ABSTRACT
Cytoplasmic mRNAs are specifically degraded in somatic cells as a part of early apoptotic response. However, no reports have been presented so far concerning mRNA fate in apoptotic gametes. In the present study, we analyzed the content of various cytoplasmic mRNAs in aging oocytes and eggs of the African clawed frog, Xenopus laevis. To circumvent large gene expression variation among the individual oocytes and eggs, single-cell monitoring of transcript levels has been implemented, using multiple cytoplasmic collections and reverse transcriptase quantitative PCR. It was found that numerous cytoplasmic mRNAs, coding for proteins classified in different functional types, are robustly degraded in apoptotic Xenopus eggs, but not in aging oocytes. mRNA degradation becomes evident in the eggs after meiotic exit at the time of cytochrome c release. A strong correlation between the length of PCR amplicon and specific transcript content was observed, suggesting endonucleolytic cleavage of mRNA. In addition, it was found that mRNA deadenylation also contributes to apoptotic mRNA degradation. Altogether, these findings indicate that the global decay of mRNA represents a hallmark of apoptosis in aging Xenopus eggs. To our knowledge, this is the first description of mRNA degradation in apoptotic gamete cells.
Introduction
Aging is associated with apoptosis of damaged and dysfunctional cells accompanied by degradation of various intracellular biopolymers. Well-characterized changes occur to proteins, lipids, RNA and DNA during apoptosis in somatic cells. In particular, cytoplasmic mRNA and rRNA are robustly degraded in mammalian cell lines as a part of early apoptotic response, preceding DNA fragmentation.Citation1 It was shown that mRNA decay occurs during classical apoptotic cell death and it depends on mitochondrial outer membrane permeabilization.Citation2 mRNA degradation was also observed in vivo, specifically during thymocyte apoptosis, suggesting its physiologic relevance.Citation1 Furthermore, rRNA and mRNA cleavage was witnessed during apoptotic cell death in plants and yeast.Citation3,4 rRNA degradation was suggested to occur by an endonucleolytic mechanism, however the identity of the ribonucleases involved has not been revealed.Citation4 These findings support the notion that RNA degradation may be a general event of apoptosis. It was proposed that RNA decay plays a role in the inhibition of protein synthesis observed in apoptotic cells.Citation5 Notably, no reports have been presented so far concerning RNA fate in aging and apoptotic gametes. Apoptosis in these cells has some uncommon features associated with their unique biologic function. Apoptotic studies of gamete cells are important because they expand our understanding of cell death by unveiling alternative physiologic mechanisms. Previously, it was demonstrated that in several species with external fertilization, such as starfish, sea urchin and frog, laid unfertilized eggs die by apoptosis outside of the animal body.Citation6-11 In the present study, we investigated the content of multiple cytoplasmic mRNAs in aging oocytes and eggs of the African clawed frog, Xenopus laevis. These cells represent a convenient experimental model for apoptotic studies because of their large size, the ease of obtaining in large numbers and biochemical tractability. They can be collected freshly from living frogs and treated after isolation like a primary cell culture. Recently, aging unfertilized Xenopus eggs were found to die by a classical apoptotic process after the spontaneous exit from meiotic arrest.Citation9-11 In the present study, to bypass large cell-to-cell variation of the transcript levels in the oocyte/egg population, single-cell gene expression analysis was implemented. Monitoring transcript levels in single living Xenopus oocytes and eggs was performed using a technique based on multiple collections of nanoliter volumes of cytoplasmic material and reverse transcriptase quantitative PCR (RT-qPCR). This technique was shown to allow reliable detection of various mRNAs expressed at the level of more than 103 copies per oocyte.Citation12
Results
Transcript profiling in Xenopus oocyte population
Grown up Xenopus oocytes contain a stock of maternal mRNA synthesized and accumulated during oogenesis. In the present study, we performed transcript profiling in individual living Xenopus oocytes using multiple collections of nanoliter amounts of the oocyte cytoplasm and RT-qPCR. This technique was shown to exert a minimal effect on oocyte viability.Citation12 The expression profiling of 18 genes, classified into different functional types, such as house-keeping, regulatory, apoptosis-related, metabolic, etc., revealed a great difference of their expression levels (). In addition, a significant degree of cell-to-cell gene expression variation was observed among individual Xenopus oocytes (). The lowest variation was observed for the highly expressed constitutive genes, such as actin, histH4 (histone H4) and gpdh (glyceraldehyde-3-phosphate dehydrogenase), which are often used as housekeeping controls in gene expression analysis. Apoptosis-related and regulatory genes showed much higher expression variations. High expression variation of apoptosis-related genes, such as bid, bak, mcl1, casp3, casp7, may help to explain a substantial heterogeneity of apoptotic response observed in unfertilized Xenopus eggs.Citation9 These results strongly suggested that monitoring transcript levels in single oocytes and eggs rather than in cell populations should be used for quantitative transcript profiling in these cells to circumvent intrinsically large gene expression heterogeneity.
