional modifications to the 4 standard RNA nucleotides range in chemical diversity from small structural changes to large, multi-step modifications. Of these modifications, pseudouridine (Ψ), a seemingly simple modification, is the most abundant, found in all 3 phylogenetic domains of life and has been referred to as the fifth nucleoside.Citation2 Many of the pseudouridine modifications in all types of RNA are generated by stand-alone enzymes called pseudouridine synthases, also called Pus enzymes in eukaryotes (in contrast to more complex H/ACA small ribonucleoproteins). Pseudouridine is a C5-glycosidic isomer of uridine that contains a C-C bond between the C1 of the ribose sugar and the C5 of uracil, rather than the usual C1-N1 bond found in uridine (). Pseudouridine is found in almost all types of non-coding RNA such as tRNA, rRNA and small nuclear RNA (snRNA) and recently also in coding RNA. This base modification is able to stabilize RNA and improve base-stacking by forming additional hydrogen bonds with water through its extra imino group.Citation3 The TΨC arm of tRNAs contains a nearly universally conserved pseudouridine at position 55 that implies a role in the basics of translation. rRNAs have numerous pseudouridine modifications near the peptidyltranferase center, the subunit interface of the ribosome and the decoding site. There are 11 pseudouridines in the Escherichia coli rRNA, 30 in yeast cytoplasmic rRNA and a single modification in mitochondrial 21S rRNA, and about 100 pseudouridines in human rRNA indicating that the extent of pseudouridylation increases with the complexity of an organism.Citation1 Deletion studies of these rRNA modifications in Saccharomyces cerevisiae revealed effects on cell growth, translation accuracy and ribosomal assembly when several pseudouridines were removed together.Citation4 Besides the ribosome, pseudouridines are also present in the second largest ribonucleoprotein machinery involved in gene expression, the spliceosome, specifically in the snRNAs U1, U2, U4, U5 and U6 as well as in the minor spliceosomal snRNA U12, U4atac and U6atac.Citation1,5,6 Of these, pseudouridylation of U2 snRNA has been the most extensively studied revealing both an important role of pseudouridines for splicing as well as inducible pseudouridine formation that is triggered by the TOR pathway.Citation7-11 Similarly, a pseudouridine in U6 snRNA is induced during filamentous growth and influences splicing efficiency of suboptimal introns.Citation5 Last but not least, pseudouridine modifications are also implicated in human diseases such as mitochondrial myopathy and sideroblastic anemia (MLASA) and Dyskeratosis congenita.Citation12,13 Together, all of these findings highlight the importance of converting uridines to seemingly very similar pseudouridines.
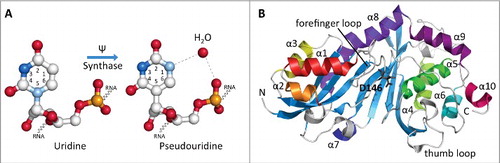
Figure 1. Structures of uridine, pseudouridine and a representative eukaryotic pseudouridine synthase. (A) Ball-and-stick structural representations of uridine and pseudouridine located within an RNA strand. Pseudouridine synthases convert the usual N-C glycosidic bond between N1 of the uracil ring and the ribose sugar into a C-C bond involving C5. N1 becomes available to form additional hydrogen bonds (dashed lines) with a water molecule (red sphere) increasing the structural stability of the RNA. Carbon atoms are shown in white, oxygen in red, phosphorous in orange and nitrogens in bright blue (N3) and light blue (N1). The structures of U12 and Ψ2604 were used as examples from the cryo-EM structure of the E. coli ribosome-EF-Tu complex (PDB ID: 5AFI). (B) Ribbon diagram of the crystal structure of the catalytic domain of human Pus1 protein shown as a representative Pus enzyme structure (PDB ID: 4IQM). The N-and C-termini are labeled with N and C, respectively. The 10 α-helices are colored and labeled according to their order in the structure. The 8-stranded mixed β-sheet is shown in blue. The catalytic aspartate residue (D146) located in the cleft of the catalytic domain is shown as a stick model and colored in black. The forefinger and thumb loops are involved in RNA recognition in the bacterial homolog TruA.
In the cell, post-transcriptional pseudouridine formation is achieved by a family of related enzymes called pseudouridine synthases that fall under one of 2 categories. The best understood are guide RNA-dependent enzymes called H/ACA sRNPs (small ribonucleoproteins) that utilize a guide RNA that is complementary to the sequence to be modified for target selection and a protein (Cbf5 in yeast, dyskerin in humans) for catalyzing the modification.Citation14 H/ACA sRNPs are found in eukaryotes and archaea, but not prokaryotes, and modify many uridines, all recruited to the enzyme by a multitude of guide RNAs. The pseudouridines found in the yeast ribosome are all modified by H/ACA sRNPs found in the nucleolus and therefore called small nucleolar RNPs (snoRNPs). In addition, H/ACA sRNPs are involved in modification of U2 snRNA and mRNA, but they also play a role in other cellular processes such as processing of pre-rRNA and stabilization of the mammalian telomerase RNA component (TERC). In this review, we focus on the second type of pseudouridine synthases found in eukaryotes as several reviews cover H/ACA sRNPs.Citation14-17
The alternative to using the RNA-guided mechanism for pseudouridine formation is a guide-independent mechanism acting through stand-alone enzymes. In eukaryotes, these enzymes are called Pus enzymes (Pseudouridine synthases), and they modify uridine by recognizing sequence and/or secondary structural elements of the RNA containing the target uridine. These enzymes are related to their bacterial counterparts, and together pseudouridine synthases are classified into 6 families (TruA, TruB, TruD, RsuA, RluA and Pus10).Citation18 However, RsuA-type pseudouridine synthases have so far only been found in bacteria while the Pus10 enzymes are only present in a few archaeal and eukaryotic organisms.Citation19 Importantly, all pseudouridine synthases, including Cbf5, share a conserved catalytic domain and presumably a conserved catalytic mechanism based on a strictly conserved catalytic aspartate residue ( and ). Despite sequence divergence between pseudouridine synthases, the conserved structure strongly suggests a common evolutionary origin for all these enzymes. In general terms, pseudouridylation involves the cleavage of the N-C glycosidic bond, rotation of uracil ring and reattachment to the ribose sugar without the requirement of ATP for catalysis. Most recently, the catalytic mechanism of pseudouridine synthases has been described to occur through deprotonation of the ribose sugar by the catalytic aspartate residue resulting in detachment of the base and a glycal intermediate.Citation20 Whereas many structures are available for prokaryotic and archaeal pseudouridine synthases, the only structures to date of eukaryotic Pus enzymes are that of the catalytic domain of human Pus1 and human Pus10.Citation21,22 Pus1 and Pus10 adopt a fold similar to the conserved catalytic domain found in prokaryotic enzymes with the central anti-parallel β sheet flanked by helices and loops (). In eukaryotes, there are 10 different types of Pus enzymes called Pus1 to Pus10 with some organisms having several paralogs of a given Pus enzyme (). Most investigations of Pus enzymes have been conducted using S. cerevisiae which has 9 Pus enzymes (Pus1 to Pus9, and ). Each Pus enzyme has either a few specific, or sometimes multiple, target uridine(s) to modify in the cell, and these modifications may take place in the nucleus, cytoplasm or mitochondria. In addition to the pseudouridines in cytoplasmic and mitochondrial tRNAs (), Pus enzymes also modify several small nuclear RNAs (snRNAs) and mitochondrial rRNA. The Pus10 enzyme which is not found in yeast, has only been studied in archaea and is responsible for the formation of pseudouridine 54 and 55 in tRNA;Citation23 notably, pseudouridine 55 in eukaryotic tRNA is (also) generated by Pus4.Citation24
Figure 2. Domain structure and localization of the Pus enzymes. (A) All Pus enzymes share a common catalytic core domain (shown in light blue), with some variations depending on which family of enzymes they belong to. Some of the enzymes have N- or C-terminal extensions beyond the catalytic domains (shown as narrow gray bars), while others have additional domains. Pus7 has an insertion in its catalytic domain (yellow), characteristic of members of the TruD family. Pus8 has an S4-like domain (green) at its N-terminus that aids in binding to RNA and a C-terminal cytidine deaminase domain (red) involved in riboflavin biosynthesis. Pus9 also contains the S4-like domain, but is preceded by an N-terminal mitochondrial targeting sequence (orange). Pus10 contains a THUMP domain (purple). Sequence numbering refers to the yeast proteins with the exception of Pus10 which is not found in yeast; in this case the numbering refers to the Pyrococcus furiosus enzyme. The total number of amino acids in each protein is shown (black) and the catalytic aspartate is indicated by an arrow and residue number above the corresponding catalytic domain (magenta). Domain location and sizes have been approximated based on sequence alignments, domain predictions and homologous structures. (B) Within a schematic yeast cell, the enzyme name is shown in each of the cellular compartments in which the enzyme is found, either in the nucleus (purple), cytoplasm (blue) or mitochondria (orange). Some enzymes are found in more than one cellular compartment. (Source: yeastgenome.org).
