 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Wobble uridines (U34) are generally modified in all species. U34 modifications can be essential in metazoans but are not required for viability in fungi. In this review, we provide an overview on the types of modifications and how they affect the physico-chemical properties of wobble uridines. We describe the molecular machinery required to introduce these modifications into tRNA posttranscriptionally and discuss how posttranslational regulation may affect the activity of the modifying enzymes. We highlight the activity of anticodon specific RNases that target U34 containing tRNA. Finally, we discuss how defects in wobble uridine modifications lead to phenotypes in different species. Importantly, this review will mainly focus on the cytoplasmic tRNAs of eukaryotes. A recent review has extensively covered their bacterial and mitochondrial counterparts.Citation1
Wobble uridine modifications are essential in evolution
Among the plethora of chemical posttranscriptional modifications that are found on tRNA, those of wobble uridine (U34) are peculiar, because U34 is almost invariably modified in any organism.Citation2,3 This phenomenon implies a strong evolutionary pressure to maintain wobble uridines modified and is further affirmed by the analysis of minimal genomes. In Mollicutes species that have drastically reduced their genome size during evolution, uridine thiolation (s2U) is part of an essential core module of translation.Citation4 Furthermore, recent attempts of synthetic biologists to generate a minimal genome based on Mycoplasma mycoides found modification systems of U34 to be required for rapid growth under laboratory conditions.Citation5 Surprisingly however, the absence of U34 modification does not cause lethality in Caenorhabditis elegans and most yeasts but is essential in some strain backgrounds of Saccharomyces cerevisiae and in mice.Citation6-12 Furthermore, several human diseases are linked to defects in U34 modifying enzymes.Citation13-15 This apparent discrepancy between essentiality during evolution and variable effects in different organisms makes wobble uridines even more worth exploring.
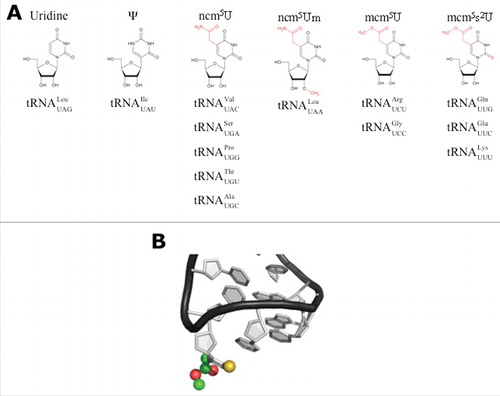
We know a lot about U34 modifications from the research of many laboratories working mainly on baker's yeast. Theoretically, 16 anticodons carry a uridine at their wobble position. UAA and UGA, 2 codons that would require U34-containing tRNAs for decoding are nonsense codons and therefore recognized by the Eukaryotic Release Factor 1 and 3 (eRF1 and eRF3) GTPase complex.Citation16,17 However, the modification machinery is of such broad specificity that U34 in suppressor is modified.Citation6
does not exist in yeast and
is unmodified.Citation18
carries a pseudouridine (Ψ34).Citation19 The remaining 11 tRNA species are decorated by 4 types of modifications: First, 5-carbamoylmethyluridine (ncm5U34), the most abundant modification, is present on
,
,
,
and
. Second,
is further 2-O-methylated to 5-carbamoylmethyl-2-O-methyluridine (ncm5U34m). Third, 5-methoxy-carbonylmethyluridine (mcm5U34) is found on
and
. Finally, 3 tRNAs are further decorated by a 2-thio group to form 5-methoxycarbonylmethyl-2-thiouridine (mcm5s2U34):
,
and
().Citation20,21
Figure 1. Modifications of wobble uridines (U34). (A) Uridine derivatives that can be found at the wobble position (U34) in anticodons of cytoplasmic tRNAs from eukaryotes. Modifications to uridine are indicated in red. (structures taken from ModomicsCitation21). Abbreviations: ncm5U: 5-carbamoylmethyluridine; ncm5Um: 5-carbamoylmethyl-2’-O-methyluridine; mcm5U: 5-methoxycarbonyl-methyluridine; mcm5s2U: 5-methoxycarbonylmethyl-2-thiouridine. (B) Detail of the structure of the anticodon stem loop (ASL) of human . mcm5s2U is highlighted by spheres. Sulfur (yellow), carbon (green), oxygen (red). Note that the nucleobases are arranged in a stacking confirmation (The structure is based on PDB 1FIR).
The physicochemical properties of wobble uridine modifications
The multitude of modifications speaks for particular requirements in the U34 position. But what are the effects of these modifications on the chemical and structural properties of uridine? In his wobble hypothesis, Francis Crick proposed that U34 recognizes A and G in the third position of the codon.Citation22 However, he could not account for the extent of modified nucleotides in tRNA as these modifications were not known. Later adaptations of the wobble hypothesis have attempted to integrate how modifications of U34 affect codon recognition.Citation20,23-26
Uridines are structurally flexible and form only weak stacking interactions with neighboring nucleosides. This is because uridines-even when decorated by 5′-modifications-adopt a C(2′) endo conformation, which is relatively flexible.Citation27,28 However, the presence of 2-thio modifications leads to the adaptation of a C(3’) endo, gauche plus [C(4’)-C(5’)], anti structure.Citation27-29 This conformation is hydrophobic, more restricted and appears to be best suited for anticodon base-stacking, therefore stabilizing the anticodon stem loop (ASL) and favoring the interaction with A in the codon.Citation27,28 The conformational stability of s2U34 is similar to U34m, which also occurs mainly in its C(3’) endo form.Citation30,31 While, s2U34 enhances the stability of the stacking of the triplet significantly over unmodified U34,Citation32,33 the stabilizing effect of the C(3’) endo conformation appears to be more important for the codon-anticodon interaction than an improvement of base stacking.Citation29,34 Furthermore, 2-thio modifications do not only restrict and stabilize the conformation of the nucleoside that carries s2 but also of a 3’-adjacent unmodified uridine.Citation35,36 Importantly, a stabilization of the interactions between 2-thiolated anticodons with their codon partners has been observed in binding models and in the context of the ribosome.Citation36-38 While the role of s2U34 modification is to restrict codon conformation, the role of xmCitation5 modifications is to open up the rigidity to stabilize U•G wobble pairing.Citation39 In general, the ribosomal grip constrains the positioning of the mRNA more than that of the ASL. Therefore, G34•U3 wobble pairs can form in unmodified anticodons, while U34•G3 pairs are much less stable, if they form at all.Citation26,40-42
Structural studies found that near-cognate tRNAs do not necessarily take up a wobble confirmation but that non-standard base pairing can retain Watson-Crick geometry.Citation43 This structural arrangement is supported by the presence of U34 modifications. Indeed, mcm5s2U34 modifications change the physicochemical behavior of the base. While s2 of mcm5s2U34 is required for proper positioning of the nucleoside, the main function of the mcmCitation5 modification is to modify the electron structure of the ring and to shift the keto-enol equilibrium toward enol thus enabling hydrogen bonding between U34•G pairs.Citation44 Therefore, while s2U rigidifies the ASL structure thereby favoring an interaction with A-ending codons, the xmCitation5 modifications relax the conformation and enable pairing with G-ending codons. However, when analyzing codon-translation speed by ribosome profiling the absence of both types of modifications seems to affect translation speed similarly.Citation45
Nevertheless, also the context of the anticodon matters. For instance, N6-threonylcarbamoyladenosine (t6A37) synergizes with U34 modifications to order the ASL such that it will bind to the correct mRNA codons. Only when U34 and A37 are fully modified, will the ASL of bind to AAA and AAG programmed ribosomesCitation41 and fulfill the function proposed by Crick.Citation22 Therefore, U34 modifications contribute to the pre-structuring of the anticodon loop to achieve optimal translation.Citation42
Wobble uridine modifications require multiple pathways
Two pathways are responsible for placing the two main classes of modifications on wobble uridine: The URM1 pathway is required for 2-thiolation and the Elongator complex needed for generating xm5U34 modifications (). Pseudouridine Synthase (Pus1) introduces pseudouridine at U34 in .Citation46,47 Finally, during the 2’-O-methylation of
tRNA Methyltransferase (Trm7) uses S-adenosylmethionine (SAM) as a methyl donor together with its cofactor Regulator of Ty1 Transposition 10 (Rtt10).Citation48,49 However, the absence of 2’-O-methylation in
does not significantly contribute to phenotypes observed in trm7▵ yeast since overexpression of
is sufficient for rescue experiments, suggesting that the physiological role of this modification in U34 is minor.Citation49 Interestingly, xm5U34 formation is independent of the presence of s2U34 while the reverse is not true. In S. cerevisiae s2U34 levels are reduced in the absence of mcmCitation5,11,50 In humans, mice and S. pombe mcmCitation5 is even strictly required for s2U34 formation.Citation51,52
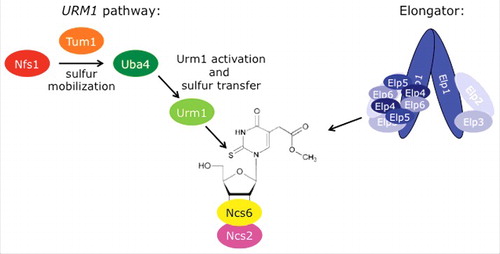
Figure 2. Model of the URM1 pathway and the Elongator complex. Schematic representation of the 2 pathways cooperating in mcm5s2U34 formation. In the URM1 pathway (left), sulfur is mobilized by Nfs1 with the help of Tum1. Uba4 activates Urm1, leading to a thiocarboxylate at Urm1's C-terminus, which acts as a sulfur carrier. Finally, the Ncs2•Ncs6 complex binds to and activates tRNA in the thiolation reaction and transfers the sulfur from Urm1 to uridine. The Elongator complex consists of twice Elp1-Elp6. Elp1 dimerizes via its C-terminus and acts as a platform for Elp2 and Elp3 binding in a wing-like structure. A ring of Elp4-Elp6 binds to one of the wings (Handedness is only partially represented in this model).Citation102,103
The URM1 pathway
Ubiquitin Related Modifier 1 (Urm1) is an ubiquitin like protein (Ubl) first described as a protein modifier in a process called urmylation.Citation53-55 It was identified through its sequence homology to bacterial sulfur-carrier proteinsCitation53 and is considered to be a molecular fossil because it is the Ubl that most closely resembles the ancestors of this class of protein modifiers.Citation56,57 Importantly, Urm1 is the only Ubl described as a protein modifier and at the same time to act in sulfur transfer, placing it at the evolutionary intersection of both pathways.Citation50,58-67 Like every Ubl, Urm1 is activated by an E1-like enzyme Ubiquitin Activating 4 (Uba4).Citation53 While the N-terminus of Uba4 contains an E1 domain, its C-terminal harbors a rhodanese homology domain (RHD).Citation68,69 The RHD has been suggested to act as an E2 domain for protein conjugation of Urm1.Citation70 However, there is no experimental support for this mechanism and the functionality of the domain rather points toward sulfur transfer. Two additional core members of the pathway are Needs Cla4 to Survive 2 and 6 (Ncs2 and Ncs6),Citation55,71 which form a complex in S. cerevisiae, C. elegans and S. pombe.Citation8,11 Difficulty in reconstituting the complex in vitro has prevented its detailed analysis. It is clear that Ncs6, an iron-sulfur (Fe/S) cluster containing protein,Citation72 and its worm homolog TUT-1 can bind to tRNA in vitroCitation11 and in vivo.Citation8 The protein carries 2 predicted zinc-finger domains and a P-loop.Citation11,73,74 Interestingly, the number of tRNA species that bind to Ncs6 in vitro exceeds the number of mcm5s2U34 targets.Citation11 Thus, Ncs2 may provide specificity to the complex. Alternatively, Ncs2 may stabilize or activate Ncs6. NCS2 is the closest homolog of NCS6 in the yeast genome. However, the critical residues required for enzymatic function are mutated. The observation that only one catalytic subunit is required in S. cerevisiae suggests a similar mechanism in enzymes that form a homo-dimer such as TtuA.Citation75 While URM1, UBA4, NCS2 and NCS6 are required for 2-thiolation, ThioUridine Modification 1 (TUM1), a gene coding for a protein with 2 RHDs, is not essential for thiolation. However, in the absence of Tum1, levels of s2U34 are significantly reduced and the ratio between modified and unmodified tRNA changes.Citation11,58,63,67
In addition to this core set of proteins, there is another group that is essential for thiolation by more generally affecting sulfur pathways. NiFS like 1 (Nfs1), a cysteine desulfurase that converts cysteine into alanine using pyridoxal phosphate as a cofactor acts upstream of several cellular sulfur pathways.Citation76 During this reaction, Nfs1 feeds sulfane sulfur to its acceptor protein. Uba4 uses the sulfur and transfers it into the downstream cascade for 2-thiolation.Citation62 The exact mechanism of Tum1 in this reaction is unclear. Tum1 receives sulfur from Nfs1 and appears to stimulate its activity.Citation63 This suggests that it acts as an enhancer of the transfer reaction or as a sulfur relay. Since Tum1 is not essential for 2-thiolation, the transfer step via Tum1 can be bypassed. Therefore, Tum1 may affect 2-thiolation through an indirect mechanism by rerouting sulfur pathways. Finally, proteins required for iron-sulfur (Fe/S) cluster biogenesis and assembly are essential for 2-thiolation. These are: Cytosolic Iron-sulfur protein Assembly 1 (Cia1), Nucleotide Binding Protein 35 (Nbp35) and Cytosolic Fe/S cluster Deficient 1 (Cfd1), which are all components of the CIA complex.Citation77,78 The CIA complex works in conjunction with IScU homologs 1 and 2 (Isu1 and Isu2), 2 proteins that reside in the mitochondrial matrix and are required for Fe/S cluster generation.Citation79 Additional proteins required for Fe/S cluster formation are, therefore, likely to affect 2-thiolation but have not been tested specifically. Whether Fe/S cluster formation affects s2U34 formation directly or indirectly via the formation of mcm5U34 will need to be determined.Citation80 Recently, a Fe/S cluster was identified in Ncs6 and its archaeal homolog TtuC.Citation72 However, its requirement for s2U34 formation remains to be demonstrated.
