Abstract
Chemical modification of nucleobases plays an important role for the control of gene expression on different levels. That includes the modulation of translation by modified tRNA-bases or silencing and reactivation of genes by methylation and demethylation of cytosine in promoter regions. Especially dynamic methylation of adenine and cytosine is essential for cells to adapt to their environment or for the development of complex organisms from a single cell. Errors in the cytosine methylation pattern are associated with most types of cancer and bacteria use methylated nucleobases to resist antibiotics. This Point of View wants to shed light on the known and potential chemistry of DNA and RNA methylation and demethylation. Understanding the chemistry of these processes on a molecular level is the first step towards a deeper knowledge about their regulation and function and will help us to find ways how nucleobase methylation can be manipulated to treat diseases.
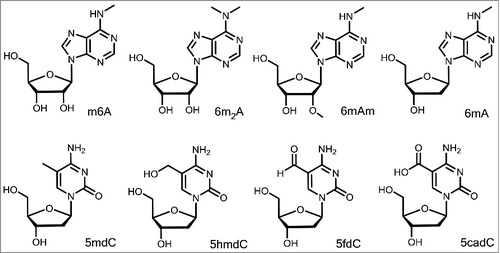
Since the discovery of (deoxy)adenosine (dA, A), (deoxy)cytidine (dC, C), (deoxy)guanosine (dG, G), (deoxy)thymidine (dT, T) and uracil (U) in the early 20th century as the information carrying building blocks, which form the basis for RNA and DNA, various modifications of these nucleosides were discovered ().Citation1,2 Particularly in transfer-RNA (tRNA) but also in rRNA (rRNA), modified bases are central elements, needed to fine tune the translation of the genetic code.Citation3-6 In rRNA of bacterial pathogens, many methylated bases are present to block binding of small molecules that work as translation inhibitors, resulting in a resistance against antibiotics such as aminoglycosides.Citation7 More recently it was discovered that also mRNA (mRNA) contains modified bases. Although it is not yet fully understood what the function of these bases are, it was revealed that the modification chemistry is to some extent reversible. This suggests that the modification and de-modification chemistry has a novel and yet unexplored regulatory function. In this regard N6-methylated adenine (m6A) is the best analyzed modification, but most recently also the reversible formation of N6,C2′-dimethyl adenine (m6Am) was discovered. According to current knowledge, reversible chemistry on modified RNA bases is limited to methyl groups, which are introduced by methyltransferases and removed by demethylases. DNA, in contrast, as the prime carrier of genetic information in the biosphere, is structurally less complex and only few modified bases are known. Most prominent is the methylated base 5-methyl deoxycytosine (5mdC). Ideas about the potential chemistry of methylations and demethylation are the focus of this review. For other aspects, the following excellent reviews can be consulted.Citation1,2
5mdC is the most abundant modified base in genomic DNA of eukaryotes and also present in the DNA of prokaryotes.Citation8-10 In mammals, 5mdC typically reaches global levels between 1 and 5% in genomic DNA.Citation11,12 Methylated adenine (6mA), which is the DNA equivalent to m6A in RNA, is another DNA modification that is under intensive investigation at the moment. Whereas 6mA is a well-characterized modification in bacterial DNA, its presence was only recently shown in several higher eukaryotic organisms.Citation13,14 In Caenorhabditis elegans, where 5mdC is not detectable, 6mA is dynamically regulated and linked to other epigenetic marksCitation15 and in early embryos of Drosophila melanogaster, 6mA levels are high, but decrease fast during development, resulting in very low 6mA levels in adult tissue.Citation16 In the unicellular green alga Chlamydomonas reinhardtii, 6mA was discovered in 84% of the genes, where it is mainly located at transcription start sites.Citation17 Recently, it was reported that mammalian DNA, including human and mouse, also contains 6mA.Citation13,18 There, 6mA seems to be distributed across the genome, but absent in gene exons,Citation19 and 6mA-demethylation in mouse embryonic stem cell (mESC) DNA was shown to correlate with ALKBH1 depletion.Citation18 These findings question the previous paradigm that DNA modifications in mammalian genome are limited to cytosine residues. However, when our group tried to confirm these results by a novel ultrasensitive UHPLC-MS method, we were not able to detect 6mA in mESC DNA or DNA from mouse tissue, whereas Chlamydomonas DNA, which served as a positive control, delivered the expected positive result.Citation20 These observations suggest that 6mA might be present at defined time points in mammalian DNA, but is not an epigenetic mark. In the coming years, the question whether 6mA is a relevant modification in mammalian DNA or not will thus certainly be under intensive investigation.
Chemistry of RNA and DNA base methylation
The addition of the methyl-group to DNA and RNA bases () is catalyzed by DNA- and RNA-methyltransferases that use S-adenosyl-methionine (SAM) as an active methyl-group donor.Citation21-23 While the methyltransferases that methylate RNA bases are now under extensive investigations, the enzymes that catalyze the methylation of dC in DNA are well characterized. In mammalian cells, 3 active DNA-methyltransferases (DNMTs: DNMT1, DNMT3a and DNMT3b) exist.Citation24,25 DNMT3a and 3b are de novo DNMTs, which methylate canonical dC bases.Citation26 In contrast, DNMT1 maintains the methylation status during cell division. DNMT1 operates on hemi-methylated DNA during replication, where the template strand is already methylated, but the newly synthesized strand is lacking methylation.Citation27 As such, DNMT1 converts the methylation of dC into an inheritable modification that can be transferred during reproduction.Citation28,29
Figure 2. Mechanism of methylation leading to the formation of het-CH3 and C-CH3 connectivities in RNA and DNA.