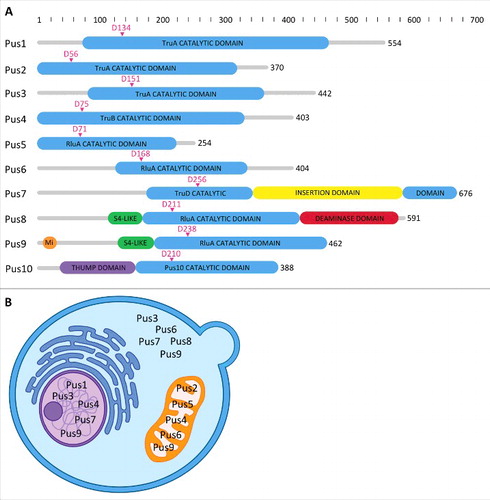
Table 1. Summary of yeast Pus enzymes. Other names for the enzymes are shown in parentheses. The family names are based on the bacterial representatives. Uridines that are modified by each enzyme are indicated by the cellular compartment, the type of RNA, and their position number. The modifications at position 35 occur in pre-tRNATyr (*) and the modification of U56 of U2 snRNA occurs only after nutrient deprivation and heat shock (#). Localization data are from the yeast genome database (yeastgenome.org) and indicated as either cytoplasmic (Cy), nuclear (Nu) or mitochondrial (Mi). The protein size is based on amino acid sequence and represented in kiloDaltons (kDa). The human homologs of the Pus enzymes are Pus1, Pus3, TruB1, TruB2, RusD1, RusD2, RusD3, RusD4, Pus7, Pus7L and Pus10.
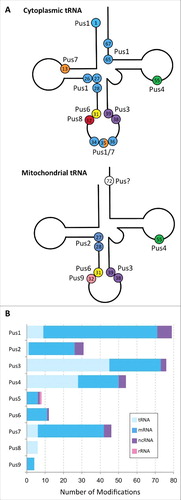
Fiure 3. Target sites of stand-alone pseudouridine synthases. (A) Cloverleaf representations of cytoplasmic and mitochondrial tRNAs showing the positions of pseudouridine modification and the Pus enzymes responsible. Cytoplasmic tRNAs can be pseudouridylated at 15 possible sites by Pus enzymes, shown as colored circles with numbers indicating their position. U35 is modified in pre-tRNATyr(GUA) by either Pus1 and Pus7. Mitochondrial tRNAs have 8 potential pseudouridines. The enzyme responsible for the modification at position 72 of mitochondrial tRNA is presently unknown. (B) General types of RNAs that are modified by each Pus enzyme under standard growth conditions. This data are based on the transcriptome-wide mapping studies by Carlile et al. and Schwartz et al. using the Pseudo-seq (or ψ-seq) technique for the identification of pseudouridines present in S. cerevisiae. The combined number of each type of RNA (tRNA, mRNA, ncRNA and rRNA) modified by each of the Pus enzymes is shown as a colored bar. Note, that alteration of growth conditions such as heat stress and nutrient deprivation leads to additional pseudouridylation events by some Pus enzymes, for example by Pus7. These additional events are not included in this figure.
Most recently, mapping of pseudouridines across the entire transcriptome of yeast and human cells has revealed their presence in mRNA transcripts.Citation25-28 By applying pseudouridine sequencing to yeast deletion strains, several Pus enzymes have been identified as the proteins responsible for the pseudouridines found in mRNA. For example in one study, Pus1 alone was found to modify over 60 mRNAs.Citation25 Notably, heat shock of yeast cells was found to result in a differential pattern of pseudouridylation compared with normal growth conditions.Citation27 Together, these studies raise the interesting hypothesis that Pus enzymes play a much bigger role in the cell than previously anticipated. By modifying not only non-coding RNAs, but also mRNAs, Pus enzymes may contribute more broadly to the regulation of gene expression than by simply modifying tRNAs, rRNAs and snRNAs. One interesting possible mechanism of Pus enzymes might be to alter translation of pseudouridylated mRNAs in response to changes in growth conditions or to adjust the stability of mRNA transcripts. While it remains unclear whether pseudouridines alter the reading of sense codons,Citation29 pseudouridine has been found to suppress translation termination of nonsense codons by enabling reading of these pseudouridylated stop codons by different tRNAs.Citation30 This alteration of stop codons to sense codons has only been observed in engineered systems and may be selected against in vivo; nevertheless, it remains a possibility that Pus enzymes could contribute to read-through of stop codons in vivo in a few specific mRNAs. Despite this recently renewed interest in Pus enzymes, the eukaryotic stand-alone pseudouridine synthases are not as well studied compared to the bacterial pseudouridine synthases and to H/ACA sRNPs. Also, most of the currently available information on Pus enzymes has been derived from studies using S. cerevisiae with only some reports on other organisms including humans. As we anticipate Pus enzymes to become the focus of research in the near future, this review therefore highlights the current knowledge on eukaryotic stand-alone pseudouridine synthases while also providing hypotheses on their cellular function.
Overview of the different Pus enzymes
First, we will begin with a description of all the major eukaryotic stand-alone pseudouridine synthases to introduce each enzyme and its characteristics based on the current literature. Due to the limited number of reports on these proteins, this overview will necessarily focus on the 9 Pus enzymes found in yeast and a brief description of Pus10. As mentioned above, these enzymes belong to 4 families named after the E. coli pseudouridine synthases (). In yeast, the TruA family, with its members Pus1, Pus2 and Pus3, is responsible for modifications at numerous sites in tRNAs as well as some positions in snRNAs and mRNAs. The highly conserved U55 of tRNA is modified by Pus4, the only member of the TruB family, which also modifies mRNA. The RluA enzyme family is the largest including Pus5, Pus6, Pus8 and Pus9 which pseudouridylate uridines in tRNAs, mitochondrial 21S rRNA and mRNA. The RluA enzymes sometimes contain an N-terminal domain that resembles S4, a ribosomal constituent protein (). Pus7 is the only member of the TruD family and modifies sites in tRNAs, rRNA and snRNA.
Pus1 – This enzyme displays a broad specificity for target uridines in tRNA that can be in non-contiguous positions.Citation31 Specifically, Pus1 modifies cytoplasmic tRNAs at positions 1 (5′ terminus of acceptor stem), 26–28 (pseudouridines at these positions at the 5′ end of anticodon arm are only found in eukaryotic tRNAs), 34–36 (anticodon positions), 65 and 67 (3′ end of TΨC arm).Citation32,33 In addition, Pus1 also acts on U44 of U2 snRNA and U28 of U6 snRNA.Citation5,6 As mentioned earlier, the pseudouridine in U6 snRNA is formed in response to environmental stress as part of the filamentation program. This study also found that Pus1 expression was increased under these conditions and spliceosome function was altered indicating that Pus1 could be involved in regulation of splicing in response to environmental conditions.Citation5 Besides forming pseudouridines, Pus1 has also been found to be involved in the export of specific tRNAs from the nucleus through its interaction with the nuclear export receptor Los1 and nucleoporin Nsp1.Citation32 In one study, tRNAIle(UAU), which is modified at 4 positions by Pus1 (U27/34/36/67), showed impaired nuclear export in a PUS1 deletion yeast strain. The authors also note that the importance of Pus1 in the nuclear export of tRNAs is supported by the fact that Pus1-like pseudouridine synthases are absent in prokaryotes. Deletion of both the PUS1 and LOS1 genes leads to reduced growth at 30°C and no growth at 37°C further supporting a role of Pus1 in the export of tRNAs from the nucleus.Citation31,32 While PUS1 deletion strains show no growth defect, a yeast double knockout of PUS1 and PUS4 results in lethality at high temperatures.Citation34 As further discussed below, Pus1 has been found to also bind to retinoic acid receptor and to modify steroid receptor RNA activator (SRA), a non-coding RNA in humans, thereby contributing to activation of nuclear receptors.Citation35 Moreover, Pus1 targets numerous other RNAs including mRNAs as recently identified by pseudouridine sequencing.Citation25,27 In summary, Pus1 emerges as a highly interesting candidate to further explore the role of pseudouridine synthases for regulation of gene expression.
