ABSTRACT
Circular (circ)RNAs have recently become a subject of great biologic interest. It is now clear that they represent a diverse and abundant class of RNAs with regulated expression and evolutionarily conserved functions. There are several mechanisms by which RNA circularization can occur in vivo. Here, we focus on the biogenesis of tRNA intronic circular RNAs (tricRNAs) in archaea and animals, and we detail their use as research tools for orthogonal, directed circRNA expression in vivo.
Background
Recent growth in the study of circular (circ)RNA processing and function has been propelled largely by advances in high-throughput sequencing technology and refined bioinformatic pipelines.Citation1,2 CircRNAs in eukaryotes were once thought of as rare molecules resulting from aberrant mRNA splicing.Citation3 It has since become clear that circRNA biogenesis in many eukaryotes proceeds through several diverse and evolutionarily conserved biochemical pathways,Citation4-7 resulting in an abundant class of RNA molecules with a host of proposed functions.Citation8,9 CircRNAs have been shown to be remarkably stable in living cells,Citation10-12 potentially due to their unique structure and lack of free termini on which the exonucleolytic RNA turnover machinery relies. Along with being generally abundant,Citation2 circRNAs can be differentially expressed in a tissue-dependent manner and can have expression profiles that differ from their parental mRNA,Citation13 suggesting regulated expression and diversity of function. CircRNAs are enriched in mammalian and Drosophila brains,Citation14-18 are positively correlated with aging,Citation16 and can be associated with disease risk.Citation19 The most well-known example of circular RNA function is their ability to act as molecular “sponges,” competing for binding of molecules that affect gene regulation. For example, the human circular transcript CDR1as/ciRS-7 contains approximately 70 seed target sequences for microRNA-7 (miR-7) and associates with exosomal Argonaute proteins in vivo in a miR-7 dependent manner.Citation17,18 CDR1as/ciRS-7 represents a striking example of the controversial “competing endogenous RNA” theory, which posits that transcripts can “compete” for binding of miRNAs and other molecules to affect gene expression.Citation20-22 This transcript has overlapping expression patterns with miR-7 in mouse brains, suggesting a potential in vivo regulatory interaction.Citation17,18 Interestingly, either inhibition of miR-7 or ectopic expression of CDR1as causes midbrain development defects in zebrafishCitation18, indicating that disruption of this type of regulation could have significant consequence.
In addition to miRNAs, circRNAs can also bind and sequester proteins. In human, mouse, and Drosophila, the RNA-binding protein Muscleblind (Mbl) has been shown to promote circularization of several transcripts, including the second exon of the Mbl pre-mRNA itself.Citation23 In all 3 species, circMbl RNA contains conserved Mbl protein binding sites, and pulldown experiments in Drosophila S2 cells revealed an in vivo interaction between Mbl protein and circMbl,Citation23 suggesting a possible feedback mechanism by which Mbl protein and circMbl interact to control splicing of the parent transcript. The relative stability of circRNAs could perhaps contribute to a unique cellular role in initiating and maintaining these types of regulatory interactions.s.
The most common pathway for RNA circularization occurs via a process termed “back-splicing.” Back-splicing proceeds via a canonical 2-step transesterification reaction, except that a downstream splice donor sequence becomes ligated to an upstream splice acceptor site, resulting in a non-canonical alternative splicing event (). Importantly, back-splicing has been shown to compete with forward splicing of certain pre-mRNA transcripts.Citation23,24 RNA elements that promote juxtaposition of splice sites close to one another in physical space are thought to promote back-splicing. For example, back-splicing of circular transcripts is associated with the presence of repetitive elements (e.g. Alu repeats in humans or different repetitive elements in other species) that flank the circularized region.Citation2,25,26 Additionally, back-splicing can be facilitated by RNA-binding proteins such as the aforementioned Mbl, QKI (quaking), hnRNPs and serine/arginine (SR) proteins.Citation23,25,27 In the following sections, we detail another conserved mode of RNA circularization occurring in archaea and eukaryotes, in which tRNA introns are spliced to form circles.
Figure 1. Two in vivo RNA circularization pathways. (A) Intron base-pairing and/or RNA-binding proteins facilitate pairing of a downstream splice site and an upstream splice site, bringing them into close proximity. Non-canonical “back-splicing” of these sites results in a circularized exon. (B) The tRNA splicing endonuclease (TSEN) complex cleaves an intron-containing pre-tRNA at the bulge-helix-bulge (BHB) motif. The exon halves are ligated, and the intron termini are also ligated to form a circle.
