ABSTRACT
Adenosine deaminases acting on RNA (ADARs) are zinc-containing enzymes that deaminate adenosine bases to inosines within dsRNA regions in transcripts. In short, structured dsRNA hairpins individual adenosine bases may be targeted specifically and edited with up to one hundred percent efficiency, leading to the production of alternative protein variants. However, the majority of editing events occur within longer stretches of dsRNA formed by pairing of repetitive sequences. Here, many different adenosine bases are potential targets but editing efficiency is usually much lower.
Recent work shows that ADAR-mediated RNA editing is also required to prevent aberrant activation of antiviral innate immune sensors that detect viral dsRNA in the cytoplasm. Missense mutations in the ADAR1 RNA editing enzyme cause a fatal auto-inflammatory disease, Aicardi–Goutières syndrome (AGS) in affected children. In addition RNA editing by ADARs has been observed to increase in many cancers and also can contribute to vascular disease. Thus the role of RNA editing in the progression of various diseases can no longer be ignored.
The ability of ADARs to alter the sequence of RNAs has also been used to artificially target model RNAs in vitro and in cells for RNA editing. Potentially this approach may be used to repair genetic defects and to alter genetic information at the RNA level.
In this review we focus on the role of ADARs in disease development and progression and on their potential use to artificially modify RNAs in a targeted manner.
A-I editing in mammals
With the introduction of high throughput methods such as RNA-Seq, a glimpse of the complexity of RNA in living organisms is being perceived. One current difficulty is that transcription and protein expression data do not correlate well.Citation1 While it is likely that there are many reasons for this lack of correlation, this is a major gap in biology that requires addressing as otherwise the monetary cost of accumulating RNA-Seq data are questionable. RNA modification can strongly influence translation.Citation2 Many reverse transcriptases currently used to produce cDNA erase modification marks thereby generating cDNA lacking important information. Instead RNA modifications require specific chemical reagents to alter them so that they can be detected by RNA-Seq.Citation3 With more than 140 different RNA modifications,Citation4 approximately 50 of which are known to occur in humans, the amount of variation present in RNA is likely to be significant and to have many pleiotropic effects. Thus, defining the landscape of RNA modification has become a pressing matter.
The deamination of adenosine to inosine is one of the most common modifications occurring in RNA. The enzymes that catalyze this reaction are the adenosine deaminases acting on RNA (ADARs).Citation5 These proteins do not recognize a specific consensus sequence surrounding the adenosine that is edited, instead they bind to and deaminate adenosine in stretches of double-stranded (ds) RNA.Citation6 Often a specific duplex RNA can form between an exon and a nearby intron, thus editing occurs co-transcriptionally in pre-mRNA.Citation7 ADAR enzymes are thought to have evolved from ADATs (adenosine deaminases that act on tRNA) enzymes as their catalytic deaminase domains are related.Citation8 The ADAR proteins acquired dsRNA binding domainsCitation9 that are N-terminal to the deaminase domain. A recent study demonstrated that inosine is present in RNA from the fungus Fusarium graminearum however no ortholog of an ADAR enzyme was found in this species.Citation10 The authors suggest that in this fungal species an ADAT enzyme may be editing the RNA. If this proves to be true it would represent the evolutionary step from an ADAT to an ADAR type of activity.
In mammals there are 3 ADAR proteins and 2 other homologous proteins; ADAD1 and ADAD2 (adenosine deaminase domain-containing protein) that are testis specific.Citation11 ADAR1 is the most highly and ubiquitously expressed of the ADARs and has 3 dsRNA binding domains.Citation12,13 It can be induced by interferon (IFN) and this results in the expression of a 150 kDa isoform that is extended at the N-terminus and is predominately cytoplasmic.Citation14 The shorter constitutively expressed p110 protein shuttlesCitation15 but is mainly nuclear. ADARp150 has 2 Z-binding domains that can bind to nucleic acids that are in Z-conformation at the amino-terminus whereas the shorter isoform has one Z-binding domain.Citation16 ADAR2, which like ADAR1 is enzymatically active, is nuclear and has 2 dsRNA binding domains.Citation17 Its expression is more restricted, it is expressed mainly in the brain and it is considered to be responsible for the majority of the site-specific editing events. The third ADAR protein; ADAR3 is brain-specific however, there is no evidence that it has any enzymatic activity.Citation18 Instead it is thought to modulate RNA editing activity by competing for binding to the same transcripts as the other ADAR enzymes.Citation19 Both ADAD1 and ADAD2 have one dsRNA binding domain and are also enzymatically inactive. They are testis-specific dsRNA binding proteins with unknown biologic functions.Citation20
A recent co-crystal of the deaminase domain of ADAR2 with an RNA substrate identified in yeast revealed how the deaminase domain flips out the adenosine base for deamination.Citation21 The structure also identified an RNA binding loop within the deaminase domain close to the active site that is evolutionarily conserved in ADAR2-type enzymes but not in ADAR1 or ADAR3. This RNA loop may explain the editing site-selectivity of the different enzymes.
Inosine is interpreted by the translational machinery, and by reverse transcriptase as a guanosine. Thus, the hallmark of RNA editing by ADARs is the replacement of adenosine in genomic DNA sequences by guanosine in cDNA sequences.Citation22 RNA-Seq data has now revealed the occurrence of millions of editing sites.Citation23,24 There are 2 classes of RNA editing sites; site-specific editing sites and promiscuous editing sites.Citation25 Site-specific editing targets a few specific adenosines within a transcript. Site-specific editing usually occurs to a high level and can even reach 100% at a particular position. If editing occurs within an exon it can result in recoding with a new amino acid being inserted at the edited position. Promiscuous editing occurs to a lower level, with different adenosines within the dsRNA region edited to approximately 10–20%. Transcripts encoding SINEs (Short Interspersed Nuclear Elements) such as Alu elements in humans are the main substrates for ADARs.Citation26 When 2 Alu elements are inserted in opposite orientations within a transcript then they can easily form a duplex that can be edited. Alu elements in humans underwent a recent expansion so inserted copies are more closely related to each other than are SINE inserts in mouse transcripts.Citation27 Moreover, SINEs constitute 10% of the human genome and consequently more double-stranded structures can be formed in the human transcriptome explaining the higher RNA editing in humans than in mice. Overall, promiscuous editing accounts for > 99% of editing by ADARs in humans.Citation28 Examination of site-specific editing events in humans have also shown that editing efficiency is frequently higher than in mice. Often there are Alu elements in the vicinity of site specific editing events in humans.Citation29 It is thought that the ADAR enzymes are attracted to the region due to the duplex formed by the inverted Alu elements thereby increasing editing efficiency at nearby sites.
