ABSTRACT
Nonsense mutations, generating premature termination codons (PTCs), account for 10% to 30% of the mutations in tumor suppressor genes. Nonsense translational suppression, induced by small molecules including gentamicin and G418, has been suggested as a potential therapy to counteract the deleterious effects of nonsense mutations in several genetic diseases and cancers. We describe here that NB124, a synthetic aminoglycoside derivative recently developed especially for PTC suppression, strongly induces apoptosis in human tumor cells by promoting high level of PTC readthrough. Using a reporter system, we showed that NB124 suppressed several of the PTCs encountered in tumor suppressor genes, such as the p53 and APC genes. We also showed that NB124 counteracted p53 mRNA degradation by nonsense-mediated decay (NMD). Both PTC suppression and mRNA stabilization contributed to the production of a full-length p53 protein capable of activating p53-dependent genes, thereby specifically promoting high levels of apoptosis. This new-generation aminoglycoside thus outperforms the only clinically available readthrough inducer (gentamicin). These results have important implications for the development of personalised treatments of PTC-dependent diseases and for the development of new drugs modifying translation fidelity.
Introduction
Nonsense mutations, creating premature termination codons (PTCs), cause the premature termination of translation, by directly interrupting translation process and by inducing transcript degradation via the nonsense-mediated decay (NMD) pathway.Citation1,2 Many cancers are linked to the appearance of a PTC in a tumor suppressor gene, resulting in the loss of the protein or the synthesis of a truncated protein unable to inhibit cell proliferation or to promote apoptosis. This is true for 2 common suppressor genes, p53 and APC, which are mutated in 50% of human cancers and 80% of colorectal cancers, respectively. PTCs account for 10% of p53 mutations and 30% of APC mutations. In the last decade, considerable interest has focused on in-frame PTCs as potential therapeutic targets. Indeed, aminoglycosides (such as gentamicin, G418, and amikacin) have been shown to promote PTC readthrough by binding to ribosomes, partly restoring the synthesis of a full-length functional protein in cultured mammalian cells and animal models.Citation3,4
This strategy has been evaluated in several genetic diseases and cancers. We have shown that the readthrough of a PTC in the p53 gene efficiently promotes the apoptosis of cancer cells.Citation5 However, since the seminal work on this topic, only a few compounds promoting readthrough have been discovered.Citation6-8 PTC124 is probably the most widely studied readthrough inducer, but its clinical benefits remain a matter of debate, despite the recent conditional approval of this drug by the EMA.Citation9 This personalised approach to treatment is currently hindered mainly because the lack of new molecules promoting the highly efficient re-expression of genes inactivated by PTCs. Gentamicin remains the molecule most widely used in this context, mainly for the proof-of-concept experiments but its long-term use leads to nephrotoxicity and ototoxicity, limiting applications in PTC suppression therapies.Citation10 Fortunately, the toxicity of aminoglycosides is not an inherent aspect of their ability to promote stop codon readthrough, but instead reflects their inhibition of mitochondrial translation.Citation11,12 It should, therefore, be possible to design aminoglycoside derivatives yielding higher levels of PTC suppression with less toxicity.Citation13,14 In this study, we tested several aminoglycoside derivatives, specially developed by us for PTC suppression, and identified one (NB124) as a potent readthrough inducer active against several of the PTCs found in the p53 and APC tumor suppressor genes. The p53 tumor suppressor gene is the cellular gatekeeper for growth and division. It encodes a transcription factor that triggers cell-cycle arrest and apoptosis in response to diverse cellular stresses, including DNA damage, oncogene activation and hypoxia.Citation15-17 The APC (adenomatous polyposis coli) tumor suppressor gene encodes a 311 kDa multidomain protein that downregulates the Wnt pathway.Citation18 We show here that NB124 restores the production of a full-length p53 protein from a gene inactivated by a PTC at position 213; the efficacy of NB124 was superior to that of gentamicin. Furthermore, this p53 protein is active, as it induces the expression of its target genes, Bax and p21, and promotes high levels of apoptosis in cancer cells. The accumulative data indicate that NB124 is a promising drug for blocking the proliferation of cancer cells with tumor suppressor genes harbouring a PTC.
Results
Synthetic aminoglycosides efficiently suppress nonsense mutations in tumor suppressor genes
The NB compounds tested here, NB74, NB84, NB122, NB124 and NB127 (Fig. S1), all contain structural components derived from paromomycin and G418, but were designed to suppress PTCs more efficiently and with lower toxicity than conventional aminoglycosides.Citation19 Several of these molecules have already been tested in various models and have displayed significant improvements in suppression activity over conventional molecules.Citation20 However, none has been tested on mutations in tumor suppressor genes. We assessed the ability of these compounds to suppress the R213X mutation, the most frequent nonsense mutation of the p53 tumor suppressor gene (accounting for 14.5% of reported nonsense mutations; UMD Database,Citation21 all curated). It has already been shown that the sequences surrounding the PTC strongly influence the suppression levels induced by these drugs.Citation22 It is, therefore, important to monitor the readthrough of the p53 R213X PTC in its natural mRNA sequence context. The stop codon of the R213X and the surrounding nucleotide context, shown in , were inserted into a dual reporter vector, for the accurate quantification of stop codon readthrough efficiency. Readthrough levels were quantified in NIH3T3 cells transiently transfected with the reporter vector, in the presence or absence of drugs. In parallel, as a control, we used conventional aminoglycosides for which the effect on readthrough efficiency had already been described: G418 and gentamicin. For each compound, we tested a range of concentrations, to identify the optimal dose yielding maximal R213X readthrough.
Table 1. nonsense mutations tested in this study.
All the molecules tested induced readthrough in a dose-dependent manner (). NB74 was the least efficient compound in this set of NB molecules tested. The maximum level of readthrough achieved with NB74 was just above 5%, a value slightly below that yielded by gentamicin at the same concentration. All the other derivatives were more efficient than gentamicin, and the most potent readthrough inducer tested was NB124. This compound was about 3 times more effective than gentamicin by increasing basal readthrough rates by a factor of 25. In addition, we note that NB124 was also significantly less cytotoxic than gentamicin and G418 in a tissue-based model of ototoxicity.Citation14 It is of note that G418 is considered as one of the strongest readthrough inducers while in parallel it is also the most toxic aminoglycoside; it is so toxic that cannot be used even as an antibiotic.Citation13 As shown in , at the highest dose compatible with cell viability (1 mg/ml for NB124 and 0.2mg/mL for G418) NB124 induced higher levels of readthrough than G418.