Pus2 – Pus1 and Pus2 are paralogs that arose from a duplication event of the gene that evolved different functions.Citation36 However, Pus2 has a different cellular localization than Pus1 as it is found in the mitochondria (). Mitochondrial tRNAs are less modified than their cytoplasmic counterparts, with modifications only observed at 8 different positions whereas cytoplasmic tRNAs are modified at 15 different positions. The Pus2 enzyme acts only on U27 and 28 of tRNAs in the mitochondria while the same uridines in cytoplasmic tRNAs are modified by Pus1. Therefore, Pus2 is specifically dedicated to mitochondrial tRNAs, despite the presence of an enzyme with specificity for the same uridines in cytoplasmic tRNAs. While Pus2 lacks a mitochondrial targeting sequence, it has been found to be associated with the insoluble fraction of disrupted yeast cells, most likely with the mitochondria.Citation37
Pus3 – This pseudouridine synthase, also called Deg1 (for Depressed Growth rate), shows significant sequence homology to yeast Pus1 and yet has different target uridines. This enzyme modifies U38/39 in both cytoplasmic and mitochondrial tRNAs in the anticodon arm, pseudouridines that are highly conserved in eukaryotes, archaea and bacteria where they are formed by TruA.Citation38 In yeast PUS3 deletion strains, cells grow at a reduced rate, in particular at higher temperatures (37°C), hence the Deg1 name.Citation39 It is tempting to speculate that the lack of pseudouridines in the anticodon stem loop of tRNAs in PUS3 deletion strains may affect the decoding of mRNA by the ribosome. When the tertiary structure of tRNAs is disrupted, modification by Pus3 is reduced suggesting its sensitivity to correct folding of the tRNA.Citation40 Yeast deletion strains of the PUS3 gene also show reduced termination read-through and frameshift efficiency, and lack of pseudouridines at positions 38 and 39 caused the reduction of stop codon read-through. Additionally, a frameshift assay showed that pseudouridine at position 39 was required for +1 frameshifts in slippery mRNA sequences.Citation41 In addition to modifying tRNA, Pus3 has many target sites in mRNA and non-coding RNA.Citation25 Specifically, Pus3 has been identified together with Pus1 to modify steroid receptor activator RNA in humans.Citation42 Like Pus1, the Pus3 enzyme has a wide target specificity which may support a broader role of Pus3 in regulation of gene expression.
Pus4 – As mentioned, the Pus4 enzyme belongs to the TruB family and is responsible for modification of the almost universally conserved U55 in the TΨC arm of tRNAs. Unlike E.coli TruB and yeast Cbf5, Pus4 seems to have no C-terminal PUA domain (pseudouridine synthase and archaeosine transglycosylase domain), a highly conserved RNA-binding motif. Chemical mapping experiments have shown that Pus4 modifies both cytoplasmic and mitochondrial tRNAs at position 55.Citation24 Pus4 has not been found to modify any other tRNA uridines suggesting a unique substrate specificity; however, new data show pseudouridylation of uridines found in mRNA by Pus4. A Pus4 consensus sequence of GUUCNANNC has been suggested which is consistent with the formation of a T-loop-type structure.Citation25
Both PUS3 and PUS4 are required in mutant yeast strains that are lacking the La gene. The La protein plays a role in tRNA stabilization, maturation and folding, yet is dispensable to yeast cells when deleted on its own. The genetic interaction between PUS3, PUS4 and the La gene suggests a redundant role for La protein and tRNA modifications during biogenesis of tRNA.Citation43 Also, Pus4 was found to bind Brome Mosaic Virus (BMV), a plant-infecting RNA virus that can replicate in yeast. Pus4 inhibits the spread of the virus in plants and prevents encapsulation of the BMV RNA, but interestingly the pseudouridylation activity of Pus4 was not required for this.Citation44 Thus, Pus4 may play a role in the cell that goes beyond its pseudouridylation activity and that depends on its ability to bind and possibly fold RNA. For example, Pus4 could acts as a tRNA chaperone similar to its bacterial homolog TruB.Citation45
Pus 5 – Very little is known about the mitochondrial pseudouridine synthase Pus5. Pus5 lacks an obvious mitochondrial targeting sequence, however global amino acid analysis classifies it as a mitochondrial enzyme.Citation46 Yeast cells with a PUS5 deletion showed no growth phenotype when grown at various temperatures and media conditions.Citation46 Originally, the only known target site of Pus5 was located in mitochondrial 21S rRNA at position U2819.Citation46 Recently however, a few Pus5-dependent pseudouridines were identified in mRNA.Citation25 Overall, Pus5 seems to be a rather specific mitochondrial pseudouridine synthase.
Pus6 – The enzyme Pus6 modifies tRNA U31 in both cytoplasmic and mitochondrial tRNA.Citation47 Despite activity of Pus6 in the mitochondria, no characteristic targeting sequence is present in the N-terminal region of the protein, similar to the observation for Pus2, Pus4 and Pus5. However, the first 30 amino acids of Pus6 contain arginine and lysine residues and also adopt an α-helical structure which are features that have been shown to direct peptides to the mitochondria. Fluorescence microscopy localization studies showed the presence of Pus6 in the cytoplasm and mitochondria, with an absence of the protein in the nucleus (), indicating modification of cytoplasmic tRNAs by Pus6 occurs after transport from the nucleus.Citation47 Since 2014, we know that Pus6 also modifies some mRNAs.Citation25
Pus7 – Being the only member of the TruD family, Pus7 was originally shown to modify U2 snRNA at position 35.Citation48 Remarkably, pseudouridylation of U2 snRNA is inducible by heat shock.Citation10 Later, Pus7 was shown to also modify tRNAs, specifically at position 13 in cytoplasmic tRNA and position 35 in pre-tRNATyr.Citation49 This finding illustrates the multi-substrate specificity of this pseudouridine synthase. Recently, Pus7 has also been shown to pseudouridylate numerous nucleotides in mRNA.Citation25,27 Since the RNA structure surrounding the target uridines varies, secondary structure is unlikely to be the major determinant of specificity albeit the presence of a stem-loop structure in the vicinity of the target uridine has been suggested.Citation50 As the predominant target feature, a recognition sequence was suggested for Pus7 target RNAs which was subsequently also identified by transcriptome-wide mapping of pseudouridines.Citation25,27,49 By recognizing a UGUAR-like (R = G > A) sequence with the second uridine being the target site for pseudouridylation, Pus7 is so far the only known pseudouridine synthase that displays a consensus sequence for substrate recognition. Interestingly, mRNA pseudouridylation by Pus7 is dramatically increased upon heat shock in yeast although both transcript and protein levels of Pus7 decrease under these conditions ().Citation27 However, the enhanced mRNA modification by Pus7 under stress is correlated with a re-localization of Pus7 from the nucleus to the cytoplasm, and mRNA pseudouridylation seems to contribute to stabilization of mRNA under heat shock. These findings point to an interesting mechanism of regulating gene expression by (somehow) altering the localization and/or activity of pseudouridine synthases targeting mRNAs with critical downstream effects, e.g. on mRNA stability. Together, all these findings render Pus7 a highly interesting enzyme with the potential to strongly influence cellular gene expression.
Table 2. Regulated expression of PUS genes. This table summarizes the PUS genes that are downregulated by at least 1.8 log2 fold change in expression on the transcriptional level under various stress conditions compared with controls. Conditions are as follows; fermentation conditions for 14 days, limiting amino acids through the use of SD medium in place of YPD medium, organic compounds through the addition of ethanol, osmotic stress through the addition of potassium chloride or sodium chloride, oxidative stress through the addition of hydrogen peroxide, pH by lowering the pH from 8.0 to 5.5, rapamycin inhibition by the addition of 200 ng/mL rapamycin to growth medium, and temperature increased to 37°C. Data from the Yeast Stress Expression Database.Citation63
Pus8 – The Pus8 enzyme (also called Rib2) modifies U32 of cytoplasmic tRNA.Citation51 As it has not (yet) been shown to modify mRNA, Pus8 appears to be a highly specific pseudouridine synthase. In addition to the N-terminal pseudouridylation domain, Pus8 has a C-terminal DRAP-deaminase domain involved in the riboflavin biosynthesis pathway.Citation51 This is the first example of an RNA modification enzyme being fused to a second enzyme with an unrelated activity. Pus8 is also the only eukaryotic stand-alone pseudouridine synthase that is essential in yeast; however, this characteristic might be attributed to its function in riboflavin biosynthesis rather than pseudouridine formation. It remains to be investigated whether the 2 activities of Pus8 influence each other.