The Elongator complex
The Elongator complex is at the heart of mcmCitation5 and ncmCitation5 side chain formation at U34. The complex has been reported to act in numerous cellular processes including transcription, DNA damage response, exocytosis, telomere gene silencing, DNA demethylation and wobble uridine modification.Citation6,81-88 Importantly, all known phenotypes of Elongator-minus yeast, except for the defect in U34 modification itself, can be rescued by overexpression of tRNA that are normally mcm5s2U34 modified.Citation12,45,51,88-95 Nevertheless, the question of whether different functions exist in other species is very persistent, in part due to the fact that tRNA overexpression experiments in metazoans are more difficult to perform than in yeast.
The Elongator complex consists of 6 subunits: Elp1-Elp6.Citation81,96,97 Each is present twice in the holo complex, which can be divided in two sub-complexes: Elp1-Elp3 and Elp4-Elp6. The latter subunits, between 30–50 kDa in size, have very similar RecA folds and assemble into a heterohexameric ring structure that resembles RecA-like ATPase complexes.Citation98 However, the subunits lack the P-loop motif, which is characteristic for ATPases.Citation98 Importantly, the Elp4-Elp6 complex binds tRNA in an ATP dependent manner, where high levels of ATP decrease the affinity of tRNA for the complex. The Elp1-Elp3 subcomplex contains Elp3, which is the catalytic subunit of the complex.Citation96,99 Elp3 carries an N-terminal radical SAM binding domain and a C-terminal histone acetyltransferase (HAT) domain.Citation96,100 Interestingly, in the archaeon Methanocaldococcus infernus a homolog of Elp3 is sufficient for U34 modification, while all other Elongator subunit genes are absent from its genome.Citation101 Elp1 and Elp2 are both WD40 domain containing proteins. Two subunits of Elp1 dimerize via their C-terminal domains, while one subunit of Elp2 and Elp3 associates with each Elp1 subunit at either side.Citation102,103 This generates a wing-like structure. Surprisingly, in the holo complex, a ring of Elp4-Elp6 is associated with the front of the left wing () resulting in an asymmetric assembly, which is in contrast to previous models.Citation98,102-105 The role of the asymmetry still needs to be determined as well as the position of the tRNA in the complex.
In addition to the Elongator complex, Kluveromyces lactis Toxin Insensitive 11–14 (Kti11-Kti14), Suppressor of Initiation of Transcription 4 (Sit4), Sit4 associated protein 185 and 190 (Sap185 and Sap190), tRNA Methyltransferase 9 and 112 (Trm9 and Trm112) are required for mcm5U34 and ncm5U34 formation. Elp1-Elp6, Kti11, Kti12, Kti14, Sit4, Sap185 and Sap190 are essential for the formation of cm5U34, which is believed to be the precursor of mcmCitation5 and ncmCitation5.Citation6,58,106 The lack of Kti13 leads to reduced levels of xmCitation5 formation.Citation6,58 Finally, the Trm9•Trm112 complex uses SAM to synthesize mcm5U34.Citation107 Whether cm5U34 or an intermediate precursor serves as the direct methylase substrate is not clear, yet. The observation that both trm9 and trm112 mutants accumulate ncm5U34 and ncm5s2U34, however, suggests the existence of an enzyme required for formation of ncm5U34 (and ncm5s2U34) from cm5U34.Citation107,108 The identity of this activity is still not known, leading to contradicting ideas of how to rationalize these later steps of the U34 modification pathway.Citation106-110
Elongator regulation by phosphorylation
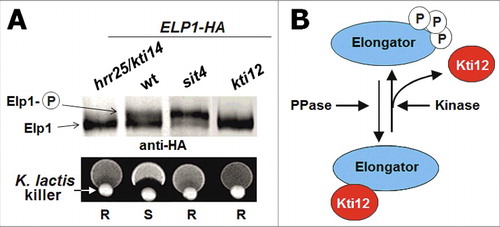
Strikingly, tRNAs that carry Elongator-dependent mcm5s2U34 can be cleaved by zymocin between anticodons position 34 and 35.Citation111-113 Zymocin is a trimeric (αβγ) tRNase toxin complex produced from K. lactis that kills yeasts including S. cerevisiae.Citation114-116 In line with this lethal mode of tRNase action, genetic studies have shown that mutations in Elongator genes trigger zymocin resistance and additional Elongator related factors (Kti11-Kti14, Sit4, Sap185 and Sap190) were genetically identified on the basis of zymocin survivor screens.Citation58,112,117-136 Rather than affecting the assembly or the integrity of the Elongator complex, these proteins appear to be regulatory. Consistent with this, a casein kinase 1 (CK1) isozyme (Kti14, also called: Hrr25), type-2A protein phosphatases (Sit4•Sap185; Sit4•Sap190) and an Elongator interactor (Kti12)Citation124,127,129 were all shown to affect the phosphorylation state of Elp1, which through dimerization assembles holo-Elongator.Citation98,102,103,137 Elp1 is present as a hypophosphorylated isoform in kti12 and hrr25 mutants and is hyperphosphorylated in sit4 mutants, while wild-type cells maintain both forms ().Citation130,131,134 This suggests that Elongator function may be phosphoregulated, which is in line with reports that tRNA modifications including mcm5s2U34 can change in response to chemical stress and cell cycle progression.Citation106,138-140
Figure 3. Phosphomodification of Elongator subunit Elp1. (A) Elp1 electrophoretic mobility shifts based on anti-HA Western blots are diagnostic for Elongator de-/phosphorylation.Citation130,134 In the kti12 and the kinase-dead hrr25/kti14 mutants, hypophosphorylated forms of Elp1-HA accumulate while sit4 phosphatase mutants induce Elp1-HA hyperphosphorylation. Wild-type (wt) cells maintain both isoforms of Elp1-HA, which mediate sensitivity (S) to growth inhibition by the tRNase toxin zymocin (killer assay: lower panel; for details see text). Zymocin resistance (R) associates with Elp1 phosphorylation defects in kti12, hrr25/kti14 and sit4 mutants. (B) Elongator phosphorylation model. Kti12 interacts with Elongator (and kinase Hrr25) thereby potentially activating Elp1 phosphorylation. In support of this, Elp1 is found to be hypophosphorylated in kti12 and hrr25/kti14 cells (see A). PPase: protein phosphatase (Sit4); Kinase: Hrr25/Kti14.
Subsequently, phosphorylation sites on Elp1 and other Elongator subunits were identified using mass spectrometry.Citation141,142 Among those mapped on Elp1, 2 (Ser-1198, Ser-1202) appear to be directly phosphorylated by Hrr25 confirming the genetic data that the CK1 isozyme has Elongator kinase activity. The analysis of phosphosite mutations revealed that Elp1 phosphorylation largely plays a positive role for Elongator activity.Citation142 Accordingly, profiling modified U34 nucleosides in tRNAs from these phosphosite mutants by LC-MS/MS showed loss of ncm5U34 and mcm5U34 formation.Citation142 The finding that normal phospholevels at Ser-1209 were detectable in a kinase-dead hrr25 mutantCitation134 implies that (at least) one additional Elongator kinase ought to exist.Citation142 Furthermore, altered interaction between Elongator, Hrr25 and Kti12 was seen in several Elp1 phosphosite mutants, in line with data showing that hrr25 mutants unable to phosphorylate Elp1 affect Elongator association with Kti12.Citation134,142 The data, therefore, suggest that normal Elongator interaction with Kti12 and proper tRNA modification are facilitated by phosphorylation of Elp1.
Regulation of s2U34 formation
Whether the URM1 pathway is regulated is not clear. Most reports describing changes in mcm5s2U34 levels in response to chemical stress do not separate the contribution of mcmCitation5 and s2 formation.Citation138,143,144 However, the analysis of 2-thiolation has shown that s2U34 levels are decreased in response to high temperatures or in growth media lacking a sulfur source.Citation140,145-148 This is not a consequence of de-thiolation but depends on active transcription of tRNA by RNA polymerase III, since the use of inhibitors or temperature sensitive alleles of RNA polymerase III prevents the decrease in s2U34.Citation140,147 Interestingly, the decrease in s2U34 is reversible when yeast is shifted back to ambient temperature, which seems like a prerequisite for active regulation. However, the kinetics for up- and downregulation of 2-thiolated tRNA are in the range of several hours.Citation140,147 This is in contrast to the idea that modification-specific changes to translation could provide a fast switch under stress conditions. The environmental stress response that elicits an extensive transcriptional change in response to various stress conditions peaks after 30 min.Citation149 A translational rewiring by reducing 2-thiolation would therefore accompany or even follow the transcriptional response to high temperatures rather than being an active driver of such a cellular transition.Citation140
But what could be mechanisms for regulation? Different high-throughput analyses have identified phosphosites in Tum1, Ncs2 and Ncs6.Citation150,151 However, to date none of these sites has been shown to affect s2U34 formation in vivo. Interestingly, Uba4 and Ncs6 were identified as targets for protein urmylationCitation65,152 and similarly ATP3BP, the human Ncs6 homolog.Citation60 However, these studies were performed using increased levels of Urm1 and oxidizing reagents. To show the in vivo relevance of urmylation it will be crucial to perform similar experiments under physiological conditions.