Figure 1. Variability of transcript contents in Xenopus oocytes. (A) Expression levels of 18 genes measured in a single Xenopus oocyte. (B) Standard deviation of gene expression in the oocyte population determined by analyzing expression levels in 16 to 22 individual oocytes. (C) Sampling design of gene expression monitoring in single aging Xenopus oocytes and eggs. Cytoplasmic collections were made from individual oocytes (O1, O2, O3, O4) before PG treatment, then from the eggs derived from these oocytes at 12, 24, 36 and 48 hours after PG addition (E1–12, E2–24, E3–36, E4–48, respectively). Cytoplasmic samples were also taken from a control untreated oocyte at the corresponding times (Oc-0, Oc-12, Oc-24, Oc-36, Oc-48). In panel (A), mRNA levels were measured in 2 to 4 replicates. SDs of replicate measurements are small and not visible in the plot.
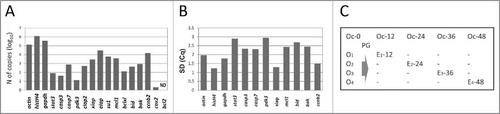
Fully grown immature Xenopus oocytes can be induced to maturation by the steroid hormone progesterone (PG). The hormone promotes oocyte transition from prophase I to metaphase II, producing fertilization-competent metaphase-arrested eggs. Previously, we have demonstrated that Xenopus oocytes can withstand multiple cytoplasmic collections, remaining intact for at least 48 hours.Citation12 In contrast, matured meiotically arrested eggs are easily activated or hyperactivated by pricking with a collection needle. They typically exit the meiotic arrest and rapidly deteriorate after the cytoplasmic collection, making impossible repeated samplings from these cells. Based on these observations, cytoplasmic collections from the single PG-treated and control Xenopus oocytes were performed according to the protocol presented in . Multiple collections (Oc-0, Oc-12, Oc-24, Oc-36, Oc-48) were made in succession from a single control PG-untreated oocyte at 12-hour intervals, providing a self-referred series of samples for quantification of various transcripts. On the other hand, cytoplasmic samplings from the PG-treated oocytes were performed only twice, before and after hormone administration at the indicated times. In this experimental format, gene expression levels determined in the individual immature oocytes before PG addition (O1, O2, O3, O4) provided the references for transcript levels in the matured or apoptotic eggs derived from these oocytes (E1–12, E2–24, E3–36, E4–48) at the indicated times ().
mRNA degradation in post-meiotic eggs
The developed protocol was applied for the analysis of age-related changes of gene expression in Xenopus oocytes and eggs. In accordance with previous reports, PG treatment had little effect on mRNA levels in oocytes at 12 hours after hormone administration (). However, a drastic fall in the content of all investigated transcripts was registered after 24 hours (). The decrease in mRNA contents has been detected when either oligo(dT) or random hexamer primers were used in the reverse transcriptase reaction. The decrease was more prominent with the oligo(dT) primer. This observation is directly related to the mechanism of mRNA degradation scrutinized further in this paper. Importantly, timing of mRNA degradation coincided with the early events of Xenopus egg apoptosis, such as meiotic exit and cytochrome c release (). After 36 hours of PG administration, only the most abundant transcripts could be detected in eggs. At that time too, mRNA degradation was more pronounced with the oligo(dT) primer (). The hallmark apoptotic events, such as caspase activation, apoptotic nuclear morphology, increase in ADP/ATP ratio, intracellular ATP depletion, were observed in the eggs by that time (). Afterwards, by 48 hours, no intact mRNA molecules could be detected in the egg cytoplasm by the applied method (data not shown). By that time, prominent intracellular acidification and egg swelling occurred, indicative of terminal stages of cell death (). Remarkably, the decrease in transcript levels was not detected in control PG-untreated oocytes that remained morphologically intact (). mRNA stability in the resting immature oocytes and mRNA degradation in the aging apoptotic eggs were additionally confirmed for the small subsets (n≤6) of analyzed transcripts in 2 other oocytes and eggs obtained from different animals (data not shown).
Figure 2. Changes of specific transcript contents in the oocytes after PG administration. In panels (A), (B), (C), specific transcript levels measured in a single Xenopus oocyte at 12, 24 and 36 hours after PG administration were referred to those measured in the same oocyte before PG treatment. Difference between the cycle threshold values of the 2 qPCR measurements (ΔCq) was plotted on the y-axis. (D) Changes of specific transcript levels were evaluated in a control oocyte after 48-hour incubation in the absence of PG. In panels (B) and (C) random hexamer (red bars) or oligo(dT) (blue bars) primers were used to generate first strand cDNA by reverse transcription. Transcript levels were measured in 2 to 4 replicates. SDs of replicate measurements are small and not visible in the panels.
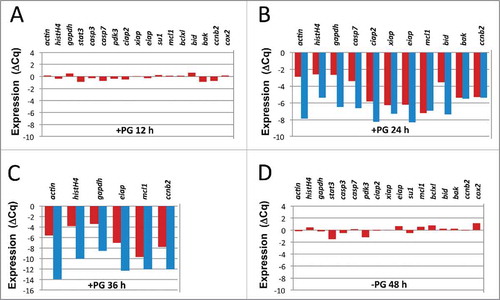
Table 1. Timing of intracellular and apoptotic events in PG-treated Xenopus oocytes.
