ABSTRACT
Asymmetric localization of mRNAs is a widespread gene regulatory mechanism that is crucial for many cellular processes. The localization of a transcript involves multiple steps and requires several protein factors to mediate transport, anchoring and translational repression of the mRNA. Specific recognition of the localizing transcript is a key step that depends on linear or structured localization signals, which are bound by RNA-binding proteins. Genetic studies have identified many components involved in mRNA localization. However, mechanistic aspects of the pathway are still poorly understood. Here we provide an overview of structural studies that contributed to our understanding of the mechanisms underlying mRNA localization, highlighting open questions and future challenges.
Introduction
The localization of mRNAs is a gene regulatory mechanism, widespread in eukaryotes, that is crucial for many processes, including patterning of embryonic axes, asymmetric cell division, cell migration and synaptic plasticity (reviewed in Holt and Bullock, 2009; Martin and Ephrussi, 2009).Citation1,Citation2 Among metazoans, the fruit fly Drosophila melanogaster has been most extensively studied, given the genetic accessibility of this system. During Drosophila oogenesis and early development, hundreds of mRNAs have been shown to localize in very defined subcellular patterns.Citation3-5 Among vertebrates, a model system that has been classically used is Xenopus laevis.Citation1,Citation6 Cell cultures of mammalian neuronal cells and fibroblasts are also widely used to explore how mRNA localization contributes to neuronal function and cell motility, respectively. Cultured cells-based systems have allowed transcriptome-wide characterization of mRNA localization. For example, several mRNAs have been shown to be localized in dendrites, in axons or in cell protrusions in these systems.Citation1,Citation6 Many mRNA-localizing factors initially identified in Drosophila have homologs in vertebrates, including Staufen,Citation7-9 Bruno,Citation10,Citation11 Bicaudal D (BicD),Citation12 ZBP1Citation13-15 and Pumilio (Pum).Citation16 In vertebrates some of the proteins that are involved in mRNA localization in Drosophila are also involved in mRNA localization, for example, mammalian Staufen and Fragile X Mental Retardation Protein (FMRP) are required for mRNA localization in neurons,Citation17-20 and the Exon Junction Complex (EJC) component eIF4AIII is involved in dendritic mRNA localization.Citation21 Genetic studies in Drosophila have provided detailed part-lists of mRNA localization components. However, our mechanistic understanding lags behind the genetic characterization of this pathway.
Here we review the contribution of structural studies to the mechanistic understanding of mRNA localization. We will discuss how target mRNAs are recognized, repressed and transported to their target site within the cell. We will only briefly touch on RNA recognition mediated by canonical RNA-binding domains (such as KH, RRM, dsRBD), for which we refer to more focused reviews.Citation22-24 We will also not address the mechanisms of motor function, mRNA localization studies in fungi, nor provide an in-depth description of pathways intersecting with mRNA localization (e.g. mRNA decay), which have been reviewed elsewhere.Citation25-28
Steps in mRNA localization
The key step in mRNA localization is the specific recognition of the transcript, which depends on cis-acting elements, generally found in 3′ untranslated regions (3′UTR) of mRNAs (reviewed in Besse and Ephrussi, 2008; Meignin and Davis, 2010; Medioni et al., 2011)Citation6,Citation29,Citation30 (). These elements are recognized by trans-acting factors, or RNA-binding proteins (RBPs). Many RBPs contain several domains, that can contribute to specific target recognition or mediate the recruitment of additional protein factors.Citation31 In the case of active localization, mRNAs are transported along the cytoskeleton either by dynein or kinesin motor proteins (mediating microtubule minus end- or plus end-directed transport, respectively) or myosins (which move along actin filaments). Alternatively, localized expression can be achieved by selective translational repression or degradation of the target mRNA in parts of the cell. Often, the localized messenger ribonucleoprotein particle (mRNP) is maintained at the destination site by anchoring to the cytoskeleton, generally actin filaments. Translational repression of the mRNP before localization is fundamental for both active and passive localization mechanisms, to prevent protein expression at ectopic sites in the cell. Such repression may persist at the destination site (e.g., during Drosophila oogenesis), until it is relieved by an extrinsic cue. This translational activation is currently one of the least understood steps of the mRNA localization pathway.
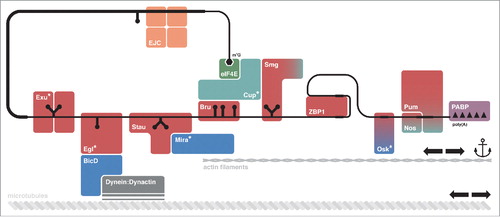
Figure 1. mRNA localization requires multiple factors. The mRNA to be localized is recognized by RNA-binding proteins (RBPs, in red) that interact with linear or structured localization signals, or cis-acting elements, on the mRNA. These RBPs can recruit adaptor proteins (in blue), which mediate anchoring or transport of the localizing messenger ribonucleoprotein complex (mRNP) along the cytoskeleton through the action of molecular motors (in gray). Other proteins (in teal) are required to maintain the mRNP in a translationally repressed state, for example by competing directly with components of the translation machinery (e.g.: Cup) or by recruiting the CCR4:NOT complex to promote shortening of the poly(A) tail on the target mRNA (e.g.: Smg, Nos, Pum). Special RBPs are represented by the Exon Junction Complex (EJC, in orange), the cap-binding protein eIF4E (in green) and the Poly(A)-Binding Protein (PABP, in purple). Asterisks (*) indicate proteins with no known ortholog in vertebrates. Not all of the proteins depicted here will associate at the same time point, or to the same mRNA.
RNA localization signals
A variety of mRNA localization signals has been identified to date, ranging from linear sequences to defined secondary structures. These signals are critical for providing recognition sites for proteins of the localization machinery.Citation32-34
Recognition of linear signals
One of the best characterized examples of a linear localization signal is the zipcode element on β-actin mRNA, recognized by the Zipcode-Binding Protein 1 (ZBP1).Citation13,Citation35 ZBP1 regulates local translation of β-actin mRNA at the base of activated dendritic spines in hippocampal neurons.Citation36 The crystal structure of human ZBP1 RNA-binding domains (hnRNP K homology, or KH domains) revealed an intramolecular pseudo-dimer arrangement that positions the RNA-binding surfaces on opposing faces of the protein. This induces the looping of the bipartite zipcode element around ZBP1 (, ). Binding of both zipcode elements enhances the affinity of ZBP1 for β-actin mRNA. Since the 2 zipcode elements can only bind if they are separated by a spacer sequence of defined length, this ensures specificity in mRNA recognition.Citation37 The occurrence of multiple domains to obtain high RNA-binding affinity and specificity, often combined with the remodelling of the target mRNA (as in the β-actin mRNA and ZBP1), is a recurrent feature of RNA-protein complexes (reviewed in Lunde et al., 2007).Citation31
Figure 2. Recognition of linear localization signals. (a) Model of RNA binding by KH3 and KH4 domains of the human ZBP1 homolog IMP1 (PDB 3KRMCitation37), showing the pseudo-dimer arrangement that positions the RNA-binding surfaces on opposing directions of the structure. The RNA molecules bound to KH3 and KH4 are derived from the structure of Gallus ZBP1 (PDB 2N8L and 2N8M, respectivelyCitation128), superposed using the cealign command in Pymol v1.7 (www.pymol.org). (b) Model of RNA binding by RRM1 and RRM2 domains of human CUGBP1 (PDB 3NNH and 3NMR, respectively). The relative orientation of the two RRM domains is modeled as described in Teplova et al., 2009Citation38. (c) Crystal structure of Drosophila Pum in complex with the Pum Recognition Element (PRE) from hunckback (hb) mRNA (PDB 5KLACitation76).