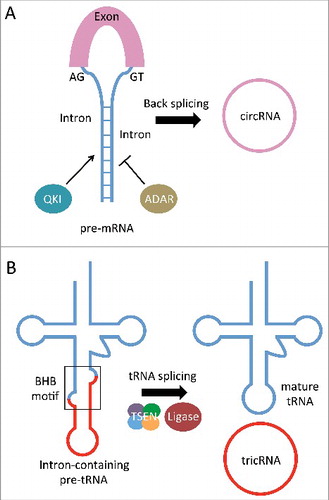
Splicing and circularization of tRNA introns in archaea
Despite the absence of the spliceosomal machinery in Bacteria and Archaea, intron-containing genes have been observed in all 3 domains of life.Citation28 Bacterial introns are of the self-splicing variety (for a review see Ref. Citation29). In Archaea, however, introns are fairly common among tRNA and rRNA genes, and their removal is catalyzed by a conserved splicing endonuclease complex.Citation30,31 Interestingly, archaeal RNA splicing typically produces a circularized intron following its removal from the primary transcript.Citation32 Splicing of pre-tRNAs is a 2-step process wherein the transcript is first cleaved at defined positions by the archaeal splicing endonuclease complex, which recognizes a bulge-helix-bulge (BHB) sequence motif ().Citation33,34 The exon halves are then ligated together to form a mature tRNA, while the intron termini are also joined to form a circle (). Although the 2 ligation events appear to use the same processing factors, the reactions can occur independently.Citation32
Circular RNAs derived from multiple tRNA isodecoder transcripts have been detected,Citation35 though it is not known if stable circles are made from every tRNA intron. To distinguish these circRNAs from their back-spliced cousins in eukaryotes, we use the term tRNA intronic circular RNAs, or tricRNAs. In the archaeon Haloferax volcanii, a tricRNA was detected in vivo from tRNATrp by RT-PCR and Northern blot analysis.Citation32 In this same study, introns derived from elongator tRNAMet appear to undergo end-joining that would be characteristic of circularization, but tricRNA expression could not be confirmed by blotting. This lack of detectability could be due to differences in overall parental tRNA expression: in Haloferax volcanii, there is only one tRNATrp gene, whereas there are 3 elongator tRNAMet genes, and only one of them contains an intron.Citation35 Thus, reduced expression of the parent tRNAMet gene might lead to lower amounts of its tricRNA that are detectable only by RT-PCR. In certain archaeal species, tricRNAs are highly expressed and/or are quite stable. For example, a whole-genome sequencing approach in Sulfolobus solfataricus revealed that among circRNAs, only those generated from 16S and 23S rRNA were more abundant than tricRNAs.Citation36 Circles were only detected for 5 of the 19 intron-containing tRNAs, but this is most likely due to an ascertainment bias, as the remaining 14 introns are quite small and would thus require specific methods of library preparation to enrich for them.
Several groups have speculated on the functions of circularized introns in archaea. Circularized rRNA introns have been found to contain putative open reading frames, and it is possible that these encode peptides.Citation11,12,37 Additionally, a functional C/D box RNA is contained in the tRNATrp intron of H. volcanii, responsible for 2′-O-methylation of its parent tRNA, though both circular and linear excised introns are capable of guiding this modification.Citation38. Similarly, the tRNATrp intron of S. solfataricus contains regions of sequence complementarity to its parent tRNA, suggesting a role for the excised intron in tRNA modification.Citation36 Whether tricRNAs from other parental tRNAs and other species also function in RNA modification remains to be determined.