This review primarily focuses the recent findings of the effect of RNA editing by ADARs on disease due to its recoding activity or when the enzyme is mutated. Due to space limitations we will not cover many topics such as the regulation of ADARs nor its role in RNA interference or in other model organisms. Some excellent reviews and research that cover these topics include.Citation30-33 We also provide a view on recent attempts to artificially redirect ADARs to novel sites to manipulate RNAs in a targeted manner.
Recoding of protein-coding sequences by ADARs
ADARs can deaminate mRNA leading to the alteration of codons and subsequent generation of novel proteins that are not encoded in the genome.Citation23,34 Moreover, splice sites can be introducedCitation35 or eliminated,Citation36 or stop codons can be eliminated. In Hepatitis delta virus, editing by ADAR can eliminate a stop codon by editing the antigenome RNA.Citation37-39 This is used by the virus to generate 2 protein products. In many cases ADAR2 is the enzyme responsible for site-specific editing events that affect protein coding regions.
Interestingly, ADAR2 can autoregulate its activity by editing its own pre-mRNA thereby generating a novel splice site that leads to an insertion of 47-nucleotide cassette in the coding region, causing a frameshift and premature termination.Citation35 Genetically modified mice which are incapable of editing ADAR2 pre-mRNA, show an increase in ADAR2 protein expression concomitant with an increase in the editing of pre-mRNAs targeted by ADAR2 but show no obvious phenotypic defects.Citation40
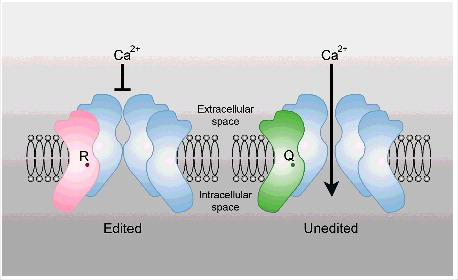
A well-studied editing target for ADAR2 is the pre-mRNA encoding glutamate receptor subunit 2 (Gria2) of aminomethylphosphonic acid (AMPA) receptors.Citation41 ADAR2 edits Gria2 at 2 exonic sites, the Q/R site located in exon 11 and the R/G site in exon 13.Citation41 In mature mRNAs the Q/R site can be edited to almost 99%. Editing changes the glutamine codon to an arginine codon which results in low Ca2+ permeability of Gria2 containing receptorsCitation41,42 (). In the absence of ADAR2 the higher Ca2+ permeability of AMPA leads to epileptic seizures and premature death in mice.Citation43 Constitutive expression of a pre-edited GRIA2 subunit rescues ADAR2 deficiency.Citation44 Lack of ADAR2 editing also leads to inefficient splicing of the nearby intron 11 causing nuclear accumulation of incompletely processed primary Gria2 transcripts and a fivefold reduction in Gria2 mRNA.Citation44 Reduced splicing levels may be caused by under-editing of an intronic hotspot in the Gria2 pre-mRNA.Citation45 In humans a decrease in editing of the GRIA2 has been found to be associated with amyotrophic lateral sclerosis (ALS).Citation46
Figure 1. Editing of protein coding targets can alter the function of the encoded protein. A critical editing site in Gria2 changes a glutamine (Q) to an arginine (R) codon. The presence of an arginine in this subunit is essential as it restricts calcium influx, thereby preventing neuronal death.Citation44
Several other mRNAs encoding receptors expressed in the brain are target for site-directed RNA editing leading to amino acids exchanges in the encoded proteins. Interestingly, overall editing of these mRNAs increases with age and is very low or absent during embryogenesis. This obviously raises the question why and when the developing nervous system seemingly requires the expression of the unedited version of these receptors.Citation47-49
Another interesting editing event changes the amino acid composition of antizyme inhibitor-1 (AZIN1). This protein inhibits cell growth by binding to and inducing degradation of tumor promoting proteins such as ornithine decarboxylase (ODC) and cyclin D1.Citation50-52 AZIN1 is an ODC homolog and shares its capacity to bind to antizyme-1 thereby competing for and stabilizing other targets of antizyme-1 by preventing their degradation.Citation50-52 The AZIN1 encoding transcript is edited by ADAR1 resulting in a serine to glycine substitution at residue 367. This amino acid exchange alters the localization from cytoplasmic to nuclear where antizyme-1, the substrate for AZIN1, is predominantly localized.Citation53 Moreover edited AZIN1 shows higher affinity for antizyme-1 therefore stabilizing cell cycle promoting proteins such as ODC. Consistently, an increase in AZIN1 editing is associated with hepatocellular carcinoma (HCC). Xenograft studies in mice, revealed that pre-edited AZIN1S367G results in high levels of tumor development when compared with wild type protein. This demonstrates a key role of RNA editing in HCC.Citation53
Control of RNA stability
One of the newly identified transcripts edited by ADAR1 encodes Cathepsin S (CTSS), a lysosomal cysteine protease which is expressed in endothelial cells and various other cell types.Citation54 CTSS participates in the degradation of antigenic peptides and extracellular matrix proteins such as laminin, fibronectin and osteocalcin and plays an important role in vasa vasorum development, angiogenesis, atherosclerotic plaque rupture and aortic aneurysm formation.Citation55-58 The 3′UTR in the mRNA harbors numerous Alu repeats which form stem loop structures, a prerequisite for RNA editing. ADAR1 was found to edit the region containing the inverted Alu repeats in CTSS transcripts in HUVEC cell lines and in patients with coronary or carotid atherosclerotic disease. Editing in the inverted Alu repeats disrupts the stem loop structure by generating mismatched wobble IU base pairs in place of the more stable AU base pairs. The incorporation of inosines destabilizes the dsRNA structure which leads to the recruitment of the stabilizing RNA-binding protein HuR, thus stabilizing the CTSS mRNA.Citation54 RNA editing levels of the 3′UTR of CTSS are significantly increased in patients with coronary or carotid atherosclerotic disease along with a concomitant increase in CTSS mRNA expression. In vitro, ADAR1 knockdown in HUVEC cells inhibits angiogenic sprouting in a 3 dimensional model of angiogenesis and in a 2D vascular network formation assay.Citation54 Thus, CTSS is an example of how editing events can have a major impact on disease progression even without changing the coding information or splice isoforms.