Endonucleolytic mechanism of mRNA degradation in apoptotic eggs
mRNA degradation in the apoptotic Xenopus eggs concerned all transcripts analyzed. However, the degree of degradation varied greatly between individual transcripts. In addition, it was more prominent with oligo(dT) than with random hexamer primers (). We hypothesized that these observations might be related to the mechanism of mRNA decay in apoptotic eggs and next investigated whether the correlations exist between the detected degree of mRNA degradation and amplicon length and position. Using random hexamer reverse transcriptase primers, no statistically significant correlations were found between ΔCq and amplicon distances from 5′ or 3′mRNA end in the analyzed transcripts at 24 and 36 hours after PG administration (), as it could be expected in the case of exonucleolytic decay. On the other hand, strong negative correlation existed between ΔCq and amplicon length both at 24 and 36 hours (), suggesting endonucleolytic mechanism of mRNA decay. Indeed, if mRNA degradation in apoptotic Xenopus eggs is endonucleolytic, the probability of transcript breakdown should be proportional to the length of a target sequence accessible for cleavage. The statistical significance of this relationship was validated by the one-tailed probability test (). The strong negative correlation between mRNA degradation degree and amplicon length (r = −0.88 at 24 hours and r = −0.87 at 36 hours), as well as the lack of significant correlation between the degradation degree and amplicon position, were additionally confirmed for the small subsets (n ≤ 6) of analyzed transcripts in 2 other eggs obtained from different animals. Furthermore, the strong negative correlation between PCR quantification cycle and amplicon length was observed in apoptotic eggs using several sets of PCR primers amplifying sequences of different length in a single gene (). Recently, similar approach was proposed for estimation of RNA integrity upon quantification of transcript number.Citation13 Altogether, our data indicate that mRNA is degraded in apoptotic Xenopus eggs by an endonucleolytic mechanism.
Figure 3. Correlations of ΔCq with amplicon length and position obtained with random hexamer RT primers. The results of pairwise correlation analysis between the decrease in transcript level and amplicon distance from the 5′ end, (A) and (D), amplicon distance from the 3′ end, (B) and (E), and amplicon length, (C) and (F), are presented. The decrease in transcript contents was determined for 11 transcripts at 24 hours after PG administration in panels (A), (B), (C) and for 6 transcripts at 36 hours in panels (D), (E), (F), as described in the legend to . First strand cDNA was generated using random hexamer primers. Calculated values of the pairwise correlation coefficient (r) and one-tailed probability test (p) are indicated in the panels.
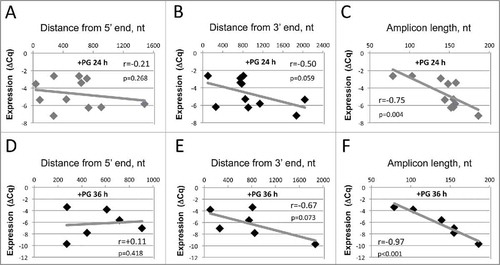
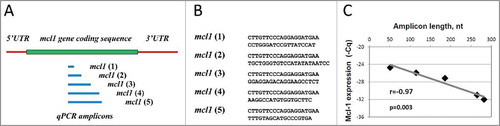
Figure 4. Correlation between Cq and amplicon length in mcl1 gene transcript observed in apoptotic eggs. Positions and sequences of 5′ qPCR primers targeting qPCR amplicons of different length in mcl1 gene transcript are presented in panels (A) and (B), respectively. First strand cDNA was generated with random hexamer primers. The results of pairwise correlation analysis between Cq and amplicon length are shown in panel (C). Calculated values of the pairwise correlation coefficient (r) and one-tailed probability test (p) are indicated in panel (C).
Poly(A)-mediated degradation of mRNA in apoptotic eggs
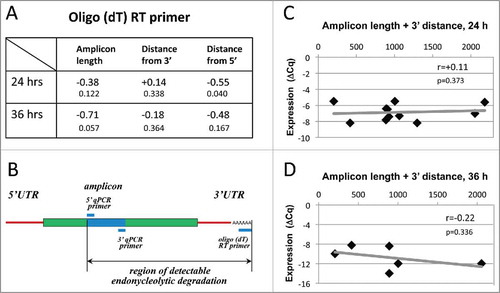
The measured degree of mRNA degradation in apoptotic eggs was universally higher when the oligo(dT) primers were used to generate first-strand DNA instead of random hexamer primers (). Importantly, the strong correlation between the amplicon length and degree of mRNA degradation observed with random primers was lost with oligo(dT) (). Instead, considering endonucleolytic mechanism of mRNA degradation and the fact that oligo(dT) anneals at the 3′ terminal of a transcript, it could be expected that correlation exists between the 3′ amplicon distance and mRNA degradation degree. However, no statistically significant relationship between the amplicon position and degradation degree has been observed (). Taking into account that an endonucleolytic event resulting in the loss of qPCR signal may occur in any place between oligo(dT) and 5′ qPCR primers (), we also analyzed the correlations between mRNA degradation and the combined parameter including amplicon length and its distance from 3′ end at 24 and 36 hours after PG administration (). In addition, to single out the influence of either amplicon length or 3′ distance, the correlations for transcripts with close amplicon length (148–157 nt) or 3′ distances (757–909 nt) were built (Fig. S1). None of the investigated relationships was statistically significant, indicating that endonucleolytic mechanism alone cannot explain the results obtained with oligo(dT), and there should exist an alternative poly(A) tail-dependent mechanism of mRNA degradation in apoptotic Xenopus eggs. The lack of significant correlation between mRNA degradation degree and amplicon length/position in oligo(dT)-generated templates was additionally confirmed for the small subsets (n≤6) of analyzed transcripts in 2 other eggs obtained from different animals.