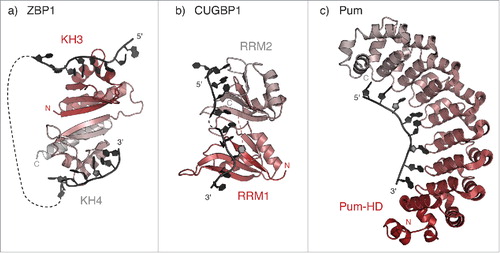
Another example is provided by the CUG-binding protein 1 (CUGBP1). The human CUGBP1 protein is a member of the CUGBP1 and ETR-like factors (CELF) protein family, which includes the Drosphila translational repressor Bruno (Bru). CELF family members share a conserved domain architecture, characterized by two adjacent RNA recognition motifs (RRMs) in the N-terminal region, and a third C-terminal RRM, which is separated from the others by a long, non-conserved, linker.Citation10 The two N-terminal RRM domains of CUGBP1 bind with similar affinities to their respective target sequences (UGUU/G). NMR studies indicate that the two domains tumble independently in solution in the absence of the RNA substrate.Citation38 However, in the presence of an RNA molecule comprising two recognition signals the two RRM domains are found in a compact and rigid arrangement (), which explains the observed binding cooperativity.Citation38 While the short linker between RRM1 and RRM2 of CUGBP1 favors cooperative binding of the two domains on adjacent sequences on the target mRNA, the longer, and probably disordered, linker that connects RRM3 could allow for recognition of a more distant sequence element, perhaps belonging to a different mRNA molecule. This mechanism could help in creating a large messenger ribonucleoprotein particle (mRNP) for transport, and would also contribute to maintaining the translational repression of the target mRNA, as has been proposed for Drosophila Bru in oskar (osk) mRNA regulation.Citation39 In addition, such unstructured regions can cooperate in RNA binding: the linker region N-terminal to the third RRM in both human CUGBP1 and Drosophila Bru extends the binding surface of the RNA-recognition motif, which increases RRM3 affinity for its target mRNA.Citation40,Citation41
Another way to achieve recognition of an RNA sequence element with high affinity and specificity is by combining multiple copies of a simple repeated structural motif within a protein domain. This approach is used by members of the Pumilio and fem-3 mRNA binding factor (Puf) protein family, of which Drosophila Pumilio (Pum) is the founder (reviewed in Wickens et al., 2002).Citation16 Puf proteins are characterized by the presence of a sequence-specific RNA-binding domain known as the Pum homology domain (Pum-HD), comprising 8 sequence repeats of three helices each, plus N- and C-terminal flanking regions.Citation42,Citation43 This helical domain is reminiscent of the Armadillo repeat domain, a protein-protein interaction module found in a wide range of proteins with diverse functions, including nucleo-cytoplasmic transport, intracellular signaling, and cytoskeletal organization (reviewed in Hatzfeld et al., 1999; Coates, 2003).Citation44,Citation45 The Pum-HD domain contacts RNA through its concave surface, where each nucleotide is recognized by a triumvirate of amino acid side chains at conserved positions within the helical repeatsCitation46 (). In addition, the Pum-HD domain can interact with the CCR4:NOT deadenylation complex and promote shortening of the poly(A) tail on the target mRNA.Citation47,Citation48 In this way, Puf proteins control stability and translation of a variety of different mRNAs, recognized through a Pum response element (PRE) in their 3′UTRs.
Recognition of structured signals
Given the difficulty in predicting tertiary or even secondary structures of RNAs with high reliability, the characterization of structured localization signals has proven more challenging and relies on experimental verification.Citation49 This is well exemplified by the Drosophila fs(1)K10 Transport and Localization Signal (K10 TLS), which mediates minus-end-directed, dynein-dependent transport along the microtubules during early Drosophila development. The structure of this localization element, derived by NMR spectroscopy, has revealed a stem-loop conformation in which purine-purine stacking within the double stranded (ds) region forces the stem to adopt an unusual A′-form conformation. This conformation has 2 widened major grooves which are oriented at 90° to one another in contrast to the standard A-form dsRNA where major and minor grooves have similar widths (). This specific arrangement (reminiscent of B-form dsDNA) is required for the localization of K10 TLS-containing mRNAs in vivoCitation33 (). Other mRNAs that are transported at various stages during Drosophila oogenesis and early development toward the minus end of microtubules also contain localization signals that could form hairpins of similar structure but of different primary sequence as the K10 TLS.Citation33 This suggests that widened major grooves in dsRNA, rather than being used for primary sequence recognition, represent an unusual structural feature that can be sensed by a common localization machinery. Indeed, several of these elements, including the K10 TLS, are recognized by the protein Egalitarian (Egl).Citation50 Egl does not contain a canonical RNA-binding domain, but rather contacts RNA through a large region including a domain that displays homology to 3′-5′ DEDD exonucleases (Exo-domain), which usually catalyze the exonucleolytic cleavage of nucleic acids.Citation50,Citation51 Mutation of the putative catalytic residues within Egl Exo-domain does not affect mRNA localization in vivo or RNA-binding activity in vitro indicating that, if the protein is a functional exonuclease, this activity is dispensable for protein function.Citation50,Citation52 Similarly, the protein Exuperantia (Exu) uses a catalytically inactive Exo-domain to bind and localize bicoid (bcd) mRNA during early oogenesis in Drosophila.Citation53 Both Egl and Exu recognize mRNA signals with defined secondary structures, though the molecular details of the interaction are not known.
Figure 3. Recognition of structured localization signals. (a-b) Solution structures of the Transport and Localization Signal from K10 mRNA (K10 TLS; a) (PDB 2KE6Citation33) and of the SOLE hairpin from osk mRNA (b) (PDB 5A17Citation70). A-U base pairs are shown in black; G-C base pairs in blue; G:A base pairs in cyan; G:U base pairs in green; unpaired residues (loops and bulges) are highlighted in red. Both stem-loops present widened major grooves (indicated by broad arrowheads) as compared with the common A-form of dsRNA helices (e). (c) Solution structure of yeast Vts1 Sterile α motif (SAM) bound to the Smg Recognition Element (SRE) (PDB 2ESECitation58). The loop residues forming a base pair are colored in blue; the specifically recognized G residue at position 3 of the loop is highlighted in red. The position of the major groove is indicated by an arrowhead. In the case of SRE RNA, the major groove has an intermediate width between that of the K10 TLS and that of an ideal A-form dsRNA and it is not involved in Smg/Vts1 recognition.Citation54 (d) Solution structure of the third dsRNA-binding domain (dsRBD3) of Drosophila Stau bound to an artificial stem-loop sequence (PDB 1EKZCitation68). The N-terminal α-helix (α1-helix), which is potentially involved in sequence-specific contacts with the RNA loop, is highlighted in red. (e) Cartoon representation of ideal dsRNA structures (generated in CootCitation129), assuming the canonical A-form (left) or the unusual A'-form conformation (right), which is reminiscent of the B-form of dsDNA. Major grooves are indicated by arrowheads (thin for A- and broad for A'-form, respectively).
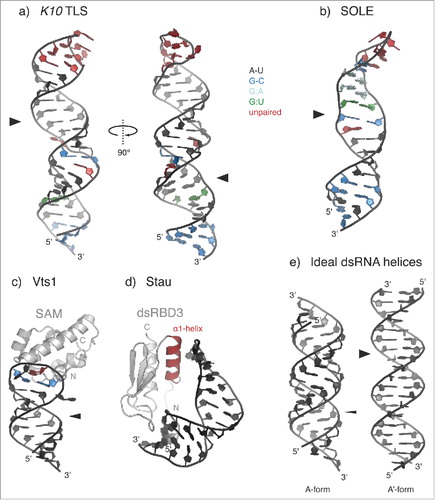
To date, there are very few examples of structured localization signals in complex with RNA-binding proteins. One is provided by the RNA-binding domain of yeast Vts1 in complex with the Smaug (Smg) Recognition Element (SRE) RNA (). Vts1 binds RNA through its Sterile α motif (SAM), a domain consisting of 5 α-helices arranged in a globular bundle, that was previously thought to be solely a protein-protein interaction motif.Citation54,Citation55 The SRE consists of a stem-loop structure, but only the 5 residues of the loop (CUGGC) are directly contacted by a shallow, positively-charged surface patch on the SAM domain. The only residue that is specifically recognized is the G at position 3 within the pentaloop, while the other protein-RNA interactions involve non-sequence specific contacts with the RNA phosphate oxygens.Citation56-58 However, the formation or stabilization of a base pair within the pentaloop upon protein binding also seems to be important, suggesting that the G3 nucleotide base within the loop is recognized within a specific structural context.Citation56,Citation57 The sequence of the stem, as long as base-pairing is preserved, does not influence Vts1 binding affinity.Citation54 Yeast Vts1 is a homolog of Drosophila Smg, a protein that contributes to anterior-posterior axis determination in the Drosophila embryo. Smg represses nanos (nos) mRNA translation everywhere in the embryo but at the posterior pole plasm. This generates a gradient of Nos protein emanating from the posterior of the embryo. Such gradient is required to trigger a series of downstream events that result in proper abdominal segmentation.Citation59 Smg also binds mRNA by its SAM domain, suggesting evolutionary conservation, but the details of this interaction are not known. A SAM-like domain is also present in Exu and mutational analysis suggests that it is important for RNA binding. However, in Exu there is an additional surface not present in Smg or Vts1 that contributes to mRNA binding.Citation53
Another conserved mRNA-localization protein, Staufen (Stau), presents a distinct RNA-binding mechanism. Members of the Stau protein family are important for mRNA localization in many organisms and contain several copies of dsRNA-binding domain (dsRBD), a domain that was first identified in the Drosophila Stau protein.Citation7 In Drosophila, Stau is required for oskar (osk) mRNA localization at the posterior pole during oogenesis, for bcd mRNA anchoring at the anterior pole in late oogenesis and early embryogenesis, and for prospero (pros) mRNA transport during neuroblast asymmetric cell division.Citation60-64,Citation65,Citation66 Structural characterization of the third dsRBD of Drosophila Staufen in complex with a non-physiologic stem-loop RNA sequence showed that the dsRBD3 interacts with the RNA sugar-phosphate backbone, without making direct contacts with the RNA bases. Some sequence specificity could be provided by the interaction of the first α-helix with the nucleotide bases in the hairpin loopCitation67,Citation68 (), although structural studies on a physiologic substrate would be required to confirm this.