Figure 1. Readthrough efficiency of various synthetic aminoglycosides for the p53 R213X mutation. Readthrough of the p53 R213X nonsense mutation was measured with a dual reporter vector after transfection of NIH3T3 cells and their treatment of 24 h. Five synthetic aminoglycoside derivatives (NB74, NB84, NB122, NB124 and NB128) were tested at 5 concentrations (0.2, 0.4, 0.6, 0.8 and 1 mg/mL), in parallel with 2 standard aminoglycosides (G418 and gentamicin) as controls. NB124 was the best readthrough inducer among all the new compounds tested. The lines in the centers of the boxes indicate the medians; the box limits indicate the 25th and 75th percentiles, as determined by R software; the whiskers extend to 1.5 times the interquartile range from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles. n = 6 samples.
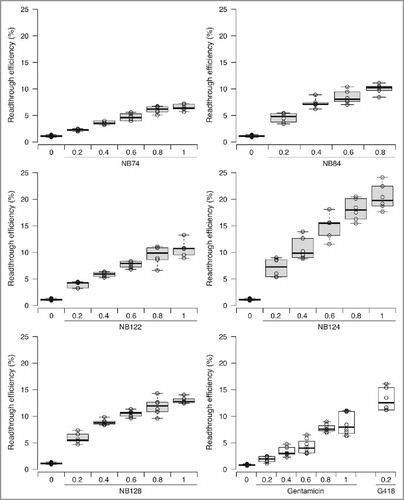
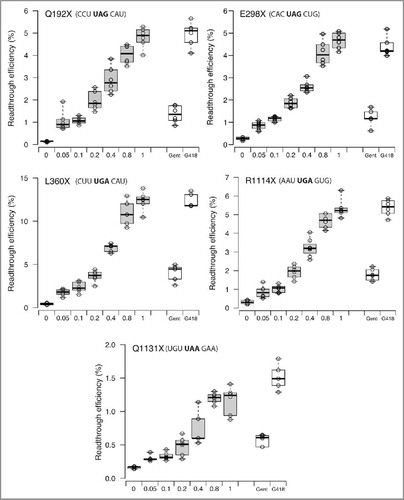
NB124, thus, clearly emerged as the most promising compound tested, and this encouraged us to further investigate its ability to suppress other nonsense mutations. The identity of a PTC and its surrounding mRNA sequence context have already been shown to have considerable influence on the ability of drugs to suppress PTCs.Citation22 It is therefore important to determine readthrough efficiency for various stop codons. We selected a set of 6 PTCs found in the APC or P53 genes of patients (). These 6 mutations correspond to the 6 PTCs most frequently encountered in the P53 and APC genes. For all the stop codons tested, NB124 clearly promoted PTC readthrough more efficiently than gentamicin (Gent), inducing 2 to 5 times more readthrough, depending on the stop codon considered (). In addition, it is of note that in all the contexts sequences tested, the effect of NB124 at the highest concentration was as strong as that obtained with G418 at 0.2 mg/mL.
Figure 2. NB124 promotes a high level of readthrough for several p53 and APC nonsense mutations. Readthrough efficiencies for 3 nonsense mutations in the P53 (Q192X, R213X, Q298X) and APC (L360X, R1114X, Q1131X) genes were determined with a dual reporter vector in NIH3T3 cells treated with gentamicin (0.8 mg/mL), G418 (0.2 mg/mL) or NB124 (0.05, 0.1, 0.2, 0.4, 0.8 and 1mg/mL) for 24 h. The lines in the centers of the boxes show the medians; box limits indicate the 25th and 75th percentiles, as determined by R software; the whiskers extend to 1.5 times the interquartile range from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles. NB124 data are indicated in gray, controls in white boxes (gentamicin and G418). n = 6 samples.
PTC readthrough levels can be quantified very efficiently with the dual reporter system, but it is also important to determine the effect of the drug on the natural gene. We next analyzed the effect of NB124 on the production of the endogenous p53 protein.
NB124 stabilises mutant p53 mRNA and restores the production of a full-length protein from the endogenous p53 R213X mRNA
We investigated whether NB124 could restore the production of a full-length p53 protein from an endogenous mutated gene, using human HDQ-P1 cells, which were established from a human primary breast carcinoma.Citation23 These cells are homozygous for the p53 R213X nonsense mutation. This mutation is located in exon 6 and generates a premature UGA stop codon more than 50 nt upstream from the last exon–exon junction. The mRNA molecule generated is thus a canonical target for degradation by the NMD pathway.Citation24
The ability of readthrough events induced by aminoglycosides, such as G418, to antagonise NMD in mammalian cells has been reported in several studies.Citation5,25 It was important to determine whether NB124 had retained this ability. Indeed, stabilization of the mutant mRNA is a prerequisite for the induction of significant protein re-expression from an endogenous nonsense mutation.
We therefore performed quantitative PCR on HDQ-P1 cells with and without NB124 treatment of 48 hours. G418 was used as a positive control (). As expected, G418 treatment increased p53 mRNA levels by a factor of 8 at the highest concentration (0.2 mg/mL). NB124 also clearly stabilised the mutant p53 mRNA in a dose-dependent manner, to a similar extent to G418. We also investigated the correlation between stabilization and readthrough properties, using apramycin, an aminoglycoside that can bind to ribosomal A-site without triggering stop codon readthrough.Citation26 No stabilization was observed with apramycin (), suggesting that only aminoglycosides able to induce readthrough stabilised PTC-containing mRNAs, by inhibiting NMD.
Figure 3. NB124 stabilises the mutant p53 mRNA and restores production of the full-length p53 protein in HDQ-P1 cancer cells. (A) HDQ-P1 cells carrying the endogenous R213X nonsense mutation were treated with NB124 (0.05, 0.1, 0.2, 0.4 and 0.8 mg/mL), G418 (0.05, 0.1 and 0.2 mg/mL) or apramycin (1 mg/mL) for 48 h. Levels of mutant p53 mRNA were determined by RT-qPCR. The results of each experiment are expressed relative to the amount of mRNA in the absence of treatment. Median values are presented, together with the SEM (n = 3). (B) HDQ-P1 cells were treated with gentamicin (0.8 mg/mL), G418 (0.2 mg/mL) or NB124 (0.05, 0.1 and 0.4 mg/mL) for 48 h. Western-blot membranes were probed with the DO-1 antibody directed against the N-terminus of p53 and an anti-actin antibody was used as the loading control. An extract from LoVo cells (p53WT) was used as a control.
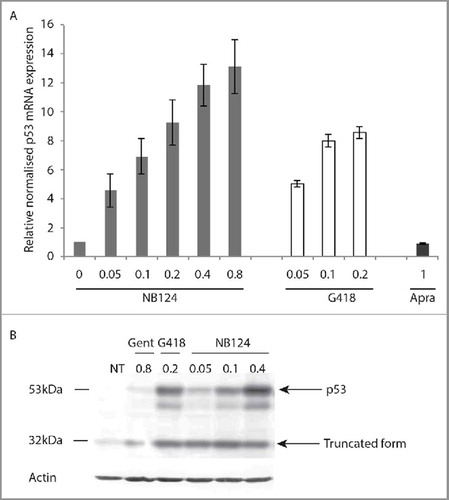
To test the production of a full-length p53 protein, HDQ-P1 cells harbouring R213X mutation were treated with NB124, gentamicin and G418 for 48 hours and the treated cells were analyzed by Western-blot (). In the absence of aminoglycoside, a faint 32 kDa band corresponding to the expected truncated protein was detected (). The weakness of this band is consistent with the low level of mutant p53 mRNA in the untreated mutant cells (). After NB124 treatment, a 53 kDa band corresponding to the full-length protein was detected, and the intensity of this band was dependent on the aminoglycoside dose used.