Splicing and circularization of tRNA introns in eukarya
Eukaryotic pre-tRNA cleavage is slightly different from archaeal in that it is performed by a complex of 4 proteins rather than one.Citation39 The heterotetrameric eukaryotic tRNA splicing endonuclease (TSEN) complex is functionally homologous to the archaeal protein; however, the RNA elements that define the sites of cleavage are different.Citation34 Many eukaryotic pre-tRNAs indeed contain a BHB-like motif, and the eukaryotic endonuclease complex has been shown to be capable of recognizing and splicing pre-tRNAs with archaeal features,Citation40 implying conservation of the structural elements underlying substrate recognition by TSEN. However, the currently accepted model of splicing by eukaryotic TSEN relies on recognition of the tRNA body, independent of a BHB, and splicing is dictated by a “ruler” mechanism whereby splice sites are chosen based on distance from the recognized tRNA body.Citation41,42 Unlike archaeal tRNA introns, which can be found at several positions in the tRNA, eukaryotic introns are nearly always found between the canonical positions 37 and 38 of the mature tRNA, adjacent to the anticodon.Citation35 This difference in intron positioning is thought to result from co-evolution of introns and their processing enzymes.Citation31
After cleavage by TSEN, tRNA halves are ligated to form a mature tRNA. There are 2 distinct ligation pathways in eukaryotes. The 5′-ligation pathway involves the multifunctional Trl1 protein and is considered to be the predominant one in fungi and plants.Citation43-45 Also known as the “healing and sealing” pathway, this process incorporates an external phosphate for use as the splice junction phosphate.Citation34 In vertebrates as well as archaea, tRNAs are ligated by the RtcB enzyme (called HSPC117 in humans) in the 3′-ligation or “direct ligation” pathway, where an external phosphate is not incorporated.Citation46,47 In RtcB knockdown experiments in Drosophila, tricRNA levels decreased significantly, indicating that tricRNA ligation is indeed dependent upon ligase activity of RtcB.Citation10
In at least one case, ligation of mRNA splice products by tRNA processing enzymes can also result in intron circularization in vitro.Citation48 The unfolded protein response in metazoans and yeast causes stress signaling, triggering an unconventional splicing of XBP1 (HAC1 in yeast) mRNA. The transcript is cleaved by IRE1 and ligated by RtcB in metazoansCitation49-51 or Trl1 in yeast.Citation52 In vitro reconstitution of these elements resulted in a circularized intron following human XBP1 splicing, and interestingly a circular intron was produced upon addition of either RtcB or the yeast Trl1 ligase,Citation48 which has homologues in some animal species. It is unclear whether this circularization occurs in vivo, if there are other mRNAs whose splicing is dependent upon tRNA ligase activity, and whether the removed introns have any cellular function.
Sequence conservation could indicate an important functional property for certain tricRNAs. In Drosophilid species, many tRNA introns are highly conserved at the sequence level. One tricRNA (tric31905) even displays compensatory mutation and preservation of base-pairing motifs, indicating a conserved secondary structure.Citation10 Additionally, small RNA sequencing analysis indicates that this circular RNA is further processed into microRNA-sized fragments.Citation10 It is currently unclear whether these small RNAs have a function in the cell or are merely degradation intermediates. Inspection of sequence and secondary structure of other Drosophilid introns does not reveal common sequence motifs or elements, suggesting that processing of tric31905 is not necessarily applicable to all tricRNAs.
Methods of ectopic circRNA expression and detection
To date, few tools exist to direct generation of RNA circles in vivo. Several groups have constructed mini-gene vectors that contain complementary sequences flanking the region of interest, designed to facilitate its circularization in a mechanistically similar fashion to endogenous exonic back-splicing.Citation17,25,53 Effective circularization has been achieved using both endogenous and designed elements, or some combination thereof. Such engineered mini-genes have also shed light on cis-acting sequences that either promote or inhibit circularization.Citation25,26,53 These ectopic circRNAs are capable of serving as a template for protein translation following inclusion of an internal ribosomal entry site (IRES) in the circularized transcript.Citation53
Back-spliced circRNA expression systems are typically driven by an RNA pol II promoter such as the one present in human cytomegalovirus (CMV).Citation25,53 Ectopic expression of tricRNAs, in contrast, relies on an RNA pol III promoter to drive transcription (). Robust expression can be achieved with an external pol III promoter, such as the one from the U6 snRNA gene.Citation10,54 Pol III transcription can exceed that of pol II, and produces more consistent levels of expression across cell types as compared with the CMV promoter.Citation55,56
Figure 2. tricRNA expression can be detected by RT-PCR. (A) Schematic of the tricRNA expression construct, showing the PCR primer binding sites (red arrows) in the intron and an external pol III promoter (blue arrow). Exon A and Exon B represent the sequences present in the mature tRNA. The dotted line indicates a variably-sized intronic region. (B) Reverse transcriptase can transcribe around a tricRNA template many times, resulting in a concatameric cDNA with numerous tandem repeats. (C) RT-PCR primers can bind to multiple sites along the cDNA concatamer, resulting in a ladder of potential PCR products. The formula for the sizes of the bands is: [size of the circle] – [distance between the divergent primers] + [size of circle]n, where n is the number of tandem repeats. (D) The ladder of PCR products can be seen for circles of several different sizes. In this experiment, the distance between the divergent primers is 105 nt, and the sizes of the circles are listed above the gel. For the 259 nt circle, the formula for the ladder is therefore: 259–105+259n. The bands detected on the gel are at 154, 413, 672, and 931 nt.