Figure 5. Correlations of ΔCq with amplicon length and position obtained using oligo(dT) RT primers. Panel (A) presents values of the pairwise correlation coefficient (big fonts) and one-tailed probability test (small fonts) determined at 24 and 36 hours after PG administration in the experiments using the oligo(dT) RT primer. Panel (B) specifies the region of detectable endonucleolytic degradation in the used experimental design. The results of pairwise correlation analysis between ΔCq and combined length of amplicon plus its distance from the 3′ mRNA end determined for 11 transcripts at 24 hours and for 6 transcripts at 36 hours after PG administration are presented in panels (C) and (D), respectively. Calculated values of the pairwise correlation coefficient (r) and one-tailed probability test (p) are indicated in the panels.
Discussion
It was reported previously that apoptosis in animal cells is accompanied by rapid shutdown of global protein synthesis,Citation14,15 preventing cellular repair mechanisms associated with de novo protein synthesis. Protein synthesis inhibitors, such as cycloheximide and anisomycin, were shown to trigger apoptosis in several cell lines,Citation16,17 suggesting that the reduction of protein synthesis may play a role in the development of cell death. In part, translation inhibition during apoptosis is caused by the caspase-mediated proteolysis of components of the cap-binding eukaryotic translation initiation factors, such as eIF4GI, eIF4B, and eIF3J.Citation14,15,18,19 Also, degradation of the poly(A)-binding protein was found to contribute to translation inhibition in apoptotic cells.Citation20 In addition, the cleavage of protein synthesis-related rRNAs occurs during apoptosis in several animal cell lines. Apoptosis-induced degradation of 28S rRNA,Citation21 18S rRNACitation22 and mitochondrial 16S rRNACitation23 has been documented. rRNA degradation was found to be site-specific and the cleavage sites of 28S rRNA have been identified in cells undergoing apoptosis.Citation24 Furthermore, mRNA degradation during apoptosis was suggested to be a contributing factor in the reduction of protein synthesis. Degradation of cytoplasmic mRNAs has been documented in several animal and plant cell lines with different degradation patterns between mRNA species.Citation1,3 Apoptosis-induced mRNA degradation is not restricted by stimulus, cell line, or species and it affects all mRNAs analyzed, suggesting that global mRNA degradation may be a general feature of cell death. It was found that mRNA decay precedes DNA degradation and correlates with the appearance of the early apoptotic marker annexin V, indicating that apoptosis-induced processing of mRNA is an early apoptotic event.Citation1 Also, the evidence was presented that a global increase in the rate of mRNA degradation occurs before the caspase-dependent cleavage of initiation factors.Citation5
In agreement with these findings, our data demonstrate that various cytoplasmic mRNAs are robustly degraded in apoptotic Xenopus eggs. mRNAs coding for proteins classified in different functional classes are prominently degraded by 24 hours of PG treatment at the time of cytochrome c release (; ). Previously, the general caspase inhibitor Z-VAD-fmk was shown to inhibit expression of all apoptotic markers including mRNA degradation in somatic mammalian cells,Citation1 suggesting that this process depends on an upstream apical caspase. However, mRNA degradation in apoptotic Xenopus eggs starts before measurable caspase activation. Indeed, highly sensitive fluorescent assay failed to detect caspase activation in the eggs at 24 hours after PG administration, whereas mRNA degradation was quite prominent at that time ( and S2A). Moreover, many of the analyzed transcripts were degraded beyond the detection limit by the time of caspase 3/7 activation and intracellular ATP depletion at 36 hours (, ). Importantly, exit from the meiotic metaphase arrest and cytochrome c release could be registered in the eggs by 24 hours of PG treatment (Fig. S2B, ), signaling the beginning of apoptosis. Massive degradation of mRNA and its temporal correlation with the early apoptotic events in aging Xenopus eggs suggest that this process may directly contribute to cell death via translational shutdown during apoptotic induction.
We further show that mRNA degradation in apoptotic Xenopus eggs is endonucleolytic. Indeed, a strong negative correlation of a high statistical significance was observed between ΔCq and qPCR amplicon length (). At present, molecular mechanisms involved in apoptotic degradation of mRNA are unknown. Normally, most mRNAs undergo decay in the cytoplasm via the deadenylation-dependent pathway.Citation25 Following deadenylation, mRNAs can be degraded by the 2 major exonucleolytic pathways, either via 3′–5′ exosome-mediated decay or via 5′–3′ decapping-dependent decay.Citation26 However, apoptotic mRNA degradation does not seem to involve these mechanisms, as it was previously found to induce a similar decay kinetics with t1/2 between 1.5 and 3 hrs for mRNAs with very different intrinsic half-lives from 20 min to more than 10 hours.Citation1 In our study too, no significant correlation have been observed between transcript content and PCR amplicon distance from 5′ or 3′transcript ends, as it could be expected if mRNA is degraded via an exonucleolytic mechanism ().