The NB124-induced p53 protein is functional
The transactivation function of the normal p53 protein underlies its function as a tumor suppressor.Citation27 We investigated whether the full-length p53 protein produced after NB124 treatment was active as a transcription factor, by transfecting HDQ-P1 cells with a reporter plasmid containing p53 binding sites upstream from the luciferase firefly gene (p53BS-luc). In this system, an active p53 protein is required to induce luciferase reporter gene expression.Citation5 Cells were left untreated or were treated with NB124 for 48 hours. G418 was used as a positive control.
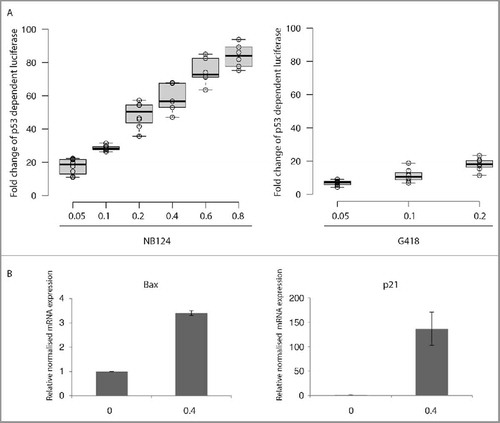
NB124 treatment strongly induced firefly luciferase expression, in a dose-dependent manner, by a factor of up to 80 with respect to that in the absence of treatment (). Treatment with G418 (at 0.2mg/mL) also increased p53-dependent firefly luciferase expression, by a factor of up to 18, but NB124 was clearly more potent than G418 at triggering the production of active p53 protein. Gentamicin was also tested at the same doses, but it had little or no effect on luciferase induction (Fig. S2). This finding is consistent with the weakness of the band corresponding to the full-length p53 protein in cells treated with gentamicin ().
Figure 4. Endogenous re-expressed full-length p53 proteins have normal transcriptional activity. (A) We assessed the transactivation capacity of the full-length p53 proteins produced after aminoglycoside treatment, by transfecting HDQ-P1 cells with a reporter plasmid containing p53 binding sites upstream from the firefly luciferase gene (p53BS-luc). We measured p53-dependent luciferase activity in the presence or absence of NB124 or G418 (0.05, 0.1 and 0.2 mg/mL) for 72 hours. The fold-change in luciferase activity between treated and untreated cells is presented. The lines in the centers of the boxes show the medians; box limits indicate the 25th and 75th percentiles, as determined by R software; the whiskers extend to 1.5 times the interquartile range from the 25th and 75th percentiles; outliers are represented by dots; data points are plotted as open circles. n = 6 samples. (B) Quantitative PCR analysis was used to assess differences in the levels of the Bax and p21 mRNAs between HDQ-P1 (p53 R213X) cells with and without NB124 (0.4 mg/ml) treatment of 72 hours. The results of each experiment are expressed relative to the amount of mRNA in the absence of treatment. Mean values are presented with the SEM (n = 3).
We then investigated whether this transcriptional activity of p53 was sufficient to promote the induction of the cellular target genes normally under the control of p53.
NB124-dependent p53 induces 2 of its main cellular targets: The BAX and p21 genes
We investigated whether the full-length p53 protein produced after NB124 treatment regulated the transcription of p53-responsive cellular genes. We focused on 2 of the most important targets of p53: the Bax gene, the expression of which is crucial for the triggering of the apoptosis pathway, and the p21 gene, which is essential for cell cycle control. We used quantitative PCR to assess differences in the expression of the Bax and p21 genes between HDQ-P1 cells with and without NB124 treatment. In each experiment, the results obtained are expressed relative to those for untreated cells (normalized to 1). Levels of Bax mRNA in HDQ-P1 cells treated with NB124 were 3.5 times higher than those in untreated cells (). The p21 mRNA was hardly detectable in the absence of NB124 but was strongly induced by NB124 treatment (induction factor of 150). In these experiments, no change in mRNA levels for any of the 3 housekeeping genes (used as qPCR reference genes) was observed in response to NB124 treatment.
Re-expressed p53 promotes apoptosis in cancer cells
The reactivation of p53 can be considered an effective anticancer treatment only if the re-expressed protein can induce the apoptosis of cancer cells. We therefore investigated whether the full-length p53 re-expressed after NB124 treatment promoted the apoptosis of a tumor cell line.
We previously showed that HDQ-P1 cells were not sensitive to the re-introduction of WT p53.Citation5 This cell line was, therefore, unsuitable for use in experiments investigating the effects of NB124-induced p53 protein. We overcame this problem by using a different cell line—H1299 (p53−/−) human cancer cells—reported to be sensitive to the re-expression of WT p53.Citation28 We improved reproducibility by constructing a cell line derived from H1299 cells (H1299-p53R213X cells) through stable transfection with a p53 cDNA carrying the R213X mutation. This new cell line was exposed to NB124 during 30 or 50 hours. We assessed apoptosis with the polymerase PARP-1, one of the main cleavage targets of caspase-3. The cleavage of the PARP protein into 89 and 116 kDa fragments is classically used as a marker for apoptotic cells.Citation29 Only the full-length form of PARP-1 was detected in untreated cells (5% of cleaved form), whereas the cleaved forms became detectable after 30 hours of NB124 (45%) treatment, and cleavage was complete after 50 hours of treatment (91%) (). We also compared the effects of NB124 and gentamicin. Gentamicin induced a weak cleavage (6% and 9% after 30 hours and 50 hours respectively) with no sign of improvement over the time ().
Figure 5. NB124 promotes p53-dependent apoptosis. (A) Induction of PARP-1 cleavage: Western blot analysis of poly (ADP-ribose) polymerase (PARP-1) cleavage in cell lysates obtained from H1299-p53R213X cells treated with NB124 (0.4 mg/ml) or gentamicin (0.8 mg/ml) for 30 or 50 hours. NT: non treated. Visible bands correspond to the full-length PARP-1 (116 kDa) and the larger fragment of the cleaved protein (89 kDa), and the β-actin used as a loading control. Percentage of the cleaved form is calculated as the ratio cleaved/ (cleaved + full length). (B) AnnexinV-PE staining: H1299-p53R213X cells were transiently transfected with a p53-targeting siRNA (p53-siRNA) or a non-targeted siRNA (NT-siRNA) or were left untransfected (−). Cells were then left untreated or treated with NB 124 (0.4 mg/ml) or gentamicin (1.2 mg/mL) for 50 hours. Percentages indicate the proportion of Annexin-PE-positive cells detected on flow cytometry, corresponding to cells undergoing apoptosis, with exposed PS (overall apoptosis).