Pus9 – Interestingly, the Pus9 enzyme catalyzes the same modification as Pus8 by modifying U32 of tRNA, but it is specific to mitochondria.Citation51 Like Pus1 and Pus2, Pus8 and Pus9 are 2 enzymes that also modify the same target uridine, but in separate compartments of the cell. Pus9 is the only yeast Pus enzyme to contain an N-terminal mitochondrial localization signal to direct the enzyme to its target compartment. In addition to pseudouridylating U32 in tRNA, Pus9 has also been reported to modify some mRNAs and is therefore somewhat less specific than Pus8.Citation25,27
Pus10 – The tenth type of stand-alone pseudouridine synthase, Pus10, remains enigmatic as it has mostly been studied in archaea, but not in eukaryotes (except for the structure of human Pus10). Pus10 is only found in certain eukaryotic organisms and is not present in S. cerevisiae. In archaea, this enzyme has been shown to modify U54 and U55 in tRNA.Citation23 In eukaryotes, a pseudouridine is only found at position 55 of tRNA which is generally formed by Pus4. It is therefore unclear whether eukaryotic Pus4 and Pus10 compete for tRNA modification or whether Pus10 has other target sites, for example in other non-coding RNAs or in mRNA.
“Novel” targets of Pus enzymes in mRNA and (long) non-coding RNA
In 2014, interesting new data was published in the field of RNA modifications when pseudouridines were discovered to be abundantly present in numerous types of RNA including mRNA.Citation25-28 Of the over one hundred possible RNA modifications known to exist, pseudouridine is one of only 6 that have been identified so far in the body of mRNA together with N6-methyladenosine, N1-methyladenosine, 5-methylcytosine, hydroxylmethylcytosine and inosine (besides formation of the mRNA cap and C-to-U editing).Citation52-55 Using a technique called Pseudo-seq (or Psi-seq), which utilizes the addition of CMC (N-cyclohexyl-N'-β-(4-methylmorpholinium) ethylcarbodiimide) groups to pseudouridines causing a stop in reverse transcription, the specific locations of these modifications in the transcriptomes of yeast, mouse and humans have been identified.Citation56 With the help of these new transcriptome-wide methods to detect pseudouridines, thousands of these modifications have been identified in many mRNAs. Although numerous H/ACA guide RNAs have unknown targets, H/ACA sRNPs seem responsible for only a moderate fraction of these mRNA modifications. Instead, deletion studies surprisingly identified many mRNA modification sites as substrates of Pus enzymes. In this context, it is important to note that the current data sets for pseudouridine sequencing in deletion strains lacking individual Pus enzymes focus on a low false-discovery rate and therefore inevitably miss numerous pseudouridines as later revealed by a modified method (CeU-Seq) which enriches pseudouridylated RNA before sequencing.Citation25-28 Hence, we know of numerous mRNA targets of Pus enzymes, but are likely to discover many more in the future. Remarkably, even with the currently available data sets, many Pus enzymes seem to have more target sites in mRNA than their “traditional” target sites in tRNA, snRNA and rRNA under standard growth conditions (). For example, combining the available data sets reveals that Pus1 modifies more than 60 sites in mRNAs, Pus2 targets at least 25 sites, Pus3 is responsible for almost 30 or more sites, Pus4 pseudouridylates 20 or more sites and Pus7 catalyzes pseudouridine formation in at least 35 positions in mRNA under normal conditions and more than 100 sites upon heat shock ().Citation25,27 Given the high cellular concentrations of the “canonical” targets of Pus enzymes such as tRNAs, the majority of cellular pseudouridylation events by Pus enzymes will still occur in these non-coding RNAs rather than in low-abundant mRNAs.
The finding of widespread mRNA pseudouridylation by Pus enzymes obviously raises the question of the importance of Pus enzymes in this context. While some of the newly discovered pseudouridylation events may simply occur by chance due to the broad specificity of Pus enzymes, it seems likely that at least some of these pseudouridines play a functional role. Notably, pseudouridines are not randomly distributed within mRNAs, but are dramatically underrepresented in the 5′UTR compared with the coding region and the 3′UTR which further indicates a functional role of these modifications.Citation28 While investigating this function, it will be crucial to keep in mind that we are likely missing many target sites for Pus enzymes, which renders it difficult to derive patterns from the targeted types of mRNAs and to generate a hypothesis for Pus enzyme function based on their targets. However, the pseudouridine sequence data provide important insight into the potential of regulating mRNA properties such as decoding, stability, protein interaction etc. by pseudouridylation, as this modification is highly dynamic in response to environmental changes. First, when yeast cells were heat shocked, additional pseudouridines were observed in mRNAs.Citation27 Many of the heat shock-induced sites were found at UGUAR Pus7 consensus sites, suggesting Pus7 is involved in the heat shock response. While it remains unclear how Pus7 affects mRNAs exactly, it was uncovered that under heat shock conditions pseudouridylated mRNAs are present in higher levels in wild-type than in ΔPUS7 cells which could be explained by an increase in mRNA stability upon pseudouridylation.Citation27 Similarly, Pus7-catalyzed pseudouridylation of U2 snRNA is induced upon heat shock.Citation10 Second, changes in pseudouridine levels in mRNA were detected during post-diauxic growth of S. cerevisiae where many additional, but also some less-modified sites were detected.Citation25 In particular, Pus1 was found to increase modifications of mRNA during post-diauxic growth. This is similar to the observation that Pus1 inducibly modifies U6 snRNA during filamentation.Citation5 Third, Pus4 was identified as the enzyme that pseudouridylates the mRNA of TEF1 (translation elongation factor 1α).Citation25,26 In one study, the authors compared several TEF1 mRNA sequences from 8 fungal species and found a conserved sequence of GUUCGA in 7 out of 8 species; of these some have a pseudouridine while others did not.Citation26 This suggests that the pseudouridine sites are specific and may be conserved across species. Interestingly, in another study, an interaction between Pus4 and elongation factor 1α was observed which raises the question whether Pus4 is somehow involved in regulating the expression and/or activity of elongation factor 1α.57
In summary, widespread pseudouridylation of mRNAs by Pus enzymes is now unambiguously proven, but we lack an understanding of both the causes and consequences of pseudouridine formation. Addressing these questions will be necessary to clearly delineate the cellular functions of Pus enzymes. The downstream effects of mRNA modification by Pus enzymes could involve re-coding of sense codons, read-through of stop codons (albeit underrepresented in current pseudouridine sequencing data sets), alteration of mRNA stability, formation or disruption of mRNA structures, binding of proteins that may act as “readers” of pseudouridines similar to the detection of N6-methyladenosine in mRNA, influencing regulation of translation or more.Citation58 It can also be envisioned that pseudouridines might be selectively removed by “eraser proteins” as part of a regulatory mechanism which has been described for N6-methyladenosine.Citation58 In addition, more knowledge is required regarding the regulation of Pus enzymes (vide infra) to further understand what is leading to the dynamic and inducible pseudouridylation of mRNA.
