ABSTRACT
Extensive work in model enterobacteria has evidenced that the RNA chaperone Hfq and several endoribonucleases, such as RNase E or RNase III, serve pivotal roles in small RNA-mediated post-transcriptional silencing of gene expression. Characterization of these protein hubs commonly provide global functional and mechanistic insights into complex sRNA regulatory networks. The legume endosymbiont Sinorhizobium meliloti is a non-classical model bacterium with a very complex lifestyle in which riboregulation is expected to play important adaptive functions. Here, we discuss current knowledge about RNA silencing in S. meliloti from the perspective of the activity of Hfq and a recently discovered endoribonuclease (YbeY) exhibiting unprecedented catalytic versatility for the cleavage of single- and double-stranded RNA molecules.
Small non-coding RNAs (sRNAs) are recognized as ubiquitous components of complex post-transcriptional regulatory networks controlling virtually any adaptive response of bacteria to environmental changes.Citation1,Citation2 The canonical activity mechanism of most RNA regulators involves base pairing to mRNAs, which may affect translation and/or turnover rates of the targeted transcripts in an either negative or positive manner.Citation3,Citation4 Complementarity between sRNAs and trans-encoded target mRNAs is typically short and imperfect, and therefore, a protein serving a chaperone function often assists these type of interactions. The widely conserved bacterial Sm-like protein Hfq has long been perceived as the major RNA binder fulfilling a dual role as sRNA stabilizer and matchmaker in trans-mRNA regulation.Citation5-7 Conversely, cis-acting antisense sRNAs (asRNAs) rely on extensive perfect complementarity to regulate mRNAs transcribed from the opposite strand and are believed to be largely protein-independent.Citation8 Although sRNA-mediated upregulation of gene expression in bacteria is not unusual, target repression is the typical outcome of sRNA activity.Citation4,Citation8 Most trans-acting sRNAs interact with their targets at the translation initiation region, thereby outcompeting ribosome binding and impairing protein synthesis. Primary translational repression is often coupled to irreversible target mRNA degradation, which is catalyzed by RNases that, in some cases, can be recruited to the sRNA-mRNA interplay by Hfq.Citation7,Citation9,Citation10 As component of the degradosome complex, the single-strand specific endoribonuclease RNase E is one Hfq-interacting partner in some bacterial species, initiating mRNA decay upon base pairing with trans-sRNAs.Citation11,Citation12 Alternatively, the prototypical double-strand endoribonuclease RNase III can initiate cleavage of some of these imperfect RNA-RNA duplexes and is a major effector of mRNA decay initiation driven by antisense transcription.Citation13,Citation14 RNA-binding proteins and ribonucleases are thus pivotal elements in riboregulatory networks and their characterization commonly provide primary functional hallmarks to complex sRNA landscapes. These paradigms about the biological roles and mechanistic principles of riboregulation mostly derive from extensive pioneering research on classical model bacteria (i.e., Escherichia coli and related enterobacteria) and clinically relevant pathogens. However, during the last two decades the availability of distantly related genomic sequences from bacteria with widely diverse lifestyles has increased exponentially. Comprehensive analysis of this overwhelming genomic information and its derived transcriptomes is expected to unveil novel insights into protein-assisted RNA regulation underlying unique ecological specializations.
Rhizobia, the nitrogen-fixing endosymbionts of legume plants, are an example of soil-dwelling microbes with complex biology and genomic architecture. During their life cycle, these bacteria shift from a free-living state in soil to a chronic intracellular residence within root nodules elicited in the plant host.Citation15 To establish competitive populations in soil, rhizobia must efficiently adapt their physiology to several abiotic and biotic variables typical of this harsh environment, e.g., oligotrophy, pH and temperature oscillations, drought, hyperosmolarity, native microbial populations or specific root exuded plant compounds like flavonoids. Upon root hair infection, nodule development and differentiation of nitrogen-fixing bacteroids are events governed by a coordinated exchange of signaling molecules between both symbiotic partners. Plant-derived signals in response to rhizobial infection include defensive reactive oxygen species, cysteine-rich peptides that promote bacteroid differentiation or microaerobiosis required by nitrogenase functioning within nodules.Citation15 These cues reprogram gene expression in rhizobia, which has been assumed to rely almost entirely on transcriptional control by protein factors, whereas post-transcriptional RNA-dependent regulation has been systematically overlooked.