DNMTs and thus cytosine methylation is essential in those multicellular organisms, where it exists. The presence or absence of 5mdC is associated with various important cellular functions, such as transcription control, X-chromosome silencing and genomic imprinting.Citation28 A global deletion of only one of the 3 DNMTs leads to severe cellular aberrations and is therefore lethal in early embryogenesis (DNMT1 and 3b) or postnatal (DNMT3a).Citation26,30 During differentiation the “methylome” is highly dynamic and a celltype-characteristic 5mdC pattern is established during this process.Citation31 While 5mdC is located to a CpG-dinucleotide context in the majority of somatic cells, non-CpG methylation is also present in embryonic stem cells, many pluripotent progenitor cells and adult brain. However, CpG-methylation is also dominating here.Citation32-34 Cytosine-methylation in vertebrates occurs in all types of DNA sequence contexts, including repetitive and regulatory sequences, genes and transposable elements; in contrast to invertebrates, where mostly repetitive sequences are methylated.Citation35 The majority of cytosines in a CpG-context, depending on the cell type up to 80%, are methylated, leaving so-called CpG islands (CGI) of actively transcribed genes as unmethylated patterns in a CpG-context.Citation36,37 CGIs are regions of high CpG frequency over a length of at least 500 base pairs compared with the bulk genomic DNA and found in 40% of promoter regions in the mammalian genome, with even higher levels (60%) in the human genome.Citation38,39 Symmetric methylation of CpG:GpC islands is consequently a hallmark of silenced genes.Citation40,41
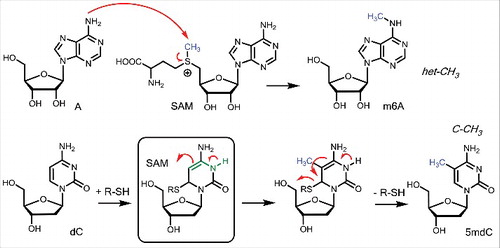
The enzymatic mechanism of how methyltransferases methylate DNA and RNA bases is shown in . Centers with a certain nucleophilicity like the amino group of the RNA base A can attack the SAM coenzyme directly leading to immediate methylation. This type of direct methylation is certainly operating for the formation of 6m2A, 4mC or m6Am. SAM as nature's “methyl iodide” is hence reactive enough to methylate even weak nucleophilic centers such as the exocyclic amino groups of A, which feature, as an sp2-hybridized N-atom only a very weak nucleophilic lone pair at the N-atom. This type of direct methylation creates bases, which possess the methyl group attached to a heteroatom establishing a het-CH3 system. This will be important in the context of active demethylation (vide infra).
In contrast to the formation of het-CH3 connections, methylation of the dC base in DNA at position C5 is far more complex. The C5-center features no nucleophilicity at all, making direct methylation impossible. Nature solves this problem by exploiting a helper nucleophile (R-SH, ). The DNMT enzymes attack the dC base first with a nucleophilic thiol in a 1,6 addition reaction. This establishes a nucleophilic enamine substructure (green in ), which can subsequently be methylated with the SAM cofactor. Importantly, the helper nucleophile is subsequently eliminated, thereby re-establishing the aromatic system. This more complex enzymatic transformation allows nature to methylate non-nucleophilic carbon atoms to create C-CH3 connectivities which feature a strong and stable C-C single bond.
Chemistry of demethylation
To establish the reversibility needed for switching biochemical processes, nature requires to remove the attached methyl groups. Removal of het-CH3 groups found predominantly in RNA was found to occur with the help of α-ketoglutarate (α-KG) dependent oxidases. These proteins contain a reactive Fe(II) center, which reacts to a strongly oxidizing Fe(IV) = O species with oxygen under concomitant decarboxylation of α-KG to succinate ().Citation42 The Fe(IV) = O species is able to abstract a H-atom from the het-CH3 group to form a het-stabilized het-CH2• radical, which reacts with the Fe-bound hydroxylradical to form a het-CH2-OH hemiaminal/acetal functionality.
Figure 3. Oxidation of m6A followed by decomposition of the hemiaminal to A and oxidation if 5mdC to stable 5hmdC.
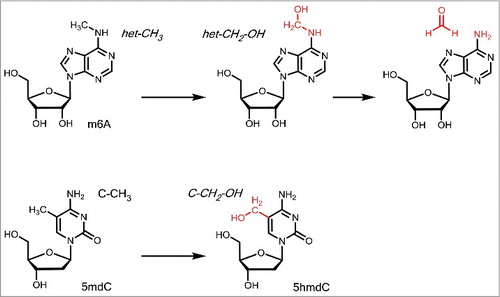
However, these structures are unstable. In water, they decompose in a spontaneous reaction under loss of formaldehyde to give the unmethylated compound. It is interesting that formaldehyde is formed as a byproduct of this reaction because it is typically a rather toxic compound. It needs to be seen how this molecule is detoxified in the context of the demethylation reaction. Particularly well studied is the removal of the N6-methyl group from m6A to revert into the canonical RNA base A. So far 2 α-KG dependent oxidases were found to catalyze the oxidation. One is the fat mass and obesity-associated protein (FTO) protein and the second is ALKBH5. It was shown, that knockdown of FTO led to increased amounts of m6A and in turn overexpression of FTO resulted in decreased m6A levels.Citation43 Alkbh5-deficient mice had a similar effect as FTO knock-down in human cells and resulted in increased m6A levels of the mRNA.Citation44 The demethylation activity of both proteins is comparable, although ALKBH5 shows direct demethylation, whereas FTO-mediated demethylation is supposed to create hm6A and f6A as intermediates.Citation42,44
In 2009 it was found that also 5mdC is further enzymatically oxidized in a stepwise fashion to give first 5-hydroxymethyldesoxycytosine (5hmdC), followed by 5-formyldesoxycytosine (5fdC) and 5-carboxydesoxycytosine (5cadC). “Ten-11 translocation” (TET) enzymes, which are Fe2+/α-KG dependent dioxygenases, were discovered to catalyze this iterative 5mdC oxidation reaction.Citation45,46 Regarding the first oxidation step that transforms 5mdC to 5hmdC, the Fe2+/α-KG catalyzed reaction generates a stable C-CH2-OH connectivity, which is as a primary alcohol stable in water (). 5hmdC is consequently a stable DNA base modification and it was suggested that the base has indeed epigenetic functions. For example, 5hmdC constitutes 0.6% of all nucleotides in Purkinje neurons, a special neural cell type of the cerebellum, and 0.032% of all nucleotides in embryonic stem (ES) cells.Citation45,47 The highest 5hmdC levels in fully differentiated tissues were found in the brain with up to 1% of all cytosines.Citation48,49 Evidence accumulates that 5hmdC in a given gene is able to accelerate transcription and it is not surprising that 5hmdC is mainly present in the promoter of actively transcribed genes.Citation50,51