Editing in cancer
Considering the link between cancer, genomic mutations and DNA methylation, there has been interest to investigate if RNA editing by ADARs correlates with particular cancers or if there is aberrant editing of specific cancer-related transcripts. Initial studies found a decrease in editing in gliomas and in pediatric astrocytoma.Citation59,60 However in other cancers the reverse was found, as for instance, an increase in editing occurs in chronic myeloid leukemia progression.Citation61
In 2015 3 in-depth studies addressed the question of whether the changes in RNA sequences generated by RNA editing are a driver or a passenger in cancer progression.Citation62-64 The major resource used in these studies was The Cancer Genome Atlas (TCGA) collection. Again these researchers found a mixed picture; in the majority of cancers there is an elevated level of editing by ADARs however there are exceptions, such as kidney renal papillary cell carcinoma tumors.Citation63 Overall, increased editing levels were associated with poor prognosis. These studies focused on promiscuous editing events in regions harboring Alu elements and they found a positive correlation between tumorigenesis and editing by ADAR1 but not by ADAR2. The study focusing on breast cancer found that Type 1 IFN response and ADAR1 copy number can explain 53% of the variation in ADAR1 expression in breast cancers.Citation64 In addition, silencing of ADAR1 in breast cancer cell lines caused a decrease in both proliferation and apoptosis.Citation64 Treatment of some cancer cell types with DNA-demethylating agents leads to IFN induction, apparently through cytoplasmic dsRNA that should also be substrate for ADAR1.Citation65 These studies suggest that RNA editing events could be used as novel biomarkers for cancer and that RNA editing by ADARs may influence the response to some therapeutic agents.
Seemingly there is no simple answer to the question whether RNA editing by ADARs is a driver or a passenger in cancer progression, as this depends on the type of cancer. While increased editing in the AZIN1 transcript is associated with hepatocellular carcinoma, suggesting ADAR1 as the driver, one has to consider that ADAR1 is induced by IFN and therefore frequently upregulated upon tumor associated inflammation.Citation64 Therefore tumor associated IFN expression can also put ADAR1 in the passenger's position. As RNA editing by ADARs can have many pleiotropic effects, adjusting editing levels as a cancer therapy may therefore have unforeseen consequences.
ADARs edit viral dsRNA and exert antiviral effects
Some of the earliest sequence-based evidence for ADAR editing came from aberrant measles viruses in brains of patients with measles inclusion body encephalitis, which showed A to G sequence changes.Citation66,67 When ADAR1 was found to have a long IFN-inducible cytoplasmic isoform that shuttles between the nucleus and the cytoplasm but accumulates in the cytoplasm, an antiviral role was proposed for ADAR1 (for reviewCitation68).
Adar1 null mutant mice were found to die by embryonic day E12.5 with aberrant IFN induction and failure of hematopoiesis in the embryonic liver.Citation69,70 In humans, heterozygous loss of function mutations in ADAR1 causes the dominantly-inherited skin disease Dyschromatosis symmetrica hereditaria 1 (DSH1).Citation71 This disorder is found predominantly in Asia. One Japanese DSH1 family also presented with a fatal dystonia due to brain calcium accumulation that resembled Aicardi–Goutières syndrome (AGS).Citation72 Characterization of the ADAR1G1007R mutant protein expressed heterozygously in this family revealed that it is a catalytically-inactive ADAR1 that binds dsRNA with a higher affinity than wildtype ADAR1.Citation73
Subsequently the group of Yanick Crow found that mutations in ADAR1 cause AGS, a fatal encephalopathy that affects infants and young children.Citation73 AGS patients have heightened levels of IFN in the serum and show induction of IFN stimulated genes (ISGs) resembling the elevated IFN signature found in ADAR1 deficient mice. The increased IFN levels lead to calcification of the brain. The Crow group found a further example of the ADAR1G1007R allele among their AGS cohort, in addition to a series of other missense mutations in ADAR1, mainly in the deaminase domain. These other mutations cause AGS when homozygous or compound heterozygous in affected children of these consanguineous Asian families living in Europe. In most DSH1 patients heterozygous mutations resulting in premature termination of ADAR1 seem to cause the milder skin disease as one functional ADAR1 gene is insufficient to suppress the pigmentation defect i.e. the dominant skin phenotype results from ADAR1 haploinsufficiency. Interestingly, a more detailed examination of ISG expression in DSH1 patients revealed that they also show an ISG signature albeit not as elevated as the one in AGS patients.Citation73 Elucidating how failure to edit dsRNA substrates leads to an autoinflammatory disease requires understanding how the innate immune system normally detects pathogenic dsRNAs.
Recognition of cytoplasmic dsRNA by the innate immune system
It is vital that vigorous antiviral responses are not activated unnecessarily as these responses are energetically very costly and damaging to the host. The cellular sensors that detect viral nucleic acids are innate immune Pattern Recognition Receptors (PRRs) and the molecular features of viral nucleic acids that activate them are Pathogen Associated Molecular Patterns (PAMPs) (for reviewCitation74). The PRRs that sense dsRNA in the cytoplasm are a family of 3 large RNA helicase-related proteins called RIG-I (retinoic acid-inducible gene I),Citation75 MDA5 (melanoma differentiation-associated protein 5)Citation76 and LGP2 (laboratory of genetics and physiology 2)Citation77 that are collectively called RIG-I-like receptors (RLRs) (). These helicase-related proteins do not melt or unwind dsRNA extensively. Instead it is thought that they use their ATPase activity to scan dsRNAs to determine if the RNA is of viral origin. RIG-I is activated by dsRNA ends that are blunt or have 3′ overhangs. The affinity of the RIG-I for dsRNA is much greater when a 5′ tri- or di-phosphate is present at the N-terminus.Citation78 Many viral RNAs have 5′ triphosphates as these are required for recognition by viral RNA polymerases. RIG-I does not require long, perfectly-paired dsRNA and it is also activated by fold-back or panhandle dsRNA structures formed during replication of single-strand RNA viruses. On the other hand, MDA5 binds long dsRNAs, coating them and forming a large nucleoprotein complex.Citation79 These different binding preferences of RIG-I and MDA5 are reflected in the type of virus each protects against, with RIG-I defending against paramyxoviruses, influenza virus and Japanese encephalitis virus that have 5′ triphosphates while MDA5 detects picornaviruses that have long dsRNA that do not have these terminal features.Citation80,81
Figure 2. Unmodified, dsRNAs can trigger an innate immune response through RIG-I, MDA5 MAVS signaling, leading to IFN signaling and the activation of ISG. This, in turn, leads to embryonic lethality. The presence of inosine in RNAs suppresses the activation of this signaling cascade. Similarly, mutations in MAVS or MDA5 can suppress the embryonic lethality displayed by Adar1 deficient animals.Citation85,87
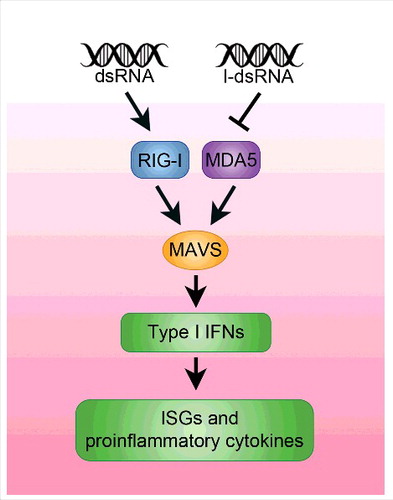
In the case of RIG-I, binding to dsRNA, leads to a conformational change in RIG-I that exposes 2 caspase-activation and recruitment domains (CARD domains) present at the N-terminus.Citation82,83 The CARD domains of RIG-I or MDA5 associate with CARD domains on the Mitochondrial Antiviral Signaling (MAVS) protein, an integral membrane protein of the mitochondrial outer membrane. Clustering of MAVS and associated nucleoprotein complexes into a signaling complex then activates the IFN response factors (IRFs).Citation84 These transcriptional regulatory proteins become phosphorylated and enter the nucleus to activate transcription of IFN that is then secreted to signal infection to neighboring cells and to trigger systemic host defenses ().