The role of phosphorylation for U34 regulation
Although the precise role for Elp1 phosphorylation is unclear, two options can be envisaged. On the one hand, phosphorylation could act as an ‘on/off’ switch for Elongator's U34 modifying activity, for example, in response to cellular stress. If translation of some mRNAs were dependent on U34 modification and hence tunable by Elongator,Citation12,90,91,153,154 this raises the possibility that Elongator is part of a translational control-mechanism that functions through its role as a U34 modifier. Such role is consistent with loss-of-function phenotypes associated with Elp1 phosphosite substitutions, kinase-dead hrr25 mutations and inhibition by ATP analogs of an analog-sensitive Hrr25-I82G kinase variant.Citation134,142 Although Hrr25 operates in many cellular functions,Citation155 which complicates the analysis of Elp1 phosphorylation signals, its kinase activity is required for full functionality of ribosomes and U34 containing tRNAs.Citation134,142,156-158 This is congruent with a role of the kinase in the regulation of a cell's capacity for proper mRNA translation and protein synthesis. Finally, hrr25 and Elongator mutants are sensitive to DNA damaging mutagens.Citation87,159 Since efficient translation of the RNR1 message coding for RiboNucleotide Reductase subunit 1, involved in the DNA damage response (DDR) requires U34-modified tRNAs,Citation160 Elp1 phosphorylation may link up to the known role the Hrr25 kinase plays in expression of other DDR genes, i.e. RNR2 and RNR3.Citation159
On the other hand, Elongator might require dynamic sequential phosphorylation and dephosphorylation cycles of Elp1 to carry out its tRNA modification reaction.Citation101,161 It was shown that phosphatase mutants like sit4 or sap185sap190 trapped Elp1 in a slower-migrating, hyperphosphorylated form whereas hrr25 kinase and kti12 mutants led to the presence of a fast-migrating, hypophosphorylated Elp1 isoform (). Both types of mutations cause loss-of-function phenotypes, suggesting that the functionality of Elongator requires sequential de-/phosphorylation of Elp1. Thus, dynamic de-/phosphorylation may impact on the catalytic activity of Elp3, its localization or even its ability to interact with accessory factors or substrate tRNAs. Importantly, the C-terminus of Elp1, which is phosphorylated, is required for dimerization and is adjacent to a basic region that is crucial for Elongator activity and tRNA binding.Citation102,103,142,162 Thus, Elp1 phosphorylation may affect holo-Elongator dimerization as well as interaction with partner proteins or its tRNA substrates.
The dynamic model is further supported by the observation that the right balance between hypo- and hyperphosphorylated Elp1 isoforms appears critical for Elongator activity (). This may explain why the presence of exclusively one of the two isoforms results in Elongator loss of function () and antagonistic de-/phosphorylation of Elp1 by phosphatase/kinase activities may indeed control its activity.Citation121,129-131,134 Although this contradicts the idea that Elp1 phosphorylation acts as an ‘on/off’ switch, data showing that loss of U34 modifications is similar in elp1, hrr25, kti12, sit4 and sap185sap190 mutants with opposite Elp1 phosphorylation states () agree with the dynamic phosphorylation model.Citation58,131 However, this would predict that mimicking constitutive phosphorylation on at least some of the Elp1 phosphosites may inhibit Elongator function. Surprisingly, all of the phosphomimetic ELP1 alleles tested in zymocin assays conferred growth arrest by the tRNase indicating proper Elongator functioning in the U34 modification pathway.Citation142 However, whether these mutations are not fully phosphomimetic and thus allowed for residual Elongator activity, has not been analyzed. Thus, although removal of phosphorylation sites provides evidence that Elp1 phosphorylation acts positively on Elongator function, a requirement for dynamic de-/phosphorylation still needs to be shown. Furthermore, it is possible that inhibitory Elp1 phosphosites exist, which were not identified by Abdel-Fattah and colleagues.Citation142 For example, although Elp1 phosphorylation at Thr-1212 was not detected by MS/MS, a substitution of this residue caused zymocin resistance suggesting that it represents a phosphosite in vivo.Citation142 To conclude, while there is clearly more to learn about Elongator phosphorylation, experimental evidence demonstrates that the kinase Hrr25 affects Elongator's tRNA modification function by phosphorylating (potentially reversible) phosphoacceptor sites in the Elp1 subunit.
Elongator regulation through Kti11, Kti12 and Kti13 proteins
Although the precise role of Kti12 is unclear, the yeast protein, its plant ortholog (DRL1/ELO4) and PSTK, a tRNA binding kinase, carry N-terminal P-loop motifs typical of nucleotide binding proteins.Citation118,163-166 Consistent with a functional role for this domain, a P-loop truncation of Kti12 triggers defects typical of Elongator mutants.Citation124 Importantly, Kti12 supports Elp1 phosphorylation and interacts with the Hrr25 kinase in an Elongator-dependent fashion.Citation121,124,125,167 kti12 knockouts abolish Hrr25 interaction with Elongator, cause loss of U34 modification and trigger the formation of hypophosphorylated Elp1 isoforms () similar to a hrr25 kinase mutant.Citation58,127,134 This led to the proposal that Kti12, through recruitment of Hrr25 to Elongator, positively acts on Elongator's U34 modification function.Citation134 Intriguingly, KTI12 overexpression triggers the accumulation of hyperphosphorylated Elp1 isoforms and suppresses zymocin sensitivity, which is typical of sit4 phosphatase mutants ().Citation118,130,133 This effect, however, is likely not caused by altered Elongator interactions in contrast to Elp1 phosphorylation defects, which enhance Kti12 association.Citation134,142 Furthermore, zymocin suppression through excess Kti12 can be rescued by overexpression of SIT4.Citation130 These genetic interactions suggest a negative role of Kti12 for Sit4 phosphatase function. In support of this idea, multi-copy KTI12 was found to suppress the rapamycin-resistance of a mutant lacking Resistant to Rapamycin Deletion 1 (Rrd1), a Sit4 activator protein and TOR pathway component, which has not been reported to directly relate to Elongator function.Citation92 It will be interesting to study whether this additional role for Kti12 can be separated from its ability to regulate Elongator phosphorylation.
Two additional Elongator regulatory factors are Kti11/Dph3 and Kti13/Ats1. Kti11 is a metal binder and electron transfer protein that copurifies with Fe/S cluster containing complexes including Elongator and Dph1•Dph2.Citation126,168,169 Moreover, it forms a dimer with Kti13 shown to promote U34 modification by Elongator and diphthamide synthesis by Dph1•Dph2.Citation132,136,170,171 Diphthamide is an exotic modification of translation Elongation Factor 2 (EF2),Citation142,172 which catalyzes ribosomal translocation during translation elongation and is, therefore, essential for protein synthesis and cell viability. Diphthamide-modified EF2 can be inhibited by cytotoxic ADP ribosylasesCitation173 including diphtheria toxin (DT).Citation172,174 As a consequence, KTI11 mutations confer resistance against DT and the zymocin tRNase.Citation123,132,172 Thus, Kti11 and Kti13 appear to partake in U34 anticodon modifications by Elongator and diphthamide synthesis on EF2174-176 presumably by providing electronsCitation170,171 to the Fe/S clusters in Elp3100 and Dph1•Dph2.Citation177 With the recent identification of a Kti11 reductase in yeast (Cbr1) that affects Elongator activity,Citation178 it will be important to clarify the precise roles of Kti11 and Kti13 and study their potential to modulate the electron flow required for both the tRNA and the EF2 modification pathways.
U34 modification dependent tRNase ribotoxins
Zymocin inhibits yeast growth through a complex mode of action that involves chitinolysis of the cell wall, tRNase toxin (γ-toxin) uptake, anticodon cleavage and depletion of tRNAs, eventually resulting in cell death.Citation111,112,114,115,117 Thus, zymocicity relies on the inhibition of mRNA translation and protein biosynthesis, reminiscent of bacterial anti-phage tRNase and colicin-type anticodon nucleases.Citation112,179-181 Importantly, up-regulating tRNA repair by overproducing the 2-component system from T4 phage (Rnl1-Pnkp) or tRNA ligase from plants (AtTRL1) suppresses anticodon cleavage and depletion of .Citation116 This suggested that the incision generated at the 3′ side of the modified wobble base is compatible with sealing and healing functions of heterologous ligases, and that tRNA repair can be an efficient antidote toward lethal anticodon damage by microbial tRNases.Citation116 This principle was further supported by findings that the inability of the yeast tRNA ligase Trl1 to repair the anticodon damage can be rectified by shuffling genetically engineered ligase constructs. Interestingly, when the domains from AtTRL1 and Trl1 were swapped, the plant ligase domain plus the yeast healing domain rescued S. cerevisiae cells against depletion of
. Thus, differences in the ligase component of the plant versus yeast enzymes likely account for tRNase antidote functions.Citation116 Intriguingly, PaT, a zymocin-related tRNase ribotoxin complex from Pichia acaciae, exploits a second site resulting in nucleotide excision rather than incision.Citation111,182 Hence, tRNA damage by PaT evades reconstitution of a functional ASL by the repair system, which is why plant AtTRL1 ligase fails to suppress PaT toxicity.Citation182,183
Other than suppressing zymocin action through direct repair, tRNA protection can also be provided by higher-than-normal levels of ,
and
, which are targeted for cleavage by the ribotoxin. In fact, a screen for zymocin insensitive mutants identified several
loci as copy-dependent suppressors of the tRNase ribotoxin.Citation118 Interestingly, this suppression can be efficiently countered by overexpressing the catalytic subunit of U34 methylase (Trm9•Trm112), suggesting that it is hypomodified
, which in excess is able to bypass zymocicity.Citation58,113 However, full resistance to zymocin is only conferred by mutations in ELP, KTI or URM1 pathway genes that trigger the loss of mcm5s2U34 modifications in tRNA.
Since the U34 modification pathways are conserved and elements of the Elongator pathway can be functionally exchanged between yeast and plants,Citation135,184 it was obvious to apply the yeast tRNase toxins to metazoans. Preliminary findings indicate that tRNase expression not only inhibits yeast growth but also affects the viability of vertebrate cells (RS & SL, data not shown). This is consistent with HeLa cell growth inhibition as well as hypersensitivity reactions in response to transient tRNase induction and tRNA cleavage in planta.Citation185,186 Similarly, bacterial PrrC-type anticodon nucleases were found to be lethal when expressed in yeast.Citation187 Importantly, using cytotoxic tRNase to study the formation of microbial biofilms suggests that tRNA cleavage may also be used in cell-cell communications.Citation181,187 Thus, it is tempting to exploit tRNase ribotoxins as anti-proliferative agents for use in biomedical interventions against infections by microbial, fungal or viral pathogens or to prevent undesired growth of tumor cells whose proliferation heavily relies on protein synthesis and therefore, proper tRNA function.Citation181,188
The origin of phenotypes of U34 defects
Exploiting U34 modifications to kill competing yeasts is a curious strategy for pathogenicity. But what are the molecular mechanisms that underlie the pleiotropic phenotypes that we observe in organisms with inappropriate levels of U34 modification? The absence of wobble uridine modifications is accompanied by increased sensitivity to biotic and abiotic stresses and defects in numerous cellular processes including transcription, DDR, exocytosis, telomere gene silencing and DNA demethylation.Citation6,81-85,87,88 In prokaryotes, s2U34 deficient tRNAs are poor substrates for aminoacyl-tRNA synthetases.Citation189 This however, does not appear to be the case in eukaryotes and can, therefore, not explain the observed phenotypes.Citation20
Most strikingly in yeast, except for the U34 modification defects, all known phenotypes can be rescued by overexpression of tRNAs that would normally be decorated by mcm5s2U34 (see above). In S. cerevisiae, overexpression of and
efficiently rescues the defects. This is in good agreement with ribosome-profiling experiments, which found that the codons CAA and AAA decoded by
and
, respectively, slow down during translation while a slow down at GAA (decoded by
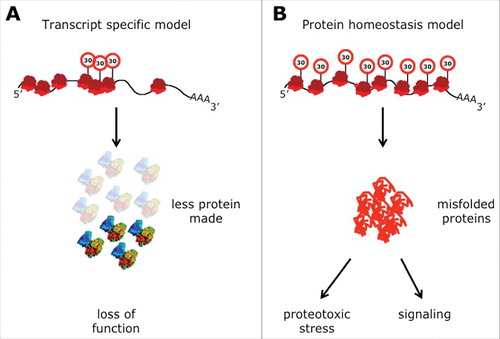
) was not detected consistently.Citation45,91 Computational attempts to identify transcripts that are enriched in A-ending codons as well as several screens and proteomics studies have reported targets that are downregulated at the protein level in U34 modification mutants.Citation143,146,160,190 Interestingly, it is possible to rescue the levels of some of these target proteins by using engineered gene constructs that have AAA codons exchanged by AAG.Citation51,191 However, in contrast to tRNA overexpression, none of these synthetic codon rescue experiments has suppressed the underlying phenotype. Failure to do so can have several reasons: First, the phenotypes may be triggered by the loss of function of a group of proteins rather than by the absence of individual proteins (). Second, instead of a loss-of-function phenotype, the effects may be triggered by a cytotoxic gain-of-function (). This concept is based on the recent findings that U34 modification mutants as well as other dysfunctional tRNA modification pathways are characterized by protein homeostasis defects.Citation45,139,154 In yeast, these lead to the aggregation of endogenous metastable proteins in the cytoplasm.Citation45 In mouse brains, the same modification defects induce the unfolded protein response in the endoplasmatic reticulum, which triggers differentiation defects in neuronal precursors, leading to microcephaly.Citation52 Thus, proteotoxic stress provides an alternative mechanism () to explain the pleiotropic phenotypes. In this scenario, codon-specific translation defects perturb the equilibrium of mRNA-translation dynamics and peptide-chain folding that has been optimized during evolution to ensure accurate protein synthesis, folding and homeostasis. As a result, proteotoxic stress alone or in combination with the standard model that favors reduction of individual proteins (), is likely the main trigger for pleiotropic defects by severely rearranging the cellular proteome or by interfering with downstream signaling of the affected cells.