Besides mRNAs, non-coding RNAs have emerged as highly interesting targets of Pus enzymes. As mentioned earlier, the first example of Pus-dependent pseudouridylation of a long non-coding RNA was the report that steroid receptor RNA activator (SRA) is modified by Pus1 and Pus3.Citation35,42 Interestingly, Pus1 forms a trimeric complex with SRA and retinoic acid receptor and is playing a role together with SRA in activating retinoic acid receptor thereby affecting gene expression of numerous nuclear-receptor activated genes.Citation42 Later, it was revealed that Pus3 can also modify SRA and thus serves as a coactivator of nuclear receptors.Citation35 The effects of Pus1 and Pus3 on SRA and nuclear receptors represent a clear example for the involvement of Pus enzymes in gene regulation which may be mediated through both modification of non-coding RNA (such as SRA) as well as through protein-protein interaction (such as the Pus1 interaction with retinoic acid receptor). Besides pseudouridylation of SRA, it was known before the pseudouridine sequencing publications that Pus1 and Pus7 are modifying the small nuclear RNAs U2 snRNA and U6 snRNA and that at least 2 of these modifications are dynamically responding to growth conditions (vide supra).Citation5,6,10,48
In the past, this impact of Pus enzymes on SRA and small nuclear non-coding RNA seemed to be an exception compared with the seemingly predominant modification of tRNAs. However, since 2014, several other interesting cases became apparent from the pseudouridine sequencing studies where Pus enzymes modify non-coding RNA. First, it is striking that almost all (if not all) Pus enzymes are modifying small nucleolar RNAs that are involved in guiding the formation of pseudouridines and the addition of 2′O-methyl groups by H/ACA and C/D snoRNPs, respectively.Citation25,27 This modification of snoRNAs may represent non-specific pseudouridylation events due to the co-localization of snoRNAs and Pus enzymes in the nucleus. However, astonishingly even Pus enzymes that are not known to reside in the nucleus such as Pus2 and Pus6, have been found to target snoRNAs which raises the question when and where in the cell this modification event will occur. Alternatively, targeting these snoRNAs for pseudouridylation by Pus enzymes may represent a feedback loop where formation of pseudouridines by Pus enzymes might affect the stability or activity of another pseudouridylation machinery, namely H/ACA snoRNPs. It is currently unclear what the function of pseudouridines in these small nucleolar RNAs might be. Presently, there are 4 more interesting examples where Pus1 and Pus7 target other non-coding RNA. First, Pus1 has been discovered to pseudouridylate the RNA component of RNase MRP in yeast.Citation25,27 RNase MRP is a catalytic RNA that processes numerous cellular RNAs, and it remains to be determined which role pseudouridylation of this RNA plays in cellular RNA homeostasis.Citation59 Pus7 appears to be even more interesting than Pus1 as it is clearly regulated by heat shock and possibly other conditions that seem to be linked to dynamic pseudouridylation.Citation27 In S. cerevisiae, Pus7 targets at least one of 4 pseudouridylation sites in telomerase RNA.Citation25 And in humans, Pus7 is responsible for modifying the long non-coding MALAT1 RNA,Citation27 a cancer-associated and well-studied long-noncoding RNA.Citation60 While the cellular function of pseudouridylating these non-coding RNAs by Pus enzymes remains to be elucidated, a recent study has uncovered a role of a pseudouridine in 7SK RNA that is generated by H/ACA snoRNP revealing that pseudouridylation modulates the function of the 7SK small nuclear ribonucleoprotein thereby affecting transcription of HIV-1 and cellular genes.Citation61 By analogy, it seems a reasonable hypothesis that Pus enzymes might affect the function of several non-coding RNAs in the cell and in doing so amplify their effects on gene regulation and other cellular processes.
Potential regulation of Pus enzymes
Should Pus enzymes have an effect on cellular gene expression in response to various stimuli, then it would be expected that their expression or activity is modulated in the cell one way or another. Here, we will discuss potential mechanisms of regulating yeast Pus enzymes based on the large data sets available for yeast genes and proteins.Citation62 Specifically, we will focus on the expression and localization of Pus enzymes, on interactions of Pus enzymes with other cellular proteins and on post-translational modifications of Pus enzymes. At least for Pus7, it has been shown that although its expression is downregulated both on the mRNA and also slightly on the protein level upon heat stress, the re-localization of Pus7 to the cytoplasm under heat stress is correlated with an increase in pseudouridylation in mRNAs by Pus7 which may represent a regulatory mechanism.Citation27 For all other Pus enzymes, the interplay of expression, localization, protein-protein interactions and post-translational modifications remains to be elucidated.
Interestingly, the yeast stress expression database reveals differential expression on the transcriptional level in particular of the PUS1 and PUS7 genes, but also PUS3, PUS4, and PUS9 under different growth conditions.Citation63 Remarkably, 8 types of stress conditions lead to downregulation of different subsets of PUS genes () which may suggest some commonalities and overlapping mechanisms in regulating the abundance of these modification enzymes. The pseudouridine sequencing data revealed changes in the pseudouridine landscape upon heat stress and nutrient deprivation.Citation25,27 Given this observation, it is notable that both elevated temperature (37°C) and different forms of nutrient deprivation (limiting amino acids or carbon source) result in reduced expression of PUS genes which could hint at a link between expression of the encoded proteins and pseudouridylation regulation. Likewise, osmotic stress induced by high salt growth conditions, oxidative stress and pH affect PUS gene expression. Last but not least, rapamycin, which targets the mTOR pathway involved in metabolism and aging,Citation64 decreases the mRNA levels of PUS1, PUS2, PUS4 and PUS7. In addition to this transcriptional regulation, expression of Pus enzymes may also be affected on the translational level, but nothing is known so far about the mechanisms of regulating Pus enzyme expression. Also, it has not been tested (yet) if any of the latter conditions affect global pseudouridine levels in coding and non-coding RNA.
As made evident by the example of Pus7, the expression levels of a given Pus enzyme might not necessarily reflect its activity which could instead be modulated by the localization of the Pus enzyme. In accordance with their tRNA targets (, ), several Pus enzymes are found in the mitochondria (Pus2, Pus4, Pus5, Pus6 and Pus9; ). Interestingly, other Pus enzymes are found in the cytoplasm, in the nucleus or in both cellular compartments. Most Pus enzymes are found in at least 2 cellular locations except for Pus1, which is strictly nuclear, and Pus2, which is only present in mitochondria. Particularly for the enzymes with multiple cellular localizations, it could be envisioned that their distribution between cellular compartments is affected by external stimuli. This further raises the issue as to where pseudouridylation of tRNAs, mRNAs and other RNAs takes place. Obviously, many questions remain to be addressed regarding localization of Pus enzymes and the modification site for various cellular RNAs.
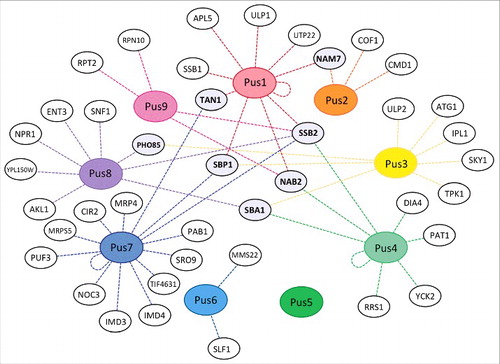
It is grossly misleading to only consider eukaryotic stand-alone pseudouridine synthases in isolation. As mentioned for Pus1, interactions of Pus enzymes with other proteins may be important for the cell such as the binding of Pus1 to nuclear receptors.Citation35 Indeed, inspecting the yeast genome database (only one of several available sources for protein-protein interactions) reveals numerous physical interactions for Pus enzymes, far too many to be discussed individually in this review ().Citation62 However, a few observations stand out regarding protein interactions of Pus enzymes. First, there are 7 yeast proteins that interact with more than one Pus enzyme (Pho85, Tan1, SBP1, SBA1, NAB2, SSB2 and NAM7). Thus, it is possible that Pus enzymes form transient or stable super-complexes containing multiple pseudouridine synthases which may facilitate substrate channeling for example during modification of tRNAs at multiple sites (). Notably, many of these proteins are in one way or another involved in cellular RNA metabolism and may help to direct pseudouridine synthases to different targets. Also, Tan1, a putative tRNA modification enzyme responsible for acetylation of tRNAs, might act in a coordinated fashion together with its binding partners Pus1 and Pus7 on select tRNAs. Second, several of the interaction partners of Pus enzymes are kinases suggesting regulation of these proteins through post-translational modifications (vide infra). For example, Pho85 is a cyclin-dependent kinase involved in cell cycle regulation that binds both to Pus3 and Pus8.Citation65 Indeed, for enzymes such as Pus3 and Pus8, a pattern of interacting with numerous kinases begins to emerge, suggesting a possible mode of regulation of Pus enzymes, for example to induce differential activity throughout the cell cycle. Third, the only pseudouridine synthase without known physical interaction partners so far is Pus5 which is strictly localized to mitochondria where it modifies rRNA. Again, this suggests that Pus5 is a highly specific enzyme with a defined task in the cell in contrast to all or most other pseudouridine synthases.
Figure 4. Physical interactions of Pus enzymes with other proteins. Each Pus enzyme is shown in a colored oval surrounded by the proteins it has been found to physically interact with, connected by dotted lines. Physical evidence includes co-immunoprecipitation, co-localization, yeast 2-hybrid system, etc. No physical interaction partners were found for Pus5. Notably, Pus1, Pus4 and Pus7 were found to interact with themselves as shown by a looped line. Proteins that interact with more than one Pus enzyme are highlighted in gray. (Source: yeastmine/yeastgenome.org).