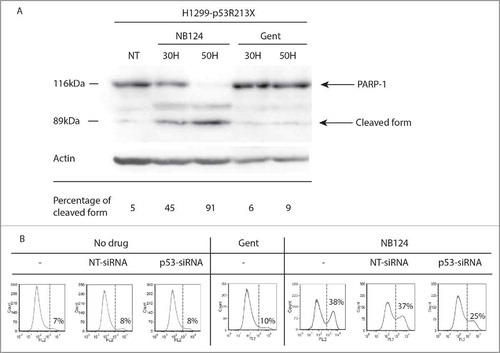
Another general marker of apoptotic cells is phosphatidylserine (PS) exposure on the cell surface. We used fluorochrome-labeled annexin V, a phospholipid-binding protein with a high affinity for PS, to detect apoptotic cells by flow cytometry. In the absence of treatment, H1299-p53R213X cells displayed a basal level of 7% apoptosis, a value typical of cultured tumor cell lines (). After 50 hours of NB124 treatment, apoptosis had been triggered in 38% of H1299-p53R213X cells. By contrast, basal levels of apoptosis were maintained in the parental H1299 cells, with only 8% of the cells displaying annexin V staining, even after 50 hours of NB124 treatment (Fig. S3). The induction of p53 production by NB124 was, therefore, essential to trigger apoptosis. We checked that the restored p53 protein was indeed responsible for apoptosis induction in NB124-treated cells, using a siRNA specifically targeting p53 mRNA. We then transfected H1299-p53R213X cells with either the p53-targeting siRNA or a siRNA not targeting the p53 mRNA with a minimal impact on known human genes. As shown in Fig. S4, the p53-targeting siRNA prevents specifically the synthesis of p53 even in presence of readthrough inducer whereas the non-targeted siRNA has no effect on p53 induction by NB124. In the presence of the p53-targeting siRNA, apoptosis rates decreased by 30%, whereas no significant change was observed with the non-specific siRNA (). This result clearly demonstrates that NB124 induced apoptosis specifically by restoring p53 expression.
Discussion
Translational nonsense suppression (TNS) strategies are very promising for the treatment of genetic diseases and cancers caused by PTCs. Unfortunately, these approaches are limited by the lack of efficient molecules. Indeed, the principal molecule currently being tested is gentamicin, which cannot be used for long-term treatments due to its well-known toxicity. Attempts have been made to identify non-aminoglycoside drugs that promote readthrough. The first such drug to be tested is ataluren (PTC124), which gave conflicting results,Citation30 despite its recent approval by the EMA.Citation31 Other drugs promoting readthrough have since been identified by high-throughput screening (RTC13 and RTC14), but these drugs are still in development to improve their efficacy.Citation6 Compounds already available in clinical practice have recently been screened, and this led to the identification of escin as the most efficient molecule for restoring CFTR function.Citation32 These results, although very promising, highlight the need for additional strategies toward the discovery of more potent and less toxic drugs. One such approach is the rational modification of aminoglycosides to achieve new structures with higher levels of PTC suppression activity and lower toxicity.Citation33,34 This approach has led to the generation of new synthetic aminoglycosides, which have been tested in several models of different genetic diseases.Citation13,19,20 However, none of these compounds has been evaluated for its capacity to re-express inactivated tumor suppressor genes to prevent the proliferation of cancer cells. Since about 15% of human cancers are associated with PTCs in a tumor suppressor gene, TNS provides potential treatment strategy of cancer. In addition, cancer treatments are, by nature, short-term treatments (lasting until the cancer cells are killed or surgically removed), so some of the issues faced in the lifelong treatment of genetic diseases are not encountered in cancer treatment. Moreover, tumor suppressor genes encode proteins with either regulatory function (such as the transcription factor p53 or APC which is a regulatory subunit of protein phosphatase) or catalytic activities (such as ATM a ser/thr kinase) that can act at low concentrations in the cells. This is a strong advantage for TNS, because less full-length protein will be required for a therapeutic benefit in comparison to genetic diseases due to the inactivation of a structural protein (like dystrophin in Duchenne muscular dystrophy).
We investigated the potential of this new class of synthetic aminoglycosides for treating cancers linked to the presence of a PTC. We first assessed the relative ability of 5 previously synthesized NB-compounds (NB74, NB84, NB122, NB124 and NB127)Citation11,35 to promote readthrough of a R213X PTC frequently found in the p53 gene. Classically, in this assay, G418 (one of the most efficient readthrough inducers known) induces 12% readthrough when used at the maximal concentration inducing moderate toxicity, whereas gentamicin yields about 8% at the highest concentration (). The NB-drugs tested here could be assigned to 3 classes: 1) NB74 was less efficient than gentamicin, providing only 7% of readthrough at its highest concentration; 2) NB84, NB122 and NB128, providing readthrough efficiencies exceeding 10% (but less than 20%); 3) NB124, providing 20% readthrough and thus significantly exceeding the performance of all the other synthetic and classical aminoglcyosides tested. The main difference between NB124 and the other NB-compounds tested is the presence of 2 chiral methyl groups as 2 important pharmacophores: (S)-5″-Me and (R)-6′-Me, on the 5-amino ribose ring (ring III) and the glucosamine ring (ring I) of paromamine, respectively. These pharmacophores ensure that the NB124 molecule has a strong preference for eukaryotic versus mitochondrial ribosomes, resulting in lower levels of toxicity and a higher level of PTC suppression.Citation11 We therefore selected this compound, NB124, for further study and characterized its effects on cancer cells.
We first addressed the question of specificity. All known readthrough inducers have been shown to have a strong bias toward certain sequences. For example, gentamicin promotes higher levels of readthrough if the PTC is preceded by a U and followed by a C.Citation22 We tested 6 PTCs from the P53 (R213X, Q192X, E298X) and APC (L360X, R1114X and Q1131X) genes. These sequences display various levels of stop codon readthrough (highest for L360X and lowest for Q1131X), but readthrough levels for all these PTCs clearly responded to NB124 in a dose-dependent manner (). Moreover, NB124 systematically outperformed gentamicin at equivalent concentration, and had a performance similar to that of G418 at its highest concentration. Q192X, E298X and R1114X are not considered as good responders with a readthrough efficiency bellow 2% in presence of 0.8mg/ml of gentamicin. However, they displayed strong levels of readthrough induction in presence of 0.8mg/ml of NB124 (higher than 4% of readthrough). This indicates that NB124 has a broader spectrum of action than gentamicin. However, a systematic analysis would be required to confirm this. The molecular mechanisms underlying such preferences remain unknown. However, it has recently been shown that eRF1 interacts not only with the stop codon, but also with the following nucleotide.Citation36 The geometry of the stop codon within the A-site seems to depend on its nucleotide context, and is an important criterion for near-cognate tRNA binding.Citation37,38 These could explain why PTC suppression varies depending the nucleotide context.