It is essential for the cell that endogenous cellular dsRNA does not trigger an immune response. It is believed that ADAR1 edits potentially immune-inducing dsRNA, altering it so that it does not aberrantly activate RLRs (). We and others showed that Adar1Δ2–13 mouse null mutant embryonic lethality is rescued in double mutant mice in which signaling from RLRs through MAVS leading to activation of IRFs and NFκB is blocked; pups are born but die within a day of birthCitation85.Citation86 The failure of Adar1Δ2–13, Mavs pups to live beyond the day of birth may be due to signaling from activated RIG-I that bypasses MAVS and has been reported to cause inflammasome activation. In addition to dsRNA editing activity the ADAR1 protein itself also appears to have significant effects on RLR signaling that may reflect sequestration of key dsRNAs or further mechanisms such as protein-protein interactions.
Mice expressing the catalytically inactive Adar1E861A allele also die embryonically but survive 2 d longer than the Adar1Δ2–13 null mutant.Citation87 However the Adar1E861A,IfiH1 double mutant lacking the Mda5 sensor only, gives complete rescue and the mice appear fully normal.Citation87,88 In contrast the Adar1Δ2–13, IfiH1 double mutants do not rescue fetal lethality of the Adar1 null mutant to live birth of pups. This significant difference between the 2 Adar1 mutants is very interesting. The delayed embryonic lethality in Adar1E861A and the complete rescue in the Adar1E861A,IfiH1 double mutant suggest that the 2 Adar1 alleles differ in the extent to which they aberrantly activate different RLRs. Aberrant activation of RIG-I may be prevented or reduced by the catalytically inactive Adar1E861A protein. On the other hand, Adar1E861A protein may not be sufficient to prevent aberrant immune activation by long dsRNA through MDA5. Therefore, in the Adar1E861A mutant the aberrant innate immune induction is more predominant due to aberrant MDA5 activation.
How does editing by ADAR1 make self dsRNA less immune-inducing? The simplest view is that I-U dsRNA has a potent suppressive effect on RLRs that is unlikely to reflect simply faster disassociation and reduced binding affinity. To address the importance of inosine, Adar1,Trp53 double mutant mouse embryo fibroblasts (MEFs) were generated that have a robust IFN activation when transfected with in vitro transcribed RNA.Citation85 A 20mer dsRNA oligonucleotide containing 2 I-U base pairs at the center was cotransfected and suppressed the aberrant immune activation. This suppression was much stronger than when cotransfecting a similar control dsRNA oligonucleotide lacking the I-U base pairs. The simple interpretation of this result is that RLR/I-U dsRNA complexes are formed that inhibit the innate immune response.
The identities of dsRNAs that cause aberrant innate immune induction in Adar1 mutant mice remain to be elucidated. There is some expectation that long dsRNAs formed by repetitive sequences such as Alu elements embedded in human transcripts are a critical type of target but there are also arguments against this. First, mice lack Alu elements and consequently have lower levels of RNA editing in their more sequence-disparate set of SINES, but there is no reason to conclude that the Adar1 mutant mouse phenotype is less severe than in humans. A more interesting possibility is that aberrant innate immune induction is triggered by some structured self RNAs conserved between human and mouse. Second, the aberrant transcription of repeat sequences in Adar1 mutant mice appears to be a secondary effect of systemic IFN activation. When this is prevented in the Adar1Δ2–13, Mavs double mutant neither IFN-induced transcripts nor repetitive sequence transcripts increase.Citation85 This indicates that loss of ADAR1 is not directly changing the dosage of these repetitive sequences.
Targeted editing
The rapidly developing CRISPR/Cas9 genomic editing methods give a new boost to gene therapeutic approaches. At the same time increasing concerns are raised about potential misuse and problems with off-target effects leading to unforeseeable consequences.Citation89 Moreover, delivery and accessibility of nuclear DNA also poses a problem for many gene therapeutic strategies. In contrast, RNA-based therapeutic strategies do not need to target nuclear DNA, can have transient effects and can also be used to modulate gene expression. Re-directing deamination of adenosines or cytosines but also other nucleic acid modifications provide good tools to alter genetic information transiently. Here, the above mentioned consequences of RNA editing, ranging from recoding, over splice site alteration to stop codon deletion and RNA destabilization can be exploited to alter, repair or inactivate existing, potentially harmful RNAs.