![Figure 2. tricRNA expression can be detected by RT-PCR. (A) Schematic of the tricRNA expression construct, showing the PCR primer binding sites (red arrows) in the intron and an external pol III promoter (blue arrow). Exon A and Exon B represent the sequences present in the mature tRNA. The dotted line indicates a variably-sized intronic region. (B) Reverse transcriptase can transcribe around a tricRNA template many times, resulting in a concatameric cDNA with numerous tandem repeats. (C) RT-PCR primers can bind to multiple sites along the cDNA concatamer, resulting in a ladder of potential PCR products. The formula for the sizes of the bands is: [size of the circle] – [distance between the divergent primers] + [size of circle]n, where n is the number of tandem repeats. (D) The ladder of PCR products can be seen for circles of several different sizes. In this experiment, the distance between the divergent primers is 105 nt, and the sizes of the circles are listed above the gel. For the 259 nt circle, the formula for the ladder is therefore: 259–105+259n. The bands detected on the gel are at 154, 413, 672, and 931 nt.](/cms/asset/f0cbeb77-50be-45fd-b6f8-21326fa7b60a/krnb_a_1317911_f0002_oc.jpg)
Accurate characterization of circRNA expression can be difficult, however, due to a wide variety of circRNA sizes. Divergent primers are used to specifically amplify circRNA-derived cDNA (). During cDNA preparation, reverse transcriptase (RT) is capable of rolling circle cDNA synthesis on a circular template RNA, resulting in concatemerized cDNA () that could lead to exaggerated measurement of circRNA levels by sequencing or RT-PCR. Indeed, concatemeric reads containing multiple copies of a tRNA intron were detected in Drosophila and C. elegans RNA-seq data sets.Citation10
For larger circles, concatemerization is less likely due to the limited processivity of reverse transcriptase. However, RT-PCR of a short circRNA can produce concatemers that are many times longer than the original transcript.Citation54 Thus, circRNA length must be carefully considered when using RT-based methods to quantify transcript abundance, and direct measurement of circRNA abundance by fluorescence or northern blotting may be preferred. We have successfully expressed ‘designer′ tricRNAs greater than 800 nt in length (), the primary transcript of which is well over 900 nt. To our knowledge, this is the longest known pol III-based transcription unit in eukaryotes. Hence, pol III processivity should not be a limiting factor for many applications aimed at expression of engineered tricRNAs in vivo.
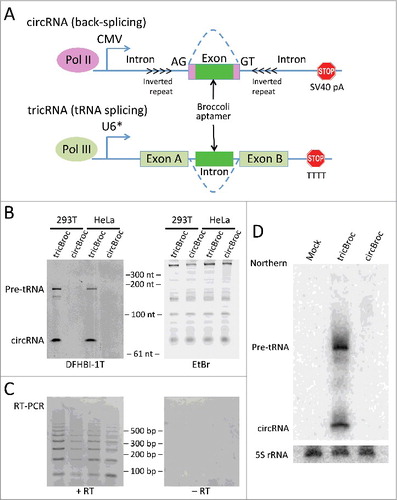
To directly compare the 2 circRNA expression systems, we generated reporter constructs using the Broccoli fluorescent RNA aptamer system ().Citation57 Each construct is designed to express an equivalently-sized aptamer-containing circle (see Methods for details). To differentiate between the 2 systems, we refer to the pol II/back-spliced Broccoli reporter as circBroc, and to the tRNA-spliced Broccoli reporter as tricBroc. Following transient transfection in human HeLa and HEK293T cells, RNA was extracted and analyzed using a convenient and powerful in-gel staining assay ().Citation54,57,58 RT-PCR (which is more sensitive, but less quantitative) was also performed (). As shown, the circBroc back-splicing reporter RNA is detectable by RT-PCR but not by in-gel fluorescence, whereas tricBroc was robustly detected by both methods. The in-gel fluorescence assay allows for detection of as little as 100 pg of Broccoli-containing RNA;Citation58 thus, we can conclude that circBroc is expressed at levels below this detection threshold. In agreement with these data, only tricBroc was detectable by RNA gel blot analysis in 293T cells (). We note that the size of the circBroc exon used here is smaller than those used in previous circRNA expression vectors (76bp compared with 300–1500 bp),Citation25,26,53 although it is certainly on par with the size of many endogenous exons (roughly 80% of human exons are shorter than 200 bp).Citation59 However, back-spliced exons average 690 bp in humansCitation2 and so it remains unclear whether back-splicing efficiency depends on exon size. Nevertheless, our results show that the pol III-based expression system provides more robust expression of circular RNAs (at least for smaller sized ones) than does the pol II-based system. Thus, the tricRNA expression system described here provides a compact and powerful method for in vivo expression of circular RNAs.