The fact that transcript degradation is more prominent when measured with the oligo(dT) RT primers, taken together with the observation that the correlation between transcript stability and qPCR amplicon length is lost in this case (), point to the existence of some additional mRNA degradation mechanism(s) targeting poly(A) 3′ tail sequence in apoptotic eggs. One plausible candidate for this is mRNA deadenylation, which can potentially attenuate oligo(dT) primer annealing and hinder transcript detection. In fact, oligo(dT)-primed RT-PCR represents a highly efficient and well-established method to detect adenylated transcripts.Citation27 Previously, microarray and RT-qPCR analyses using oligo(dT) primed cDNA synthesis were used for transcriptional profiling of mRNAs deadenylation in Xenopus tropicalis oocytes and eggs upon maturation, fertilization and aging.Citation28,29 Based on the evaluation of reverse transcription efficiency for mRNAs, containing poly(A)-tails of different size, it was estimated that the threshold for detection of poly(A)-tailed transcripts lies between 5 and 10 adenine nucleotides.Citation28 This experimental approach helped to reveal 9 categories of maternal mRNA with distinct adenylation behavior in egg maturation and fertilization. In direct relation to our present study, it has been reported that aging of Xenopus tropicalis eggs is accompanied by deadenylation of a specific set of maternal mRNAs.Citation29 Of note, the difference in the degree of mRNA degradation measured with random and oligo(dT) primers in our study was especially prominent in the case of housekeeping genes, such as actin, histH4 and gapdh (). In this connection, the global analysis of poly(A) lengths in multiple eukaryotic organisms demonstrated that housekeeping genes tend to have shorter poly(A) tails.Citation30 The existence of an apoptotic deadenylation-mediated mRNA degradation mechanism proposed by our present study is consistent with this report. Notably, deadenylation may not be necessarily correlated with exhaustive mRNA degradation, because there is no decapping enzyme in Xenopus oocytes and eggs, and most deadenylated mRNAs in dividing Xenopus embryos remain stable until the mid-blastula transition, when the decapping enzyme is synthesized and maternal mRNAs are degraded.
The observations of apoptotic mRNA degradation raise a question about identity of the enzymes involved in this process. It is highly unlikely that a caspase-activated enzyme is involved in apoptotic processing of mRNA, as it is the case with the caspase-activated DNase (CAD) responsible for DNA fragmentation and chromatin disassembly during apoptosis.Citation31,32 As discussed above, mRNA degradation in apoptotic Xenopus eggs starts before measurable caspase activation. Correspondingly, apoptotic nuclear morphology acquired due to chromatin disassembly cannot be observed at that time (). Of note, previously reported degradation patterns were dominated by the specific intermediates of a defined length, rather than a smear of degradation products.Citation1,3,4 This fact points to the existence of designated cleavage sites and suggests the involvement of site-specific endoribonuclease(s). The ubiquitous interferone-inducible RNase L has been implicated in the degradation of 28S rRNA during apoptosis in mammalian cells.Citation33 It is quite plausible however that degradation of mRNA and rRNA is executed by different enzymes. Another candidate for an apoptotic RNase may be the mitochondrial endonuclease G, which is released from the mitochondrial intermembrane space during apoptosis and forms complexes with AIF and Hsp70.Citation34 The enzyme was shown to digest RNA more favorably than DNA at physiologic conditions, however its direct involvement in apoptotic degradation of mRNA has not been demonstrated.
Concerning possible mechanisms of the apoptotic deadenylation, it was found that apoptosis in U937 cells is associated with dephosphorylation, proteolysis and reduced activity of poly(A) polymerase.Citation35 Similar modifications have also been observed in other cell lines. Also, degradation of poly(A)-binding protein was found to contribute to translation inhibition in apoptotic cells.Citation20 Most recently, generation of 3′ truncated mRNA decay intermediates with nontemplated uridylate-rich tails was observed during apoptosis in cultured mammalian cells.Citation2 The uridylate-rich tails are added by the terminal uridylyl transferases ZCCHC6 and ZCCCH11, then the uridylated transcripts are degraded by the 3′ to 5′ exonuclease DIS3L22. It is interesting to investigate whether the enzymes and molecular mechanisms involved in mRNA deadenylation in mammalian cells are also employed in apoptotic Xenopus eggs.
In sum, our study demonstrates that robust and global decay of mRNA occurs in aging Xenopus eggs early in apoptosis via an endonucleolytic process. Deadenylation also contributes to the apoptotic mRNA degradation. Temporal correlation of mRNA degradation with the onset of apoptosis implies that it may directly contribute to cell death via translational shutdown.