Most mRNAs carrying a PTC are rapidly degraded by nonsense-mediated decay (NMD). PTC translational suppression has been reported to antagonise NMD, preventing mRNA degradation.Citation5,39 From a therapeutic standpoint, this would be highly beneficial, as greater mRNA stability should make it possible to produce more protein. We addressed this question, using HDQ-P1 cells carrying the endogenous R213X mutation in the P53 gene. The amount of P53 mRNA clearly increases as a function of the concentration of G418 or NB124, whereas no such relationship was observed for apramycin, another aminoglycoside that strongly binds to the eukaryotic ribosome and inhibits protein synthesis but does not promote PTC readthrough ().Citation26 The observed lack of mRNA stabilization by apramycin clearly points on the necessity of PTC readthrough ability by an aminoglycoside as an important characteristic for efficient inhibition of NMD and stabilization of mRNA. The extent of this ability was consistent with the larger amount of p53 observed on western blots (). Indeed, treatment resulted in increases in both the full-length and truncated p53 forms, but at different concentrations. The truncated form was visible from the lowest (0.05 mg/mL) to the highest (0.8 mg/mL) dose, with no obvious difference between concentrations, despite the accumulation of the mRNA. At higher NB124 concentrations, PTC readthrough increases considerably, resulting in a significantly higher proportion of the full-length protein. Indeed, large increase in levels of the full-length p53 was observed at a concentration of 0.4 mg/mL (). These results clearly show that NB124 treatment leads to the production of large amounts of full-length p53.
The re-expression of a full-length protein is an essential step, but there is no guarantee that this protein will be functional. Indeed, we have previously shown in yeast that at least 3 different amino acids are incorporated at the PTC during readthrough.Citation40 Little is currently known about the amino acids incorporated at stop codon in human cells. However, given the conservation of the tRNA pool, the set of amino acids incorporated is likely to be identical to that in yeast. In the absence of predictive rules, it is important to determine the activity of the restored full-length protein. We performed 4 different assays to determine whether p53 was functional. The first was based on the expression of a luciferase gene under the control of a P53 promoter.Citation5 We tested this system in HDQ-P1 cells carrying an endogenous PTC in the P53 gene. As expected, luciferase activity increased with increasing G418 or NB124 concentration (), suggesting that the restored full-length p53 was active. We then determined using RT-qpCR whether the full-length p53 induced by NB124 could also promote transcription of the Bax and p21 genes, 2 major targets of p53. Both genes were significantly expressed in the presence of NB124, confirming the ability of the p53 protein produced to activate the transcription of its target genes (). These 2 assays demonstrate that the restored full-length p53 was able to activate the genes under its control in natural conditions. However, it remained unclear whether this induction would be sufficient to induce the apoptosis of cancer cells. We investigated this aspect by first determining whether PARP-1 was cleaved by caspase-3, an early sign of apoptosis. This assay requires cells with a functional p53-dependent apoptosis pathway. We, therefore, stably inserted a p53 cDNA carrying a PTC (R213X) into H1299 cells, which have been reported to be sensitive to the re-expression of WT P53.Citation28 As expected, the parental cell line (H1299) displayed no signs of PARP-1 cleavage, whereas such cleavage was clearly observed in cells expressing p53-R213X in the presence of NB124, but not in the presence of gentamicin (). This result clearly confirms that NB124 is more effective than gentamicin for promoting the production of a functional p53. We confirmed this result by flow cytometry to detect apoptosis. After 50 hours of treatment with NB124, 38% were in an apoptotic state, whereas the parental cell line displayed no induction of apoptosis (Fig. S3). Moreover, using a siRNA specifically targeting p53 mRNA we observed a clear decrease, but not a total inhibition, of apoptosis in the presence of NB124 (). We know that the remaining 25% of apoptosis are due to the siRNA that is probably not 100% efficient at inhibiting p53 synthesis, because NB124 does not promote apoptosis in the parental cell line lacking functional p53 (figs. S3 and S4).
The accumulative data clearly demonstrate the advantage of NB124 over gentamicin for counteracting tumor development. NB124 is the first molecule shown to be as potent as G418 for inducing the re-expression of a functional protein by PTC suppression but without the toxicity of G418. The recent determination of the X-ray structure of the yeast ribosome in the presence of G418 has provided important insight into the binding of aminoglycosides within the A-site,Citation3 making it possible to develop promising new leads by rational design strategy.Citation33 The race to develop more potent readthrough inducers has only just begun. We now know more about the mode of action of aminoglycosides, but their specificity for certain nucleotide contexts remains a major obstacle to the development of drugs with a broader range of action.
Materials and methods
Cell lines and cell culture
All cells were cultured in DMEM plus GlutaMAX (Invitrogen), except for H1299 cells, which were cultured in RPMI plus GlutaMAX (Invitrogen). The medium was supplemented with 10% foetal calf serum (FCS, Invitrogen) and 100 U/ml penicillin/streptomycin. Cells were kept in a humidified atmosphere containing 5.5% CO2, at 37°C. NIH3T3 cells are embryonic mouse fibroblasts. H1299 is a p53-null cell line established from a human lung carcinoma (provided by the ATCC). H1299-p53R213X cells are H1299 cells stably transfected with the p53 R213X cDNA under the control of the CMV promoter. HDQ-P1 is homozygous for a nonsense mutation at codon 213 (CGA to TGA) in the p53 gene. This cell line was established from a human primary breast carcinomaCitation23 and was provided by DSMZ-German collection of Microorganisms and cell cultures. LoVo (WT p53) cells are epithelial cells derived from a human colorectal adenocarcinoma provided by the ATCC.
Readthrough quantification in cell culture
Complementary oligonucleotides corresponding to nonsense mutations embedded in their natural context (sequences in ) were annealed and ligated into the pAC99 dual reporter plasmid, as described previously.Citation41 This dual reporter can be used to quantify stop-codon readthrough through the measurement of luciferase and β-galactosidase (internal calibration) activities, as described previously.Citation42 Readthrough levels for nonsense mutations were analyzed in the presence or absence of gentamicin, G418 or NB124. NIH3T3 and H1299 cells were used to seed a 96-well plate. The next day, the cells were transfected with the reporter plasmid in the presence of JetPei reagent (Ozyme). The following day, they were rinsed and fresh medium, with or without gentamicin, G418, or NB124, was added. In these experiments, no cell toxicity was observed for any of the doses used. Cells were harvested 24 hours later, with trypsin–EDTA (Invitrogen), lysed with Glo lysis buffer (Promega) and β-galactosidase and luciferase activities were assayed as described previously.Citation42 Readthrough efficiency was estimated by calculating the ratio of luciferase activity to β-galactosidase activity obtained with the test construct, with normalization against an in-frame control construct. At least 3 independent transfection experiments were performed for each construct.