Figure 3. Testing in vivo methods of RNA circularization using the Broccoli RNA aptamer as a reporter. (A) Schematic of the 2 constructs, where the bright green box indicates the placement of the Broccoli aptamer sequence. The dotted arcs indicate the splice junctions. The U6* promoter includes the first 27 nucleotides of U6 snRNA in the transcriptional unit. This promotes 5′ cappingCitation60 and enhances stability of the expressed RNA, resulting in a higher yield of tricRNAs.Citation10 (B) In-gel imaging following transient transfection of the reporter constructs into HeLa and 293T cells. The left hand image is the DFHBI-1T stain, which binds to all Broccoli-containing RNAs. In this image, the top band is the pre-tRNA and the lower band is the circular RNA. The doublet of bands in the first lane is most likely due to transcription beginning from both the external U6 promoter, which has a longer 5′ leader sequence, and the internal tRNA promoter. The right hand image is the ethidium bromide stain of the same gel, which marks total RNA. (C) RT-PCR was performed on cDNA generated from the RNA used in . Diverging PCR primers were used (similar to ), such that they only generate products of the appropriate size from a circularized template (see also ). In this experiment, the lengths for tricBroc and circBroc are 77 nt and 76 nt, respectively. (D) Northern blot analysis was performed to quantitatively assess circRNA expression in 293T cells.
Concluding remarks
Despite their conservation across great evolutionary distances, the function of tRNA introns has, for the most part, yet to be elucidated. Many interesting questions remain regarding their role in tRNA biosynthesis (as DNA elements), as well as their fate and function in the cell (as tricRNAs) following removal during processing. As with many non-coding RNAs, characterization of circular RNAs has been greatly aided by the progress of detection technologies and molecular genetic approaches. As detailed here, tricRNA vectors provide an effective means for the orthogonal expression of a wide variety of genetically engineered circRNAs.
Materials and methods
Cloning circBroc reporter constructs
Annealed oligonucleotides were ligated into the pCircGFP plasmid (Zefeng Wang, PICB Shanghai) following digestion with BamHI and SacII to generate a new vector, called pCircBroc. Top oligo: ggtctctttcttccagGAGCGGCCGCGAGACGGTCGGGTCCAGATATTCGTATCTGTCGAGTAGAGTGTGGGCTCGTGGCCGCGGTGTTGAGgtaagtctcgacg, bottom oligo: gatccgtcgagacttacctcaacaccgcggccacgagcccacactctactcgacagatacgaatatctggacccgaccgtctcgcggccgctcctggaagaaagagaccgc. For the top strand oligo, the exon is shown in uppercase letters and the intronic sequence is in lower case. The nucleotides corresponding to Broccoli are underlined.
Cell transfection and RNA isolation
All cells were grown at 37°C with 5% CO2 in DMEM (Gibco) supplemented with 10% fetal bovine serum and 1% penicillin and streptomycin. One million HEK293T or HeLa cells were plated in T-25 flasks and incubated at 37°C. After 24 hours, cells were transfected with equimolar amounts of either circBroc or tricBroc expression vector using Fugene HD transfection reagent (Promega cat#E2311). After 72 hours, total RNA was isolated from cells using Trizol reagent (Thermo Fisher Scientific cat#15596026).
RT-PCR
Total RNA was treated with Turbo DNase (Thermo Fisher Scientific cat#AM2238) and used to prepare cDNA using SuperScript III (Invitrogen cat#18080051) with random hexamer primers. Divergent primers used for circBroc PCR amplification were as follows: F-GTCGAGTAGAGTGTGGGCTCGT, R-GATACGAATATCTGGACCCGACCGTC.
In-gel fluorescence imaging
Ten micrograms of total RNA from transfected cells was run on a Novex 10% TBE-Urea gel (Thermo Fisher Scientific cat#EC68572BOX) for 40 minutes at 300V. After washing 3 times with ddH20, the gel was stained with DFHBI-1T staining solution (40 mM HEPES pH 7.4, 100 mM KCl, 1 mM MgCl2, 10 μm DFHBI-1T [Lucerna cat#410–1 mg]) and fluorescence imaged using a GE Typhoon Trio variable mode imager. To visualize total RNA, the gel was then stained with ethidium bromide and fluorescence imaged.