TET enzymes are in this sense required to orchestrate the transcriptional activity of genes. In vertebrates, TET proteins exist in 3 different types (TET1 – TET3) that do not differ regarding their chemistry, but seem to have different spatio-temporal activity. Whereas TET1 is mostly expressed in stem cells, TET3 is upregulated during differentiation and the most abundant TET enzyme in fully differentiated cells.Citation52-54 A global TET3-knockout is lethal in embryogenesis, because it prevents epigenetic reprogramming during differentiation.Citation55 It is interesting, that the presence of 5hmdC in mammalian DNA was described first already in 1972.Citation56 It took more than 30 y to confirm that 5hmdC is really present in substantial amounts that are highly depending on the cell and tissue type.Citation57
The further oxidized bases 5fdC and especially 5cadC () could not be associated yet with distinct cellular functions, but for 5fdC it was reported that it might have regulatory purposes and is also a stable epigenetic mark.Citation58 In accordance with these previous findings, a recently reported single-cell 5fdC-sequencing method called CLEVER-seq revealed that the generation of 5fdC in promoter regions precedes the upregulation of gene expression.Citation59 Despite this faint evidence for epigenetic functions, 5fdC and 5cadC are currently mainly considered to be intermediates on the way of an active DNA demethylation process. DNA demethylation is a crucial process of cell development. Especially during fertilization (paternal part of the genome), early embryogenesis (maternal part of the genome) and the development of germ cells, DNA demethylation takes place in a genome-wide manner, allowing a broad reprogramming of the fertilized oocyte and the cells in the early embryo.Citation60-63 But not only during development, also in fully differentiated cells, it occurs at specific sites of the genome. In brain, for example, locus-specific DNA demethylation and de novo methylation is induced by neural activation, arguing that DNA demethylation is important for normal brain function, including memory formation and learning.Citation64-66 DNA demethylation can take place either actively, which means replication-independent, or passively when DNMT1 does not methylate the nascent DNA strand in hemi-methylated DNA after replication. Passive demethylation occurs, when DNMT1 is absent or blocked during the replication process, which happens for example during early embryogenesis to ensure the demethylation of the maternal genome.Citation67 Interestingly, 6mA demethylation in Drosophila is catalyzed by Drosophila's TET homolog (DMAD or dTet). DMAD depletion results in higher 6mA levels, but unchanged 5mdC patterns, and is lethal at pupa stage or shortly after.Citation16 DMAD and TET possess similar catalytic active Cys-rich and DSBH domains, however, 6mA-demethylation activity was not observed yet for mammalian TET enzymes.Citation16
Figure 4. Potential mechanism of chemically induced active demethylation with a potential immediate re-methylation.
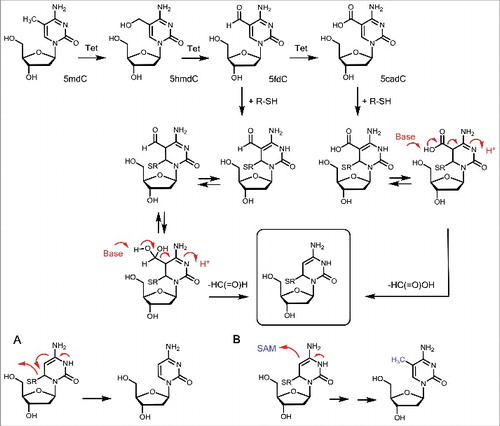
Although oxidation of 5hmdC to 5fdC and 5cadC creates stable molecules due to the lack of a het-atom in β-position, it is discussed that both could be turned into unstable structures upon further chemical manipulation. A chemically attractive mechanism requires that 5fdC and 5cadC are attacked by a helper nucleophile, preferentially a thiol group at the C6 position, in a Michael-type reaction (). Hydratization of 5fdC and tautomerization of the reacted 5fdC and 5cadC allows us to formulate a “β-imino-type” substructure that is prone to deformylation and decarboxylation (red arrows in ). Indeed, we could show that reaction of 5fdC and 5cadC with a thiol-nucleophile leads to spontaneous deformylation and decarboxylation showing that the suggested chemistry is feasible. There is currently no evidence that this type of chemistry occurs in vivo but we could show that stem cell lysates feature a decarboxylating activity.Citation68 Interesting is the observation that deformylation and decarboxylation of 5fdC and 5cadC after reaction with a thiol nucleophile leads to a reaction intermediate (boxed in and ) that is the key intermediate observed already during methylation of dC to 5mdC by the DNMTs. It is therefore tempting to speculate that DNMT enzymes are involved in the deformylation and decarboxylation maybe followed by immediate re-methylation. Although this reaction sequence would follow chemical logic, it needs to clarified in the near future, if such reactions occur indeed in nature. It was, however, shown that C5-DNA-methyltransferases are indeed able to remove formaldehyde from 5hmdC, converting 5hmdC directly to dC, therefore supporting these ideas.Citation69
In this context, it is interesting to note that 5hmC and 5fC were also discovered in RNA. In human cells at tRNA position C34, the oxidation of the corresponding RNA base 5mrC to 5frC is catalyzed by the Fe2+/α-KG dependent enzyme ALKBH1, which is also responsible for m1 A demethylation in mammalian tRNA.Citation70,71 Interestingly, 5hmrC was not detected as an intermediate in the ALKBH1-dependent 5mrC oxidation.Citation70 In Drosophila, 5hmrC was discovered in polyadenylated RNA and is associated with enhanced mRNA-translation efficiency back to normal level, when 5mrC has lowered the efficiency.Citation72 Surprisingly, the oxidation reaction is catalyzed by Drosophila's TET homolog dTet that is also responsible for 6mA demethylation, but does not oxidize 5mdC.Citation16,72 Moreover, there is evidence that TET enzymes are also responsible for 5mrC oxidation,Citation73,74 but at the moment it is not clear whether TET-mediated 5hmrC or 5frC formation are stable or rather transient modifications.