To this end 2 strategies have been developed to target the catalytic activity of ADARs to specific sites. The group of Rosenthal, for instance tagged the ADAR2 deaminase domain with a 22 aa long λ- N protein.Citation90 λ- N binds the 17 nucleotide long box B RNA. A box B fusion with an antisense guide RNA can therefore guide the catalytic activity of the deaminase domain to specific sites of interest that become deaminated with reasonable selectivityCitation90 (). In vitro, this system can reach editing levels of more than 90 % but also shows off-target effects. As a proof of principle this approach was used to correct the premature stop codon in the cystic fibrosis transmembrane conductance regulator (CFTR) in Xenopus oocytes. To achieve this, the substrate RNA harboring the CFTR W496X mutation was microinjected alongside with an RNA encoding the λ- N-deaminase fusion with the targeting oligonucleotide. In oocytes, editing levels of about 30% could be achieved and most importantly, CFTR activity could be restored (). Also genetically encoded λ- N-deaminase constructs could be used to repair premature stop codons in reporter constructs when transfected with a repair-oligo in tissue culture cellsCitation90 (). This system has the potential to be further optimized. Point mutations in the deaminase domain and improved tethering by an increase of λ- N domains and box B sites in the targeting oligo have been used to boost editing efficiencies in cells from 11 to 70%.Citation91
Figure 3. RNA editing enzymes can be repurposed to correct genetic defects at the RNA level. Several approaches have been taken to retarget ADARs to novel sites. (A) An antisense RNA (pink) harboring a Box B (a Lambda-N binding site) is used to target the deaminase domain of ADAR2 (DEAM) expressed as a fusion with a Lambda-N domain to an mRNA (black). (B) Alternatively, chemically modified oligos (pink) are used to target a deaminase-SNAP tag fusion to novel editing sites. (C) Complex guide RNAs (pink) that resemble endogenous ADAR targets can be used to attract endogenous ADARs to novel sites in endogenous RNAs (black). (D) These systems have been used to eliminate stop codons, but can conceptually also be used to introduce other A to I mediated codon exchanges.
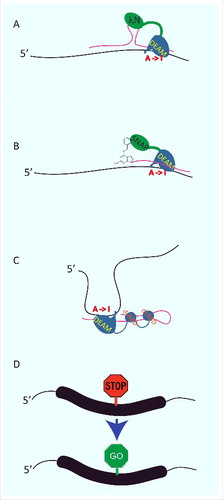
A conceptually similar, yet different approach was developed in the Stafforst laboratory, where 5′-O-benzylguanine (BG) modified antisense guide RNAs were used to target SNAP tagged deaminase domains to regions of interest (). This approach was successfully used in vitro to reach editing levels of 60–90%.Citation92 Chemical modifications of the guide RNAs helped to improve stability, membrane permeability, and substrate specificity to allow application of the system in cells with an efficiency of up to 70%.Citation93 Also, protecting the BG modification with a photoinactivatable cage did allow specific photoactivation of editing in cells and polychaete embryos.Citation94
In trying to avoid the need for the transfection of activated oligonucleotides the Stafforst group has also aimed at retargeting endogenous ADARs to novel sites. Here, guide RNAs were designed to basepair with candidate editing substrates in a way to resemble known editing stems. By systematically probing the length of complementarity, the optimal basepair composition and relative position to the site to be edited specific adenosines could be optimized to target various substrates with up to 30% efficiency in cells (). At present this system still relies on the ectopic expression of ADAR2, a reporter construct and a guide RNA but has also been used to repair a premature stop codon in ectopically expressed PINK195 and to target several endogenous RNAs.Citation95 However, attempts are on the way to also retarget the ubiquitously expressed ADAR1 which should eliminate the need for enzyme expression and would only require expression or delivery of a well designed guide RNA.Citation96
Such a single component system would have applications for gene therapy but could also be used to modulate and alter gene function, RNA stability or splicing in model systems. Moreover, retargeting of nucleic acid modifying enzymes may have applications way beyond adenosine deaminations. Retargeting modifications such adenosine methylation, pseudouridinylation, or ribose methylation may be used to modify RNA stability, turnover, or translation, to name a few.Citation97-100
The initial studies on RNA editing by ADARs focused on recoding and the generation of diverse isoforms of proteins that were not genetically encoded. While this is still an active area of research, the field has expanded to ADAR's role in cancer and other disease. The essential role of ADAR1 in innate immunity in the RLR pathway has come as a surprise. Now there is a new twist whereby ADARs enzymatic activity may be harness for gene therapy and modulation of gene function. Seemingly, 30 y after the discovery of A to I editing ADARs are still offering new perspectives.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
M.A. O'C and L. K. have received funding from the European Union's Seventh Framework Program for research, technological development and demonstration under grant agreement No 621368.” Work in the laboratory of MFJ is funded by the Austrian Science Foundation grant numbers SFB F4313, P26845 and P26882. PB is funded by the Austrian Science Foundation doctoral program W1207.
References
- Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 2012; 13:227-32; PMID:22411467; http://dx.doi.org/10.1038/nrg3185
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, He C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015; 161:1388-99; PMID:26046440; http://dx.doi.org/10.1016/j.cell.2015.05.014
- Behm-Ansmant I, Helm M, Motorin Y. Use of specific chemical reagents for detection of modified nucleotides in RNA. J Nucleic Acids 2011; 2011:408053; PMID:21716696; http://dx.doi.org/10.4061/2011/408053