Figure 4. Two models to explain phenotypes of U34 modification mutants. (A) Specific mRNA enriched in codons that depend on tRNA modifications are translated at lower rates. This leads to reduced levels of the protein encoded by this transcript triggering a loss-of-function phenotype. (B) Ribosomes slow down when translating codons that depend on tRNA modifications. The slowdown perturbs the optimized equilibrium between speed of protein synthesis and protein folding. The increased rate of protein stress leads to a systemic failure in protein homeostasis and the aggregation of endogenous proteins that associates with it in a toxic gain-of-function scenario. This can either affect viability of the cells directly or by changing cellular signaling (Street signs with “30” indicate slow speed of ribosomes).
Importantly, the underlying mechanism of U34 defects has significant consequences for our options to remedy phenotypes, particularly in the context of human disease: The loss-of-function model suggests that the identification of undertranslated mRNAs will lead to treatment options by enhancing the activity of their encoded proteins. In contrast, the protein-homeostasis model suggests that instead of repairing individual proteins, our response rather has to focus on alleviating proteotoxicity or signaling output that is induced downstream of proteotoxic stress.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank all scientists in the field who have worked since many years to provide insights into wobble uridine modification systems and owing to space limitations, apologize for not having cited all their findings that have contributed to this review topic. We thank Drs Monica Montero-Lomeli (Universidade Federal do Rio de Janeiro, Brazil), Roland Klassen and Wael Abdel-Fattah (both Universität Kassel, Germany) and Michael J.R. Stark (University of Dundee, Scotland, UK) for thematic suggestions and/or critically reading the manuscript. Project and cooperation support by the Deutsche Forschungsgemeinschaft (DFG) to RS (SCHA 750/15, SCHA 750/18 and SCHA 750/19) and funds through their Priority Programs SPP1784 Chemical Biology of Native Nucleic Acid Modifications to RS (SCHA 750/20) and SAL (LE 3260/2) and SPP1927 Iron-Sulfur for Life: Cooperative Function of Iron-Sulfur Centers in Assembly, Biosynthesis, Catalysis and Disease to RS (SCHA 750/21) and by the European Research Council to SAL (ERC-2012-StG 310489-tRNAmodi) are gratefully acknowledged.
References
- Armengod ME, Meseguer S, Villarroya M, Prado S, Moukadiri I, Ruiz-Partida R, Garzón MJ, Navarro-González C, Martínez-Zamora A. Modification of the wobble uridine in bacterial and mitochondrial tRNAs reading NNA/NNG triplets of 2-codon boxes. RNA Biol 2015; 11:1495-507; http://dx.doi.org/10.4161/15476286.2014.992269
- Jackman JE, Alfonzo JD. Transfer RNA modifications: nature's combinatorial chemistry playground. WIREs RNA 2012; 4:35-48; PMID:23139145; http://dx.doi.org/10.1002/wrna.1144
- Helm M, Alfonzo JD. Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland. Chem Biol 2014; 21:174-85; PMID:24315934; http://dx.doi.org/10.1016/j.chembiol.2013.10.015
- Grosjean H, Breton M, Sirand-Pugnet P, Tardy F, Thiaucourt F, Citti C, Barré A, Yoshizawa S, Fourmy D, de Crécy-Lagard V, et al. Predicting the Minimal Translation Apparatus: Lessons from the Reductive Evolution of Mollicutes. PLoS Genet 2014; 10:e1004363; PMID:24809820; http://dx.doi.org/10.1371/journal.pgen.1004363
- Hutchison CA, Chuang R-Y, Noskov VN, Assad-Garcia N, Deerinck TJ, Ellisman MH, Gill J, Kannan K, Karas BJ, Ma L, et al. Design and synthesis of a minimal bacterial genome. Science 2016; 351:aad6253; PMID:27013737; http://dx.doi.org/10.1126/science.aad6253
- Huang B, Johansson MJO, Byström AS. An early step in wobble uridine tRNA modification requires the Elongator complex. RNA 2005; 11:424-36; PMID:15769872; http://dx.doi.org/10.1261/rna.7247705
- Bjork GR, Huang B, Persson OP, Byström AS. A conserved modified wobble nucleoside (mcm5s2U) in lysyl-tRNA is required for viability in yeast. RNA 2007; 13:1245-55; PMID:17592039; http://dx.doi.org/10.1261/rna.558707
- Dewez M, Bauer F, Dieu M, Raes M, Vandenhaute J, Hermand D. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci USA 2008; 105:5459-64; PMID:18391219; http://dx.doi.org/10.1073/pnas.0709404105
- Chen C, Tuck S, Byström AS. Defects in tRNA modification associated with neurological and developmental dysfunctions in Caenorhabditis elegans elongator mutants. PLoS Genet 2009; 5:e1000561; PMID:19593383; http://dx.doi.org/10.1371/journal.pgen.1000561
- Chen Y-T, Hims MM, Shetty RS, Mull J, Liu L, Leyne M, Slaugenhaupt SA. Loss of mouse Ikbkap, a subunit of elongator, leads to transcriptional deficits and embryonic lethality that can be rescued by human IKBKAP. Mol Cell Biol 2009; 29:736-44; PMID:19015235; http://dx.doi.org/10.1128/MCB.01313-08
- Leidel S, Pedrioli PGA, Bucher T, Brost RL, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature 2009; 458:228-32; PMID:19145231; http://dx.doi.org/10.1038/nature07643
- Klassen R, Grunewald P, Thüring KL, Eichler C, Helm M, Schaffrath R. Loss of Anticodon Wobble Uridine Modifications Affects tRNALys Function and Protein Levels in Saccharomyces cerevisiae. PLoS ONE 2015; 10:e0119261; PMID:25747122; http://dx.doi.org/10.1371/journal.pone.0119261
- Torres AG, Batlle E, Ribas de Pouplana L. Role of tRNA modifications in human diseases. Trends Mol Med 2014; 20:306-14; PMID:24581449; http://dx.doi.org/10.1016/j.molmed.2014.01.008
- Sarin LP, Leidel SA. Modify or die? - RNA modification defects in metazoans. RNA Biol 2014; 11:1555-67; PMID:25692999; http://dx.doi.org/10.4161/15476286.2014.992279
- Kojic M, Wainwright B. The Many Faces of Elongator in Neurodevelopment and Disease. Front Mol Neurosci 2016; 9:1-10; PMID:26834556; http://dx.doi.org/10.3389/fnmol.2016.00115
- Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, Paushkin SV, Nierras CR, Cox BS, Ter-Avanesyan MD, Tuite MF. The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 1995; 14:4365-73; PMID:7556078
- Zhouravleva G, Frolova L, Le Goff X, Le Guellec R, Inge-Vechtomov S, Kisselev L, Philippe M. Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 1995; 14:4065-72; PMID:7664746
- Randerath E, Gupta RC, Chia LL, Chang SH, Randerath K. Yeast tRNA Leu UAG. Purification, properties and determination of the nucleotide sequence by radioactive derivative methods. Eur J Biochem 1979; 93:79-94; PMID: 374075; http://dx.doi.org/10.1111/j.1432-1033.1979.tb12797.x
- Szweykowska-Kulinska Z, Senger B, Keith G, Fasiolo F, Grosjean H. Intron-dependent formation of pseudouridines in the anticodon of Saccharomyces cerevisiae minor tRNAIle. EMBO J 1994; 13:4636-44; PMID:7925304
- Johansson MJO, Esberg A, Huang B, Bjork GR, Byström AS. Eukaryotic wobble uridine modifications promote a functionally redundant decoding system. Mol Cell Biol 2008; 28:3301-12; PMID:18332122; http://dx.doi.org/10.1128/MCB.01542-07
- Dunin-Horkawicz S, Czerwoniec A, Gajda MJ, Feder M, Grosjean H, Bujnicki JM. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res 2006; 34:D145-9; PMID:16381833; http://dx.doi.org/10.1093/nar/gkj084
- Crick FH. Codon-anticodon pairing: the wobble hypothesis. J Mol Biol 1966; 19:548-55; PMID:5969078; http://dx.doi.org/10.1016/S0022-2836(66)80022-0
- Agris PF. Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie 1991; 73:1345-9; PMID:1799628; http://dx.doi.org/10.1016/0300-9084(91)90163-U
- Yokoyama S, Nishimura S. Modified Nucleosides and Codon Recognition. In: tRNA: Structure, Biosynthesis, and Function, Soll D and RajBhandary UL (eds); American Society for Microbiology, Washington 1995; 207-223; http://dx.doi.org/10.1128/9781555818333.ch12
- Agris PF. Decoding the genome: a modified view. Nucleic Acids Res 2004; 32:223-38; PMID:14715921; http://dx.doi.org/10.1093/nar/gkh185
- Murphy FV, Ramakrishnan V, Malkiewicz A, Agris PF. The role of modifications in codon discrimination by tRNALysUUU. Nat Struct Mol Biol 2004; 11:1186-91; PMID:15558052; http://dx.doi.org/10.1038/nsmb861
- Sierzputowska H, Sochacka E, Malkiewicz A, Kuo KC, Gehrke CW, Agris PF. Chemistry and Structure of Modified Uridines in the Anticodon, Wobble Position of Transfer-RNA Are Determined by Thiolation. J Am Chem Soc 1987; 109:7171-7; http://dx.doi.org/10.1021/ja00257a044
- Agris PF, Sierzputowska H, Smith W, Malkiewicz A, Sochacka E, Nawrot B. Thiolation of Uridine Carbon-2 Restricts the Motional Dynamics of the Transfer-RNA Wobble Position Nucleoside. J Am Chem Soc 1992; 114:2652-6; http://dx.doi.org/10.1021/ja00033a044
- Yokoyama S, Yamaizumi Z, Nishimura S, Miyazawa T. 1H NMR studies on the conformational characteristics of 2-thiopyrimidine nucleotides found in transfer RNAs. Nucleic Acids Res 1979; 6:2611-26; PMID:379825; http://dx.doi.org/10.1093/nar/6.7.2611
- Kawai G, Yamamoto Y, Kamimura T, Masegi T, Sekine M, Hata T, Iimori T, Watanabe T, Miyazawa T, Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry 1992; 31:1040-6; PMID:1310418; http://dx.doi.org/10.1021/bi00119a012
- Yamamoto Y, Yokoyama S, Miyazawa T, Watanabe K, Higuchi S. NMR analyses on the molecular mechanism of the conformational rigidity of 2-thioribothymidine, a modified nucleoside in extreme thermophile tRNAs. FEBS Lett 1983; 157:95-9; PMID:6190675; http://dx.doi.org/10.1016/0014-5793(83)81123-5
- Testa SM, Disney MD, Turner DH, Kierzek R. Thermodynamics of RNA-RNA duplexes with 2- or 4-thiouridines: implications for antisense design and targeting a group I intron. Biochemistry 1999; 38:16655-62; PMID:10600128; http://dx.doi.org/10.1021/bi991187d