For other localized mRNAs, the protein factors required for the recognition of their structured localization signals are still unidentified. This is the case of the Spliced osk Localization Element (SOLE)Citation69,Citation70 (). The SOLE is generated upon splicing of the first intron of osk and it is required for plus end-directed, posterior localization of osk mRNA during oogenesis in Drosophila.Citation69 Osk localization also requires the deposition of the EJC upstream of the first exon-exon junction. The isolated SOLE assumes a stem-loop structure capped by a 5 nucleotide loop, and, like the K10 TLS, displays a widened major groove.Citation70 A protein binding partner of the SOLE has yet to be identified.
Combinatorial regulation mechanisms (The importance of a bigger picture)
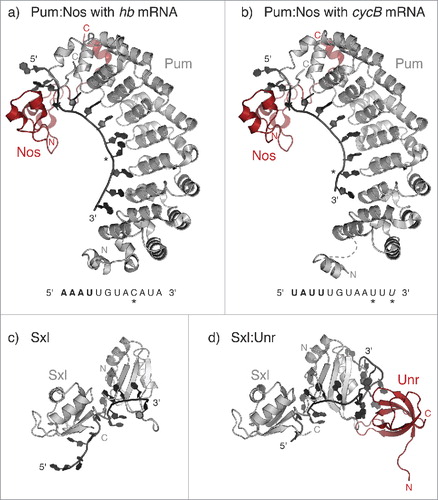
In many of the examples described above, the specificity of the RNA-binding proteins (such as Vts156, Egl,Citation50 Stau,Citation23 ExuCitation53) appears to be less stringent in vitro than in vivo.Citation31,Citation71 This could be due to the use of isolated RBDs in in vitro assays. The inclusion of multiple domains, where present, would be important in future studies to better approximate in vivo conditions. In other cases, the binding affinity and specificity of an RNA-binding protein (RBP) for its target mRNA is modulated by the interaction with a protein partner. Such cases highlight the importance of structural studies focusing on multiprotein complexes together with their bound mRNA targets. The effects of partner proteins on mRNA binding could be mediated by partner-dependent stabilization of RBPs, as has been proposed for Drosophila Egl and Bicaudal D (BicD).Citation50,Citation72,Citation73 Alternatively, binding partners could alter RBP binding specificity either by promoting conformational changes or by contributing additional surfaces for binding. A striking example is provided by the complex of Pum and Nanos (Nos). These proteins co-regulate several mRNAs in Drosophila, including repression of maternal hunchback (hb) mRNA at the posterior pole of the embryo, and of Cyclin B (CycB) mRNA in primordial germ cells and germline stem cells.Citation74,Citation75 The crystal structure of the Pum:Nos complex bound to hb and CycB recognition elements revealed that Nos not only induces conformational changes in Pum that increase its affinity for RNA, but also directly contacts the RNA upstream of the Pum canonical recognition sequenceCitation76 (). In this way, Nos adds upstream recognition specificity, thereby relaxing the requirement for a perfect Pum-binding consensus sequence. The Pum:Nos complex can thus regulate a broader range of mRNA targets than Pum alone. Cooperativity is also exhibited by the proteins Sex-lethal (Sxl) and Upstream of N-Ras (Unr). In Drosophila females, the two proteins bind adjacent sequences in the 3′UTR of male-specific lethal 2 (msl2) mRNA, which encodes for the limiting component of the dosage compensation complex and repress its translation.Citation77 The crystal structure of the Sxl:Unr:msl2 mRNA ternary complex showed how the 5′ end of the RNA sequence is recognized by Sxl, while the 3′ end is sandwiched by both proteins. The formation of such a “triple zipper” allows additional protein:RNA contacts that would not be possible with either protein aloneCitation78 ().
Figure 4. Examples of combinatorial control. (a-b) Crystal structure of the Drosophila Pum:Nos complex bound to the Pum Recognition Element (PRE) from hunckback (hb) (a) (PBD 5KL1) or cyclinB (cycB) mRNA (b) (PDB 5KL8Citation76). The RNA sequence is written under the corresponding structure, with the nucleotides recognized by Nos in bold; asterisks indicate nucleotides that deviate from the PRE consensus. The U residue at the 3′ end of cycB PRE (in italic) is not visible in the structure. (c) Crystal structure of the two RRM domains of Sxl in complex with the Sxl-binding element from transformer (tra) mRNA (PDB 1B7FCitation130). (d) Crystal structure of the ternary complex between Sxl, Unr and the RNA recognition element from male-specific lethal 2 (msl2) mRNA (PDB 4QQBCitation78). Unr binds RNA through its Cold Shock domain.
Transport and anchoring
Long-range active transport usually depends on microtubules, with kinesin and dynein motors mediating plus end- and minus-end directed movement, respectively. There are very few instances where all biochemical links between the localizing mRNA and the molecular motor have been identified. These include the complex linking ASH1 mRNA to myosin 4 motor during localization to the bud of Saccaromyces cerevisiae (reviewed in Niedner et al., 2014)Citation26 and Drosophila BicD, a protein linking localizing mRNAs to the dynein motor during minus end-directed mRNA localization (reviewed in Hoogenraad and Akhmanova, 2016)Citation79 (). BicD is the founder of a conserved family of motor adaptor proteins, which, in addition to coupling the dynein motor to various cargoes, stimulates processive dynein motility by stabilizing dynein interaction with its constitutive cofactor dynactin.Citation80-82 While the N-terminal 2 coiled-coil domains of BicD associate simultaneously with dynein and dynactin, stabilizing their interaction,Citation82 the C-terminal part of BicD can recognize different protein partners, that in turn mediate the transport of specific cargoes.Citation83 In Drosophila, one of the most important partners of BicD is Egl, which is required for dynein-dependent localization of a variety of transcripts.Citation50 Another BicD-associated factor is FMRP, which has an important role in mRNA localization in neurons and is mutated in the most common inherited form of cognitive deficiency in humans.Citation84 All existing data point to BicD binding to one partner at a time, and in some cases through overlapping interaction surfaces.Citation83 Though we can draw a link between localizing mRNPs and dynein-mediated transport, little is known about kinesin-mediated transport.
Figure 5. Transport and anchoring. (a) Schematic representation of Egl-BicD mediated transport of K10 mRNA. Structural information is currently available for K10 TLS (PDB 2KE6Citation33); for the first and second coiled coil regions of BicD in complex with the Dynein:Dynactin molecular motor (PDB 5AFUCitation82); and for the C-terminal coiled coil region of BicD, with the Egl-binding site defined by mutation analysis (PDB 4BL6Citation83). Still unknown are the molecular details of Egl interaction with the RNA and with BicD, as well as the connection between BicD N-terminal and C-terminal coiled coil domains. (b) Crystal structure of a fragment of Mira coiled coil region bound to the fifth dsRBD (dsRBD5) of Stau (PDB 5CFFCitation91). BicD and Mira both act as homodimers.