- Machnicka MA, Milanowska K, Osman Oglou O, Purta E, Kurkowska M, Olchowik A, Januszewski W, Kalinowski S, Dunin-Horkawicz S, Rother KM, et al. MODOMICS: a database of RNA modification pathways–2013 update. Nucleic Acids Res 2013; 41:D262-7; PMID:23118484; http://dx.doi.org/10.1093/nar/gks1007
- Bass BL, Nishikura K, Keller W, Seeburg PH, Emeson RB, O'Connell MA, Samuel CE, Herbert A. A standardized nomenclature for adenosine deaminases that act on RNA. RNA 1997; 3:947-9; PMID:9292492
- Higuchi M, Single FN, Kohler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell 1993; 75:1361-70; PMID:8269514; http://dx.doi.org/10.1016/0092-8674(93)90622-W
- Ryman K, Fong N, Bratt E, Bentley DL, Ohman M. The C-terminal domain of RNA Pol II helps ensure that editing precedes splicing of the GluR-B transcript. Rna 2007; 13:1071-8; PMID:17525170; http://dx.doi.org/10.1261/rna.404407
- Gerber AP, Keller W. RNA editing by base deamination: more enzymes, more targets, new mysteries. Trends Biochem Sci 2001; 26:376-84; PMID:11406411; http://dx.doi.org/10.1016/S0968-0004(01)01827-8
- St Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Natl Acad Sci USA 1992; 89:10979-83
- Wang C, Xu JR, Liu H. A-to-I RNA editing independent of ADARs in filamentous fungi. RNA Biol 2016; 13:940-5; http://dx.doi.org/10.1080/15476286.2016.1215796
- Schumacher JM, Lee K, Edelhoff S, Braun RE. Distribution of Tenr, an RNA-binding protein, in a lattice-like network within the spermatid nucleus in the mouse. Biol Reprod 1995; 52:1274-83; PMID:7543294
- Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNAs for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc Natl Acad Sci USA 1994; 91:11457-61; PMID:7972084
- O'Connell MA, Krause S, Higuchi M, Hsuan JJ, Totty NF, Jenny A, Keller W. Cloning of cDNAs encoding mammalian double-stranded RNA-specific adenosine deaminase. Mol Cell Biol 1995; 15:1389-97; PMID:7862132
- Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol 1995; 15:5376-88
- Strehblow A, Hallegger M, Jantsch MF. Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol Biol Cell 2002; 13:3822-35; PMID:12429827; http://dx.doi.org/10.1091/mbc.E02-03-0161
- Herbert A, Schade M, Lowenhaupt K, Alfken J, Schwartz T, Shlyakhtenko LS, Lyubchenko YL, Rich A. The Zalpha domain from human ADAR1 binds to the Z-DNA conformer of many different sequences. Nucleic Acids Res 1998; 26:3486-93; PMID:9671809
- Melcher T, Maas S, Higuchi M, Keller W, Seeburg PH. Editing of AMPA receptor GluR-B pre-mRNA in vitro reveals site-selective adenosine to inosine conversion. J Biol Chem 1995; 270:8566-70; PMID:7721757; http://dx.doi.org/10.1074/jbc.270.15.8566
- Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. RED2, a brain specific member of the RNA-specific adenosine deaminase family. J Biol Chem 1996; 271:31795-8
- Chen CX, Cho DS, Wang Q, Lai F, Carter KC, Nishikura K. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single- and double-stranded RNA binding domains. Rna 2000; 6:755-67; PMID:10836796
- Connolly CM, Dearth AT, Braun RE. Disruption of murine Tenr results in teratospermia and male infertility. Dev Biol 2005; 278:13-21; PMID:15649457; http://dx.doi.org/10.1016/j.ydbio.2004.10.009
- Matthews MM, Thomas JM, Zheng Y, Tran K, Phelps KJ, Scott AI, Havel J, Fisher AJ, Beal PA. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat Struct Mol Biol 2016; 23:426-33; PMID:27065196; http://dx.doi.org/10.1038/nsmb.3203
- Bass BL, Weintraub H, Cattaneo R, Billeter MA. Biased hypermutation of viral RNA genomes could be due to unwinding/modification of double-stranded RNA. Cell 1989; 56:331
- Ramaswami G, Zhang R, Piskol R, Keegan LP, Deng P, O'Connell MA, Li JB. Identifying RNA editing sites using RNA sequencing data alone. Nat Methods 2013; 10:128-32; PMID:23291724; http://dx.doi.org/10.1038/nmeth.2330
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 2014; 24:365-76; PMID:24347612; http://dx.doi.org/10.1101/gr.164749.113
- Lehmann KA, Bass BL. The importance of internal loops within RNA substrates of ADAR1. J Mol Biol 1999; 291:1-13
- Levanon EY, Eisenberg E, Yelin R, Nemzer S, Hallegger M, Shemesh R, Fligelman ZY, Shoshan A, Pollock SR, Sztybel D, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat Biotechnol 2004; 22:1001-5; PMID:15258596; http://dx.doi.org/10.1038/nbt996
- Neeman Y, Levanon EY, Jantsch MF, Eisenberg E. RNA editing level in the mouse is determined by the genomic repeat repertoire. RNA 2006; 12:1802-9; PMID:16940548;; http://dx.doi.org/10.1261/rna.165106
- Pinto Y, Cohen H, Levanon E. Mammalian conserved ADAR targets comprise only a small fragment of the human editosome. Genome Biol 2014; 15:R5
- Daniel C, Silberberg G, Behm M, Ohman M. Alu elements shape the primate transcriptome by cis-regulation of RNA editing. Genome Biol 2014; 15:R28
- Alon S, Garrett SC, Levanon EY, Olson S, Graveley BR, Rosenthal JJ, Eisenberg E. The majority of transcripts in the squid nervous system are extensively recoded by A-to-I RNA editing. eLife 2015; 4; PMID:25569156; http://dx.doi.org/10.7554/eLife.05198
- Mannion N, Arieti F, Gallo A, Keegan LP, O'Connell MA. New Insights into the Biological Role of Mammalian ADARs; the RNA Editing Proteins. Biomolecules 2015; 5:2338-62; PMID:26437436; http://dx.doi.org/10.3390/biom5042338
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu Rev Biochem 2009; 79:321-49
- Paro S, Li X, O'Connell MA, Keegan LP. Regulation and functions of ADAR in Drosophila. Current Topics in Microbiology and Immunology 2012; 353:221-36; PMID:21761288; http://dx.doi.org/10.1007/82_2011_152
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 2009; 324:1210-3; PMID:19478186; http://dx.doi.org/10.1126/science.1170995
- Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature 1999; 399:75-80
- Ferreira PG, Oti M, Barann M, Wieland T, Ezquina S, Friedlander MR, Rivas MA, Esteve-Codina A, GEUVADIS Consortium, Rosenstiel P, Strom TM, et al. Sequence variation between 462 human individuals fine-tunes functional sites of RNA processing. Sci Rep 2016; 6:32406; PMID:27617755; http://dx.doi.org/10.1038/srep32406
- Casey JL. RNA editing in hepatitis delta virus. Curr Top Microbiol Immunol 2006; 307:67-89; PMID:16903221
- Casey JL, Gerin JL. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J Virol 1995; 69:7593-600