In contrast to the chemical mechanism of active demethylation discussed above, strong evidence exists that active demethylation via formation of 5fdC and 5cadC is also linked to base excision repair (BER), which repairs also mismatches caused by deamination of 5hmdC to 5hmdU (). This mechanism includes excision of 5dfC or 5cadC and subsequent activation of BER. The dG/dT mismatch specific thymine DNA glycosylase (TDG) recognizes dG/dT mismatches, but with an even higher activity it excises 5fdC or 5cadC, but not 5mdC and 5hmdC, in vitro.Citation75 This reactivity was not observed for other DNA glycosylases. Evidence that TDG excises 5fdC and 5cadC also in vivo is given by the fact that 5fdC and 5cadC levels are 5–10 times increased in TDG-deficient ES cells compared with the wildtype.Citation76 However, TET/TDG-mediated demethylation is very unlikely to be the only demethylation mechanism. It rather occurs at defined promoter regions in the genome than in a genome-wide manner. First, TDG-activity causes abasic sites.Citation77 If this happened genome-wide, it may impair genomic stability, which is crucial for correct development. Second, TDG knockout starts to be lethal not before embryonic day 12.5 and TDG levels are very low in the zygote, where the paternal genome is demethylated.Citation78,79
Figure 5. Active demethylation via base excision repair. Two possibilities are discussed: A direct removal of 5fdC and 5cadC in xdC:dG base pairs or removal of a deaminated 5hmdU in a 5hmdU:dG mismatch by BER glycosylase.
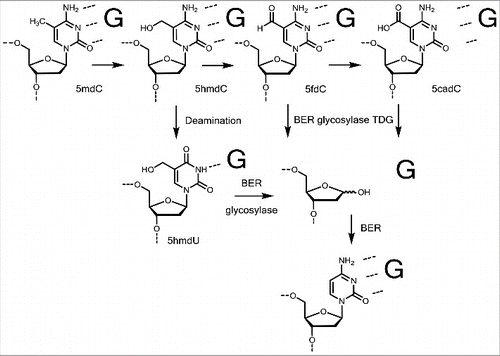
Most recently it was suggested that nature may not need to oxidize 5mdC to 5fdC and 5cadC for demethylation and that a third TET-independent pathway has to exist. In the zygote, the most drastic demethylation occurs when 5mdC is globally erased from the paternal part of the genome, while the maternal part is shielded from demethylation. DNA-demethylation of the paternal pro-nuclei is replication- and TET-independent, since 5hmdC levels increase after 5mdC levels have dropped and global demethylation can be detected in Tet3-deficient zygotes.Citation80 It might be that deamination of genomic 5mdC to dT and subsequent dT/dG mismatch repair are the mechanism behind this observation.Citation81 However, this would also impair genomic stability.
Implication of misguided methylation and demethylation
Whereas the distribution of 5mdC and 5hmdC is tightly regulated to ensure the anticipated functionality of a cell and its response to DNA damage, one hallmark of cancer cells is their completely different methylation and hydroxyl-methylation pattern.Citation82,83 In many cancer types, the global methylation levels are decreased, while promoter regions of important regulatory and tumor suppressor genes are hypermethylated and therefore silenced.Citation84 One example is the hypermethylation of the promoter region of HIC1, which is a transcriptional repressor of SIRT1, a survival protein (proto-oncogene) that is consequently upregulated.Citation85 It was also shown that certain CGIs are coordinately methylated in some tumor cells, which is called “CpG island methylator phenotype.”Citation86-88 Since TET1 was initially discovered in 2002 as a fusion protein to MLL1 H3K4 methyltransferase in patients with acute lymphoblastic leukemia, which is characterized by mutations in the MLL1 protein, it was considered to be an oncogene.Citation89 Only when the biologic function of the TET enzymes was elucidated in 2009, it was proven a few years later that TET enzymes are actually tumor suppressors that are silenced in various types of tumors. Decreased levels and activity of TET1 and therefore reduced 5hmdC levels are associated with haematopoietic malignancies, colon, breast, prostate, liver and lung cancers, which show greater levels of proliferation and for breast cancer increased invasion rates as a direct consequence of TET1 downregulation.Citation90-93 TET2 mutations and consequently decreased 5hmdC levels occur in various myeloid malignancies, including chronic myelomonocytic leukemia, myeloid proliferative neoplasm and acute myeloid leukemia.Citation82,94,95 Mutations in the genes of isocitrate dehydrogenase (IDH) 1 and 2 lead to the production of D-2-hydroxyglutarate (D-2HG), a metabolite that inhibits TET2 activity for example in AML and MPN, but also in malignant gliomas, resulting in a dramatic decrease of 5hmdC levels.Citation96 Interestingly, IDH1/2 and TET2 mutations seem to be mutually exclusive in these types of tumor, with IDH1/2 mutations being the ones with higher oncogenic potential.Citation97,98 Recent results show that not only mutations in TET genes or their inhibition by cancer metabolites are important for tumorigenesis, but also tumor hypoxia is responsible for reduced TET activity.Citation99
There is more and more evidence that epigenetics and metabolism are closely connected not only via D-2HG in cancer metabolism, but also in normal cells.Citation100,101 As an intermediate in the tri-citric acid (TCA) cycle and part of nitrogen catabolism through deamination of glutamate, α-KG is one of the key metabolites. Since it is the co-substrate of TET enzymes and other dioxygenases involved in epigenetic regulation, such as histone lysine demethylase, it links epigenetics directly to metabolism. Levels of α-KG are rate limiting for TET activity and higher α-KG levels result in higher TET activity with direct impact on differentiation processes.Citation102 Depending on the cell type and status, α-KG can either promote self-renewal or induce differentiation.Citation103 In brown adipose tissue (BAT) development, for example, TET3 mediates cell commitment to BAT by demethylating the Prdm16 promoter. AMP activated protein kinase α1 (AMPKα1) influences α-KG levels positively and therefore increase TET3 activity.Citation104 Glutamate and glutamine metabolism increases α-KG levels, leading to self-renewal in pluripotent mouse embryonic stem cells, while succinate supply leads to differentiation.Citation102 Additionally, succinate and also fumarate, another 2 intermediates of TCA cycle, show an inhibitory effect on TET enzymes in vitro.Citation105
In the future, it will be challenging not only to prove the existence, but to reveal the distinct biologic functions of the various DNA and RNA modifications that exist. The role of the modified bases in mRNA are currently under extensive investigation and for DNA, especially the functions of 5hmdC in regulatory and learning processes in brain, but also during development and in cancer cells are of great interest.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
FRT thanks the Boehringer Ingelheim Fonds for a PhD fellowship. We thank the Deutsche Forschungsgemeinschaft for financial support through the programs: SFBs 646, 749 and 1032, as well as the SPP1784. Further support is acknowledged from the Excellence Cluster CiPSM (Center for Integrated Protein Science).