- Larsen AT, Fahrenbach AC, Sheng J, Pian J, Szostak JW. Thermodynamic insights into 2-thiouridine-enhanced RNA hybridization. Nucleic Acids Res 2015; 43:7675-87; PMID:26240387; http://dx.doi.org/10.1093/nar/gkv761
- Davis DR, Durant PC. Nucleoside modifications affect the structure and stability of the anticodon of tRNALys,3. Nucleosides Nucleotides 1999; 18:1579-81; PMID:10474235; http://dx.doi.org/10.1080/07328319908044790
- Smith WS, Sierzputowska-Gracz H, Sochacka E, Malkiewicz A, Agris PF. Chemistry and Structure of Modified Uridine Dinucleosides are Determined by Thiolation. J Am Chem Soc 1992; 114:7989-97; http://dx.doi.org/10.1021/ja00047a005
- Kumar RK, Davis DR. Synthesis and studies on the effect of 2-thiouridine and 4-thiouridine on sugar conformation and RNA duplex stability. Nucleic Acids Res 1997; 25:1272-80; PMID:9092639; http://dx.doi.org/10.1093/nar/25.6.1272
- Houssier C, Degée P, Nicoghosian K, Grosjean H. Effect of uridine dethiolation in the anticodon triplet of tRNA(Glu) on its association with tRNA(Phe). J Biomol Struct Dyn 1988; 5:1259-66; PMID:2482764; http://dx.doi.org/10.1080/07391102.1988.10506468
- Ashraf SS, Sochacka E, Cain R, Guenther R, Malkiewicz A, Agris PF. Single atom modification (O->S) of tRNA confers ribosome binding. RNA 1999; 5:188-94; PMID:10024171; http://dx.doi.org/10.1017/S1355838299981529
- Kurata S, Weixlbaumer A, Ohtsuki T, Shimazaki T, Wada T, Kirino Y, Takai K, Watanabe K, Ramakrishnan V, Suzuki T. Modified Uridines with C5-methylene Substituents at the First Position of the tRNA Anticodon Stabilize U:G Wobble Pairing during Decoding. J Biol Chem 2008; 283:18801-11; PMID:18456657; http://dx.doi.org/10.1074/jbc.M800233200
- Ogle JM, Murphy FV, Tarry MJ, Ramakrishnan V. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 2002; 111:721-32; PMID:12464183; http://dx.doi.org/10.1016/S0092-8674(02)01086-3
- Yarian C, Marszalek M, Sochacka E, Malkiewicz A, Guenther R, Miskiewicz A, Agris PF. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry 2000; 39:13390-5; PMID:11063576; http://dx.doi.org/10.1021/bi001302g
- Grosjean H, Westhof E. An integrated, structure- and energy-based view of the genetic code. Nucleic Acids Res 2016; 44:8020-40; PMID:27448410; http://dx.doi.org/10.1093/nar/gkw608
- Demeshkina N, Jenner L, Westhof E, Yusupov M, Yusupova G. A new understanding of the decoding principle on the ribosome. Nature 2012; 484:256-9; PMID:22437501; http://dx.doi.org/10.1038/nature10913
- Vendeix FAP, Murphy FV, Cantara WA, Leszczynska G, Gustilo EM, Sproat B, Malkiewicz A, Agris PF. Human tRNALys3UUU is pre-structured by natural modifications for cognate and wobble codon binding through keto-enol tautomerism. J Mol Biol 2012; 416:467-85; PMID:22227389; http://dx.doi.org/10.1016/j.jmb.2011.12.048
- Nedialkova DD, Leidel SA. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell 2015; 161:1606-18; PMID:26052047; http://dx.doi.org/10.1016/j.cell.2015.05.022
- Simos G, Tekotte H, Grosjean H, Segref A, Sharma K, Tollervey D, Hurt EC. Nuclear pore proteins are involved in the biogenesis of functional tRNA. EMBO J 1996; 15:2270-84; PMID:8641292
- Motorin Y, Keith G, Simon C, Foiret D, Simos G, Hurt E, Grosjean H. The yeast tRNA:pseudouridine synthase Pus1p displays a multisite substrate specificity. RNA 1998; 4:856-69; PMID:9671058; http://dx.doi.org/10.1017/S1355838298980396
- Pintard L, Lecointe F, Bujnicki JM, Bonnerot C, Grosjean H, Lapeyre B. Trm7p catalyses the formation of two 2′-O-methylriboses in yeast tRNA anticodon loop. EMBO J 2002; 21:1811-20; PMID:11927565; http://dx.doi.org/10.1093/emboj/21.7.1811
- Guy MP, Podyma BM, Preston MA, Shaheen HH, Krivos KL, Limbach PA, Hopper AK, Phizicky EM. Yeast Trm7 interacts with distinct proteins for critical modifications of the tRNAPhe anticodon loop. RNA 2012; 18:1921-33; PMID:22912484; http://dx.doi.org/10.1261/rna.035287.112
- Nakai Y, Nakai M, Hayashi H. Thio-modification of yeast cytosolic tRNA requires a ubiquitin-related system that resembles bacterial sulfur transfer systems. J Biol Chem 2008; 283:27469-76; PMID:18664566; http://dx.doi.org/10.1074/jbc.M804043200
- Fernández-Vázquez J, Vargas-Pérez I, Sansó M, Buhne K, Carmona M, Paulo E, Hermand D, Rodríguez-Gabriel M, Ayté J, Leidel S, et al. Modification of tRNALysUUU by elongator is essential for efficient translation of stress mRNAs. PLoS Genet 2013; 9:e1003647; PMID:23874237; http://dx.doi.org/10.1371/journal.pgen.1003647
- Laguesse S, Creppe C, Nedialkova DD, Prévot P-P, Borgs L, Huysseune S, Franco B, Duysens G, Krusy N, Lee G, et al. A Dynamic Unfolded Protein Response Contributes to the Control of Cortical Neurogenesis. Dev Cell 2015; 35:553-67; PMID:26651292; http://dx.doi.org/10.1016/j.devcel.2015.11.005
- Furukawa K, Mizushima N, Noda T, Ohsumi Y. A protein conjugation system in yeast with homology to biosynthetic enzyme reaction of prokaryotes. J Biol Chem 2000; 275:7462-5; PMID:10713047; http://dx.doi.org/10.1074/jbc.275.11.7462
- Goehring AS, Rivers DM, Sprague GF. Attachment of the ubiquitin-related protein Urm1p to the antioxidant protein Ahp1p. Eukaryotic Cell 2003; 2:930-6; PMID:14555475; http://dx.doi.org/10.1128/EC.2.5.930-936.2003
- Goehring AS, Rivers DM, Sprague GF. Urmylation: a ubiquitin-like pathway that functions during invasive growth and budding in yeast. Mol Biol Cell 2003; 14:4329-41; PMID:14551258; http://dx.doi.org/10.1091/mbc.E03-02-0079
- Singh S, Tonelli M, Tyler RC, Bahrami A, Lee MS, Markley JL. Three-dimensional structure of the AAH26994.1 protein from Mus musculus, a putative eukaryotic Urm1. Protein Sci 2005; 14:2095-102; PMID:16046629; http://dx.doi.org/10.1110/ps.051577605
- Xu J, Zhang J, Wang L, Zhou J, Huang H, Wu J, Zhong Y, Shi Y. Solution structure of Urm1 and its implications for the origin of protein modifiers. Proc Natl Acad Sci USA 2006; 103:11625-30; PMID:16864801; http://dx.doi.org/10.1073/pnas.0604876103
- Huang B, Lu J, Byström AS. A genome-wide screen identifies genes required for formation of the wobble nucleoside 5-methoxycarbonylmethyl-2-thiouridine in Saccharomyces cerevisiae. RNA 2008; 14:2183-94; PMID:18755837; http://dx.doi.org/10.1261/rna.1184108
- Pedrioli PGA, Leidel S, Hofmann K. Urm1 at the crossroad of modifications. “Protein Modifications: Beyond the Usual Suspects” Review Series. EMBO Rep 2008; 9:1196-202; PMID:19047990; http://dx.doi.org/10.1038/embor.2008.209
- Schlieker CD, Van der Veen AG, Damon JR, Spooner E, Ploegh HL. A functional proteomics approach links the ubiquitin-related modifier Urm1 to a tRNA modification pathway. Proc Natl Acad Sci USA 2008; 105:18255-60; PMID: 19017811; http://dx.doi.org/10.1073/pnas.0808756105
- Schmitz J, Chowdhury MM, Hänzelmann P, Nimtz M, Lee E-Y, Schindelin H, Leimkuhler S. The sulfurtransferase activity of Uba4 presents a link between ubiquitin-like protein conjugation and activation of sulfur carrier proteins. Biochemistry 2008; 47:6479-89; PMID:18491921; http://dx.doi.org/10.1021/bi800477u
- Nakai Y, Umeda N, Suzuki T, Nakai M, Hayashi H, Watanabe K, Kagamiyama H. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J Biol Chem 2004; 279:12363-8; PMID:14722066; http://dx.doi.org/10.1074/jbc.M312448200
- Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res 2009; 37:1335-52; PMID:19151091; http://dx.doi.org/10.1093/nar/gkn1023
- Petroski MD, Salvesen GS, Wolf DA. Urm1 couples sulfur transfer to ubiquitin-like protein function in oxidative stress. Proc Natl Acad Sci USA 2011; 108:1749-50; PMID:21245332; http://dx.doi.org/10.1073/pnas.1019043108
- Van der Veen AG, Schorpp K, Schlieker C, Buti L, Damon JR, Spooner E, Ploegh HL, Jentsch S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc Natl Acad Sci USA 2011; 108:1763-70; PMID:21209336; http://dx.doi.org/10.1073/pnas.1014402108
- Van der Veen AG, Ploegh HL. Ubiquitin-Like Proteins. Annu Rev Biochem 2012; 81:323-57; PMID:22404627; http://dx.doi.org/10.1146/annurev-biochem-093010-153308
- Jüdes A, Bruch A, Klassen R, Helm M, Schaffrath R. Sulfur transfer and activation by ubiquitin-like modifier system Uba4•Urm1 link protein urmylation and tRNA thiolation in yeast. Microb Cell 2016; 423-433; http://dx.doi.org/10.15698/mic2016.11.539
- Hofmann K, Bucher P, Kajava AV. A model of Cdc25 phosphatase catalytic domain and Cdk-interaction surface based on the presence of a rhodanese homology domain. J Mol Biol 1998; 282:195-208; PMID:9733650
- Bordo D, Bork P. The rhodanese/Cdc25 phosphatase superfamily. Sequence-structure-function relations. EMBO Rep 2002; 3:741-6; PMID:12151332; http://dx.doi.org/10.1093/embo-reports/kvf150
- Hochstrasser M. Evolution and function of ubiquitin-like protein-conjugation systems. Nat Cell Biol 2000; 2:E153-7; PMID:10934491; http://dx.doi.org/10.1038/35019643
- Goehring AS, Mitchell DA, Tong AHY, Keniry ME, Boone C, Sprague GF. Synthetic lethal analysis implicates Ste20p, a p21-activated potein kinase, in polarisome activation. Mol Biol Cell 2003; 14:1501-16; PMID:12686605
- Liu Y, Vinyard DJ, Reesbeck ME, Suzuki T, Manakongtreecheep K, Holland PL, Brudvig GW, Soll D. A [3Fe-4S] cluster is required for tRNA thiolation in archaea and eukaryotes. Proc Natl Acad Sci USA 2016; published online 24.10.2016; PMID:27791189; http://dx.doi.org/10.1073/pnas.1615732113
- Kambampati R, Lauhon CT. MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 2003; 42:1109-17; PMID:12549933; http://dx.doi.org/10.1021/bi026536+