Lastly, we need to consider post-translational modification of Pus enzymes as a means to regulate their activity and/or localization. In fact, the majority of these proteins with the exception of Pus2, Pus5 and Pus8 are known to be modified either by ubiquitination (Pus1, Pus2 and Pus7), sumoylation (Pus1), phosphorylation (Pus1, Pus3, Pus4, Pus6, Pus9) or succinylation (Pus7).Citation66-70 The enzymes responsible for these post-translational modifications are not yet known, nor the effects of the modifications on the respective Pus enzyme. Consequently, it remains a promising hypothesis that Pus enzymes are regulated by post-translational modifications which in turn may affect gene expression in the cell depending on the cellular condition.
Pus enzymes and disease
The importance of stand-alone pseudouridine synthases is further underlined by the fact that mutations in the human PUS1 and PUS3 genes are associated with inherited diseases. MLASA stands for mitochondrial myopathy, lactic acidosis and sideroblastic anemia, an autosomal recessive disease in humans. It is an oxidative phosphorylation and iron metabolism disorder in skeletal muscle and bone marrow and is caused by different mutations, for example in the mitochondrial tyrosyl-tRNA synthetase gene, but also in PUS1.Citation12 In 2 families, a homozygous missense mutation was found that results in a R116W substitution in the Pus1 protein; this arginine is located very close to the catalytic aspartate in the conserved catalytic core of the protein suggesting potential effects on pseudouridylation by this Pus1 variant. Indeed, patients were found to be lacking the modifications of U27 and U28 in selected tRNAs.Citation71 Alternatively, MLASA can also be caused by a stop mutation in PUS1 that deletes almost half of the amino acid residues of the Pus1 protein.Citation72 Recently, a long-surviving patient with sideroblastic anemia and mild cognitive impairment with 2 novel mutations in PUS1 was described.Citation73 Together, the discovery of multiple disease-causing mutations in PUS1 underlines the importance of PUS1, in particular for mitochondria, even though the Pus1 enzyme is localized to the nucleus in yeast. Obviously, we are still lacking a detailed understanding of the molecular pathology of MLASA. A new Pus1-deficient mouse model may help to further elucidate the physiologic function of the Pus1 enzyme and suggests an effect of PUS1 mutations on muscle metabolism related to mitochondria.Citation74 In this regard it is noteworthy that mutations in the PUS1 gene also affect cellular differentiation in the haploid protozoan parasite Toxoplasma gondii that can cause a lifelong, chronic infection in humans.Citation75 Very recently, a homozygous, truncated form of Pus3 has been found to be present in patients with an intellectual disability which is accompanied by reduced levels of U39 pseudouridylation in tRNA in these patients.Citation76 Taken together the 2 examples of diseases caused by PUS1 and PUS3 mutations indicates that these enzymes are particularly important for mitochondrial function in muscle as well as for the brain; possibly, in both cases the affected tissue may have increased sensitivity to translational deficiency due to reduced levels of pseudouridylation in tRNA, but it cannot be excluded that other pseudouridylation targets of these enzymes are implicated in the diseases.
Future perspectives
Eukaryotic stand-alone pseudouridine synthases introduce apparently simple modifications by converting uridines to their isomers, pseudouridine; however, it is now clear that these enzymes target a complex set of RNAs modifying a surprisingly large number of uridines. Moreover, transcriptome-wide mapping of pseudouridines, but also expression studies, identification of interaction partners and post-translational modifications of Pus enzymes all support the hypothesis that the activity of these enzymes is likely regulated in response to different types of cellular stress and/or depending on the cell cycle. Together with the disease mutations found in human PUS1 and PUS3 genes, we are beginning to appreciate the complex function and cellular importance of this enzyme family.
Obviously, we are lacking significant information for all of these pseudouridine synthases as well as detailed studies on their individual functions as each enzyme is characterized by a distinct specificity for its target RNAs as well as its protein interaction partners. Several important questions have to be addressed to dissect the presumably complex role of pseudouridines in health and disease.
First, it is mandatory to dissect the upstream effectors influencing the availability and activity of Pus enzymes to understand how these proteins are tied into cellular signal transduction networks and how they are influenced by the cellular state. Therefore, the following questions will have to be addressed: How is expression of PUS genes regulated? How are Pus enzymes localized to the different areas of the cell? Which interaction partners contribute to this localization? When and how is localization of Pus enzymes altered depending on the cellular state? In which location do the Pus enzymes modify which target RNA and how does localization affect enzyme activity? What are the functions of post-translational modifications of Pus enzymes? Under which conditions are they modified? Is the post-translational modification dynamic in response to external stimuli? What are the modifying enzymes? Does modification affect the pseudouridylation activity of the Pus enzymes?
Second, we need to better understand the downstream effects of RNA modification by pseudouridine synthases in the cell. This will include answering the following questions for numerous RNA targets: What are the consequences of pseudouridylating the different types of RNA ranging from mRNAs to short and long non-coding RNAs? Is translation affected? How is RNA stability impacted? Are there effects on protein-RNA interactions?
Last, but not least and on a more general level, it is crucial to enhance our knowledge about the mechanisms and evolution of Pus enzymes in eukaryotes as well as about their tissue-specific effects: How is the RNA specificity of each enzyme defined? Which RNA structure or sequence elements are recognized by each Pus enzyme? How conserved are the characteristics of Pus enzymes among eukaryotes? What are the functions of paralogs found in humans? What are the molecular mechanisms underlying diseases that are resulting from mutations in PUS genes? Why are only certain tissues affected by the mutations?
This long list of outstanding questions regarding pseudouridine synthases clearly indicates that we are only at the very beginning of a promising research endeavor. Clearly, many interesting characteristics of pseudouridine synthases are likely to be discovered in the coming years that will help us to not only understand the function of the Pus enzymes. Rather, by studying this enzyme family, general principles of RNA metabolism and regulation of gene expression are likely to be identified. Pseudouridine is only one of very few modifications found in mRNA so far, and all current evidence suggests that pseudoouridines are more than mere byproducts of unspecific modification activity by pseudouridine synthases. Judging by our knowledge of the function of N6-methyladenosine in mRNA,Citation77 it seems highly probable that pseudouridines and therefore Pus enzymes likewise play an important role in cellular RNA metabolism and gene expression in general.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by Alberta Innovates Technology Futures under Grant number G201400254; and the Natural Sciences and Engineering Research Council of Canada under Grant RGPIN-2014–05954.
References
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41(Database issue):D262-7; PMID:23118484; http://dx.doi.org/10.1093/nar/gks1007
- Spenkuch F, Motorin Y, Helm M. Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol 2014; 11(12):1540-54; PMID:25616362; http://dx.doi.org/10.4161/15476286.2014.992278
- Arnez JG, Steitz TA. Crystal structure of unmodified tRNA(Gln) complexed with glutaminyl-tRNA synthetase and ATP suggests a possible role for pseudo-uridines in stabilization of RNA structure. Biochemistry 1994; 33(24):7560-7567; PMID:8011621
- Liang XH, Liu Q, Fournier MJ. Loss of rRNA modifications in the decoding center of the ribosome impairs translation and strongly delays pre-rRNA processing. RNA 2009; 15(9):1716-1728; PMID:19628622; http://dx.doi.org/10.1261/rna.1724409
- Basak A, Query CC. A pseudouridine residue in the spliceosome core is part of the filamentous growth program in yeast. Cell Rep 2014; 8(4):966-973; PMID:25127136; http://dx.doi.org/10.1016/j.celrep.2014.07.004
- Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C. Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase Pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol 1999; 19(3):2142-2154; PMID:10022901
- Wu G, Adachi H, Ge J, Stephenson D, Query CC, Yu YT. Pseudouridines in U2 snRNA stimulate the ATPase activity of Prp5 during spliceosome assembly. EMBO J 2016; 35(6):654-667; PMID:26873591; http://dx.doi.org/10.15252/embj.201593113
- Yang C, McPheeters DS, Yu YT. Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem 2005; 280(8):6655-6662; PMID:15611063
- Zhao X, Yu YT. Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo. Nucleic Acids Res 2007; 35(2):550-558; PMID:17169984
- Wu G, Xiao M, Yang C, Yu YT. U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP. EMBO J 2011; 30(1):79-89; PMID:21131909; http://dx.doi.org/10.1038/emboj.2010.316
- Wu G, Radwan MK, Xiao M, Adachi H, Fan J, Yu YT. The TOR signaling pathway regulates starvation-induced pseudouridylation of yeast U2 snRNA. RNA 2016; 22(8):1146-1152; http://dx.doi.org/10.1261/rna.056796.116.