Northern blotting
Ten micrograms of total RNA was run on a 10% TBE-Urea gel for 40 minutes at 300V and transferred to a nylon membrane at 10 mA overnight. The membrane was dried and UV-crosslinked, followed by pre-hybridization in Rapid-hyb Buffer (GE Healthcare Life Sciences cat#RPN1635) at 42°C for 30 minutes. Oligonucleotide probes were radiolabeled using T4 PNK (NEB) and then purified using Illustra Microspin G-50 columns (GE Healthcare Life Sciences cat#27–5330–71). They were then boiled for 5 minutes and cooled on ice for 2 minutes, and then added to the Rapid-hyb Buffer. The membrane was hybridized at 42°C for one hour with rocking, and then washed at ambient temperature twice for 15 minutes with 2X SSC with 0.1% w/v SDS and twice for 15 minutes with 0.1X SSC with 0.1% w/v SDS at 42°C before overnight PhosphorImager screen exposure. The blot was then imaged using a GE Typhoon Trio variable mode imager.
Probe sequences: anti-Broccoli-5′-gagcccacactctactcgacagatacgaatatctggacccgaccgtctc-3′, anti-5S rRNA- 5′-ccctgcttagcttccgagatcagacgagat-3′.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Zefeng Wang for providing the pCircGFP vector. C.A.S. was supported in part by a National Scientific Foundation Graduate Research Fellowship, DGE-1144081. This work was supported by a Grant from the National Institutes of Health, R01-GM118636, to A.G.M.
References
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453-61; PMID:24811520; https://doi.org/10.1038/nbt.2890
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013; 19:141-57; PMID:23249747; https://doi.org/10.1261/rna.035667.112
- Coqcuerelle C, Mascrez B, Hetuin D, Bailleul B. Mis-splicing yields circular RNA molecules. FASEB J 1993; 7:155-60; PMID:7678559; https://doi.org/10.1261/rna.047126.114
- Lasda E, Parker R. Circular RNAs: Diversity of form and function. RNA 2014; 20:1829-42; PMID:25404635; https://doi.org/10.1261/rna.047126.114
- Petkovic S, Müller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res 2015; 43:2454-65; PMID:25662225; https://doi.org/10.1093/nar/gkv045
- Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol 2016; PMID:27982727; https://doi.org/10.1080/15476286.2016.1271524
- Fischer JW, Leung AKL. CircRNAs: A regulator of cellular stress. Crit Rev Biochem Mol Biol 2017; 9238:1-17; PMID:28095716; https://doi.org/10.1080/10409238.2016.1276882
- Salzman J. Circular RNA expression: Its potential regulation and function. Trends Genet 2016; 32:309-16; PMID:27050930; https://doi.org/10.1016/j.tig.2016.03.002
- Qu S, Zhong Y, Shang R, Zhang X, Song W, Kjems J, Li H. The emerging landscape of circular RNA in life processes. RNA Biol 2016; PMID:27617908; https://doi.org/10.1080/15476286.2016.1220473
- Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Matera AG. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 2015; 21:1554-65; PMID:26194134; https://doi.org/10.1261/rna.052944.115
- Dalgaard JZ, Garrett RA. Protein-coding introns from the 23S rRNA-encoding gene form stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene 1992; 121:103-10; PMID:1427083; https://doi.org/10.1016/0378-1119(92)90167-N
- Kjems J, Garrett RA. Novel splicing mechanism for the ribosomal RNA intron in the archaebacterium desulfurococcus mobilis. Cell 1988; 54:693-703; PMID:3136929; https://doi.org/10.1016/S0092-8674(88)80014-X
- Chen L. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 2016; 17:205-11; PMID:26908011; https://doi.org/10.1038/nrm.2015.32
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-Type specific features of circular RNA expression. PLoS Genet 2013; 9:pgen.1003777; PMID:24039610; https://doi.org/10.1371/journal.pgen.1003777
- Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 2014; 58:870-85; PMID:25921068; https://doi.org/10.1016/j.molcel.2015.03.027
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014; 9:1966-80; PMID:25544350; https://doi.org/10.1016/j.celrep.2014.10.062
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384-8; PMID:23446346; https://doi.org/10.1038/nature11993
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333-8; PMID:23446348; https://doi.org/10.1038/nature11928
- Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010; 6:1-15; PMID:21151960; https://doi.org/10.1371/journal.pgen.1001233
- Seitz H. Redefining MicroRNA targets. Curr Biol 2009; 19:870-3; PMID:19375315; https://doi.org/10.1016/j.cub.2009.03.059
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The rosetta stone of a hidden RNA language? Cell 2011; 146:353-58; PMID:21802130; https://doi.org/10.1016/j.cell.2011.07.014
- Tay Y, Kats L, Salmena L, Weiss D, Tan SM, Ala U, Karreth F, Poliseno L, Provero P, Di Cunto F, et al. Coding-independent regulation of the tumor suppressor PTEN by competing endogenous mRNAs. Cell 2011; 147:344-57; PMID:22000013; https://doi.org/10.1016/j.cell.2011.09.029
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. CircRNA Biogenesis competes with Pre-mRNA splicing. Mol Cell 2014; 56:55-66; PMID:25242144; https://doi.org/10.1016/j.molcel.2014.08.019
- Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A. Exon circularization requires canonical splice signals. Cell Rep 2015; 10:103-11; PMID:25543144; https://doi.org/10.1016/j.celrep.2014.12.002
- Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 2015; 29:2168-82; PMID:26450910; https://doi.org/10.1101/gad.270421.115
- Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28:2233-47; PMID:25281217; https://doi.org/10.1101/gad.251926.114
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015; 160:1125-34; PMID:25768908; https://doi.org/10.1016/j.cell.2015.02.014
- Johnson PJ. Spliceosomal introns in a deep-branching eukaryote: The splice of life. Proc Natl Acad Sci USA 2002; 99:3359-61; PMID:11904397; https://doi.org/10.1073/pnas.072084199
- Woodson SA. Ironing out the kinks: Splicing and translation in bacteria. Genes Dev 1998; 12:1243-7; PMID:9573040; https://doi.org/10.1101/gad.12.9.1243
- Calvin K, Li H. RNA-splicing endonuclease structure and function. Cell Mol Life Sci 2008; 65:1176-85; PMID:18217203; https://doi.org/10.1007/s00018-008-7393-y
- Tocchini-Valentini GD, Fruscoloni P, Tocchini-Valentini GP. Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea. Proc Natl Acad Sci USA 2005; 102:15418-22; PMID:16221764; https://doi.org/10.1073/pnas.0506750102
- Salgia SR, Singh SK, Gurha P, Gupta R. Two reactions of Haloferax volcanii RNA splicing enzymes: joining of exons and circularization of introns. RNA 2003; 9:319-30; PMID:12592006; https://doi.org/10.1261/rna.2118203
- Xue S, Calvin K, Li H. RNA recognition and cleavage by a splicing endonuclease. Science 2006; 312:906-10; PMID:16690865; https://doi.org/10.1126/science.1126629
- Yoshihisa T. Handling tRNA introns, archaeal way and eukaryotic way. Front Genet 2014; 5:1-16; PMID:24567736; https://doi.org/10.3389/fgene.2014.00213
- Chan PP, Lowe TM. GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res 2009; 37:93-7; PMID:18984615; https://doi.org/10.1093/nar/gkn787
- Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res 2012; 40:3131-42; PMID:22140119; https://doi.org/10.1093/nar/gkr1009
- Burggraf S, Larsen N, Woese CR, Stetter KO. An intron within the 16S ribosomal RNA gene of the archaeon Pyrobaculum aerophilum. Proc Natl Acad Sci U S A 1993; 90:2547-50; PMID:8460170; https://doi.org/10.1073/pnas.90.6.2547
- Singh SK, Gurha P, Tran EJ, Maxwell ES, Gupta R. Sequential 2′-O-methylation of archaeal pre-tRNA Trp nucleotides is guided by the intron-encoded but trans-acting box C/D ribonucleoprotein of pre-tRNA. J Biol Chem 2004; 279:47661-71; PMID:15347671; https://doi.org/10.1074/jbc.M408868200
- Trotta CR, Miao F, Arn EA, Stevens SW, Ho CK, Rauhut R, Abelson JN. The yeast tRNA splicing endonuclease: a tetrameric enzyme with two active site subunits homologous to the archaeal tRNA endonucleases. Cell 1997; 89:849-58; PMID:9200603; https://doi.org/10.1016/S0092-8674(00)80270-6
- Di Segni G, Borghese L, Sebastiani S, Tocchini-Valentini GP. A pre-tRNA carrying intron features typical of Archaea is spliced in yeast. RNA 2005; 11:70-6; PMID:15574514; https://doi.org/10.1261/rna.7138805