- Mueller EG, Palenchar PM. Using genomic information to investigate the function of ThiI, an enzyme shared between thiamin and 4-thiouridine biosynthesis. Protein Sci 1999; 8:2424-7; PMID:10595545; http://dx.doi.org/10.1110/ps.8.11.2424
- Nakagawa H, Kuratani M, Goto-Ito S, Ito T, Katsura K, Terada T, Shirouzu M, Sekine S-I, Shigi N, Yokoyama S. Crystallographic and mutational studies on the tRNA thiouridine synthetase TtuA. Proteins 2013; 81:1232-44; PMID:23444054; http://dx.doi.org/10.1002/prot.24273
- Kolman C, Soll D. SPL1-1, a Saccharomyces cerevisiae mutation affecting tRNA splicing. J Bacteriol 1993; 175:1433-42; PMID:8444805; http://dx.doi.org/10.1128/jb.175.5.1433-1442.1993
- Nakai Y, Nakai M, Lill R, Suzuki T, Hayashi H. Thio modification of yeast cytosolic tRNA is an iron-sulfur protein-dependent pathway. Mol Cell Biol 2007; 27:2841-7; PMID:17283054; http://dx.doi.org/10.1128/MCB.01321-06
- Paul VD, Lill R. Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim Biophys Acta 2015; 1853:1528-39; PMID:25583461; http://dx.doi.org/10.1016/j.bbamcr.2014.12.018
- Gerber J, Neumann K, Prohl C, Mühlenhoff U, Lill R. The yeast scaffold proteins Isu1p and Isu2p are required inside mitochondria for maturation of cytosolic Fe/S proteins. Mol Cell Biol 2004; 24:4848-57; PMID:15143178; http://dx.doi.org/10.1128/MCB.24.11.4848-4857.2004
- Inigo S, Durand AN, Ritter A, Le Gall S, Termathe M, Klassen R, Tohge T, De Coninck B, Van Leene J, De Clercq R, et al. Glutaredoxin GRXS17 Associates with the Cytosolic Iron-Sulfur Cluster Assembly Pathway. Plant Physiol 2016; 172:858-73; PMID:27503603; http://dx.doi.org/10.1104/pp.16.00261
- Otero G, Fellows J, Li Y, de Bizemont T, Dirac AM, Gustafsson CM, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator, a multisubunit component of a novel RNA polymerase II holoenzyme for transcriptional elongation. Mol Cell 1999; 3:109-18; PMID:10024884; http://dx.doi.org/10.1016/S1097-2765(00)80179-3
- Winkler GS, Petrakis TG, Ethelberg S, Tokunaga M, Erdjument-Bromage H, Tempst P, Svejstrup JQ. RNA polymerase II elongator holoenzyme is composed of two discrete subcomplexes. J Biol Chem 2001; 276:32743-9; PMID:11435442; http://dx.doi.org/10.1074/jbc.M105303200
- Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo. Proc Natl Acad Sci USA 2002; 99:3517-22; PMID:11904415; http://dx.doi.org/10.1073/pnas.022042899
- Li Y, Takagi Y, Jiang Y, Tokunaga M, Erdjument-Bromage H, Tempst P, Kornberg RD. A multiprotein complex that interacts with RNA polymerase II elongator. J Biol Chem 2001; 276:29628-31; PMID:11390369; http://dx.doi.org/10.1074/jbc.C100274200
- Rahl PB, Chen CZ, Collins RN. Elp1p, the yeast homolog of the FD disease syndrome protein, negatively regulates exocytosis independently of transcriptional elongation. Mol Cell 2005; 17:841-53; PMID:15780940; http://dx.doi.org/10.1016/j.molcel.2005.02.018
- Svejstrup JQ. Elongator complex: how many roles does it play? Curr Opin Cell Biol 2007; 19:331-6; PMID:17466506; http://dx.doi.org/10.1016/j.ceb.2007.04.005
- Li Q, Fazly AM, Zhou H, Huang S, Zhang Z, Stillman B. The elongator complex interacts with PCNA and modulates transcriptional silencing and sensitivity to DNA damage agents. PLoS Genet 2009; 5:e1000684; PMID:19834596; http://dx.doi.org/10.1371/journal.pgen.1000684
- Chen C, Huang B, Eliasson M, Rydén P, Byström AS. Elongator complex influences telomeric gene silencing and DNA damage response by its role in wobble uridine tRNA modification. PLoS Genet 2011; 7:e1002258; PMID:21912530; http://dx.doi.org/10.1371/journal.pgen.1002258
- Esberg A, Huang B, Johansson MJO, Byström AS. Elevated levels of two tRNA species bypass the requirement for elongator complex in transcription and exocytosis. Mol Cell 2006; 24:139-48; PMID:17018299; http://dx.doi.org/10.1016/j.molcel.2006.07.031
- Bauer F, Hermand D. A coordinated codon-dependent regulation of translation by Elongator. Cell Cycle 2012; 11; 4524-9; PMID: 23165209; http://dx.doi.org/10.4161/cc.22689
- Zinshteyn B, Gilbert WV. Loss of a conserved tRNA anticodon modification perturbs cellular signaling. PLoS Genet 2013; 9:e1003675; PMID:23935536; http://dx.doi.org/10.1371/journal.pgen.1003675
- Scheidt V, Jüdes A, Bär C, Klassen R. Loss of wobble uridine modification in tRNA anticodons interferes with TOR pathway signaling. Microb Cell 2014; 1:416-426; http://dx.doi.org/10.15698/mic2014.12.179
- Tigano M, Ruotolo R, Dallabona C, Fontanesi F, Barrientos A, Donnini C, Ottonello S. Elongator-dependent modification of cytoplasmic tRNALysUUU is required for mitochondrial function under stress conditions. Nucleic Acids Res 2015; 43:8368-80; PMID:26240381; http://dx.doi.org/10.1093/nar/gkv765
- Tükenmez H, Xu H, Esberg A, Byström AS. The role of wobble uridine modifications in +1 translational frameshifting in eukaryotes. Nucleic Acids Res 2015; 43:9489-99; PMID:26283182; http://dx.doi.org/10.1093/nar/gkv832
- Klassen R, Bruch A, Schaffrath R. Independent suppression of ribosomal +1 frameshifts by different tRNA anticodon loop modifications. RNA Biol 2016; published online 12.12.2016; PMID:27937809; http://dx.doi.org/10.1080/15476286.2016.1267098
- Wittschieben BO, Otero G, de Bizemont T, Fellows J, Erdjument-Bromage H, Ohba R, Li Y, Allis CD, Tempst P, Svejstrup JQ. A novel histone acetyltransferase is an integral subunit of elongating RNA polymerase II holoenzyme. Mol Cell 1999; 4:123-8; PMID:10445034; http://dx.doi.org/10.1016/S1097-2765(00)80194-X
- Krogan NJ, Greenblatt JF. Characterization of a six-subunit holo-elongator complex required for the regulated expression of a group of genes in Saccharomyces cerevisiae. Mol Cell Biol 2001; 21:8203-12; PMID:11689709; http://dx.doi.org/10.1128/MCB.21.23.8203-8212.2001
- Glatt S, Létoquart J, Faux C, Taylor NMI, Séraphin B, Müller CW. The Elongator subcomplex Elp456 is a hexameric RecA-like ATPase. Nat Struct Mol Biol 2012; 19:314-20; PMID:22343726; http://dx.doi.org/10.1038/nsmb.2234
- Wittschieben BO, Fellows J, Du W, Stillman DJ, Svejstrup JQ. Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo. EMBO J 2000; 19:3060-8; PMID:10856249; http://dx.doi.org/10.1093/emboj/19.12.3060
- Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol 2006; 59:795-806; PMID:16420352; http://dx.doi.org/10.1111/j.1365-2958.2005.04989.x
- Selvadurai K, Wang P, Seimetz J, Huang RH. Archaeal Elp3 catalyzes tRNA wobble uridine modification at C5 via a radical mechanism. Nat Chem Biol 2014; 10:810-2; PMID:25151136; http://dx.doi.org/10.1038/nchembio.1610
- Setiaputra DT, Cheng DT, Lu S, Hansen JM, Dalwadi U, Lam CH, To JL, Dong M-Q, Yip CK. Molecular architecture of the yeast Elongator complex reveals an unexpected asymmetric subunit arrangement. EMBO Rep 2016; Published online 21.11.2016; PMID:27872205; http://dx.doi.org/10.15252/embr.201642548
- Dauden MI, Kosinski J, Kolaj-Robin O, Desfosses A, Ori A, Faux C, Hoffmann NA, Onuma OF, Breunig KD, Beck M, et al. Architecture of the yeast Elongator complex. EMBO Rep 2016; 18: 264-279; PMID:27974378; http://dx.doi.org/10.15252/embr.201643353
- Glatt S, Séraphin B, Müller CW. Elongator: transcriptional or translational regulator? Transcription 2012; 3:273-6; PMID:22889844; http://dx.doi.org/10.4161/trns.21525
- Glatt S, Müller CW. Structural insights into Elongator function. Curr Opin Struct Biol 2013; 23:235-42; PMID:23510783; http://dx.doi.org/10.1016/j.sbi.2013.02.009
- Gu C, Begley TJ, Dedon PC. tRNA modifications regulate translation during cellular stress. FEBS Lett 2014; 588:4287-96; PMID:25304425; http://dx.doi.org/10.1016/j.febslet.2014.09.038
- Mazauric M-H, Dirick L, Purushothaman SK, Bjork GR, Lapeyre B. Trm112p is a 15-kDa zinc finger protein essential for the activity of two tRNA and one protein methyltransferases in yeast. J Biol Chem 2010; 285:18505-15; PMID:20400505; http://dx.doi.org/10.1074/jbc.M110.113100
- Chen C, Huang B, Anderson JT, Byström AS. Unexpected accumulation of ncm5U and ncm5s2U in a trm9 mutant suggests an additional step in the synthesis of mcm5U and mcm5s2U. PLoS ONE 2011; 6:e20783; PMID:21687733; http://dx.doi.org/10.1371/journal.pone.0020783
- Kalhor HR, Clarke S. Novel methyltransferase for modified uridine residues at the wobble position of tRNA. Mol Cell Biol 2003; 23:9283-92; PMID:14645538; http://dx.doi.org/10.1128/MCB.23.24.9283-9292.2003
- Deng W, Babu IR, Su D, Yin S, Begley TJ, Dedon PC. Trm9-Catalyzed tRNA Modifications Regulate Global Protein Expression by Codon-Biased Translation. PLoS Genet 2015; 11:e1005706; PMID:26670883; http://dx.doi.org/10.1371/journal.pgen.1005706
- Lu J, Huang B, Esberg A, Johansson MJO, Byström AS. The Kluyveromyces lactis γ-toxin targets tRNA anticodons. RNA 2005; 11:1648-54; PMID:16244131; http://dx.doi.org/10.1261/rna.2172105
- Jablonowski D, Schaffrath R. Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem Soc Trans 2007; 35:1533-7; PMID:18031261; http://dx.doi.org/10.1042/BST0351533
- Jablonowski D, Zink S, Mehlgarten C, Daum G, Schaffrath R. tRNAGlu wobble uridine methylation by Trm9 identifies Elongator's key role for zymocin-induced cell death in yeast. Mol Microbiol 2006; 59:677-88; PMID:16390459; http://dx.doi.org/10.1111/j.1365-2958.2005.04972.x
- Meinhardt PDF, Schaffrath DR. Extranuclear Inheritance: Cytoplasmic linear double-stranded DNA killer elements of the dairy yeast Kluyveromyces lactis. In: Progress in Botany, Esser K, Lüttge U, Kadereit JW, Beyschlag W (eds); Springer Verlag, Berlin Heidelberg New York; 62:51-70