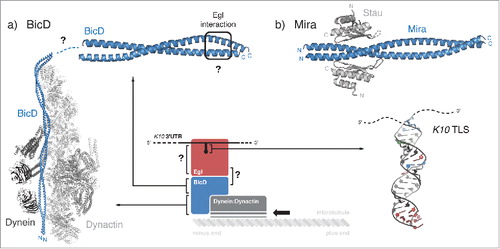
Another key adaptor protein in mRNA localization is Miranda (Mira). Mira is required for the segregation of the cell fate determinants Brain tumor (Brat), Prospero (Pros) and Numb to the basal cortex during the asymmetric cell division of neuroblasts in Drosophila.Citation66,Citation85-88,Citation89,Citation90 In addition, Mira directly interacts with the dsRNA-binding protein Stau to localize pros mRNA in an actin-dependent manner both in neuroblasts and epithelial cellsCitation66,Citation88,Citation91 (). In this case, the connection with the cytoskeletal motor is still unknown.
The mechanism by which osk mRNA is anchored at the posterior cortex during Drosophila oogenesis and early embryogenesis is also poorly understood. At the posterior pole, two Osk protein isoforms are translated from alternative start codons: Short Osk is necessary and sufficient to induce pole cell formation and posterior patterning in the embryo, while Long Osk is required for proper anchoring of both osk mRNA and Short Osk to the posterior cortex of the oocyte.Citation92-95 Short Osk recruits the DEAD box helicase Vasa through its N-terminal LOTUS domain to initiate germ plasm assembly.Citation94,Citation96 In addition, short Osk associates with germ plasm localized mRNAs, including its own and nos mRNA.Citation96,Citation97 Analogous to Egl and Exu, Osk binds RNA through an enzyme fold: the C-terminal OSK domain, which resembles a SGNH hydrolase, but lacks the catalytic residues.Citation96,Citation97 The RNA sequence specificity of Osk has not been yet determined. In addition to the LOTUS and OSK domains, Long Osk contains an N-terminal extension, which is required, but not sufficient, for the posterior anchoring of osk mRNA.Citation95 Despite the presence of the LOTUS domain, Long Osk cannot recruit Vasa, suggesting that the N-terminal extension is somehow modulating the function of the other Osk domains.Citation92,Citation94,Citation96 Instead, Long Osk stimulates clathrin-mediated endocytosis and contributes to the organization of the actin cytoskeleton at the posterior of the oocyte, promoting its own and Short Osk maintenance at the posterior pole through a poorly understood mechanism.Citation98
Translational repression and selective degradation
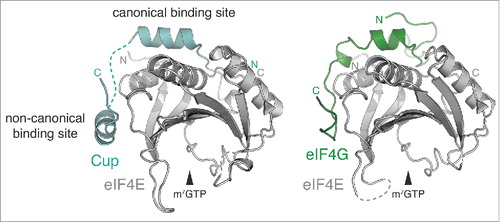
Translational repression mechanisms frequently target translation initiation, which is generally rate-limiting. During initiation, the scaffolding factor eIF4G interacts with the cap-binding protein eIF4E and the Poly(A)-Binding Protein (PABP), to connect 5′ and 3′ end of the mRNA in a closed loop conformation that is thought to stimulate translation; in addition, eIF4G interacts with eIF3, which in turn recruits the small ribosomal subunit (reviewed in Jackson et al., 2010).Citation99 The eIF4E:eIF4G interaction is targeted by a conserved class of translational repressors, the eIF4E binding proteins (4E-BPs) (reviewed in Topisirovic et al., 2011).Citation100 A member of this family is the Drosophila protein Cup, which is required during oogenesis and early development for repressing translation of the axis-determining factors osk, nos, and gurken (grk). Cup does not contact RNA, but is recruited to its mRNA targets by different RBPs (Bru in the case of osk and grk; Smg for nos).Citation101-104 The crystal structure of an N-terminal fragment of Cup in complex with eIF4E revealed how Cup interacts through two binding motifs (one canonical and one non-canonical), and engages the same surfaces of eIF4E responsible for interaction with eIF4GCitation102,Citation104-106,Citation107,Citation108 (). In addition, Cup binding stabilizes eIF4E and increases its grip on the target mRNA, thus protecting it from decapping and degradation.Citation107,Citation108 Alternatively, translation initiation can be inhibited by preventing the recruitment of the large ribosomal subunit, as described for ZBP1.Citation36
Figure 6. Translational repression. Crystal structure of Drosophila eIF4E in complex with a peptide from Cup (left) (PDB 4AXGCitation107) or eIF4G (right) (PDB 5T47Citation131).
Other proteins repress translation of their target mRNA by recruiting the CCR4:NOT deadenylation complex: examples include Cup, Bicaudal C (BicC), Smg, Pum and Nos.Citation109,Citation47,Citation108,Citation110 For the recruitment of the CCR4:NOT complex, at least some of these proteins rely on short linear motifs embedded in peptide regions of predicted disorder (e.g., NosCitation111). Interestingly, many RNA- and DNA-binding proteins show a significant enrichment in low complexity regions when compared with the entire proteome.Citation112 In addition to providing quickly-evolving interaction surfaces, these regions have the potential for forming reversible amyloid-like fibers in vitroCitation113; in vivo, the same sequences could mediate oligomerization and assembly of large particles that would reinforce translational repression by steric exclusion of the ribosomes, as proposed for osk mRNA.Citation39
Indeed, to ensure efficient repression of the localizing mRNA, translation is usually inhibited at multiple steps. Complete repression of nos expression, for instance, requires both Cup-mediated inhibition at the initiation stepCitation104 and Smg-mediated deadenylation.Citation109,Citation114
Translational activation
How mRNA recognition factors, translational repressors and localization machinery could dissociate from the localized mRNP is a critical but poorly understood step in the mRNA localization pathway. Release could be mediated by targeting the localizing factor for degradation when it is no longer needed, as has been shown for Mira.Citation88 Another mechanism involves dissociation of the RBP from the recognition element on the mRNA. For example, the majority of nos mRNA is evenly distributed throughout the Drosophila embryo and translationally repressed in a Smg-dependent manner.Citation104,Citation109,Citation114 However, at the posterior pole, Osk protein prevents Smg binding to nos mRNA, thereby allowing its translation.Citation114-116 The molecular details of this regulation are still unclear. Osk could bind Smg directly, and perhaps alter its affinity for RNACitation115; alternatively, Osk and Smg could compete for the same binding site on nos mRNA, as suggested by the discovery that Osk is also an RNA-binding protein.Citation96,Citation97
Post-translational modifications, such as phosphorylation, also seem to play an important role. The affinity of 4E-BPs for eIF4E, for example, is decreased by the phosphorylation of 4E-BPs at multiple sites.Citation117,Citation118 Analogously, Src-dependent phosphorylation of ZBP1 relieves translational repression of β-actin mRNA by disrupting ZBP1:RNA interaction,Citation36 though the molecular details of this regulation are still unclear.
Future perspectives
Structural studies have greatly advanced our understanding of the mechanisms underlying mRNA localization. However, many issues remain. The challenge for the future is to understand the assembly and dynamics of the large multiprotein-RNA complexes involved in mRNA localization.
First, structural studies will have to tackle larger multimolecular assemblies. RBPs often contain multiple domains that combine protein- and RNA-binding activities, and, due to their complexity and flexibility, offer a challenge for structural studies. Moreover, many localizing mRNAs assemble in multimeric complexes, through both protein- (e.g., Exu, Bru, Osk, BicD, Mira) and RNA-mediated dimerization (e.g., bcd). The increasing number of examples in which combinations of structural motifs, protein domains and protein interaction partners are exploited to modulate RNA-binding affinity and specificity, however, prompts for an effort in the characterization of more physiologic complexes. It will be challenging to characterize stable assemblies and to purify them in suitable amounts for structural studies. Cryo-EM approaches hold promise to solve large assemblies at high resolution and with the need of less material. However, inherent flexibility and heterogeneous composition of the targets remains an issue across techniques when tackling large assemblies.