References
- Breiling A, Lyko F. Epigenetic regulatory functions of DNA modifications: 5-methylcytosine and beyond. Epigenetics Chromatin 2015; 8:24; PMID:26195987; https://doi.org/10.1186/s13072-015-0016-6
- Chen K, Zhao BS, He C. Nucleic acid modifications in regulation of gene expression. Cell Chem Biol 2016; 23:74-85; PMID:26933737; https://doi.org/10.1016/j.chembiol.2015.11.007
- Duechler M, Leszczynska G, Sochacka E, Nawrot B. Nucleoside modifications in the regulation of gene expression: Focus on tRNA. Cell Mol Life Sci 2016; 73:3075-95; PMID:27094388; https://doi.org/10.1007/s00018-016-2217-y
- Penzo M, Galbiati A, Trere D, Montanaro L. The importance of being (slightly) modified: The role of rRNA editing on gene expression control and its connections with cancer. Biochim Biophys Acta 2016; 1866:330-8; PMID:27815156; https://doi.org/10.1016/j.bbcan.2016.10.007
- Sloan KE, Warda AS, Sharma S, Entian KD, Lafontaine DL, Bohnsack MT. Tuning the ribosome: The influence of rRNA modification on eukaryotic ribosome biogenesis and function. RNA Biol 2016:1-16; PMID:27911188; https://doi.org/10.1080/15476286.2016.1259781
- El Yacoubi B, Bailly M, de Crecy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet 2012; 46:69-95; PMID:22905870; https://doi.org/10.1146/annurev-genet-110711-155641
- Lioy VS, Goussard S, Guerineau V, Yoon E-J, Courvalin P, Galimand M, Grillot-Courvalin C. Aminoglycoside resistance 16S rRNA methyltransferases block endogenous methylation, affect translation efficiency and fitness of the host. RNA 2014; 20:382-91; PMID:24398977; https://doi.org/10.1261/rna.042572.113
- Ehrlich M, Wang RY-H. 5-Methylcytosine in eukaryotic DNA. Science 1981; 212:1350-7; PMID:6262918; https://doi.org/10.1126/science.6262918
- Kahramanoglou C, Prieto AI, Khedkar S, Haase B, Gupta A, Benes V, Fraser GM, Luscombe NM, Seshasayee AS. Genomics of DNA cytosine methylation in escherichia coli reveals its role in stationary phase transcription. Nat Commun 2012; 3:886; PMID:22673913; https://doi.org/10.1038/ncomms1878
- Ehrlich M, Gama-Sosa MA, Carreira LH, Ljungdahl LG, Kuo KC, Gehrke CW. DNA methylation in thermophylic bacteria: N4-methylcytosine, 5-methylcytosine, and N6-methyladenine. Nucleic Acids Res 1985; 13:1399-412; PMID:4000939; https://doi.org/10.1093/nar/13.4.1399
- Ehrlich M, Gama-Sosa MA, Huang L-H, Midgett RM, Kuo KC, McCune RA, Gehrke C. Amount and distribution of 5-methylcytosine in human DNA from different types of tissues or cells. Nucleic Acids Res 1982; 10:2709-21; PMID:7079182; https://doi.org/10.1093/nar/10.8.2709
- Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 2010; 5:e15367; PMID:21203455; https://doi.org/10.1371/journal.pone.0015367
- Koziol MJ, Bradshaw CR, Allen GE, Costa AS, Frezza C, Gurdon JB. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol 2016; 23:24-30; PMID:26689968; https://doi.org/10.1038/nsmb.3145
- Liu J, Zhu Y, Luo GZ, Wang X, Yue Y, Wang X, Zong X, Chen K, Yin H, Fu Y. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat Commun 2016; 7:13052; PMID:27713410; https://doi.org/10.1038/ncomms13052
- Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizabal-Corrales D, Hsu CH, Aravind L, He C, Shi Y. DNA methylation on N6-Adenine in C. elegans. Cell 2015; 161:868-78; PMID:25936839; https://doi.org/10.1016/j.cell.2015.04.005
- Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J, et al. N6-methyladenine DNA modification in drosophila. Cell 2015; 161:893-906; PMID:25936838; https://doi.org/10.1016/j.cell.2015.04.018
- Fu Y, Luo GZ, Chen K, Deng X, Yu M, Han D, Hao Z, Liu J, Lu X, Doré LC, et al. N6-methyldeoxyadenosine marks active transcription start sites in chlamydomonas. Cell 2015; 161:879-92; PMID:25936837; https://doi.org/10.1016/j.cell.2015.04.010
- Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature 2016; 532:329-33; PMID:27027282; https://doi.org/10.1038/nature17640
- Koziol MJ, Bradshaw CR, Allen GE, Costa AS, Frezza C, Gurdon JB. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol 2016; 23:24-30; PMID:26689968; https://doi.org/10.1038/nsmb.3145
- Schiffers S, Ebert C, Rahimoff R, Kosmatchev O, Steinbacher J, Bohne A-V, Spada F, Michalakis S, Nickelsen J, Müller M, et al. Quantitative LC–MS provides no evidence for m6da or m4dc in the genome of mouse embryonic stem cells and tissues. Angew Chem Int Ed 2017; PMID:28371147; https://doi.org/10.1002/anie.201700424
- Kumar S, Cheng X, Klimasauskas S, Mi S, Postfai J, Roberts RJ, Wilson GG. The DNA (cytosine-5) methyltransferases. Nucleic Acids Res 1993; 22:1-10; PMID:8127644; https://doi.org/10.1093/nar/22.1.1
- Smith SS, Kaplan BE, Sowers LC, Newman EM. Mechanism of human methyl-directed DNA methyltransferases and the fidelity of cytosine methylation. Proc Natl Acad Sci USA 1992; 89:4744-8; PMID:1584813; https://doi.org/10.1073/pnas.89.10.4744
- Hori H. Methylated nucleosides in tRNA and tRNA methyltransferases. Front Genet 2014; 5:144; PMID:24904644; https://doi.org/10.3389/fgene.2014.00144
- Bestor T, Laudano A, Mattaliano R, Ingram V. Cloning and sequencing of a cDNA encoding dna methyltransferase of mouse cells. J Mol Biol 1988; 203:971-83; PMID:3210246; https://doi.org/10.1016/0022-2836(88)90122-2
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet 1998; 19:219-20; PMID:9662389; https://doi.org/10.1038/890