- Lu J, Esberg A, Huang B, Byström AS. Kluyveromyces lactis γ-toxin, a ribonuclease that recognizes the anticodon stem loop of tRNA. Nucleic Acids Res 2008; 36:1072-80; PMID:18096622; http://dx.doi.org/10.1093/nar/gkm1121
- Nandakumar J, Schwer B, Schaffrath R, Shuman S. RNA repair: an antidote to cytotoxic eukaryal RNA damage. Mol Cell 2008; 31:278-86; PMID:18657509; http://dx.doi.org/10.1016/j.molcel.2008.05.019
- Schaffrath R, Breunig KD. Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet Biol 2000; 30:173-90; PMID:11035939; http://dx.doi.org/10.1006/fgbi.2000.1221
- Butler AR, White JH, Folawiyo Y, Edlin A, Gardiner D, Stark MJ. Two Saccharomyces cerevisiae genes which control sensitivity to G1 arrest induced by Kluyveromyces lactis toxin. Mol Cell Biol 1994; 14:6306-16; PMID:8065362; http://dx.doi.org/10.1128/MCB.14.9.6306
- Kishida M, Tokunaga M, Katayose Y, Yajima H, Kawamura-Watabe A, Hishinuma F. Isolation and genetic characterization of pGKL killer-insensitive mutants (iki) from Saccharomyces cerevisiae. Biosci Biotechnol Biochem 1996; 60:798-801; PMID:8704309; http://dx.doi.org/10.1271/bbb.60.798
- Yajima H, Tokunaga M, Nakayama-Murayama A, Hishinuma F. Characterization of IKI1 and IKI3 genes conferring pGKL killer sensitivity on Saccharomyces cerevisiae. Biosci Biotechnol Biochem 1997; 61:704-9; PMID:9145530; http://dx.doi.org/10.1271/bbb.61.704
- Frohloff F, Fichtner L, Jablonowski D, Breunig KD, Schaffrath R. Saccharomyces cerevisiae Elongator mutations confer resistance to the Kluyveromyces lactis zymocin. EMBO J 2001; 20:1993-2003; PMID:11296232; http://dx.doi.org/10.1093/emboj/20.8.1993
- Frohloff F, Jablonowski D, Fichtner L, Schaffrath R. Subunit communications crucial for the functional integrity of the yeast RNA polymerase II elongator (γ-toxin target (TOT)) complex. J Biol Chem 2003; 278:956-61; PMID:12424236; http://dx.doi.org/10.1074/jbc.M210060200
- Fichtner L, Schaffrath R. KTI11 and KTI13, Saccharomyces cerevisiae genes controlling sensitivity to G1 arrest induced by Kluyveromyces lactis zymocin. Mol Microbiol 2002; 44:865-75; PMID:11994165; http://dx.doi.org/10.1046/j.1365-2958.2002.02928.x
- Fichtner L, Frohloff F, Bürkner K, Larsen M, Breunig KD, Schaffrath R. Molecular analysis of KTI12/TOT4, a Saccharomyces cerevisiae gene required for Kluyveromyces lactis zymocin action. Mol Microbiol 2002; 43:783-91; PMID:11929532; http://dx.doi.org/10.1046/j.1365-2958.2002.02794.x
- Fichtner L, Frohloff F, Jablonowski D, Stark MJR, Schaffrath R. Protein interactions within Saccharomyces cerevisiae Elongator, a complex essential for Kluyveromyces lactis zymocicity. Mol Microbiol 2002; 45:817-26; PMID:12139626; http://dx.doi.org/10.1046/j.1365-2958.2002.03055.x
- Fichtner L, Jablonowski D, Schierhorn A, Kitamoto HK, Stark MJR, Schaffrath R. Elongator's toxin-target (TOT) function is nuclear localization sequence dependent and suppressed by post-translational modification. Mol Microbiol 2003; 49:1297-307; PMID:12940988; http://dx.doi.org/10.1046/j.1365-2958.2003.03632.x
- Mehlgarten C, Schaffrath R. Mutant casein kinase I (Hrr25p/Kti14p) abrogates the G1 cell cycle arrest induced by Kluyveromyces lactis zymocin in budding yeast. Mol Genet Genomics 2003; 269:188-96; PMID:12756531; http://dx.doi.org/10.1007/s00438-003-0807-5
- Jablonowski D, Frohloff F, Fichtner L, Stark MJ, Schaffrath R. Kluyveromyces lactis zymocin mode of action is linked to RNA polymerase II function via Elongator. Mol Microbiol 2001; 42:1095-105; PMID:11737649; http://dx.doi.org/10.1046/j.1365-2958.2001.02705.x
- Jablonowski D, Butler AR, Fichtner L, Gardiner D, Schaffrath R, Stark MJ. Sit4p protein phosphatase is required for sensitivity of Saccharomyces cerevisiae to Kluyveromyces lactis zymocin. Genetics 2001; 159:1479-89; PMID:11779790
- Jablonowski D, Fichtner L, Stark MJR, Schaffrath R. The yeast elongator histone acetylase requires Sit4-dependent dephosphorylation for toxin-target capacity. Mol Biol Cell 2004; 15:1459-69; PMID:14718557; http://dx.doi.org/10.1091/mbc.E03-10-0750
- Jablonowski D, Täubert J-E, Bär C, Stark MJR, Schaffrath R. Distinct subsets of Sit4 holophosphatases are required for inhibition of Saccharomyces cerevisiae growth by rapamycin and zymocin. Eukaryotic Cell 2009; 8:1637-47; PMID:19749176; http://dx.doi.org/10.1128/EC.00205-09
- Bär C, Zabel R, Liu S, Stark MJR, Schaffrath R. A versatile partner of eukaryotic protein complexes that is involved in multiple biological processes: Kti11/Dph3. Mol Microbiol 2008; 69:1221-33; PMID:18627462; http://dx.doi.org/10.1111/j.1365-2958.2008.06350.x
- Mehlgarten C, Zink S, Rutter J, Schaffrath R. Dosage suppression of the Kluyveromyces lactis zymocin by Saccharomyces cerevisiae ISR1 and UGP1. FEMS Yeast Res 2007; 7:722-30; PMID:17367514; http://dx.doi.org/10.1111/j.1567-1364.2007.00216.x
- Mehlgarten C, Jablonowski D, Breunig KD, Stark MJR, Schaffrath R. Elongator function depends on antagonistic regulation by casein kinase Hrr25 and protein phosphatase Sit4. Mol Microbiol 2009; 73:869-81; PMID:19656297; http://dx.doi.org/10.1111/j.1365-2958.2009.06811.x
- Mehlgarten C, Jablonowski D, Wrackmeyer U, Tschitschmann S, Sondermann D, Jager G, Gong Z, Byström AS, Schaffrath R, Breunig KD. Elongator function in tRNA wobble uridine modification is conserved between yeast and plants. Mol Microbiol 2010; 76:1082-94; PMID:20398216; http://dx.doi.org/10.1111/j.1365-2958.2010.07163.x
- Zabel R, Bär C, Mehlgarten C, Schaffrath R. Yeast alpha-tubulin suppressor Ats1/Kti13 relates to the Elongator complex and interacts with Elongator partner protein Kti11. Mol Microbiol 2008; 69:175-87; PMID:18466297; http://dx.doi.org/10.1111/j.1365-2958.2008.06273.x
- Xu H, Lin Z, Li F, Diao W, Dong C, Zhou H, Xie X, Wang Z, Shen Y, Long J. Dimerization of elongator protein 1 is essential for Elongator complex assembly. Proc Natl Acad Sci USA 2015; 112:10697-702; PMID: 26261306; http://dx.doi.org/10.1073/pnas.1502597112
- Chan CTY, Dyavaiah M, Demott MS, Taghizadeh K, Dedon PC, Begley TJ. A Quantitative Systems Approach Reveals Dynamic Control of tRNA Modifications during Cellular Stress. PLoS Genet 2010; 6:e1001247; PMID:21187895; http://dx.doi.org/10.1371/journal.pgen.1001247
- Patil A, Chan CTY, Dyavaiah M, Rooney JP, Dedon PC, Begley TJ. Translational infidelity-induced protein stress results from a deficiency in Trm9-catalyzed tRNA modifications. RNA Biol 2012; 9:990-1001; PMID:22832247; http://dx.doi.org/10.4161/rna.20531
- Alings F, Sarin LP, Fufezan C, Drexler HCA, Leidel SA. An evolutionary approach uncovers a diverse response of tRNA 2-thiolation to elevated temperatures in yeast. RNA 2015; 21:202-12; PMID:25505025; http://dx.doi.org/10.1261/rna.048199.114
- Soulard A, Cremonesi A, Moes S, Schütz F, Jenö P, Hall MN. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell 2010; 21:3475-86; PMID:20702584; http://dx.doi.org/10.1091/mbc.E10-03-0182
- Abdel-Fattah W, Jablonowski D, Di Santo R, Thüring KL, Scheidt V, Hammermeister A, Have Ten S, Helm M, Schaffrath R, Stark MJR. Phosphorylation of Elp1 by Hrr25 is required for elongator-dependent tRNA modification in yeast. PLoS Genet 2015; 11:e1004931; PMID:25569479; http://dx.doi.org/10.1371/journal.pgen.1004931
- Chan CTY, Pang YLJ, Deng W, Babu IR, Dyavaiah M, Begley TJ, Dedon PC. Reprogramming of tRNA modifications controls the oxidative stress response by codon-biased translation of proteins. Nat Commun 2012; 3:937; PMID:22760636; http://dx.doi.org/10.1038/ncomms1938
- Chan CTY, Deng W, Li F, Demott MS, Babu IR, Begley TJ, Dedon PC. Highly Predictive Reprogramming of tRNA Modifications Is Linked to Selective Expression of Codon-Biased Genes. Chem Res Toxicol 2015; 28:978-88; PMID:25772370; http://dx.doi.org/10.1021/acs.chemrestox.5b00004
- Han L, Kon Y, Phizicky EM. Functional importance of Ψ38 and Ψ39 in distinct tRNAs, amplified for tRNAGln(UUG) by unexpected temperature sensitivity of the s2U modification in yeast. RNA 2015; 21:188-201; PMID:25505024; http://dx.doi.org/10.1261/rna.048173.114
- Tyagi K, Pedrioli PGA. Protein degradation and dynamic tRNA thiolation fine-tune translation at elevated temperatures. Nucleic Acids Res 2015; 43:4701-12; PMID:25870413; http://dx.doi.org/10.1093/nar/gkv322
- Damon JR, Pincus D, Ploegh HL. tRNA thiolation links translation to stress responses in Saccharomyces cerevisiae. Mol Biol Cell 2015; 26:270-82; PMID:25392298; http://dx.doi.org/10.1091/mbc.E14-06-1145
- Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. Sulfur Amino Acids Regulate Translational Capacity and Metabolic Homeostasis through Modulation of tRNA Thiolation. Cell 2013; 154:416-29; PMID:23870129; http://dx.doi.org/10.1016/j.cell.2013.06.043
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 2000; 11:4241-57; PMID:11102521; http://dx.doi.org/10.1091/mbc.11.12.4241
- Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. PhosphoPep–a database of protein phosphorylation sites in model organisms. Nat Biotechnol 2008; 26:1339-40; PMID:19060867; http://dx.doi.org/10.1038/nbt1208-1339
- Stark C, Su T-C, Breitkreutz A, Lourenco P, Dahabieh M, Breitkreutz B-J, Tyers M, Sadowski I. PhosphoGRID: a database of experimentally verified in vivo protein phosphorylation sites from the budding yeast Saccharomyces cerevisiae. Database (Oxford) 2010; 2010:bap026; PMID:20428315; http://dx.doi.org/10.1093/database/bap026
- Jüdes A, Ebert F, Bär C, Thüring KL, Harrer A, Klassen R, Helm M, Stark MJR, Schaffrath R. Urmylation and tRNA thiolation functions of ubiquitin-like Uba4·Urm1 systems are conserved from yeast to man. FEBS Lett 2015; 589:904-9; PMID:25747390; http://dx.doi.org/10.1016/j.febslet.2015.02.024