RNA extraction and quantitative PCR analysis
For the analysis of mRNA levels for p53 and its transcriptional target genes, Bax and p21, we extracted total RNA from HDQ-P1 cells that had or had not been treated with G418 (0.05, 0.1, 0.2, 0.4 mg/mL) apramycin (1 mg/mL) or NB124 (0.05, 0.1, 0.2, 0.4 and 0.8 mg/mL) for 48 h (RNeasy Mini Kit, Qiagen). The RNA was treated with DNAse I (RNase-free DNase) and quantified with a Nanodrop spectrometer (ThermoScientific). The absence of RNA degradation was confirmed by agarose gel electrophoresis. The first-strand cDNA was synthesized from 2μg of total RNA, with random primers and the SuperScript II Reverse Transcriptase (Invitrogen), as recommended by the manufacturer. Quantitative PCR was then performed on equal amounts of the various cDNAs, with a CFX96 thermocycler (Biorad), and the accumulation of products was monitored with the intercalating dye FastStart Universal SYBRGreen Master (ROX) reagent (Roche). We quantified mRNA levels relative to 3 reference mRNAs: RPL32, Hprt1 and HMBS. In each experiment, results are expressed relative to those for untreated cells, for which the value obtained was set to 1. Relative levels of gene expression were calculated at early stages of PCR, when the amplification was exponential and might, therefore, be correlated with the initial number of copies of the transcript. The specificity of quantitative PCR was checked by agarose gel electrophoresis, which showed that a single product of the desired length was produced for each gene. A melting curve analysis was also performed. Single product-specific melting temperatures were identified for each gene. For the quantification of each mRNA, 3 independent experiments (from biologic replicates) were performed in triplicate. We used the following oligonucleotides pairs for amplification: HPRT1 forward: 5′GACCAGTCAACAGGG GACAT 3′ and reverse: 5′AACACTTCGTGGGTCCTTTTC 3′; HMBS forward: 5′GGCAATGCG GCTGCAA 3′ and reverse: 5′GGGTACCCACGCGAATCAC3′; RPL32 forward: 5′ GCATTGACAAC AGGGTTCGTAG 3′ and reverse: 5′ GCGGTTCTTGGAGGAAACATTG 3′; p53 forward: 5′CCGCAGT CAGATCCTAGCG 3′ and reverse: 5′CCATTGCTTGGGACGGCAAGG 3′; p21 forward: 5′CAAGCT CTACCTTCCCACGG3′ and reverse 5′GCCAGGGTATGTACATGAGG 3′; Bax forward: 5′GCTGTTG GGCTGGATCCAAG 3′ and reverse 5′ TCAGCCCATCTTCTTCCAGA 3′.
Western-blot analysis
HDQ-P1 cells (R213X) were treated with G418 (0.2 mg/mL), gentamicin (0.8 mg/mL) or NB124 (0.05, 0.1 and 0.4 mg/mL) for 48 h. Cells were harvested by treatment with trypsin–EDTA (Invitrogen), lysed in 350 mM NaCl, 50 mM Tris–HCl pH 7.5, 1% NP-40, and protease inhibitor cocktail (Roche) and disrupted by passage through a syringe. Total proteins were quantified with Bradford reagent (Biorad) and extracts were denatured by incubation in Laemmli buffer for 5 minutes at 90°C. We subjected 30µg of total protein from HDQ-1 cells to SDS–PAGE in 4/10% Bis–Tris gels. Proteins were transferred onto nitrocellulose membranes, according to the manufacturer's instructions (Biorad). Membranes were saturated by overnight incubation in 5% skimmed milk powder in PBS, and incubated for 1 hour with the primary monoclonal antibody, DO-1 (N-terminal epitope mapping between amino acid residues 11 and 25 of p53; Santa Cruz Biotechnologies, 1/400) or a monoclonal antibody against mouse actin (Millipore, 1/2000). After 3 washes in PBS supplemented with 0.1% Tween, the membranes were incubated with the secondary antibody [horseradish peroxidase-conjugated anti-mouse IgG (1/ 2500) for 45 minutes. The membranes were washed 5 times and chemiluminescence was detected with ECL Prime Western Blotting Detection Reagents (Amersham, GE Healthcare). The signal was quantified with ImageJ software. For western-blot analysis of siRNA effect on p53 expression in H1299-p53R213X, we used the same protocol except that the cells were transfected or not with siRNA and treated or not with NB124 (0.4 mg/ml) for 50 hours. For western-blot analysis of poly (ADP-ribose) polymerase I (PARP-1) cleavage in cell lysates obtained from H1299-p53R213X cells, we used the same protocol except that the cells were treated with NB124 (0.4 mg/ml) or gentamicin (0.8 mg/ml) for 30 or 50 hours. The antibody used was a mouse anti-PARP-1 (Ab-2) mAb from Calbiochem.
siRNA transfections
The siRNAs used in this study are double-stranded chemically synthesized oligonucleotides provided by Thermo Scientific. The siRNA targeting p53 is referred to as 3329–14–0020 and the non-targeting siRNA is referred to as 1810–01–05. H1299-p53R213X cells were transfected, with the siRNA in the presence of jetPRIME from Polyplus (1µg of siRNA per well of a 6-well plate). 22 hours after transfection the medium was renewed and cells were incubated with or without NB124 (0.4 mg/ml) for 50 hours.
Protein activity assays
We investigated the transcriptional activity of the p53 protein in HDQ-P1 cells, which carry the endogenous p53 R213X nonsense mutation. We added G418, gentamicin or NB124 to the medium 24 h before transfection and then again 2 d later (total of 72 hours of treatment). Cells were cotransfected, by the JetPei method, with the p53BS-luc reporter plasmid containing the firefly luciferase gene downstream from 7 p53 binding sites and pCMVLacZ, and were left untreated or incubated with one of the 3 drugs tested. Protein extracts were prepared 24 h after transfection, and enzymatic activities were measured. Transfection with pCMVLacZ was used to normalize transfection efficiency, cell viability and protein extraction. At least 4 independent transfection experiments were performed for each set of conditions.
Flow cytometry analysis
H1299 and the stably transfected clone H1299-p53R213X cells were used to seed 6-well plates at a density of 250,000 cells per well and were incubated for 24 h. The cells were then left untreated or were treated for 50 hours with gentamicin (1 mg/mL) or NB124 (0.4 mg/mL).