- Polson AG, Bass BL, Casey JL. RNA editing of hepatitis delta antigenome by dsRNA-adenosine deaminase. Nature (London) 1996; 380:454-6; PMID:8602246; http://dx.doi.org/10.1038/380454a0
- Feng Y, Sansam CL, Singh M, Emeson RB. Altered RNA editing in mice lacking ADAR2 autoregulation. Mol Cell Biol 2006; 26:480-8; PMID:16382140; http://dx.doi.org/10.1128/MCB.26.2.480-488.2006
- Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell 1991; 67:11-9
- Burnashev N, Monyer, H, Seeburg, PH, Sakmann, B. Divalent ion permeability of AMPA receptor channels is dominated by the edited form of a single subunit. Neuron 1992; 8:189-98; PMID:1370372
- Brusa R, Zimmermann F, Koh DS, Feldmeyer D, Gass P, Seeburg PH, Sprengel R. Early-onset epilepsy and postnatal lethality associated with an editing-deficient GluR-B allele in mice. Science 1995; 270:1677-80; PMID:750208
- Higuchi M, Maas S, Single F, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 2000; 406:78-81
- Schoft VK, Schopoff S, Jantsch MF. Regulation of glutamate receptor B pre-mRNA splicing by RNA editing. Nucleic Acids Res 2007; 35:3723-32; PMID:17517775
- Kawahara Y, Ito K, Sun H, Aizawa H, Kanazawa I, Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature 2004; 427:801
- Veno M, Bramsen JB, Bendixen C, Panitz F, Holm I, Ohman M, Kjems J. Spatio-temporal regulation of ADAR editing during development in porcine neural tissues. RNA biology 2012; 9:1054-65; PMID:22858680; http://dx.doi.org/10.4161/rna.21082
- Ring H, Boije H, Daniel C, Ohlson J, Ohman M, Hallbook F. Increased A-to-I RNA editing of the transcript for GABAA receptor subunit alpha3 during chick retinal development. Vis Neurosci 2010; 27:149-57
- Daniel C, Wahlstedt H, Ohlson J, Bjork P, Ohman M. A-to-I RNA editing affects trafficking of the {gamma}-aminobutyric acid type A (GABAA) receptor. J Biol Chem 2010
- Bercovich Z, Kahana C. Degradation of antizyme inhibitor, an ornithine decarboxylase homologous protein, is ubiquitin-dependent and is inhibited by antizyme. J Biol Chem 2004; 279:54097-102; PMID:15491992; http://dx.doi.org/10.1074/jbc.M410234200
- Kahana C. Antizyme and antizyme inhibitor, a regulatory tango. Cell Mol Life Sci 2009; 66:2479-88
- Newman RM, Mobascher A, Mangold U, Koike C, Diah S, Schmidt M, Finley D, Zetter BR. Antizyme targets cyclin D1 for degradation. A novel mechanism for cell growth repression. J Biol Chem 2004; 279:41504-11; PMID:15277517; http://dx.doi.org/10.1074/jbc.M407349200
- Chen L, Li Y, Lin CH, Chan TH, Chow RK, Song Y, Liu M, Yuan YF, Fu L, Kong KL, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat Med 2013; 19:209-16
- Stellos K, Gatsiou A, Stamatelopoulos K, Perisic Matic L, John D, Lunella FF, Jaé N, Rossbach O, Amrhein C, Sigala F, et al. Adenosine-to-inosine RNA editing controls cathepsin S expression in atherosclerosis by enabling HuR-mediated post-transcriptional regulation. Nat Med 2016; 22:1140-50; PMID:27595325; http://dx.doi.org/10.1038/nm.4172
- Shi GP, Sukhova GK, Kuzuya M, Ye Q, Du J, Zhang Y, Pan JH, Lu ML, Cheng XW, Iguchi A, et al. Deficiency of the cysteine protease cathepsin S impairs microvessel growth. Circ Res 2003; 92:493-500; PMID:12600886;http://dx.doi.org/10.1161/01.RES.0000060485.20318.96
- Wang B, Sun J, Kitamoto S, Yang M, Grubb A, Chapman HA, Kalluri R, Shi GP. Cathepsin S controls angiogenesis and tumor growth via matrix-derived angiogenic factors. J Biol Chem 2006; 281:6020-9
- Riese RJ, Mitchell RN, Villadangos JA, Shi GP, Palmer JT, Karp ER, De Sanctis GT, Ploegh HL, Chapman HA. Cathepsin S activity regulates antigen presentation and immunity. J Clin Invest 1998; 101:2351-63; PMID:9616206
- Reiser J, Adair B, Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest 2010; 120:3421-31; PMID:20921628; http://dx.doi.org/10.1172/JCI42918
- Cenci C, Barzotti R, Galeano F, Corbelli S, Rota R, Massimi L, Di Rocco C, O'Connell MA, Gallo A. Down-regulation of RNA editing in pediatric astrocytomas: ADAR2 editing activity inhibits cell migration and proliferation. J Biol Chem 2008; 283:7251-60; PMID:18178553; http://dx.doi.org/10.1074/jbc.M708316200
- Maas S, Patt S, Schrey M, Rich A. Underediting of glutamate receptor GluR-B mRNA in malignant gliomas. Proc Natl Acad Sci U S A 2001; 98:14687-92
- Jiang Q, Crews LA, Barrett CL, Chun HJ, Court AC, Isquith JM, Zipeto MA, Goff DJ, Minden M, Sadarangani A, et al. ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U S A 2013; 110:1041-6; PMID:23275297; http://dx.doi.org/10.1073/pnas.1213021110
- Paz-Yaacov N, Bazak L, Buchumenski I, Porath HT, Danan-Gotthold M, Knisbacher BA, Eisenberg E, Levanon EY. Elevated RNA Editing Activity Is a Major Contributor to Transcriptomic Diversity in Tumors. Cell Rep 2015; 13:267-76; PMID:26440895; http://dx.doi.org/10.1016/j.celrep.2015.08.080
- Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner HM, Eterovic AK, Yuan Y, et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer Cell 2015; 28:515-28; PMID:26439496; http://dx.doi.org/10.1016/j.ccell.2015.08.013
- Fumagalli D, Gacquer D, Rothe F, Lefort A, Libert F, Brown D, Kheddoumi N, Shlien A, Konopka T, Salgado R, et al. Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome. Cell reports 2015; 13:277-89
- Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell 2016; 164:1073; PMID:27064190; http://dx.doi.org/10.1016/j.cell.2015.10.020
- Cattaneo R, Kaelin K, Baczko K, Billeter MA. Measles virus editing provides an additional cysteine-rich protein. Cell 1989; 56:759-64; PMID:2924348
- Cattaneo R, Schmid A, Eschle D, Baczko K, ter Meulen V, Billeter MA. Biased hypermuation and other genetic changes in defective measles viruses in human brain infections. Cell 1988; 55:255-65; PMID:3167982
- Samuel CE. Adenosine deaminases acting on RNA (ADARs) are both antiviral and proviral. Virology 2011; 411:180-93
- Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 2004; 279:4952-61; PMID:14613934; http://dx.doi.org/10.1074/jbc.M310162200
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 2004; 279:4894-902