- Bykhovskaya Y, Casas K, Mengesha E, Inbal A. N Fischel-Ghodsian. Missense mutation in pseudouridine synthase 1 (PUS1) causes mitochondrial myopathy and sideroblastic anemia (MLASA). Am J Hum Genet 2004; 74(6):1303-1308; PMID:15108122; http://dx.doi.org/10.1086/421530
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 1998; 19(1):32-38; PMID:9590285; http://dx.doi.org/10.1038/ng0598-32
- Yu YT, Meier UT. RNA-guided isomerization of uridine to pseudouridine–pseudouridylation. RNA Biol 2014; 11(12):1483-1494; PMID:25590339; http://dx.doi.org/10.4161/15476286.2014.972855
- Kiss T, Fayet-Lebaron E, Jady BE. Box H/ACA small ribonucleoproteins. Mol Cell 2010; 37(5):597-606; PMID:20227365; http://dx.doi.org/10.1016/j.molcel.2010.01.032
- Hamma T, Ferre-D'Amare AR. The box H/ACA ribonucleoprotein complex: interplay of RNA and protein structures in post-transcriptional RNA modification. J Biol Chem 2010; 285(2):805-809; PMID:19917616; http://dx.doi.org/10.1074/jbc.R109.076893
- Meier UT. The many facets of H/ACA ribonucleoproteins. Chromosoma 2005; 114(1):1-14; PMID:15770508; http://dx.doi.org/10.1007/s00412-005-0333-9
- Hamma T, Ferre-D'Amare AR. Pseudouridine synthases. Chem Biol 2006; 13(11):1125-1135; PMID:17113994; PMID:10871366
- Watanabe Y, Gray MW. Evolutionary appearance of genes encoding proteins associated with box H/ACA snoRNAs: cbf5p in Euglena gracilis, an early diverging eukaryote, and candidate Gar1p and Nop10p homologs in archaebacteria. Nucleic Acids Res 2000; 28(12):2342-2352
- Veerareddygari GR, Singh SK, Mueller EG. The Pseudouridine Synthases Proceed through a Glycal Intermediate. J Am Chem Soc 2016; 138(25):7852-7855; PMID:27292228; http://dx.doi.org/10.1021/jacs.6b04491
- Czudnochowski N, Wang AL, Finer-Moore J, Stroud RM. In human pseudouridine synthase 1 (hPus1), a C-terminal helical insert blocks tRNA from binding in the same orientation as in the Pus1 bacterial homologue TruA, consistent with their different target selectivities. J Mol Biol 2013; 425(20):3875-3887; PMID:23707380; http://dx.doi.org/10.1016/j.jmb.2013.05.014
- McCleverty CJ, Hornsby M, Spraggon G, Kreusch A. Crystal structure of human Pus10, a novel pseudouridine synthase. J Mol Biol 2007; 373(5):1243-1254; PMID:17900615
- Gurha P, Gupta R. Archaeal Pus10 proteins can produce both pseudouridine 54 and 55 in tRNA. RNA 2008; 14(12):2521-2527; PMID:18952823; http://dx.doi.org/10.1261/rna.1276508
- Becker HF, Motorin Y, Planta RJ, Grosjean H. The yeast gene YNL292w encodes a pseudouridine synthase (Pus4) catalyzing the formation of psi55 in both mitochondrial and cytoplasmic tRNAs. Nucleic Acids Res 1997; 25(22):4493-4499; PMID:9358157
- Carlile TM, Rojas-Duran MF, Zinshteyn B, Shin H, Bartoli KM, Gilbert WV. Pseudouridine profiling reveals regulated mRNA pseudouridylation in yeast and human cells. Nature 2014; 515(7525):143-146; PMID:25192136; http://dx.doi.org/10.1038/nature13802
- Lovejoy AF, Riordan DP, Brown PO. Transcriptome-wide mapping of pseudouridines: pseudouridine synthases modify specific mRNAs in S. cerevisiae. PLoS One 2014; 9(10):e110799; PMID:25353621; http://dx.doi.org/10.1371/journal.pone.0110799
- Schwartz S, Bernstein DA, Mumbach MR, Jovanovic M, Herbst RH, León-Ricardo BX, Engreitz JM, Guttman M, Satija R, Lander ES, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell 2014; 159(1):148-162; PMID:25219674; http://dx.doi.org/10.1016/j.cell.2014.08.028
- Li X, Zhu P, Ma S, Song J, Bai J, Sun F, Yi C. Chemical pulldown reveals dynamic pseudouridylation of the mammalian transcriptome. Nat Chem Biol 2015; 11(8):592-597; PMID:26075521; http://dx.doi.org/10.1038/nchembio.1836
- Parisien M, Yi C, Pan T. Rationalization and prediction of selective decoding of pseudouridine-modified nonsense and sense codons. RNA 2012; 18(3):355-367; PMID:22282339; http://dx.doi.org/10.1261/rna.031351.111
- Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 2011; 474(7351):395-398; PMID:21677757; http://dx.doi.org/10.1038/nature10165
- Motorin Y, Keith G, Simon C, Foiret D, Simos G, Hurt E, Grosjean H. The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA 1998; 4(7):856-869; PMID:9671058
- Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt EC. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J 1996; 15(9):2270-2284; PMID:8641292
- Behm-Ansmant I, Massenet S, Immel F, Patton JR, Motorin Y, Branlant C. A previously unidentified activity of yeast and mouse RNA:pseudouridine synthases 1 (Pus1p) on tRNAs. RNA 2006; 12(8):1583-1593; PMID:16804160; http://dx.doi.org/10.1261/rna.100806
- Grosshans H, Lecointe F, Grosjean H, Hurt E, Simos G. Pus1p-dependent tRNA pseudouridinylation becomes essential when tRNA biogenesis is compromised in yeast. J Biol Chem 2001; 276(49):46333-46339; PMID:11571299; http://dx.doi.org/10.1074/jbc.M107141200
- Zhao X, Patton JR, Davis SL, Florence B, Ames SJ, Spanjaard RA. Regulation of nuclear receptor activity by a pseudouridine synthase through posttranscriptional modification of steroid receptor RNA activator. Mol Cell 2004; 15(4):549-558; PMID:15327771; http://dx.doi.org/10.1016/j.molcel.2004.06.044
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res 2005; 15(10):1456-1461; PMID:16169922
- Behm-Ansmant I, Branlant C, Motorin Y. The Saccharomyces cerevisiae Pus2 protein encoded by YGL063w ORF is a mitochondrial tRNA:Psi27/28-synthase. RNA 2007; 13(10):1641-1647; PMID:17684231
- Cortese R, Kammen HO, Spengler SJ, Ames BN. Biosynthesis of pseudouridine in transfer ribonucleic acid. J Biol Chem 1974; 249(4):1103-1108; PMID:4592259
- Carbone ML, Solinas M, Sora S, Panzeri L. A gene tightly linked to CEN6 is important for growth of Saccharomyces cerevisiae. Curr Genet 1991; 19(1):1-8; PMID:2036682
- Lecointe F, Simos G, Sauer A, Hurt EC, Motorin Y, Grosjean H. Characterization of yeast protein Deg1 as pseudouridine synthase (Pus3) catalyzing the formation of psi 38 and psi 39 in tRNA anticodon loop. J Biol Chem 1998; 273(3):1316-1323; PMID:9430663
- Lecointe F, Namy O, Hatin I, Simos G, Rousset JP, Grosjean H. Lack of pseudouridine 38/39 in the anticodon arm of yeast cytoplasmic tRNA decreases in vivo recoding efficiency. J Biol Chem 2002; 277(34):30445-30453; PMID:12058040; http://dx.doi.org/10.1074/jbc.M203456200
- Zhao X, Patton JR, Ghosh SK, Fischel-Ghodsian N, Shen L, Spanjaard RA. Pus3p- and Pus1p-dependent pseudouridylation of steroid receptor RNA activator controls a functional switch that regulates nuclear receptor signaling. Mol Endocrinol 2007; 21(3):686-699; PMID:17170069
- Copela LA, Chakshusmathi G, Sherrer RL, Wolin SL. The La protein functions redundantly with tRNA modification enzymes to ensure tRNA structural stability. RNA 2006; 12(4):644-654; PMID:16581807
- Zhu J, Gopinath K, Murali A, Yi G, Hayward SD, Zhu H, Kao C. RNA-binding proteins that inhibit RNA virus infection. Proc Natl Acad Sci U S A 2007; 104(9):3129-3134; PMID:17360619
- Keffer-Wilkes LC, Veerareddygari GR, Kothe U. RNA modification enzyme TruB is a tRNA chaperone. Proc Natl Acad Sci U S A 2016; http://dx.doi.org/10.1073/pnas.1607512113.