Localizing mRNP composition changes at the different steps of gene expression. Studies of single molecule dynamics are particularly promising to gain mechanistic insights of mRNA localization at high resolution. These studies provide a direct readout of the spatial and temporal details of localization. It will be particularly challenging to integrate information from these kinds of studies with the static information derived from structural studies. In this respect, structural snapshots of the same component/complex in different cellular states are very informative although technically very challenging.
A third avenue will be to integrate the wealth of genetic information with structural and single molecule studies. Genetic studies, especially in Drosophila, provide much data on the identity and hierarchy of the factors required for mRNA localization, and represent an excellent starting point for a more detailed biochemical characterization. Furthermore, novel genome editing techniques will facilitate the in vivo validation of functional hypotheses from structural data in reverse genetic approaches.
It will also be essential to chart the complement of RBPs and RNA target motives by large-scale studies. New methods of RNA-protein interactionCitation119,Citation120 including CRAC,Citation121 CLIP,Citation122 PAR-CLIP,Citation123 iCLIP,Citation124 hiCLIPCitation125 provide a rich source of RBPs and RNA targets. Those will be the starting points for structural studies of complexes to understand RNA binding modes.
The variety of canonical and non-canonical RNA-binding domains used by RBPs, together with the divergence in RNA signal sequences, makes it difficult to derive rules for target mRNA selection. Bioinformatics predictions and studies on isolated RNA elements can provide valuable information. However, experimental validation of the protein:RNA interaction remains essential; this is especially true for cases in which the RNA secondary structure changes following post-transcriptional modifications of the nucleotide bases (e.g., N6-methyladenosine-dependent RNA structural switchesCitation126), or interaction with the protein partner (e.g., structural rearrangements in the E3 localization element from yeast ASH1 mRNA bound to the localization machineryCitation127).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We would like to thank Atlanta Cook and Gáspár Jékely for critical reading of the manuscript. The authors are funded by the Max Planck Gesellshaft, the European Research Council under the European Union's Seventh Framework Program (FP7/2007-2013), ERC grant agreement no. 310957 and the Deutsche Forschungsgemeinschaft (BO3588/2-1 to F.B.).
We apologize to those investigators whose work was inadvertently overlooked or omitted for space constraints.
References
- Holt CE, Bullock SL. Subcellular mRNA Localization in Animal Cells and Why It Matters. Science 2009; 326:1212-6; PMID:19965463; https://doi.org/10.1126/science.1176488
- Martin KC, Ephrussi A. mRNA localization: Gene expression in the spatial dimension. Cell 2009; 136:719-30; PMID:19239891; https://doi.org/10.1016/j.cell.2009.01.044
- Lécuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007; 131:174-87; PMID:17923096; https://doi.org/10.1016/j.cell.2007.08.003
- Jambor H, Surendranath V, Kalinka AT, Mejstrik P, Saalfeld S, Tomancak P. Systematic imaging reveals features and changing localization of mRNAs in Drosophila development. eLife 2015; 4:e05003; PMID:25838129; https://doi.org/10.7554/eLife.05003
- Wilk R, Hu J, Blotsky D, Krause HM. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev 2016; 30:594-609; PMID:26944682; https://doi.org/10.1101/gad.276931.115
- Medioni C, Mowry K, Besse F. Principles and roles of mRNA localization in animal development. Development 2012; 139:3263-76; PMID:22912410; https://doi.org/10.1242/dev.078626
- St Johnston D, Brown NH, Gall JG, Jantsch M. A conserved double-stranded RNA-binding domain. Proc Nati Acad Sci U S A 1992; 89:10979-83; PMID:1438302; https://doi.org/10.1073/pnas.89.22.10979
- Wickham L, Duchaîne T, Luo M, Nabi IR, DesGroseillers L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol Cell Biol 1999; 19:2220-30; PMID:10022909; https://doi.org/10.1128/MCB.19.3.2220
- Marión RM, Fortes P, Beloso A, Dotti C, Ortín J. A human sequence homologue of Staufen is an RNA-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol Cell Biol 1999; 19:2212-9; PMID:10022908; https://doi.org/10.1128/MCB.19.3.2212
- Good PJ, Chen Q, Warner SJ, Herring DC. A family of human RNA-binding proteins related to the Drosophila Bruno translational regulator. J Biol Chem 2000; 275:28583-92; PMID:10893231; https://doi.org/10.1074/jbc.M003083200
- Suzuki H, Maegawa S, Nishibu T, Sugiyama T, Yasuda K, Inoue K. Vegetal localization of the maternal mRNA encoding an EDEN-BP//Bruno-like protein in zebrafish. Mech Dev 2000; 93:205-9; PMID:10781958; https://doi.org/10.1016/S0925-4773(00)00270-7
- Schlager MA, Kapitein LC, Grigoriev I, Burzynski GM, Wulf PS, Keijzer N, de Graaff E, Fukuda M, Shepherd IT, Akhmanova A, et al. Pericentrosomal targeting of Rab6 secretory vesicles by Bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J 2010; 29:1637-51; PMID:20360680; https://doi.org/10.1038/emboj.2010.51
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH. Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol 1997; 17:2158-65; PMID:9121465; https://doi.org/10.1128/MCB.17.4.2158
- Deshler JO, Highett MI, Schnapp BJ. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science 1997; 276:1128-31; PMID:9148809; https://doi.org/10.1126/science.276.5315.1128
- Boylan KLM, Mische S, Li M, Marqués G, Morin X, Chia W, Hays TS. Motility screen identifies Drosophila IGF-II mRNA-binding protein–zipcode-binding protein acting in oogenesis and synaptogenesis. PLoS Genet 2008; 4:36; PMID:18282112; https://doi.org/10.1371/journal.pgen.0040036
- Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3'UTR regulation as a way of life. Trends Genet 2002; 18:150-7; PMID:11858839; https://doi.org/10.1016/S0168-9525(01)02616-6
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol 2000; 20:8536-47; PMID:11046149; https://doi.org/10.1128/MCB.20.22.8536-8547.2000
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, et al. The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol 2003; 13:286-96; PMID:12593794; https://doi.org/10.1016/S0960-9822(03)00064-2
- Dictenberg JB, Swanger SA, Antar LN, Singer RH, Bassell GJ. A direct role for FMRP in activity-dependent dendritic mRNA transport links filopodial-spine morphogenesis to fragile X syndrome. Dev Cell 2008; 14:926-39; PMID:18539120; https://doi.org/10.1016/j.devcel.2008.04.003
- Vessey JP, Amadei G, Burns SE, Kiebler MA, Kaplan DR, Miller FD. An asymmetrically localized Staufen2-dependent RNA complex regulates maintenance of mammalian neural stem cells. Cell Stem Cell 2012; 11:517-28; PMID:22902294; https://doi.org/10.1016/j.stem.2012.06.010
- Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell 2007; 130:179-91; PMID:17632064; https://doi.org/10.1016/j.cell.2007.05.028