Materials and methods
Animals and cells
Adult female frogs of Xenopus laevis were obtained from Hamamatsu Seibutsu Kyozai (Hamamatsu, Japan). Animal handling was performed in accordance with the guidelines of the Kobe University Animal Experimentation Regulations. All experimental protocols concerning the animals were approved by the Institutional Animal Care and Use Committee of Kobe University (permission number KEN-12). The experiments with oocytes and eggs were conducted at ambient temperature 21–23°C. To isolate oocytes, the frogs were anesthetized, then the ovaries were surgically removed and placed into OR-2 solution containing 82.5 mM NaCl, 2.5 mM KCl, 1 mM CaCl2, 1 mM MgCl2, 1 mM Na2HPO4, 5 mM HEPES, pH 7.6. The chemicals used for buffer preparation were purchased from Wako (Osaka, Japan), Nacalai Tesque (Kyoto, Japan) and Sigma (St. Louis, MO). The ovaries were manually dissected into clumps of 50–100 oocytes and extensively washed with OR-2 solution. Oocytes were treated with 0.5 mg/ml collagenase (280 U/mg; Wako) in OR-2 at 21°C for 3 hours by shaking at 60 rpm, extensively washed in OR-2 solution and left for stabilization over 4 hours. Undamaged defolliculated oocytes of stage VI were manually selected and used in experiments. Oocyte maturation was induced by addition of 10 μM progesterone (Sigma) and monitored by the appearance of a white spot on the animal hemisphere of oocytes. Microscopic observations of oocyte and egg morphology were performed using a stereomicroscope Leica S8APO (Leica Microsystems, Wetzlar, Germany). Oocytes and eggs were bench-top aged in 110 mm dishes for up to 48 hours.
Cytoplasmic collections
In detail, the technique of multiple cytoplasmic collections from Xenopus oocytes was described previously.Citation12 Briefly, capillaries for cytopasmic collections were prepared using a dual-stage glass micropipette puller from Narishige (Tokyo, Japan). The capillary was positioned for cytoplasmic samplings using a 3-axis micromanipulator under microscopic observation. Cytoplasmic collections were made with the pulse-directed nanoliter injector system Nanoject II (Drummond, Broomall, PA). The capillary tip was inserted into the oocyte to a depth of 300–500 μm followed by collection of a 10-nl volume of cytoplasmic material. The sample was withdrawn into 2 μl of nanopure water containing 1 U/μl of RNasin Plus ribonuclease inhibitor (Promega, Fitchburg, WI). The collected samples were frozen in liquid nitrogen and kept at −80°C till following gene expression analysis.
RT-qPCR
Quantification of gene-specific transcripts was performed in the samples using RT-qPCR technique. Total RNA was extracted from the collected samples using an RNeasy Mini purification kit from Qiagen (Valencia, CA). Reverse transcription was performed using an AccuScript High Fidelity 1st strand cDNA Synthesis Kit from Agilent (Santa Clara, CA), 50 ng of total RNA and poly(dT) or random hexamer primers. Gene–specific nucleotide sequences were detected with a SYBR Green detection system from Roche (Penzberg, Germany). qPCR was performed in a final volume of 20 μl using a LightCycler 480 Instrument (Roche) under the following cycling conditions: 95°C for 2 min, 45 cycles at 95°C for 10 sec, 60°C for 10 sec, 72°C for 10 sec. After cycling, the melting curve was recorded between 65°C and 95°C with a ramp rate of 0.11°C/sec. The detection primers were designed with the help of Primer3 softwareCitation36 [http://biotools.umassmed.edu/bioapps/primer3_www.cgi]. Primer sequences are presented in Table 1S. The primers were purchased from Invitrogen (Carlsbad, CA) and used in PCR at a final concentration of 0.5 μM. The primers produced a single peak in the derivative of the melting curve and did not amplify non-template controls.
Data analysis and statistics
The data of qPCR experiments were analyzed with the built-in LightCycler 480 Software. Measured Cq values of qPCR replicates were averaged. Absolute quantification of the transcript numbers for studied genes was performed using calibration curves obtained with the corresponding double-stranded PCR products on the assumption that efficiency of reverse transcription is 100%. The experimentally determined efficiency of PCR reactions for studied gene products was ≥95%, as calculated by the software used. No PCR efficiency correction has been applied to the data. For relative comparisons of gene expression, the levels of individual transcripts in aging oocytes and eggs were referenced to those in freshly obtained oocytes. Correlations of mRNA content with amplicon length and position were evaluated by calculating Pearson's pairwise correlation coefficients. Statistical significance of the correlation coefficients was determined by calculating one-tailed probability values, given the values of correlation coefficient and sample size, with a 0.95 confidence level. Calculations of both correlation coefficients and p-values were performed using the online statistics calculators available at http://www.danielsoper.com/statcalc3/.