- Ansmant I, Massenet S, Grosjean H, Motorin Y, Branlant C. Identification of the Saccharomyces cerevisiae RNA:pseudouridine synthase responsible for formation of psi(2819) in 21S mitochondrial ribosomal RNA. Nucleic Acids Res 2000; 28(9):1941-1946; PMID:10756195
- Ansmant I, Motorin Y, Massenet S, Grosjean H, Branlant C. Identification and characterization of the tRNA:Psi 31-synthase (Pus6p) of Saccharomyces cerevisiae. J Biol Chem 2001; 276(37):34934-24940; PMID:11406626; http://dx.doi.org/10.1074/jbc.M103131200
- Ma X, Zhao X, Yu YT. Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J 2003; 22(8):1889-1897; PMID:12682021; http://dx.doi.org/10.1093/emboj/cdg191
- Behm-Ansmant I, Urban A, Ma X, Yu YT, Motorin Y, Branlant C. The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA 2003; 9(11):1371-1382; PMID:14561887
- Urban A, Behm-Ansmant I, Branlant C, Motorin Y. RNA sequence and two-dimensional structure features required for efficient substrate modification by the Saccharomyces cerevisiae RNA:{Psi}-synthase Pus7p. J Biol Chem 2009; 284(9):5845-5858; PMID:19114708; http://dx.doi.org/10.1074/jbc.M807986200
- Behm-Ansmant I, Grosjean H, Massenet S, Motorin Y, Branlant C. Pseudouridylation at position 32 of mitochondrial and cytoplasmic tRNAs requires two distinct enzymes in Saccharomyces cerevisiae. J Biol Chem 2004; 279(51):52998-53006; PMID:15466869
- Gilbert WV, Bell TA, Schaening C. Messenger RNA modifications: Form, distribution, and function. Science 2016; 352(6292):1408-1412; PMID:27313037; http://dx.doi.org/10.1126/science.aad8711
- Nishikura K. A-to-I editing of coding and non-coding RNAs by ADARs. Nat Rev Mol Cell Biol 2016; 17(2):83-96; PMID:26648264; http://dx.doi.org/10.1038/nrm.2015.4
- Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, et al. RNA biochemistry. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 2016; 351(6270):282-285; PMID:26816380; http://dx.doi.org/10.1126/science.aac5253
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016; 530(7591):441-446; PMID:26863196; http://dx.doi.org/10.1038/nature16998
- Zaringhalam M, Papavasiliou FN. Pseudouridylation meets next-generation sequencing. Methods 2016; 107:63-72; PMID:26968262; http://dx.doi.org/10.1016/j.ymeth.2016.03.001
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature 2006; 440(7084):631-636; PMID:16429126
- Maity A, Das B. N6-methyladenosine modification in mRNA: machinery, function and implications for health and diseases. FEBS J 2016; 283(9):1607-1630; PMID:26645578; http://dx.doi.org/10.1111/febs.13614
- Hernandez-Cid A, Aguirre-Sampieri S, Diaz-Vilchis A, Torres-Larios A. Ribonucleases P/MRP and the expanding ribonucleoprotein world. IUBMB Life 2012; 64(6):521-628; PMID:22605678; http://dx.doi.org/10.1002/iub.1052
- Ren D, Li H, Li R, Sun J, Guo P, Han H, Yang Y, Li J. Novel insight into MALAT-1 in cancer: Therapeutic targets and clinical applications. Oncol Lett 2016; 11(3):1621-1630; PMID:26998053; http://dx.doi.org/10.3892/ol.2016.4138
- Zhao Y, Karijolich J, Glaunsinger B, Zhou Q. Pseudouridylation of 7SK snRNA promotes 7SK snRNP formation to suppress HIV-1 transcription and escape from latency. EMBO Rep 2016; http://dx.doi.org/10.15252/embr.201642682
- Cherry JM, Hong EL, Amundsen C, Balakrishnan R, Binkley G, Chan ET, Christie KR, Costanzo MC, Dwight SS, Engel SR, et al. Saccharomyces Genome Database: the genomics resource of budding yeast. Nucleic Acids Res 2012; 40(Database issue):D700-5; PMID:22110037; http://dx.doi.org/10.1093/nar/gkr1029
- Wanichthanarak K, Nookaew I, Petranovic D. yStreX: yeast stress expression database. Database (Oxford) 2014; 2014; http://dx.doi.org/10.1093/database/bau068
- Kennedy BK, Lamming DW. The Mechanistic Target of Rapamycin: The Grand ConducTOR of Metabolism and Aging. Cell Metab 2016; 23(6):990-1003; PMID:27304501; http://dx.doi.org/10.1016/j.cmet.2016.05.009
- Jimenez J, Ricco N, Grijota-Martinez C, Fado R, Clotet J. Redundancy or specificity? The role of the CDK Pho85 in cell cycle control. Int J Biochem Mol Biol 2013; 4(3):140-149; PMID:24049669
- Swaney DL, Beltrao P, Starita L, Guo A, Rush J, Fields S, Krogan NJ, Villen J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat Methods 2013; 10(7):676-682; PMID:23749301; http://dx.doi.org/10.1038/nmeth.2519
- Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 2008; 7(7):1389-1396; PMID:18407956; http://dx.doi.org/10.1074/mcp.M700468-MCP200
- Weinert BT, Scholz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep 2013; 4(4):842-851; PMID:23954790; http://dx.doi.org/10.1016/j.celrep.2013.07.024
- Sung MK, Lim G, Yi DG, Chang YJ, Yang EB, Lee K, Huh WK. Genome-wide bimolecular fluorescence complementation analysis of SUMO interactome in yeast. Genome Res 2013; 23(4):736-746; PMID:23403034; http://dx.doi.org/10.1101/gr.148346.112
- Duttler S, Pechmann S, Frydman J. Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell 2013; 50(3):379-393; PMID:23583075; http://dx.doi.org/10.1016/j.molcel.2013.03.010
- Patton JR, Bykhovskaya Y, Mengesha E, Bertolotto C, Fischel-Ghodsian N. Mitochondrial myopathy and sideroblastic anemia (MLASA): missense mutation in the pseudouridine synthase 1 (PUS1) gene is associated with the loss of tRNA pseudouridylation. J Biol Chem 2005; 280(20):19823-19828; PMID:15772074
- Fernandez-Vizarra E, Berardinelli A, Valente L, Tiranti V, Zeviani M. Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA). J Med Genet 2007; 44(3):173-180; PMID:17056637
- Cao M, Donà M, Valentino ML, Semplicini C, Maresca A, Cassina M, Torraco A, Galletta E, Manfioli V, Sorarù G, et al. Clinical and molecular study in a long-surviving patient with MLASA syndrome due to novel PUS1 mutations. Neurogenetics 2016; 17(1):65-70; PMID:26556812; http://dx.doi.org/10.1007/s10048-015-0465-x
- Mangum JE, Hardee JP, Fix DK, Puppa MJ, Elkes J, Altomare D, Bykhovskaya Y, Campagna DR, Schmidt PJ, Sendamarai AK, et al. Pseudouridine synthase 1 deficient mice, a model for Mitochondrial Myopathy with Sideroblastic Anemia, exhibit muscle morphology and physiology alterations. Sci Rep 2016; 6:26202; PMID:27197761; http://dx.doi.org/10.1038/srep26202
- Anderson MZ, Brewer J, Singh U, Boothroyd JC. A pseudouridine synthase homologue is critical to cellular differentiation in Toxoplasma gondii. Eukaryot Cell 2009; 8(3):398-409; PMID:19124578; http://dx.doi.org/10.1128/EC.00329-08
- Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM, Alkuraya FS. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet 2016; 135(7):707-713; PMID:27055666; http://dx.doi.org/10.1007/s00439-016-1665-7
- Yue Y, Liu J, He C. RNA N6-methyladenosine methylation in post-transcriptional gene expression regulation. Genes Dev 2015; 29(13):1343-1355; PMID:26159994; http://dx.doi.org/10.1101/gad.262766.115