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99:247-57; PMID:10555141; https://doi.org/10.1016/S0092-8674(00)81656-6
- Pradhan S, Bacolla A, Wells RD, Roberts RJ. Recombinant human DNA (Cytosine-5) methyltransferase. J Biol Chem 1999; 274:33002-10; PMID:10551868; https://doi.org/10.1074/jbc.274.46.33002
- Klose RJ, Bird AP. Genomic DNA methylation: The mark and its mediators. Trends Biochem Sci 2006; 31:89-97; PMID:16403636; https://doi.org/10.1016/j.tibs.2005.12.008
- Spada F, Haemmer A, Kuch D, Rothbauer U, Schermelleh L, Kremmer E, Carell T, Längst G, Leonhardt H. DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells. J Cell Biol 2007; 176:565-71; PMID:17312023; https://doi.org/10.1083/jcb.200610062
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992; 69:915-26; PMID:1606615; https://doi.org/10.1016/0092-8674(92)90611-F
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 2011; 480:490-5; PMID:22170606; https://doi.org/10.1038/nature10716
- Ramsahoye BH, Biniszkiewicz D, Lyko F, Clark V, Bird AP, Jaenisch R. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc Natl Acad Sci USA 2000; 97:5237-42; PMID:10805783; https://doi.org/10.1073/pnas.97.10.5237
- Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009; 462:315-22; PMID:19829295; https://doi.org/10.1038/nature08514
- Xie W, Barr CL, Kim A, Yue F, Lee AY, Eubanks J, Dempster EL, Ren B. Base-resolution analyses of sequence and parent-of-origin dependent DNA methylation in the mouse genome. Cell 2012; 148:816-31; PMID:22341451; https://doi.org/10.1016/j.cell.2011.12.035
- Suzuki MM, Bird A. DNA methylation landscapes: Provocative insights from epigenomics. Nat Rev Genet 2008; 9:465-76; PMID:18463664; https://doi.org/10.1038/nrg2341
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell 1985; 40:91-9; PMID:2981636; https://doi.org/10.1016/0092-8674(85)90312-5
- Bird AP. CpG-rich islands and the function of DNA methylation. Nature 1986; 321:209-13; PMID:2423876; https://doi.org/10.1038/321209a0
- Antequera F, Bird A. Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci USA 1993; 90:11995-9; PMID:7505451; https://doi.org/10.1073/pnas.90.24.11995
- Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA. Footprinting of mammalian promoters: Use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res 2005; 33:e176; PMID:16314307; https://doi.org/10.1093/nar/gni180
- Boyes J, Bird A. DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell 1991; 64:1123-34; PMID:2004419; https://doi.org/10.1016/0092-8674(91)90267-3
- Siegfried Z, Eden S, Mendelsohn M, Feng X, Tsuberi B-Z, Cedar H. DNA methylation represses transcription in vivo. Nat Genet 1999; 22:203-6; PMID:10369268; https://doi.org/10.1038/9727
- Zou S, Toh JDW, Wong KHQ, Gao Y-G, Hong W, Woon ECY. N(6)-methyladenosine: A conformational marker that regulates the substrate specificity of human demethylases FTO and ALKBH5. Sci Rep 2016; 6:25677; PMID:27156733; https://doi.org/10.1038/srep25677
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol 2011; 7:885-7; PMID:22002720; https://doi.org/10.1038/nchembio.687
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vågbø CB, Shi Y, Wang WL, Song SH, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell 2013; 49:18-29; PMID:23177736; https://doi.org/10.1016/j.molcel.2012.10.015
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 2009; 324:930-5; PMID:19372391; https://doi.org/10.1126/science.1170116
- Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, He C, Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011; 333:1300-3; PMID:21778364; https://doi.org/10.1126/science.1210597
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in purkinje neurons and the brain. Science 2009; 324:929-30; PMID:19372393; https://doi.org/10.1126/science.1169786
- Bachman M, Uribe-Lewis S, Yang X, Williams M, Murrell A, Balasubramanian S. 5-Hydroxymethylcytosine is a predominantly stable DNA modification. Nat Chem 2014; 6:1049-55; PMID:25411882; https://doi.org/10.1038/nchem.2064
- Wagner M, Steinbacher J, Kraus TF, Michalakis S, Hackner B, Pfaffeneder T, Perera A, Müller M, Giese A, Kretzschmar HA, et al. Age-dependent levels of 5-methyl-, 5-hydroxymethyl-, and 5-formylcytosine in human and mouse brain tissues. Angew Chem Int Ed Engl 2015; 54:12511-4; PMID:26137924; https://doi.org/10.1002/anie.201502722
- Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell 2012; 151:1417-30; PMID:23260135; https://doi.org/10.1016/j.cell.2012.11.022
- Perera A, Eisen D, Wagner M, Laube SK, Künzel AF, Koch S, Steinbacher J, Schulze E, Splith V, Mittermeier N, et al. TET3 is recruited by REST for context-specific hydroxymethylation and induction of gene expression. Cell Rep 2015; 11:283-94; PMID:25843715; https://doi.org/10.1016/j.celrep.2015.03.020
- Costa Y, Ding J, Theunissen TW, Faiola F, Hore TA, Shliaha PV, Fidalgo M, Saunders A, Lawrence M, Dietmann S, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 2013; 495:370-4; PMID:23395962; https://doi.org/10.1038/nature11925
- Koh KP, Yabuuchi A, Rao S, Huang Y, Cunniff K, Nardone J, Laiho A, Tahiliani M, Sommer CA, Mostoslavsky G, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell 2011; 8:200-13; PMID:21295276; https://doi.org/10.1016/j.stem.2011.01.008
- Szwagierczak A, Bultmann S, Schmidt CS, Spada F, Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res 2010; 38:e181; PMID:20685817; https://doi.org/10.1093/nar/gkq684
- Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, Xie ZG, Shi L, He X, Jin SG, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 2011; 477:606-10; PMID:21892189; https://doi.org/10.1038/nature10443
- Penn NW, Suwalski R, O'Riley C, Bojanowski K, Yura R. The presence of 5-hydroxymethylcytosine in animal deoxyribonucleic acid. Biochem J 1972; 126:781-90; PMID:4538516; https://doi.org/10.1042/bj1260781
- Globisch D, Münzel M, Müller M, Michalakis S, Wagner M, Koch S, Brückl T, Biel M, Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 2010; 5:e15367; PMID:21203455; https://doi.org/10.1371/journal.pone.0015367
- Bachman M, Uribe-Lewis S, Yang X, Burgess HE, Iurlaro M, Reik W, Murrell A, Balasubramanian S. 5-Formylcytosine can be a stable DNA modification in mammals. Nat Chem Biol 2015; 11:555-7; PMID:26098680; https://doi.org/10.1038/nchembio.1848
- Zhu C, Gao Y, Guo H, Xia B, Song J, Wu X, Zeng H, Kee K, Tang F, Yi C. Single-cell 5-formylcytosine landscapes of mammalian early embryos and escs at single-base resolution. Cell Stem Cell 2017; 20:720-731.e5; PMID:28343982; https://doi.org/10.1016/j.stem.2017.02.013
- Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Embryogenesis: Demethylation of the zygotic paternal genome. Nature 2000; 403:501-2; PMID:10676950; https://doi.org/10.1038/35000656
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol 2000; 10:475-8; PMID:10801417; https://doi.org/10.1016/S0960-9822(00)00448-6
- Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science 2010; 330:622-7; PMID:21030646; https://doi.org/10.1126/science.1190614
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: Reprogramming and beyond. Nat Rev Genet 2008; 9:129-40; PMID:18197165; https://doi.org/10.1038/nrg2295
- Guo JU, Ma DK, Mo H, Ball MP, Jang M-H, Bonaguidi MA, Balazer JA, Eaves HL, Xie B, Ford E, et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat Neurosci 2011; 14:1345-51; PMID:21874013; https://doi.org/10.1038/nn.2900
- Morris MJ, Monteggia LM. Role of DNA methylation and the DNA methyltransferases in learning and memory. Dialogues Clin Neurosci 2014; 16:359-71; PMID:25364286
- Ma DK, Jang M-H, Guo JU, Kitabatake Y, Chang M-l, Pow-anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity–induced gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science 2009; 323:1074-7; PMID:19119186; https://doi.org/10.1126/science.1166859
- Seisenberger S, Peat JR, Reik W. Conceptual links between DNA methylation reprogramming in the early embryo and primordial germ cells. Curr Opin Cell Biol 2013; 25:281-8; PMID:23510682; https://doi.org/10.1016/j.ceb.2013.02.013
- Schiesser S, Hackner B, Pfaffeneder T, Müller M, Hagemeier C, Truss M, Carell T. Mechanism and stem-cell activity of 5-carboxycytosine decarboxylation determined by isotope tracing. Angew Chem Int Ed Engl 2012; 51:6516-20; PMID:22644704; https://doi.org/10.1002/anie.201202583
- Liutkevičiūtė Z, Lukinavičius G, Masevičius V, Daujotytė D, Klimaašauskas S. Cytosine-5-methyltransferases add aldehydes to DNA. Nat Chem Biol 2009; 5:400-2; PMID:19430486; https://doi.org/10.1038/nchembio.172
- Haag S, Sloan KE, Ranjan N, Warda AS, Kretschmer J, Blessing C, Hübner B, Seikowski J, Dennerlein S, Rehling P, et al. NSUN3 and ABH1 modify the wobble position of mt-tRNA Met to expand codon recognition in mitochondrial translation. EMBO J 2016; 35:2104-19; PMID:27497299; https://doi.org/10.15252/embj.201694885
- Liu F, Clark W, Luo G, Wang X, Fu Y, Wei J, Wang X, Hao Z, Dai Q, Zheng G, et al. ALKBH1-mediated tRNA demethylation regulates translation. Cell 2016; 167:816-28.e16; PMID:27745969; https://doi.org/10.1016/j.cell.2016.09.038
- Delatte B, Wang F, Ngoc LV, Collignon E, Bonvin E, Deplus R, Calonne E, Hassabi B, Putmans P, Awe S, et al. Transcriptome-wide distribution and function of RNA hydroxymethylcytosine. Science 2016; 351:282-5; PMID:26816380; https://doi.org/10.1126/science.aac5253
- Fu L, Guerrero CR, Zhong N, Amato NJ, Liu Y, Liu S, Cai Q, Ji D, Jin SG, Niedernhofer LJ, et al. Tet-mediated formation of 5-hydroxymethylcytosine in RNA. J Am Chem Soc 2014; 136:11582-5; PMID:25073028; https://doi.org/10.1021/ja505305z
- Zhang H-Y, Xiong J, Qi B-L, Feng Y-Q, Yuan B-F. The existence of 5-hydroxymethylcytosine and 5-formylcytosine in both DNA and RNA in mammals. Chem Commun 2016; 52:737-40; PMID:26562407; https://doi.org/10.1039/C5CC07354E
- Maiti A, Drohat AC. Thymine DNA glycosylase can rapidly excise 5-formylcytosine and 5-carboxylcytosine: Potential implications for active demethylation of CpG sites. J Biol Chem 2011; 286:35334-8; PMID:21862836; https://doi.org/10.1074/jbc.C111.284620
- He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, Ding J, Jia Y, Chen Z, Li L, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011; 333:1303-7; PMID:21817016; https://doi.org/10.1126/science.1210944
- Schomacher L, Han D, Musheev MU, Arab K, Kienhofer S, von Seggern A, Niehrs C. Neil DNA glycosylases promote substrate turnover by Tdg during DNA demethylation. Nat Struct Mol Biol 2016; 23:116-24; PMID:26751644; https://doi.org/10.1038/nsmb.3151
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 2011; 146:67-79; PMID:21722948; https://doi.org/10.1016/j.cell.2011.06.020
- Cortazar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 2011; 470:419-23; PMID:21278727; https://doi.org/10.1038/nature09672