Other methods
The total content of isolated RNA in the collected cytoplasmic samples was determined using a NanoDrop ND-1000 spectrophotometer (Nano-Drop Technologies, Wilmington, DE, USA). Measurements of Cdk1 activity, MAPK activation, cytochrome c release, caspase 3/7 activity, detection of nuclear morphology, assessment of intracellular ATP and ADP contents, egg diameter and intracellular pH were performed as described previously.Citation9
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Supplemental_Data.pptx
Download MS Power Point (151.9 KB)Funding
This work was supported by the Research Fund for Foreign Visiting Professor from Kobe University (to A.A.T.) and the Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (25440023 to A.A.T., 21770142 to T.I., 15K07083 to K-I.S. and 21570225 to Y.F.).
References
- Del Prete MJ, Robles MS, Guáo A, Martínez-A C, Izquierdo M, Garcia-Sanz JA. Degradation of cellular mRNA is a general early apoptosis-induced event. FASEB J 2002; 16:2003-5; PMID:12397088; http://dx.doi.org/10.1096/fj.02-0392fje
- Thomas MP, Liu X, Whangbo J, McCrossan G, Sanborn KB, Basar E, Walch M, Lieberman J. Apoptosis triggers specific, rapid, and global mRNA decay with 3′ uridylated intermediates degraded by DIS3L2. Cell Rep 2015; 11:1079-89; PMID:25959823; http://dx.doi.org/10.1016/j.celrep.2015.04.026
- Hoat TX, Nakayashiki H, Tosa Y, Mayama S. Specific cleavage of ribosomal RNA and mRNA during victorin-induced apoptotic cell death in oat. Plant J 2006; 46:922-33; PMID:16805727; http://dx.doi.org/10.1111/j.1365-313X.2006.02752.x
- Mroczek S, Kufel J. Apoptotic signals induce specific degradation of ribosomal RNA in yeast. Nucleic Acids Res 2008; 36:2874-88; PMID:18385160; http://dx.doi.org/10.1093/nar/gkm1100
- Bushell M, Stoneley M, Sarnow P, Willis AE. Translation inhibition during the induction of apoptosis: RNA or protein degradation? Biochem Soc Trans 2004; 32:606-10; PMID:15270687; http://dx.doi.org/10.1042/BST0320606
- Sasaki K, Chiba K. Fertilization blocks apoptosis of starfish eggs by inactivation of the MAP kinase pathway. Dev Biol 2001; 237:18-28; PMID:11518502; http://dx.doi.org/10.1006/dbio.2001.0337
- Yuce O, Sadler KC. Postmeiotic unfertilized starfish eggs die by apoptosis. Dev Biol 2001; 237:29-44; PMID:11518503; http://dx.doi.org/10.1006/dbio.2001.0361
- Voronina E, Wessel GM. Apoptosis in sea urchin oocytes, eggs, and early embryos. Mol Reprod Dev 2001; 60:553-61; PMID:11746966; http://dx.doi.org/10.1002/mrd.1120
- Tokmakov AA, Iguchi S, Iwasaki T, Fukami Y. Unfertilized frog eggs die by apoptosis following meiotic exit. BMC Cell Biol 2011; 12:56; PMID:22195698; http://dx.doi.org/10.1186/1471-2121-12-56
- Pasquier DD, Dupre A, Jessus C. Unfertilized Xenopus eggs die by Bad-dependent apoptosis under the control of Cdk1 and JNK. PLoS One 2011; 6(8):e23672; PMID:21858202; http://dx.doi.org/10.1371/journal.pone.0023672
- Iguchi S, Iwasaki T, Fukami Y, Tokmakov AA. Unlaid Xenopus eggs degrade by apoptosis in the genital tract. BMC Cell Biol 2013; 14:11; PMID:23452868; http://dx.doi.org/10.1186/1471-2121-14-11
- Tokmakov AA, Hashimoto T, Hasegawa Y, Iguchi S, Iwasaki T, Fukami Y. Monitoring gene expression in a single Xenopus oocyte using multiple cytoplasmic collections and quantitative RT-PCR. FEBS J 2014; 281:104-14; PMID:24165194; http://dx.doi.org/10.1111/febs.12576
- Brisco MJ, Morley AA. Quantification of RNA integrity and its use for measurements of transcript number. Nucleic Acids Res 2012; 40:e144; PMID:22735698; http://dx.doi.org/10.1093/nar/gks588
- Marissen WE, Lloyd RE. Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol Cell Biol 1998; 18:7565-74; PMID:9819442; http://dx.doi.org/10.1128/MCB.18.12.7565
- Clemens MJ, Bushell M, Jeffrey IW, Pain VM, Morley SJ. Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ 2000; 7:603-15; PMID:10889505; http://dx.doi.org/10.1038/sj.cdd.4400695
- Martin SJ, Lennon SV, Bonham AM, Cotter TG. Induction of apoptosis (programmed cell death) in human leukemic HL-60 cells by inhibition of RNA or protein synthesis. J Immunol 1990; 145:1859-67; PMID:2167911.