- Daubner GM, Cléry A, Allain FHT. RRM-RNA recognition: NMR or crystallography…and new findings. Curr Opin Struct Biol 2013; 23:100-8; PMID:23253355; https://doi.org/10.1016/j.sbi.2012.11.006
- Masliah G, Barraud P, Allain FHT. RNA recognition by double-stranded RNA binding domains: A matter of shape and sequence. Cell Mol Life Sci 2013; 70:1875-95; PMID:22918483; https://doi.org/10.1007/s00018-012-1119-x
- Nicastro G, Taylor IA, Ramos A. KH-RNA interactions: Back in the groove. Curr Opin Struct Biol 2015; 30:63-70; PMID:25625331; https://doi.org/10.1016/j.sbi.2015.01.002
- Bullock SL. Messengers, motors and mysteries: Sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem Soc Trans 2011; 39:1161-5; PMID:21936782; https://doi.org/10.1042/BST0391161
- Niedner-Boblenz A, Edelmann FT, Niessing D. Of social molecules: The interactive assembly of ASH1 mRNA-transport complexes in yeast. RNA Biol 2014; 11:998-1009; PMID:25482892; https://doi.org/10.4161/rna.29946
- Singer-Krüger B, Jansen RP. Here, there, everywhere. mRNA localization in budding yeast. RNA Biol 2014; 11:1031-9; PMID:25482891; https://doi.org/10.4161/rna.29945
- Temme C, Simonelig M, Wahle E. Deadenylation of mRNA by the CCR4-NOT complex in Drosophila: Molecular and developmental aspects. Front Genet 2014; 5:143; PMID:24904643; https://doi.org/10.3389/fgene.2014.00143
- Besse F, Ephrussi A. Translational control of localized mRNAs: Restricting protein synthesis in space and time. Nat Rev Mol Cell Biol 2008; 9:971-80; PMID:19023284; https://doi.org/10.1038/nrm2548
- Meignin C, Davis I. Transmitting the message: Intracellular mRNA localization. Curr Opin Cell Biol 2010; 22:112-9; PMID:20022233; https://doi.org/10.1016/j.ceb.2009.11.011
- Lunde BM, Moore C, Varani G. RNA-binding proteins: Modular design for efficient function. Nat Rev Mol Cell Biol 2007; 8:479-90; PMID:17473849; https://doi.org/10.1038/nrm2178
- Singer RH. RNA zipcodes for cytoplasmic addresses. Curr Bio 1993; 3:719-21; PMID:15335871; https://doi.org/10.1016/0960-9822(93)90079-4
- Bullock SL, Ringel I, Ish-Horowicz D, Lukavsky PJ. A′-form RNA helices are required for cytoplasmic mRNA transport in Drosophila. Nat Publ Group 2010; 17:703-9; PMID:20473315; https://doi.org/10.1038/nsmb.1813. Epub 2010 May 16
- Pratt CA, Mowry KL. Taking a cellular road-trip: mRNA transport and anchoring. Curr Opin Cell Biol 2013; 25:99-106; PMID:23200723; https://doi.org/10.1016/j.ceb.2012.08.015
- Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol 1994; 127:441-51; PMID:7929587; https://doi.org/10.1083/jcb.127.2.441
- Hüttelmaier S, Zenklusen D, Lederer M, Dictenberg J, Lorenz M, Meng X, Bassell GJ, Condeelis J, Singer RH. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 2005; 438:512-5; PMID:16306994; https://doi.org/10.1038/nature04115
- Chao JA, Patskovsky Y, Patel V, Levy M, Almo SC, Singer RH. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes Dev 2010; 24:148-58; PMID:20080952; https://doi.org/10.1101/gad.1862910
- Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural insights into RNA recognition by the alternate-splicing regulator CUG-Binding protein 1. Structure 2010; 18:1364-77; PMID:20947024; https://doi.org/10.1016/j.str.2010.06.018
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 2006; 124:521-33; PMID:16469699; https://doi.org/10.1016/j.cell.2006.01.031
- Lyon AM, Reveal BS, Macdonald PM, Hoffman DW. Bruno protein contains an expanded RNA recognition motif. Biochemistry 2009; 48:12202-12; PMID:19919093; https://doi.org/10.1021/bi900624j
- Tsuda K, Kuwasako K, Takahashi M, Someya T, Inoue M, Terada T, Kobayashi N, Shirouzu M, Kigawa T, Tanaka A, et al. Structural basis for the sequence-specific RNA-recognition mechanism of human CUG-BP1 RRM3. Nucleic Acids Res 2009; 37:5151-66; PMID:19553194; https://doi.org/10.1093/nar/gkp546
- Edwards TA, Pyle SE, Wharton RP, Aggarwal AK. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 2001; 105:281-9; PMID:11336677; https://doi.org/10.1016/S0092-8674(01)00318-X
- Wang X, Zamore PD, Hall TM. Crystal structure of a Pumilio homology domain. Mol Cell 2001; 7:855-65; PMID:11336708; https://doi.org/10.1016/S1097-2765(01)00229-5
- Hatzfeld M. The armadillo family of structural proteins. Int Rev Cytol 1999; 186:179-224; PMID:9770300; https://doi.org/10.1016/S0074-7696(08)61054-2
- Coates JC. Armadillo repeat proteins: Beyond the animal kingdom. Trends Biol 2003; 13:463-71; PMID:12946625; https://doi.org/10.1016/S0962-8924(03)00167-3
- Wang X, McLachlan J, Zamore PD, Hall TMT. Modular recognition of RNA by a human pumilio-homology domain. Cell 2002; 110:501-12; PMID:12202039; https://doi.org/10.1016/S0092-8674(02)00873-5
- Goldstrohm AC, Hook BA, Seay DJ, Wickens M. PUF proteins bind Pop2p to regulate messenger RNAs. Nat Struct Mol Biol 2006; 13:533-9; PMID:16715093; https://doi.org/10.1038/nsmb1100
- Goldstrohm AC, Seay DJ, Hook BA, Wickens M. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J Biol Chem 2007; 282:109-14; PMID:17090538; https://doi.org/10.1074/jbc.M609413200
- Weeks KM. Advances in RNA structure analysis by chemical probing. Curr Opin Struct Biol 2010; 20:295-304; PMID:20447823; https://doi.org/10.1016/j.sbi.2010.04.001
- Dienstbier M, Boehl F, Li X, Bullock SL. Egalitarian is a selective RNA-binding protein linking mRNA localization signals to the dynein motor. Genes Dev 2009; 23:1546-15; PMID:19515976; https://doi.org/10.1101/gad.531009
- Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res 1997; 25:5110-8; PMID:9396823; https://doi.org/10.1093/nar/25.24.5110
- Navarro C, Puthalakath H, Adams JM, Strasser A, Lehmann R. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat Cell Biol 2004; 6:427-35; PMID:15077115; https://doi.org/10.1038/ncb1122
- Lazzaretti D, Veith K, Kramer K, Basquin C, Urlaub H, Irion U, Bono F. The bicoid mRNA localization factor Exuperantia is an RNA-binding pseudonuclease. Nat Struct Mol Biol 2016; 23:705-13; PMID:27376588; https://doi.org/10.1038/nsmb.3254
- Aviv T, Lin Z, Lau S, Rendl LM, Sicheri F, Smibert CA. The RNA-binding SAM domain of Smaug defines a new family of post-transcriptional regulators. Nat Struct Biol 2003; 10:614-21; PMID:12858164; https://doi.org/10.1038/nsb956
- Green JB, Gardner CD, Wharton RP, Aggarwal AK. RNA recognition via the SAM domain of Smaug. Mol Cell 2003; 11:1537-48; PMID:12820967; https://doi.org/10.1016/S1097-2765(03)00178-3
- Aviv T, Lin Z, Ben-Ari G, Smibert CA, Sicheri F. Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p. Nat Struct Mol Biol 2006; 13:168-76; PMID:16429151; https://doi.org/10.1038/nsmb1053
- Johnson PE, Donaldson LW. RNA recognition by the Vts1p SAM domain. Nat Struct Mol Biol 2006; 13:177-8; PMID:16429155; https://doi.org/10.1038/nsmb1039
- Oberstrass FC, Lee A, Stefl R, Janis M, Chanfreau G, Allain FH. Shape-specific recognition in the structure of the Vts1p SAM domain with RNA. Nat Struct Mol Biol 2006; 13:160-7; PMID:16429156; https://doi.org/10.1038/nsmb1038
- Gavis ER, Lehmann R. Translational regulation of nanos by RNA localization. Nature 1994; 369:315-8; PMID:7514276; https://doi.org/10.1038/369315a0