- Zhang XJ, He PP, Li M, He CD, Yan KL, Cui Y, Yang S, Zhang KY, Gao M, Chen JJ, et al. Seven novel mutations of the ADAR gene in Chinese families and sporadic patients with dyschromatosis symmetrica hereditaria (DSH). Hum Mutat 2004; 23:629-30; PMID:15146470; http://dx.doi.org/10.1002/humu.9246
- Tojo K, Sekijima Y, Suzuki T, Suzuki N, Tomita Y, Yoshida K, Hashimoto T, Ikeda S. Dystonia, mental deterioration, and dyschromatosis symmetrica hereditaria in a family with ADAR1 mutation. Mov Disord 2006; 21:1510-3
- Rice GI, Kasher PR, Forte GM, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA, et al. Mutations in ADAR1 cause Aicardi-Goutieres syndrome associated with a type I interferon signature. Nat Genetics 2012; 44:1243-8; PMID:23001123; http://dx.doi.org/10.1038/ng.2414
- Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell 2010; 140:805-20
- Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol 2004; 5:730-7; PMID:15208624; http://dx.doi.org/10.1038/ni1087
- Kang DC, Gopalkrishnan RV, Wu Q, Jankowsky E, Pyle AM, Fisher PB. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci U S A 2002; 99:637-42; PMID:11805321; http://dx.doi.org/10.1073/pnas.022637199
- Satoh T, Kato H, Kumagai Y, Yoneyama M, Sato S, Matsushita K, Tsujimura T, Fujita T, Akira S, Takeuchi O. LGP2 is a positive regulator of RIG-I- and MDA5-mediated antiviral responses. Proc Natl Acad Sci U S A 2010; 107:1512-7; PMID:20080593; http://dx.doi.org/10.1073/pnas.0912986107
- Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 2009; 31:25-34; PMID:19576794; http://dx.doi.org/10.1016/j.immuni.2009.05.008
- Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S. Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 2013; 152:276-89; PMID:23273991; http://dx.doi.org/10.1016/j.cell.2012.11.048
- Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 2006; 441:101-5; PMID:16625202
- Loo YM, Fornek J, Crochet N, Bajwa G, Perwitasari O, Martinez-Sobrido L, Akira S, Gill MA, García-Sastre A, Katze MG, et al. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J Virol 2008; 82:335-45; PMID:17942531
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell 2011; 147:423-35; PMID:22000019; http://dx.doi.org/10.1016/j.cell.2011.09.039
- Luo D, Ding SC, Vela A, Kohlway A, Lindenbach BD, Pyle AM. Structural insights into RNA recognition by RIG-I. Cell 2011; 147:409-22; PMID:22000018; http://dx.doi.org/10.1016/j.cell.2011.09.023
- Cai X, Chen J, Xu H, Liu S, Jiang QX, Halfmann R, Chen ZJ. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 2014; 156:1207-22; PMID:24630723; http://dx.doi.org/10.1016/j.cell.2014.01.063
- Mannion NM, Greenwood SM, Young R, Cox S, Brindle J, Read D, Nellåker C, Vesely C, Ponting CP, McLaughlin PJ, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell reports 2014; 9:1482-94; PMID:25456137; http://dx.doi.org/10.1016/j.celrep.2014.10.041
- Pestal K, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB. Isoforms of RNA-Editing Enzyme ADAR1 Independently Control Nucleic Acid Sensor MDA5-Driven Autoimmunity and Multi-organ Development. Immunity 2015; 43:933-44; PMID:26588779; http://dx.doi.org/10.1016/j.immuni.2015.11.001
- Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 2015; 349:1115-20; PMID:26275108; http://dx.doi.org/10.1126/science.aac7049
- Liddicoat BJ, Chalk AM, Walkley CR. ADAR1, inosine and the immune sensing system: distinguishing self from non-self. Wiley Interdiscip Rev RNA 2016; 7:157-72; PMID:26692549; http://dx.doi.org/10.1002/wrna.1322
- Barrangou R, Doudna JA. Applications of CRISPR technologies in research and beyond. Nat Biotechnol 2016; 34:933-41; PMID:27606440; http://dx.doi.org/10.1038/nbt.3659
- Montiel-Gonzalez MF, Vallecillo-Viejo I, Yudowski GA, Rosenthal JJ. Correction of mutations within the cystic fibrosis transmembrane conductance regulator by site-directed RNA editing. Proc Natl Acad Sci U S A 2013; 110:18285-90; PMID:24108353; http://dx.doi.org/10.1073/pnas.1306243110
- Montiel-Gonzalez MF, Vallecillo-Viejo IC, Rosenthal JJ. An efficient system for selectively altering genetic information within mRNAs. Nucleic Acids Res 2016; 44:e157; PMID:27557710; http://dx.doi.org/10.1093/nar/gkw738
- Stafforst T, Schneider MF. An RNA-deaminase conjugate selectively repairs point mutations. Angew Chem Int Ed Engl 2012; 51:11166-9; PMID:23038402; http://dx.doi.org/10.1002/anie.201206489
- Vogel P, Schneider MF, Wettengel J, Stafforst T. Improving site-directed RNA editing in vitro and in cell culture by chemical modification of the guideRNA. Angew Chem Int Ed Engl 2014; 53:6267-71; PMID:24890431; http://dx.doi.org/10.1002/anie.201402634
- Hanswillemenke A, Kuzdere T, Vogel P, Jekely G, Stafforst T. Site-Directed RNA Editing in Vivo Can Be Triggered by the Light-Driven Assembly of an Artificial Riboprotein. J Am Chem Soc 2015; 137:15875-81; PMID:26594902; http://dx.doi.org/10.1021/jacs.5b10216
- Wettengel J, Reautschnig P, Geisler S, Kahle PJ, Stafforst T. Harnessing human ADAR2 for RNA repair - Recoding a PINK1 mutation rescues mitophagy. Nucleic Acids Res 2016; PMID:27907896; http://dx.doi.org/10.1093/nar/gkw911
- Heep M, Mach P, Reautschnig P, Wettengel J, Stafforst T. Applying Human ADAR1p110 and ADAR1p150 for Site-Directed RNA Editing-G/C Substitution Stabilizes GuideRNAs against Editing. Genes (Basel) 2017; 8:E34; PMID:28098820; http://dx.doi.org/10.3390/genes8010034
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 2015; 347:1002-6; PMID:25569111; http://dx.doi.org/10.1126/science.1261417
- Dominissini D, Nachtergaele S, Moshitch-Moshkovitz S, Peer E, Kol N, Ben-Haim MS, Dai Q, Di Segni A, Salmon-Divon M, Clark WC, et al. The dynamic N(1)-methyladenosine methylome in eukaryotic messenger RNA. Nature 2016; 530:441-6; PMID:26863196; http://dx.doi.org/10.1038/nature16998
- Vaidyanathan PP, AlSadhan I, Merriman DK, Al-Hashimi H, Herschlag D. Pseudouridine and N-6 methyladenosine modifications weaken PUF protein/RNA interactions. RNA 2017; PMID:28138061; http://dx.doi.org/10.1261/rna.060053.116
- Karijolich J, Yu YT. Converting nonsense codons into sense codons by targeted pseudouridylation. Nature 2011; 474:395-8; PMID:21677757; http://dx.doi.org/10.1038/nature10165