PS is exposed at the surface of cells undergoing apoptosis. It was detected by incubation with Annexin V conjugated to the fluorochrome phycoerythrin (PE), from BD PharMingen, in accordance with the manufacturer's instructions. We analyzed the proportion of apoptotic cells by flow cytometry. For each experiment, at least 4 independent transfection experiments were performed.
Disclosure of potential conflicts of interest
Timor Baasov acknowledges that he is used by Technion who owns the patents relating to NB compounds discussed in this study, that were licensed to a third party.
Author contributions
L.B and O.B performed cell experiments, V.B and T.B designed and synthesized aminoglycoside derivatives. L.B, O.B, T.B and O.N designed the experiments. All the authors interpreted the data and participated in the writing of the manuscript.
Acknowledgments
We thank Alex Edelman & Associates for correcting English usage. We would like to thank our collaborators from Dundee University, especially Virginia Appleyard, Jean-Christophe Bourdon and Alastair Thompson, for their help and discussions during this project.
Funding
This project is supported by the French foundation ARC [SFI20101201647 and PJA 20131200234]; and the French association ‘Ligue Nationale contre le cancer’ [3FI10167LVCY and, grants awarded to O.N.].
References
- Linde L, Kerem B. Introducing sense into nonsense in treatments of human genetic diseases. Trends Genet 2008; 24(11):552-63; PMID:18937996; http://dx.doi.org/10.1016/j.tig.2008.08.010
- Isken O, Maquat LE. The multiple lives of NMD factors: balancing roles in gene and genome regulation. Nat Rev Genet 2008; 9(9):699-712; PMID:18679436; http://dx.doi.org/10.1038/nrg2402
- Garreau de Loubresse N, Prokhorova I, Holtkamp W, Rodnina MV, Yusupova G, Yusupov M. Structural basis for the inhibition of the eukaryotic ribosome. Nature 2014; 513(7519):517-22; PMID:25209664; http://dx.doi.org/10.1038/nature13737
- Keeling KM, Xue X, Gunn G, Bedwell DM. Therapeutics based on stop codon readthrough. Annual Rev Genomics Hum Genetics 2014; 15:371-94; PMID:24773318; http://dx.doi.org/10.1146/annurev-genom-091212-153527
- Floquet C, Deforges J, Rousset JP, Bidou L. Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res 2011; 39(8):3350-62; PMID:21149266; http://dx.doi.org/10.1093/nar/gkq1277
- Du L, Jung ME, Damoiseaux R, Completo G, Fike F, Ku JM, Nahas S, Piao C, Hu H, Gatti RA. A new series of small molecular weight compounds induce read through of all three types of nonsense mutations in the ATM gene. Mol Ther 2013; 21(9):1653-60; PMID:23774824; http://dx.doi.org/10.1038/mt.2013.150
- Du L, Damoiseaux R, Nahas S, Gao K, Hu H, Pollard JM, Goldstine J, Jung ME, Henning SM, Bertoni C, et al. Nonaminoglycoside compounds induce readthrough of nonsense mutations. J Exp Med 2009; 206(10):2285-97; PMID:19770270; http://dx.doi.org/10.1084/jem.20081940
- Welch EM, Barton ER, Zhuo J, Tomizawa Y, Friesen WJ, Trifillis P, Paushkin S, Patel M, Trotta CR, Hwang S, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature 2007; 447(7140):87-91. Epub 2007/04/24; PMID:17450125; http://dx.doi.org/10.1038/nature05756
- Haas M, Vlcek V, Balabanov P, Salmonson T, Bakchine S, Markey G, Weise M, Schlosser-Weber G, Brohmann H, Yerro CP, et al. European Medicines Agency review of ataluren for the treatment of ambulant patients aged 5 years and older with Duchenne muscular dystrophy resulting from a nonsense mutation in the dystrophin gene. Neuromuscul Disord 2015; 25(1):5-13; PMID:25497400; http://dx.doi.org/10.1016/j.nmd.2014.11.011
- Forge A, Schacht J. Aminoglycoside antibiotics. Audiol Neurootol 2000; 5(1):3-22; PMID:10686428; http://dx.doi.org/10.1159/000013861
- Kandasamy J, Atia-Glikin D, Shulman E, Shapira K, Shavit M, Belakhov V, Baasov T. Increased selectivity toward cytoplasmic versus mitochondrial ribosome confers improved efficiency of synthetic aminoglycosides in fixing damaged genes: a strategy for treatment of genetic diseases caused by nonsense mutations. J Med Chem 2012; 55(23):10630-43; PMID:23148581; http://dx.doi.org/10.1021/jm3012992
- Matt T, Ng CL, Lang K, Sha SH, Akbergenov R, Shcherbakov D, Meyer M, Duscha S, Xie J, Dubbaka SR, et al. Dissociation of antibacterial activity and aminoglycoside ototoxicity in the 4-monosubstituted 2-deoxystreptamine apramycin. Proc Natl Acad Sci U S A 2012; 109(27):10984-9; PMID:22699498; http://dx.doi.org/10.1073/pnas.1204073109
- Shulman E, Belakhov V, Wei G, Kendall A, Meyron-Holtz EG, Ben-Shachar D, Schacht J, Baasov T. Designer aminoglycosides that selectively inhibit cytoplasmic rather than mitochondrial ribosomes show decreased ototoxicity: a strategy for the treatment of genetic diseases. J Biol Chem 2014; 289(4):2318-30; PMID:24302717; http://dx.doi.org/10.1074/jbc.M113.533588
- Xue X, Mutyam V, Tang L, Biswas S, Du M, Jackson LA, Dai Y, Belakhov V, Shalev M, Chen F, et al. Synthetic aminoglycosides efficiently suppress cystic fibrosis transmembrane conductance regulator nonsense mutations and are enhanced by ivacaftor. Am J Respir Cell Mol Biol 2014; 50(4):805-16; PMID:24251786; http://dx.doi.org/10.1165/rcmb.2013-0282OC
- Asker C, Wiman KG, Selivanova G. p53-induced apoptosis as a safeguard against cancer. Biochem Biophys Res Commun 1999; 265(1):1-6; PMID:10548481; http://dx.doi.org/10.1006/bbrc.1999.1446
- Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev 1998; 12(19):2973-83; PMID:9765199; http://dx.doi.org/10.1101/gad.12.19.2973
- Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev 1996; 10(9):1054-72; PMID:8654922; http://dx.doi.org/10.1101/gad.10.9.1054
- Kolligs FT, Bommer G, Goke B. Wnt/beta-catenin/tcf signaling: a critical pathway in gastrointestinal tumorigenesis. Digestion 2002; 66(3):131-44; PMID:12481159; http://dx.doi.org/10.1159/000066755