- Ephrussi A, Dickinson LK, Lehmann R. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 1991; 66:37-50; PMID:2070417; https://doi.org/10.1016/0092-8674(91)90137-N
- Kim-Ha J, Smith JL, Macdonald PM. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 1991; 66:23-35; PMID:2070416; https://doi.org/10.1016/0092-8674(91)90136-M
- St Johnston D, Driever W, Berleth T, Richstein S, Nüsslein-Volhard C. Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 1989; 107 Suppl:13-9; PMID:2483989
- St Johnston D, Beuchle D, Nüsslein-Volhard C. Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 1991; 66:51-63; PMID:1712672; https://doi.org/10.1016/0092-8674(91)90138-O
- Ferrandon D, Elphick L, Nüsslein-Volhard C, St Johnston D. Staufen protein associates with the 3'UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 1994; 79:1221-32; PMID:8001156; https://doi.org/10.1016/0092-8674(94)90013-2
- Broadus J, Fuerstenberg S, Doe CQ. Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter-cell fate. Nature 1998; 391:792-5; PMID:9486649; https://doi.org/10.1038/35861
- Schuldt AJ, Adams JH, Davidson CM, Micklem DR, Haseloff J, St Johnston D, Brand AH. Miranda mediates asymmetric protein and RNA localization in the developing nervous system. Genes Dev 1998; 12:1847-57; PMID:9637686; https://doi.org/10.1101/gad.12.12.1847
- Ramos A, Bayer P, Varani G. Determination of the structure of the RNA complex of a double-stranded RNA-binding domain from Drosophila Staufen protein. Biopolymers 1999; 52:181-96; PMID:11295750; https://doi.org/10.1002/1097-0282(1999)52:4%3c181::AID-BIP1003%3e3.0.CO;2-5
- Ramos A, Grünert S, Adams J, Micklem DR, Proctor MR, Freund S, Bycroft M, St Johnston D, Varani G. RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J 2000; 19:997-1009; PMID:10698941; https://doi.org/10.1093/emboj/19.5.997
- Ghosh S, Marchand V, Gaspar I, Ephrussi A. Control of RNP motility and localization by a splicing-dependent structure in oskar mRNA. Nat Publ Group 2012; 19:441-9; PMID:22426546; https://doi.org/10.1038/nsmb.2257
- Simon B, Masiewicz P, Ephrussi A, Carlomagno T. The structure of the SOLE element of oskar mRNA. RNA 2015; 21:1444-53; PMID:26089324; https://doi.org/10.1261/rna.049601.115
- Jankowsky E, Harris ME. Specificity and nonspecificity in RNA-protein interactions. Nat Rev Mol Cell Biol 2015; 16:533-44; PMID:26285679; https://doi.org/10.1038/nrm4032
- Dix CI, Soundararajan HC, Dzhindzhev NS, Begum F, Suter B, Ohkura H, Stephens E, Bullock SL. Lissencephaly-1 promotes the recruitment of dynein and dynactin to transported mRNAs. J Cell Biol 2013; 202:479-94; PMID:23918939; https://doi.org/10.1083/jcb.201211052
- Vazquez-Pianzola P, Schaller B, Colombo M, Beuchle D, Neuenschwander S, Marcil A, Bruggmann R, Suter B. The mRNA transportome of the BicD/Egl transport machinery. RNA Biology 2017; 14:73-89; PMID:27801632; https://doi.org/10.1080/15476286.2016.1251542
- Wreden C, Verrotti AC, Schisa JA, Lieberfarb ME, Strickland S. Nanos and pumilio establish embryonic polarity in Drosophila by promoting posterior deadenylation of hunchback mRNA. Development 1997; 124:3015-23; PMID:9247343.
- Asaoka-Taguchi M, Yamada M, Nakamura A, Hanyu K, Kobayashi S. Maternal pumilio acts together with Nanos in germline development in Drosophila embryos. Nat Cell Biol 1999; 1:431-7; PMID:10559987; https://doi.org/10.1038/15666
- Weidmann CA, Qiu C, Arvola RM, Lou TF, Killingsworth J, Campbell ZT, Tanaka Hall TM, Goldstrohm AC. Drosophila Nanos acts as a molecular clamp that modulates the RNA-binding and repression activities of Pumilio. eLife 2016; 5:1948; PMID:27482653; https://doi.org/10.7554/eLife.17096
- Graindorge A, Militti C, Gebauer F. Posttranscriptional control of X-chromosome dosage compensation. Wiley Interdiscip Rev 2011; 2:534-45; PMID:21957042; https://doi.org/10.1002/wrna.75
- Hennig J, Militti C, Popowicz GM, Wang I, Sonntag M, Geerlof A, Gabel F, Gebauer F, Sattler M. Structural basis for the assembly of the Sxl-Unr translation regulatory complex. Nature 2014; 515:287-90; PMID:25209665; https://doi.org/10.1038/nature13693
- Hoogenraad CC, Akhmanova A. Bicaudal D family of motor adaptors: Linking dynein motility to cargo binding. Trends Biol 2016; 26:327-40; PMID:26822037; https://doi.org/10.1016/j.tcb.2016.01.001
- Schlager MA, Serra-Marques A, Grigoriev I, Gumy LF, Esteves da Silva M, Wulf PS, Akhmanova A, Hoogenraad CC. Bicaudal d family adaptor proteins control the velocity of Dynein-based movements. Cell Rep 2014; 8:1248-56; PMID:25176647; https://doi.org/10.1016/j.celrep.2014.07.052
- McKenney RJ, Huynh W, Tanenbaum ME, Bhabha G, Vale RD. Activation of cytoplasmic dynein motility by dynactin-cargo adapter complexes. Science 2014; 345:337-41; PMID:25035494; https://doi.org/10.1126/science.1254198
- Urnavicius L, Zhang K, Diamant AG, Motz C, Schlager MA, Yu M, Patel NA, Robinson CV, Carter AP. The structure of the dynactin complex and its interaction with dynein. Science 2015; 347:1441-6; PMID:25814576; https://doi.org/10.1126/science.aaa4080
- Liu Y, Salter HK, Holding AN, Johnson CM, Stephens E, Lukavsky PJ, Walshaw J, Bullock SL. Bicaudal-D uses a parallel, homodimeric coiled coil with heterotypic registry to coordinate recruitment of cargos to dynein. Genes Dev 2013; 27:1233-46; PMID:23723415; https://doi.org/10.1101/gad.212381.112
- Bianco A, Dienstbier M, Salter HK, Gatto G, Bullock SL. Bicaudal-D regulates fragile X mental retardation protein levels, motility, and function during neuronal morphogenesis. Curr Biol 2010; 20:1487-92; PMID:20691595; https://doi.org/10.1016/j.cub.2010.07.016
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 1997; 390:625-9; PMID:9403694; https://doi.org/10.1038/37641
- Shen CP, Jan LY, Jan YN. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 1997; 90:449-58; PMID:9267025; https://doi.org/10.1016/S0092-8674(00)80505-X
- Matsuzaki F, Ohshiro T, Ikeshima-Kataoka H, Izumi H. miranda localizes staufen and prospero asymmetrically in mitotic neuroblasts and epithelial cells in early Drosophila embryogenesis. Development 1998; 125:4089-98; PMID:9735369.
- Fuerstenberg S, Peng CY, Alvarez-Ortiz P, Hor T, Doe CQ. Identification of Miranda protein domains regulating asymmetric cortical localization, cargo binding, and cortical release. Mol Cell Neurosci 1998; 12:325-39; PMID:9888987; https://doi.org/10.1006/mcne.1998.0724
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev Cell 2006; 10:441-9; PMID:16549393; https://doi.org/10.1016/j.devcel.2006.01.017
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell 2006; 124:1241-53; PMID:16564014; https://doi.org/10.1016/j.cell.2006.01.038
- Jia M, Shan Z, Yang Y, Liu C, Li J, Luo ZG, Zhang M, Cai Y, Wen W, Wang W. The structural basis of Miranda-mediated Staufen localization during Drosophila neuroblast asymmetric division. Nat Commun 2015; 6:8381; PMID:26423004; https://doi.org/10.1038/ncomms9381
- Markussen FH, Michon AM, Breitwieser W, Ephrussi A. Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development 1995; 121:3723-32; PMID:8582284
- Rongo C, Gavis ER, Lehmann R. Localization of oskar RNA regulates oskar translation and requires Oskar protein. Development 1995; 121:2737-46; PMID:7555702.