- Reyes VM, Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell 1988; 55:719-30; PMID:3141064; https://doi.org/10.1016/0092-8674(88)90230-9
- Greer CL, Soll D, Willis I. Substrate recognition and identification of splice sites by the tRNA-splicing endonuclease and ligase from Saccharomyces cerevisiae. Mol Cell Biol 1987; 7:76-84; PMID:3550427; https://doi.org/10.1128/MCB.7.1.76
- Phizicky EM, Schwartz RC, Abelson J. Saccharomyces cerevisiae tRNA ligase. Purification of the protein and isolation of the structural gene. J Biol Chem 1986; 261:2978-86; PMID:3512545
- Englert M, Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res 2005; 33:388-99; PMID:15653639; https://doi.org/10.1093/nar/gki174
- Wang LK, Schwer B, Englert M, Beier H, Shuman S. Structure-function analysis of the kinase-CPD domain of yeast tRNA ligase (Trl1) and requirements for complementation of tRNA splicing by a plant Trl1 homolog. Nucleic Acids Res 2006; 34:517-27; PMID:16428247; https://doi.org/10.1093/nar/gkj441
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lührmann R, Söll D, et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science 2011; 331:760-4; PMID:21311021; https://doi.org/10.1126/science.1197847
- Englert M, Sheppard K, Aslanian A, Yates JR, Söll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc Natl Acad Sci USA 2011; 108:1290-5; PMID:21209330; https://doi.org/10.1073/pnas.1018307108
- Peschek J, Acosta-Alvear D, Mendez AS, Walter P. A conformational RNA zipper promotes intron ejection during non-conventional XBP1 mRNA splicing. EMBO Rep 2015; 16:1688-98; PMID:26483401; https://doi.org/10.15252/embr.201540955
- Jurkin J, Henkel T, Nielsen AF, Minnich M, Popow J, Kaufmann T, Heindl K, Hoffmann T, Busslinger M, Martinez J. The mammalian tRNA ligase complex mediates splicing of XBP1 mRNA and controls antibody secretion in plasma cells. EMBO J 2014; 33:2922-36; PMID:25378478; https://doi.org/10.15252/embj.201490332
- Kosmaczewski SG, Edwards TJ, Han SM, Eckwahl MJ, Meyer BI, Peach S, Hesselberth JR, Wolin SL, Hammarlund M. The RtcB RNA ligase is an essential component of the metazoan unfolded protein response. EMBO Rep 2014; 15:1278-85; PMID:25366321; https://doi.org/10.15252/embr.201439531
- Lu Y, Liang FX, Wang X. A synthetic biology approach identifies the mammalian UPR RNA ligase RtcB. Mol Cell 2014; 55:758-70; PMID:25087875; https://doi.org/10.1016/j.molcel.2014.06.032
- Sidrauski C, Cox JS, Walter P. tRNA ligase is required for regulated mRNA splicing in the unfolded protein response. Cell 1996; 87:405-13; PMID:8898194; https://doi.org/10.1016/S0092-8674(00)81361-6
- Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA 2015; 21:172-9; PMID:25449546; https://doi.org/10.1261/rna.048272.114
- Schmidt CA, Noto JJ, Filonov GS, Matera AG. A method for expressing and imaging abundant, stable, circular RNAs in vivo using tRNA splicing. Methods Enzymol 2016; 572:215-36. PMID:27241756; https://doi.org/10.1016/bs.mie.2016.02.018
- Lebbink RJ, Lowe M, Chan T, Khine H, Wang X, McManus MT. Polymerase II promoter strength determines efficacy of microRNA adapted shRNAs. PLoS One 2011; 6:e26213; PMID:22031824; https://doi.org/10.1371/journal.pone.0026213
- Qin JY, Zhang L, Clift KL, Hulur I, Xiang AP, Ren BZ, Lahn BT. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One 2010; 5:e10611; PMID:20485554; https://doi.org/10.1371/journal.pone.0010611
- Filonov GS, Moon JD, Svensen N, Jaffrey SR. Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J Am Chem Soc 2014; 136:16299-308; PMID:25337688; https://doi.org/10.1021/ja508478x
- Filonov GS, Kam CW, Song W, Jaffrey SR. In-gel imaging of RNA processing using broccoli reveals optimal aptamer expression strategies. Chem Biol 2015; 22:649-60; PMID:26000751; https://doi.org/10.1016/j.chembiol.2015.04.018
- Sakharkar MK, Chow VTK, Kangueane P. Distributions of exons and introns in the human genome. In Silico Biol 2004; 4:387-93; PMID:15217358
- Good P, Krikos A, Li S, Bertrand E, Lee N, Giver L, Ellington A, Zaia J, Rossi J, Engelke D. Expression of small, therapeutic RNAs in human cell nuclei. Gene Ther 1997; 4:45-54; PMID:9068795; https://doi.org/10.1038/sj.gt.3300354