- Shalev M, Baasov T. When proteins start to make sense: Fine-tuning aminoglycosides for PTC suppression therapy. Medchemcomm 2014; 5(8):1092-105; PMID:25147726; http://dx.doi.org/10.1039/C4MD00081A
- Goldmann T, Overlack N, Moller F, Belakhov V, van Wyk M, Baasov T, Wolfrum U, Nagel-Wolfrum K. A comparative evaluation of NB30, NB54 and PTC124 in translational read-through efficacy for treatment of an USH1C nonsense mutation. EMBO Mol Med 2012; 4(11):1186-99; PMID:23027640; http://dx.doi.org/10.1002/emmm.201201438
- Beroud C, Hamroun D, Collod-Beroud G, Boileau C, Soussi T, Claustres M. UMD (Universal Mutation Database): 2005 update. Hum Mutat 2005; 26(3):184-91; PMID:16086365; http://dx.doi.org/10.1002/humu.20210
- Floquet C, Hatin I, Rousset JP, Bidou L. Statistical analysis of readthrough levels for nonsense mutations in mammalian cells reveals a major determinant of response to gentamicin. PLoS Genet 2012; 8(3):e1002608. Epub 2012/04/06; PMID:22479203; http://dx.doi.org/10.1371/journal.pgen.1002608
- Wang CS, Goulet F, Lavoie J, Drouin R, Auger F, Champetier S, Germain L, Têtu B. Establishment and characterization of a new cell line derived from a human primary breast carcinoma. Cancer Genet Cytogenet 2000; 120(1):58-72; PMID:10913678; http://dx.doi.org/10.1016/S0165-4608(99)00253-8
- Popp MW, Maquat LE. Organizing principles of mammalian nonsense-mediated mRNA decay. Annu Rev Genet 2013; 47:139-65; PMID:24274751; http://dx.doi.org/10.1146/annurev-genet-111212-133424
- Allamand V, Bidou L, Arakawa M, Floquet C, Shiozuka M, Paturneau-Jouas M, et al. Drug-induced readthrough of premature stop codons leads to the stabilization of laminin alpha2 chain mRNA in CMD myotubes. J Gene Med 2008; 10(2):217-24; PMID:18074402; http://dx.doi.org/10.1002/jgm.1140
- Kondo J, Hainrichson M, Nudelman I, Shallom-Shezifi D, Barbieri CM, Pilch DS, et al. Differential selectivity of natural and synthetic aminoglycosides towards the eukaryotic and prokaryotic decoding A sites. Chembiochem 2007; 8(14):1700-9; PMID:17705310; http://dx.doi.org/10.1002/cbic.200700271
- Gotz C, Montenarh M. P53 and its implication in apoptosis (review). Int J Oncol 1995; 6(5):1129-35; PMID:21556650
- Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev 1996; 10(19):2438-51; PMID:8843196; http://dx.doi.org/10.1101/gad.10.19.2438
- Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem 1998; 273(50):33533-9; PMID:9837934; http://dx.doi.org/10.1074/jbc.273.50.33533
- Sheridan C. Doubts raised over ‘read-through’ Duchenne drug mechanism. Nat Biotechnol 2013; 31(9):771-3; PMID:24022133; http://dx.doi.org/10.1038/nbt0913-771
- Mullard A. EMA reconsiders ‘read-through’ drug against Duchenne muscular dystrophy following appeal. Nat Biotechnol 2014; 32(8):706; PMID:25101726; http://dx.doi.org/10.1038/nbt0814-706
- Mutyam V, Du M, Xue X, Keeling KM, White EL, Bostwick JR, Rasmussen L, Liu B, Mazur M, Hong JS, et al. Discovery of clinically approved agents that promote suppression of CFTR nonsense mutations. Am J Respir Crit Care Med 2016; 194(9):1092-1103; PMID:27104944; http://dx.doi.org/10.1164/rccm.201601-0154OC
- Sabbavarapu NM, Shavit M, Degani Y, Smolkin B, Belakhov V, Baasov T. Design of novel aminoglycoside derivatives with enhanced suppression of diseases-causing nonsense mutations. ACS Med Chem Lett 2016; 7(4):418-23; PMID:27096052; http://dx.doi.org/10.1021/acsmedchemlett.6b00006
- Nudelman I, Rebibo-Sabbah A, Cherniavsky M, Belakhov V, Hainrichson M, Chen F, Schacht J, Pilch DS, Ben-Yosef T, Baasov T. Development of novel aminoglycoside (NB54) with reduced toxicity and enhanced suppression of disease-causing premature stop mutations. J Med Chem 2009; 52(9):2836-45; PMID:19309154; http://dx.doi.org/10.1021/jm801640k
- Nudelman I, Glikin D, Smolkin B, Hainrichson M, Belakhov V, Baasov T. Repairing faulty genes by aminoglycosides: development of new derivatives of geneticin (G418) with enhanced suppression of diseases-causing nonsense mutations. Bioorg Med Chem 2010; 18(11):3735-46. Epub 2010/04/23; PMID:20409719; http://dx.doi.org/10.1016/j.bmc.2010.03.060
- Brown A, Shao S, Murray J, Hegde RS, Ramakrishnan V. Structural basis for stop codon recognition in eukaryotes. Nature 2015; 524(7566):493-6; PMID:26245381; http://dx.doi.org/10.1038/nature14896
- Rozov A, Demeshkina N, Khusainov I, Westhof E, Yusupov M, Yusupova G. Novel base-pairing interactions at the tRNA wobble position crucial for accurate reading of the genetic code. Nat Commun 2016; 7:10457; PMID:26791911; http://dx.doi.org/10.1038/ncomms10457
- Rozov A, Demeshkina N, Westhof E, Yusupov M, Yusupova G. Structural insights into the translational infidelity mechanism. Nat Commun 2015; 6:7251; PMID:26037619; http://dx.doi.org/10.1038/ncomms8251
- Linde L, Boelz S, Nissim-Rafinia M, Oren YS, Wilschanski M, Yaacov Y, Virgilis D, Neu-Yilik G, Kulozik AE, Kerem E, et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest 2007; 117(3):683-92. Epub 2007/02/10; PMID:17290305; http://dx.doi.org/10.1172/JCI28523
- Blanchet S, Cornu D, Argentini M, Namy O. New insights into the incorporation of natural suppressor tRNAs at stop codons in Saccharomyces cerevisiae. Nucleic Acids Res 2014; 42(15):10061-72; PMID:25056309; http://dx.doi.org/10.1093/nar/gku663
- Bidou L, Hatin I, Perez N, Allamand V, Panthier JJ, Rousset JP. Premature stop codons involved in muscular dystrophies show a broad spectrum of readthrough efficiencies in response to gentamicin treatment. Gene Ther 2004; 11(7):619-27; PMID:14973546; http://dx.doi.org/10.1038/sj.gt.3302211
- Stahl G, Bidou L, Rousset JP, Cassan M. Versatile vectors to study recoding: conservation of rules between yeast and mammalian cells. Nucleic Acids Res 1995; 23(9):1557-60; PMID:7784210; http://dx.doi.org/10.1093/nar/23.9.1557