- Dedon PC, Begley TJ. A System of RNA Modifications and Biased Codon Use Controls Cellular Stress Response at the Level of Translation. Chem Res Toxicol 2014; 27:330-7; PMID:24422464; http://dx.doi.org/10.1021/tx400438d
- Klassen R, Ciftci A, Funk J, Bruch A, Butter F, Schaffrath R. tRNA anticodon loop modifications ensure protein homeostasis and cell morphogenesis in yeast. Nucleic Acids Res 2016; PMID:27496282; http://dx.doi.org/10.1093/nar/gkw705
- Cheong JK, Virshup DM. Casein kinase 1: Complexity in the family. Int J Biochem Cell Biol 2011; 43:465-9; PMID:21145983; http://dx.doi.org/10.1016/j.biocel.2010.12.004
- Schäfer T, Maco B, Petfalski E, Tollervey D, Böttcher B, Aebi U, Hurt E. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature 2006; 441:651-5; PMID:16738661; http://dx.doi.org/10.1038/nature04840
- Ray P, Basu U, Ray A, Majumdar R, Deng H, Maitra U. The Saccharomyces cerevisiae 60 S ribosome biogenesis factor Tif6p is regulated by Hrr25p-mediated phosphorylation. J Biol Chem 2008; 283:9681-91; PMID:18256024; http://dx.doi.org/10.1074/jbc.M710294200
- Ghalei H, Schaub FX, Doherty JR, Noguchi Y, Roush WR, Cleveland JL, Stroupe ME, Karbstein K. Hrr25/CK1δ-directed release of Ltv1 from pre-40S ribosomes is necessary for ribosome assembly and cell growth. J Cell Biol 2015; 208:745-59; PMID:25778921; http://dx.doi.org/10.1083/jcb.201409056
- Ho Y, Mason S, Kobayashi R, Hoekstra M, Andrews B. Role of the casein kinase I isoform, Hrr25, and the cell cycle-regulatory transcription factor, SBF, in the transcriptional response to DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 1997; 94:581-6; PMID:9012827; http://dx.doi.org/10.1073/pnas.94.2.581
- Begley U, Dyavaiah M, Patil A, Rooney JP, DiRenzo D, Young CM, Conklin DS, Zitomer RS, Begley TJ. Trm9-catalyzed tRNA modifications link translation to the DNA damage response. Mol Cell 2007; 28:860-70; PMID:18082610; http://dx.doi.org/10.1016/j.molcel.2007.09.021
- Glatt S, Zabel R, Kolaj-Robin O, Onuma OF, Baudin F, Graziadei A, Taverniti V, Lin T-Y, Baymann F, Séraphin B, et al. Structural basis for tRNA modification by Elp3 from Dehalococcoides mccartyi. Nat Struct Mol Biol 2016; 23:794-802; PMID:27455459; http://dx.doi.org/10.1038/nsmb.3265
- Di Santo R, Bandau S, Stark MJR. A conserved and essential basic region mediates tRNA binding to the Elp1 subunit of the Saccharomyces cerevisiae Elongator complex. Mol Microbiol 2014; 92:1227-42; PMID:24750273; http://dx.doi.org/10.1111/mmi.12624
- Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol 2002; 317:41-72; PMID:11916378; http://dx.doi.org/10.1006/jmbi.2001.5378
- Nelissen H, Clarke JH, De Block M, De Block S, Vanderhaeghen R, Zielinski RE, Dyer T, Lust S, Inzé D, Van Lijsebettens M. DRL1, a homolog of the yeast TOT4/KTI12 protein, has a function in meristem activity and organ growth in plants. Plant Cell 2003; 15:639-54; PMID:12615938; http://dx.doi.org/10.1105/tpc.007062
- Carlson BA, Xu X-M, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci USA 2004; 101:12848-53; PMID:15317934; http://dx.doi.org/10.1073/pnas.0402636101
- Jun SE, Cho K-H, Hwang J-Y, Abdel-Fattah W, Hammermeister A, Schaffrath R, Bowman JL, Kim G-T. Comparative analysis of the conserved functions of Arabidopsis DRL1 and yeast KTI12. Mol Cells 2015; 38:243-50; PMID:25518926; http://dx.doi.org/10.14348/molcells.2015.2297
- Petrakis TG. Physical and Functional Interaction between Elongator and the Chromatin-associated Kti12 Protein. J Biol Chem 2005; 280:19454-60; PMID:15772087; http://dx.doi.org/10.1074/jbc.M413373200
- Proudfoot M, Sanders SA, Singer A, Zhang R, Brown G, Binkowski A, Xu L, Lukin JA, Murzin AG, Joachimiak A, et al. Biochemical and structural characterization of a novel family of cystathionine beta-synthase domain proteins fused to a Zn ribbon-like domain. J Mol Biol 2008; 375:301-15; PMID:18021800; http://dx.doi.org/10.1016/j.jmb.2007.10.060
- Dong M, Su X, Dzikovski B, Dando EE, Zhu X, Du J, Freed JH, Lin H. Dph3 is an electron donor for Dph1-Dph2 in the first step of eukaryotic diphthamide biosynthesis. J Am Chem Soc 2014; 136:1754-7; PMID:24422557; http://dx.doi.org/10.1021/ja4118957
- Glatt S, Zabel R, Vonkova I, Kumar A, Netz DJ, Pierik AJ, Rybin V, Lill R, Gavin A-C, Balbach J, et al. Structure of the Kti11/Kti13 heterodimer and its double role in modifications of tRNA and eukaryotic elongation factor 2. Structure 2015; 23:149-60; PMID:25543256; http://dx.doi.org/10.1016/j.str.2014.11.008
- Kolaj-Robin O, McEwen AG, Cavarelli J, Séraphin B. Structure of the Elongator cofactor complex Kti11/Kti13 provides insight into the role of Kti13 in Elongator-dependent tRNA modification. FEBS J 2015; 282:819-33; PMID:25604895; http://dx.doi.org/10.1111/febs.13199
- Liu S, Milne GT, Kuremsky JG, Fink GR, Leppla SH. Identification of the proteins required for biosynthesis of diphthamide, the target of bacterial ADP-ribosylating toxins on translation elongation factor 2. Mol Cell Biol 2004; 24:9487-97; PMID:15485916; http://dx.doi.org/10.1128/MCB.24.21.9487-9497.2004
- Deng Q, Barbieri JT. Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu Rev Microbiol 2008; 62:271-88; PMID:18785839; http://dx.doi.org/10.1146/annurev.micro.62.081307.162848
- Schaffrath R, Abdel-Fattah W, Klassen R, Stark MJR. The diphthamide modification pathway from Saccharomyces cerevisiae - revisited. Mol Microbiol 2014; 94:1213-26; PMID:25352115; http://dx.doi.org/10.1111/mmi.12845
- Schaffrath R, Stark MJR. Decoding the biosynthesis and function of diphthamide, an enigmatic modification of translation elongation factor 2 (EF2). Microb Cell 2014; 1:203-5; http://dx.doi.org/10.15698/mic2014.06.151
- Su X, Lin Z, Lin H. The biosynthesis and biological function of diphthamide. Crit Rev Biochem Mol Biol 2013; 48:515-21; PMID:23971743; http://dx.doi.org/10.3109/10409238.2013.831023
- Zhu X, Dzikovski B, Su X, Torelli AT, Zhang Y, Ealick SE, Freed JH, Lin H. Mechanistic understanding of Pyrococcus horikoshii Dph2, a [4Fe-4S] enzyme required for diphthamide biosynthesis. Mol Biosyst 2011; 7:74-81; PMID:20931132; http://dx.doi.org/10.1039/C0MB00076K
- Lin Z, Dong M, Zhang Y, Lee EA, Lin H. Cbr1 is a Dph3 reductase required for the tRNA wobble uridine modification. Nat Chem Biol 2016; 12:995-7; PMID:27694803; http://dx.doi.org/10.1038/nchembio.2190
- Kaufmann G. Anticodon nucleases. Trends Biochem Sci 2000; 25:70-4; PMID:10664586; http://dx.doi.org/10.1016/S0968-0004(99)01525-X
- Satwika D, Klassen R, Meinhardt F. Anticodon nuclease encoding virus-like elements in yeast. Appl Microbiol Biotechnol 2012; 96:345-56; PMID:22899498; http://dx.doi.org/10.1007/s00253-012-4349-9
- Ogawa T. tRNA-targeting ribonucleases: molecular mechanisms and insights into their physiological roles. Biosci Biotechnol Biochem 2016; 80:1037-45; PMID:26967967; http://dx.doi.org/10.1080/09168451.2016.1148579
- Meineke B, Kast A, Schwer B, Meinhardt F, Shuman S, Klassen R. A fungal anticodon nuclease ribotoxin exploits a secondary cleavage site to evade tRNA repair. RNA 2012; 18:1716-24; PMID:22836353; http://dx.doi.org/10.1261/rna.034132.112
- Klassen R, Paluszynski JP, Wemhoff S, Pfeiffer A, Fricke J, Meinhardt F. The primary target of the killer toxin from Pichia acaciae is tRNAGln. Mol Microbiol 2008; 69:681-97; PMID:18532979; http://dx.doi.org/10.1111/j.1365-2958.2008.06319.x
- Chen Z, Zhang H, Jablonowski D, Zhou X, Ren X, Hong X, Schaffrath R, Zhu J-K, Gong Z. Mutations in ABO1/ELO2, a subunit of holo-Elongator, increase abscisic acid sensitivity and drought tolerance in Arabidopsis thaliana. Mol Cell Biol 2006; 26:6902-12; PMID:16943431; http://dx.doi.org/10.1128/MCB.00433-06
- Kheir E, Bär C, Jablonowski D. Cell growth control by tRNase ribotoxins from bacteria and yeast. In: Méndez-Vilas A, editor. Science and Technology against Microbial Pathogens: Research, Development and Evaluation. London (United Kingdom): World Scientific Publishing Co. Pte. Ltd; 2011:398-402.
- Leitner J, Retzer K, Malenica N, Bartkeviciute R, Lucyshyn D, Jager G, Korbei B, Byström AS, Luschnig C. Meta-regulation of Arabidopsis auxin responses depends on tRNA maturation. Cell Rep 2015; 11:516-26; PMID:25892242; http://dx.doi.org/10.1016/j.celrep.2015.03.054
- Meineke B, Schwer B, Schaffrath R, Shuman S. Determinants of eukaryal cell killing by the bacterial ribotoxin PrrC. Nucleic Acids Res 2011; 39:687-700; PMID:20855293; http://dx.doi.org/10.1093/nar/gkq831
- Uthman S, Kheir E, Bär C, Jablonowski D. Growth inhibition strategies based on antimicrobial microbes/toxins. In: Méndez-Vilas A, editor. Science against Microbial Pathogens: Communicating Current Research and Technological Advances. Vol. 2. Badajoz (Spain): FORMATEX; 2011:1321-29.
- Sylvers LA, Rogers KC, Shimizu M, Ohtsuka E, Soll D. A 2-thiouridine derivative in tRNAGlu is a positive determinant for aminoacylation by Escherichia coli glutamyl-tRNA synthetase. Biochemistry 1993; 32:3836-41; PMID:8385989; http://dx.doi.org/10.1021/bi00066a002
- Rezgui VAN, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PGA. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci USA 2013; 110:12289-94; PMID:23836657; http://dx.doi.org/10.1073/pnas.1300781110
- Bauer F, Matsuyama A, Candiracci J, Dieu M, Scheliga J, Wolf DA, Yoshida M, Hermand D. Translational Control of Cell Division by Elongator. CellReports 2012; 1:424-33; PMID:22768388; http://dx.doi.org/10.1016/j.celrep.2012.04.001