- King KL, Jewell CM, Bortner CD, Cidlowski JA. 28S ribosome degradation in lymphoid cell apoptosis: evidence for caspase and Bcl-2-dependent and -independent pathways. Cell Death Differ 2000; 7:994-1001; PMID:11279546; http://dx.doi.org/10.1038/sj.cdd.4400731
- Bushell M, McKendrick L, Jänicke RU, Clemens MJ, Morley SJ. Caspase-3 is necessary and sufficient for cleavage of protein synthesis eukaryotic initiation factor 4G during apoptosis. FEBS Lett 1999; 451:332-6; PMID:10371215; http://dx.doi.org/10.1016/S0014-5793(99)00614-6
- Bushell M, Poncet D, Marissen WE, Flotow H, Lloyd RE, Clemens MJ, Morley SJ. Cleavage of polypeptide chain initiation factor eIF4GI during apoptosis in lymphoma cells: characterisation of an internal fragment generated by caspase-3-mediated cleavage. Cell Death Differ 2000; 7:628-36; PMID:10889507; http://dx.doi.org/10.1038/sj.cdd.4400699
- Marissen WE, Triyoso D, Younan P, Lloyd RE. Degradation of poly(A)-binding protein in apoptotic cells and linkage to translation regulation. Apoptosis 2004; 9:67-75; PMID:14739600; http://dx.doi.org/10.1023/B:APPT.0000012123.62856.20
- Houge G, Døskeland SO, Bøe R, Lanotte M. Selective cleavage of 28S rRNA variable regions V3 and V13 in myeloid leukemia cell apoptosis. FEBS Lett 1993; 315:16-20; PMID:8416804; http://dx.doi.org/10.1016/0014-5793(93)81123-H
- Lafarga M, Lerga A, Andres MA, Polanco JI, Calle E, Berciano MT. Apoptosis induced by methylazoxymethanol in developing rat cerebellum: organization of the cell nucleus and its relationship to DNA and rRNA degradation. Cell Tissue Res 1997; 289:25-38; PMID:9182598; http://dx.doi.org/10.1007/s004410050849
- Crawford DR, Lauzon RJ, Wang Y, Mazurkiewicz JE, Schools GP, Davies KJ. 16S mitochondrial ribosomal RNA degradation is associated with apoptosis. Free Radic Biol Med 1997; 22:1295-300; PMID:9098105; http://dx.doi.org/10.1016/S0891-5849(96)00544-8
- Houge G, Robaye B, Eikhom TS, Golstein J, Mellgren G, Gjertsen BT, Lanotte M, Døskeland SO. Fine mapping of 28S rRNA sites specifically cleaved in cells undergoing apoptosis. Mol Cell Biol 1995; 15:2051-62; PMID:7891700; http://dx.doi.org/10.1128/MCB.15.4.2051
- Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol 2007; 8:113-26; PMID:17245413; http://dx.doi.org/10.1038/nrm2104
- Ghosh S, Jacobson A. RNA decay modulates gene expression and controls its fidelity. Wiley Interdiscip Rev RNA 2010; 1:351-61; PMID:21132108; http://dx.doi.org/10.1002/wrna.25
- Slomovic S, Portnoy V, Schuster G. Detection and characterization of polyadenylated RNA in Eukarya, Bacteria, Archaea, and organelles. Methods Enzymol 2008; 447:501-20; PMID:19161858; http://dx.doi.org/10.1016/S0076-6879(08)02224-6
- Graindorge A, Thuret R, Pollet N, Osborne HB, Audic Y. Identification of post-transcriptionally regulated Xenopus tropicalis maternal mRNAs by microarray. Nucleic Acid Res 2006; 34:986-95; PMID:16464828; http://dx.doi.org/10.1093/nar/gkj492
- Kosubek A, Klein-Hitpass L, Rademacher K, Horsthemke B, Ryffel GU. Aging of Xenopus tropicalis eggs leads to deadenylation of a specific set of maternal mRNAs and loss of developmental potential. PLoS One 2010; 5(10):e13532; PMID:21042572; http://dx.doi.org/10.1371/journal.pone.0013532
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 2014; 508:66-71; PMID:24476825; http://dx.doi.org/10.1038/nature13007
- Widlak P, Garrard WT. Discovery, regulation, and action of the major apoptotic nucleases DFF40/CAD and endonuclease G. J Cell Biochem 2005; 94:1078-87; PMID:15723341; http://dx.doi.org/10.1002/jcb.20409
- Larsen BD, Sørensen CS. The caspase-activated DNase: apoptosis and beyond. FEBS J 2016; Nov 19; PMID:27865056; http://dx.doi.org/10.1111/febs.13970
- Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, et al. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J 1997; 16:6355-63; PMID:9351818; http://dx.doi.org/10.1093/emboj/16.21.6355
- Kalinowska M, Garncarz W, Pietrowska M, Garrard WT, Widlak P. Regulation of human apoptotic DNase/RNase endonuclease G: involvement of Hsp70 and ATP. Apoptosis 2005; 10:821-30; PMID:16133872; http://dx.doi.org/10.1007/s10495-005-0410-9
- Atabasides H, Tsiapalis CM, Havredaki M. Dephosphorylation, proteolysis, and reduced activity of poly(A) polymerase associated with U937 cell apoptosis. Exp Cell Res 1998; 244:433-40; PMID:9806793; http://dx.doi.org/10.1006/excr.1998.4231
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and biologists programmers. Methods Mol Biol 2000; 132:365-86; PMID:10547847