- Breitwieser W, Markussen FH, Horstmann H, Ephrussi A. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev 1996; 10:2179-88; PMID:8804312; https://doi.org/10.1101/gad.10.17.2179
- Vanzo NF, Ephrussi A. Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development 2002; 129:3705-14; PMID:12117819
- Jeske M, Bordi M, Glatt S, Müller S, Rybin V, Müller CW, Ephrussi A. The crystal structure of the drosophila germline inducer oskar identifies two domains with distinct vasa Helicase- and RNA-binding activities. Cell Rep 2015; 12:587-98; PMID:26190108; https://doi.org/10.1016/j.celrep.2015.06.055
- Yang N, Yu Z, Hu M, Wang M, Lehmann R, Xu RM. Structure of drosophila oskar reveals a novel RNA binding protein. Proc Nati Acad Sci U S A 2015; 112:11541-6; PMID:26324911; https://doi.org/10.1073/pnas.1515568112
- Vanzo N, Oprins A, Xanthakis D, Ephrussi A, Rabouille C. Stimulation of endocytosis and actin dynamics by Oskar polarizes the Drosophila oocyte. Dev Cell 2007; 12:543-55; PMID:17419993; https://doi.org/10.1016/j.devcel.2007.03.002
- Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 2010; 11:113-27; PMID:20094052; https://doi.org/10.1038/nrm2838
- Topisirovic I, Svitkin YV, Sonenberg N, Shatkin AJ. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev 2011; 2:277-98; PMID:21957010; https://doi.org/10.1002/wrna.52
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol 2003; 163:1197-204; PMID:14691132; https://doi.org/10.1083/jcb.200309088
- Nakamura A, Sato K, Hanyu-Nakamura K. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell 2004; 6:69-78; PMID:14723848; https://doi.org/10.1016/S1534-5807(03)00400-3
- Zappavigna V, Piccioni F, Villaescusa JC, Verrotti AC. Cup is a nucleocytoplasmic shuttling protein that interacts with the eukaryotic translation initiation factor 4E to modulate Drosophila ovary development. Proc Nati Acad Sci U S A 2004; 101:14800-5; PMID:15465908; https://doi.org/10.1073/pnas.0406451101
- Nelson MR, Leidal AM, Smibert CA. Drosophila Cup is an eIF4E-binding protein that functions in Smaug-mediated translational repression. EMBO J 2004; 23:150-9; PMID:14685270; https://doi.org/10.1038/sj.emboj.7600026
- Marcotrigiano J, Gingras AC, Sonenberg N, Burley SK. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 1999; 3:707-16; PMID:10394359; https://doi.org/10.1016/S1097-2765(01)80003-4
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 2003; 115:739-50; PMID:14675538; https://doi.org/10.1016/S0092-8674(03)00975-9
- Kinkelin K, Veith K, Grunwald M, Bono F. Crystal structure of a minimal eIF4E-Cup complex reveals a general mechanism of eIF4E regulation in translational repression. RNA 2012; 18:1624-34; PMID:22832024; https://doi.org/10.1261/rna.033639.112
- Igreja C, Izaurralde E. CUP promotes deadenylation and inhibits decapping of mRNA targets. Genes Dev 2011; 25:1955-67; PMID:21937713; https://doi.org/10.1101/gad.17136311
- Semotok JL, Cooperstock RL, Pinder BD, Vari HK, Lipshitz HD, Smibert CA. Smaug recruits the CCR4/POP2/NOT deadenylase complex to trigger maternal transcript localization in the early Drosophila embryo. Curr Biol 2005; 15:284-94; PMID:15723788; https://doi.org/10.1016/j.cub.2005.01.048
- Van Etten J, Schagat TL, Hrit J, Weidmann CA, Brumbaugh J, Coon JJ, Goldstrohm AC. Human Pumilio proteins recruit multiple deadenylases to efficiently repress messenger RNAs. J Biol Chem 2012; 287:36370-83; PMID:22955276; https://doi.org/10.1074/jbc.M112.373522
- Raisch T, Bhandari D, Sabath K, Helms S, Valkov E, Weichenrieder O, Izaurralde E. Distinct modes of recruitment of the CCR4-NOT complex by Drosophila and vertebrate Nanos. EMBO J 2016; 35:974-90; PMID:26968986; https://doi.org/10.15252/embj.201593634
- Michelitsch MD, Weissman JS. A census of glutamine/asparagine-rich regions: Implications for their conserved function and the prediction of novel prions. Proc Nati Acad Sci U S A 2000; 97:11910-5; PMID:11050225; https://doi.org/10.1073/pnas.97.22.11910
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 2012; 149:753-67; PMID:22579281; https://doi.org/10.1016/j.cell.2012.04.017
- Zaessinger S, Busseau I, Simonelig M. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 2006; 133:4573-83; PMID:17050620; https://doi.org/10.1242/dev.02649
- Dahanukar A, Walker JA, Wharton RP. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell 1999; 4:209-18; PMID:10488336; https://doi.org/10.1016/S1097-2765(00)80368-8
- Jeske M, Moritz B, Anders A, Wahle E. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. EMBO J 2010; 30:90-103; PMID:21081899; https://doi.org/10.1038/emboj.2010.283
- Gingras AC, Gygi SP, Raught B, Polakiewicz RD, Abraham RT, Hoekstra MF, Aebersold R, Sonenberg N. Regulation of 4E-BP1 phosphorylation: A novel two-step mechanism. Genes Dev 1999; 13:1422-37; PMID:10364159; https://doi.org/10.1101/gad.13.11.1422
- Gingras AC, Raught B, Gygi SP, Niedzwiecka A, Miron M, Burley SK, Polakiewicz RD, Wyslouch-Cieszynska A, Aebersold R, Sonenberg N. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev 2001; 15:2852-64. PMID:11691836; https://doi.org/10.1101/gad.912401
- Hentze MW, Preiss T. The REM phase of gene regulation. Trends Biochem Sci 2010; 35:423-6; PMID:20554447; https://doi.org/10.1016/j.tibs.2010.05.009
- Castello A, Horos R, Strein C, Fischer B, Eichelbaum K, Steinmetz LM, Krijgsveld J, Hentze MW. System-wide identification of RNA-binding proteins by interactome capture. Nat Protoc 2013; 8:491-500; PMID:23411631; https://doi.org/10.1038/nprot.2013.020
- Granneman S, Kudla G, Petfalski E, Tollervey D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Nati Acad Sci U S A 2009; 106:9613-8; PMID:19482942; https://doi.org/10.1073/pnas.0901997106
- Wang Z, Tollervey J, Briese M, Turner D, Ule J. CLIP: Construction of cDNA libraries for high-throughput sequencing from RNAs cross-linked to proteins in vivo. Methods 2009; 48:287-93; PMID:19272451; https://doi.org/10.1016/j.ymeth.2009.02.021
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp AC, Munschauer M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 2010; 141:129-41; PMID:20371350; https://doi.org/10.1016/j.cell.2010.03.009
- König J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Publ Group 2010; 17:909-15; PMID:20601959; https://doi.org/10.1038/nsmb.1838
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D'Ambrogio A, Luscombe NM, Ule J. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 2015; 519:491-4; PMID:25799984; https://doi.org/10.1038/nature14280
- Liu N, Pan T. Probing RNA modification status at single-nucleotide resolution in total RNA. Methods Enzymol 2015; 560:149-59. PMID:26253970; https://doi.org/10.1016/bs.mie.2015.03.005
- Edelmann FT, Schlundt A, Heym RG, Jenner A, Niedner-Boblenz A, Syed MI, Paillart JC, Stehle R, Janowski R, Sattler M, et al. Molecular architecture and dynamics of ASH1 mRNA recognition by its mRNA-transport complex. Nat Publ Group 2017; 24:152-61; PMID:28092367; https://doi.org/10.1038/nsmb.3351
- Nicastro G, Candel AM, Uhl M, Oregioni A, Hollingworth D, Backofen R, Martin SR, Ramos A. Mechanism of β-actin mRNA Recognition by ZBP1. Cell Rep 2017; 18:1187-99; PMID:28147274; https://doi.org/10.1016/j.celrep.2016.12.091
- Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sec D Biol Crystallogr 2004; 60:2126-32; PMID:15572765; https://doi.org/10.1107/S0907444904019158
- Handa N, Nureki O, Kurimoto K, Kim I, Sakamoto H, Shimura Y, Muto Y, Yokoyama S. Structural basis for recognition of the tra mRNA precursor by the Sex-lethal protein. Nature 1999; 398:579-85; PMID:10217141; https://doi.org/10.1038/19242
- Grüner S, Peter D, Weber R, Wohlbold L, Chung MY, Weichenrieder O, Valkov E, Igreja C, Izaurralde E. The structures of eIF4E-eIF4G complexes reveal an extended interface to regulate translation initiation. Mol Cell 2016; 64:467-79; PMID:27773676; https://doi.org/10.1016/j.molcel.2016.09.020