- Amouroux R, Nashun B, Shirane K, Nakagawa S, Hill PW, D'Souza Z, Nakayama M, Matsuda M, Turp A, Ndjetehe E, et al. De novo DNA methylation drives 5hmC accumulation in mouse zygotes. Nat Cell Biol 2016; 18:225-33; PMID:26751286; https://doi.org/10.1038/ncb3296
- Santos F, Peat J, Burgess H, Rada C, Reik W, Dean W. Active demethylation in mouse zygotes involves cytosine deamination and base excision repair. Epigenetics Chromatin 2013; 6:39; PMID:24279473; https://doi.org/10.1186/1756-8935-6-39
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010; 468:839-43; PMID:21057493; https://doi.org/10.1038/nature09586
- Esteller M. Epigenetics in Cancer. N Engl J Med 2008; 358:1148-59; PMID:18337604; https://doi.org/10.1056/NEJMra072067
- Jones PA, Baylin SB. The epigenomics of cancer. Cell 2007; 128:683-92; PMID:17320506; https://doi.org/10.1016/j.cell.2007.01.029
- Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell 2005; 123:437-48; PMID:16269335; https://doi.org/10.1016/j.cell.2005.08.011
- Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB. Issa J-PJ. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999; 96:8681-6; PMID:10411935; https://doi.org/10.1073/pnas.96.15.8681
- Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006; 38:787-93; PMID:16804544; https://doi.org/10.1038/ng1834
- Issa J-P. CpG island methylator phenotype in cancer. Nat Rev Cancer 2004; 4:988-93; PMID:15573120; https://doi.org/10.1038/nrc1507
- Ono R, Taki T, Taketani T, Taniwaki M, Kobayashi H, Hayashi Y. LCX, leukemia-associated protein with a CXXC domain, is fused to MLL in acute myeloid leukemia with trilineage dysplasia having t(10;11)(q22;q23). Cancer Res 2002; 62:4075-80; PMID:12124344
- Yang H, Liu Y, Bai F, Zhang JY, Ma SH, Liu J, Xu ZD, Zhu HG, Ling ZQ, Ye D, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 2013; 32:663-9; PMID:22391558; https://doi.org/10.1038/onc.2012.67
- Neri F, Dettori D, Incarnato D, Krepelova A, Rapelli S, Maldotti M, Parlato C, Paliogiannis P, Oliviero S. TET1 is a tumour suppressor that inhibits colon cancer growth by derepressing inhibitors of the WNT pathway. Oncogene 2015; 34:4168-76; PMID:25362856; https://doi.org/10.1038/onc.2014.356
- Hsu C-H, Peng K-L, Kang M-L, Chen Y-R, Yang Y-C, Tsai C-H, Chu CS, Jeng YM, Chen YT, Lin FM, et al. TET1 suppresses cancer invasion by activating the tissue inhibitors of metalloproteinases. Cell Rep 2012; 2:568-79; PMID:22999938; https://doi.org/10.1016/j.celrep.2012.08.030
- Liu C, Liu L, Chen X, Shen J, Shan J, Xu Y, Yang Z, Wu L, Xia F, Bie P, et al. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One 2013; 8:e62828; PMID:23671639; https://doi.org/10.1371/journal.pone.0062828
- Langemeijer SMC, Kuiper RP, Berends M, Knops R, Aslanyan MG, Massop M, Stevens-Linders E, van Hoogen P, van Kessel AG, Raymakers RA, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet 2009; 41:838-42; PMID:19483684; https://doi.org/10.1038/ng.391
- Lemonnier F, Couronné L, Parrens M, Jaïs J-P, Travert M, Lamant L, Tournillac O, Rousset T, Fabiani B, Cairns RA, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 2012; 120:1466-9; PMID:22760778; https://doi.org/10.1182/blood-2012-02-408542
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 2011; 19:17-30; PMID:21251613; https://doi.org/10.1016/j.ccr.2010.12.014
- Inoue S, Lemonnier F, Mak TW. Roles of IDH1/2 and TET2 mutations in myeloid disorders. Int J Hematol 2016; 103:627-33; PMID:26980223; https://doi.org/10.1007/s12185-016-1973-7
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Cancer 2012; 12:599-612; PMID:22898539; https://doi.org/10.1038/nrc3343
- Thienpont B, Steinbacher J, Zhao H, D'Anna F, Kuchnio A, Ploumakis A, Ghesquière B, Van Dyck L, Boeckx B, Schoonjans L, et al. Tumour hypoxia causes DNA hypermethylation by reducing TET activity. Nature 2016; 537:63-8; PMID:27533040; https://doi.org/10.1038/nature19081
- Yang H, Lin H, Xu H, Zhang L, Cheng L, Wen B, Shou J, Guan K, Xiong Y, Ye D. TET-catalyzed 5-methylcytosine hydroxylation is dynamically regulated by metabolites. Cell Res 2014; 24:1017-20; PMID:24971736; https://doi.org/10.1038/cr.2014.81
- Sciacovelli M, Goncalves E, Johnson TI, Zecchini VR, da Costa AS, Gaude E, Drubbel AV, Theobald SJ, Abbo SR, Tran MG, et al. Fumarate is an epigenetic modifier that elicits epithelial-to-mesenchymal transition. Nature 2016; 537:544-7; PMID:27580029; https://doi.org/10.1038/nature19353
- Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2015; 518:413-6; PMID:25487152; https://doi.org/10.1038/nature13981
- TeSlaa T, Chaikovsky AC, Lipchina I, Escobar SL, Hochedlinger K, Huang J, Graeber TG, Braas D, Teitell MA. alpha-ketoglutarate accelerates the initial differentiation of primed human pluripotent stem cells. Cell Metab 2016; 24:485-93; PMID:27476976; https://doi.org/10.1016/j.cmet.2016.07.002
- Yang Q, Liang X, Sun X, Zhang L, Fu X, Rogers CJ, Berim A, Zhang S, Wang S, Wang B, et al. AMPK/alpha-ketoglutarate axis dynamically mediates dna demethylation in the prdm16 promoter and brown adipogenesis. Cell Metab 2016; 24:542-54; PMID:27641099; https://doi.org/10.1016/j.cmet.2016.08.010
- Laukka T, Mariani CJ, Ihantola T, Cao JZ, Hokkanen J, Kaelin WG Jr., Godley LA, Koivunen P. Fumarate and succinate regulate expression of hypoxia-inducible genes via TET enzymes. J Biol Chem 2016; 291:4256-65; PMID:26703470; https://doi.org/10.1074/jbc.M115.688762