The group of root-nodule bacteria includes species mostly belonging to the Rhizobiales order within the larger α–subgroup of proteobacteria. Among them, Sinorhizobium meliloti, the partner of the forage legume alfalfa (Medicago sativa) and the model plant M. truncatula, has served as main genetically tractable model species to study riboregulation in rhizobia.Citation16-18 The closely related S. meliloti reference strains Rm1021 and Rm2011 have highly syntenic composite genomes consisting of 3 megareplicons; the chromosome (3.54 Mb) and the so-called symbiotic plasmids pSymA (1.35 Mb) and pSymB (1.68 Mb), the latter exhibiting chromosome-like features.Citation19,Citation20 Computational comparative genomics and RNAseq-based screens have uncovered a large and heterogeneous inventory of non-coding transcripts expressed by this bacterium.Citation21-25 The challenge now is deciphering the function and activity mechanisms of this plethora of newly discovered S. meliloti sRNAs in the regulation of the adaptive responses imposed by both the saprophytic life in soil and the symbiotic transitions. Here, we discuss insights into RNA-based regulation in S. meliloti derived from RNA networks of two proteins functionally linked to sRNA activity; the RNA chaperone Hfq (henceforth SmHfq) and the novel recently discovered endoribonuclease YbeY (SmYbeY).Citation26,Citation27
Discrete contribution of SmHfq to riboregulation
Hfq has been positioned at the core of post-transcriptional regulatory networks in bacteria. Accordingly, the identity of Hfq regulated genes and binding transcripts typically draft major pathways relying on sRNA-based regulation.Citation28-30 In S. meliloti, lack of Hfq compromises adaptation of free-living bacteria to abiotic stress and the overall symbiotic performance on legume roots.Citation31-34 Profiling of the SmHfq regulon and RNA ligands by a diversity of genome-wide screens has traced the pathways specifying this phenotypic outcome.Citation27,Citation31,Citation33-35 These independent studies converged in the identification of nutrient uptake, energy metabolism, stress response, biosynthesis of surface macromolecules, quorum sensing (QS) and symbiotic nitrogen-fixation as major processes influenced by Hfq in S. meliloti. Most SmHfq-associated RNA species derive from or are full-length mRNAs, which collectively represent ∼18% of the S. meliloti protein coding genes. More than third of these genes have been reported to be differentially expressed in hfq knockout mutants. In contrast, SmHfq binds and stabilize only small fractions of the trans-acting and asRNAs expressed by this bacterium (14% and 2%, respectively).Citation27,Citation36
Almost half of the mRNAs concurrently bound and regulated by SmHfq encode components of ABC transport systems and cytoplasmic metabolic enzymes.Citation27 Remarkably, SmHfq negatively affects the steady-state levels of a large array of mRNA partners that encode periplasmic substrate binding proteins with affinity for a diversity of nitrogen-containing compounds (e.g., amino acids, peptides or polyamines), a feature common to many bacterial Hfq homologs characterized to date. In enterobacteria, the Hfq-dependent sRNA GcvB post-transcriptionally silences many of these mRNAs, which integrate one of the largest sRNA regulons described in bacteria.Citation37 Similarly, computer predictions in S. meliloti delivered a plethora of putative targets of the SmHfq-dependent homologous AbcR1 and AbcR2 sRNAs within the set of SmHfq mRNA ligands encoding proteins for the uptake of nitrogen sources.Citation27,Citation31,Citation38 Further genetic reporter assays in vivo confirmed AbcR1/2-mediated downregulation of some of these mRNAs, most likely by a canonical silencing mechanism involving primary occlusion of the respective ribosome binding site.Citation27,Citation38 Similar regulatory roles and activity mechanisms have been described for AbcR homologs in the related α–proteobacteria Agrobacterium tumefaciens and Brucella abortus.Citation39,Citation40 Rhizobial genomes encode exceptionally large inventories of ABC transport systems (e.g., 200 in S. meliloti compared with 67 in E. coli) for the uptake of widely diverse growth limiting nutrients in soil and specific plant-derived compounds in the rhizosphere and endosymbiotic compartments.Citation19,Citation41 Selective silencing of unnecessary simultaneous transcription of transporter gene sets, which is commonly driven by analogous compounds, would contribute to optimize rhizobial metabolism throughout the symbiotic interaction.
From a methodological point of view, these findings provide another proof of principle validating the atlas of Hfq-binding transcripts as a reliable source of functional sRNA-mRNA regulatory pairs in bacteria. However, since most S. meliloti sRNAs are Hfq independent, mining of this data set would render a rather partial view of RNA regulation in this bacterium. Indeed, it has been recently reported that the Hfq-independent trans-RNAs RcsR1 and EcpR1 regulate other symbiotically relevant processes such as QS or cell cycle.Citation42,Citation43 In line with this notion, post-genomic research on phylogenetically distant bacterial species increasingly suggests that the contribution of Hfq to riboregulation is more limited than expected from the early studies in classical model bacteria. Even in Salmonella, Hfq-binding sRNAs represent only a third of the non-coding transcripts expressed by this bacterium.Citation44 On the other hand, almost half of bacterial species lack an hfq gene and in others, this is fully dispensable for riboregulation.Citation8 These evidences have prompted searches for alternative bacterial RNA-binding proteins fulfilling Hfq-like roles as novel nodes of post-transcriptional regulatory networks.
SmYbeY is a novel endoribonuclease with unprecedented metal-dependent catalytic versatility
A few years ago, it was proposed that the YbeY protein might functionally replace Hfq for RNA regulation in S. meliloti.Citation45 YbeY belongs to the UPF0054 family of metallo-hydrolases and is almost ubiquitous in bacteria. Indeed, its coding gene is part of the minimal prokaryotic genome set, thus anticipating that it serves a fundamental function probably affecting a wide range of cellular processes.Citation46 Consequently, YbeY has been shown to be essential in some bacterial species, whereas in others its loss-of-function leads to severe pleiotropic phenotypes that in S. meliloti resemble those of the hfq mutants.Citation45,Citation47-50 Structural modeling of SmYbeY unveiled a positively charged cavity reminiscent of the MID domain of the Argonaute (AGO) proteins that assist RNA silencing in eukaryotes.Citation45 Further supporting a likely similar competence of bacterial YbeY homologs for RNA binding and/or regulation, recent studies in S. meliloti, E. coli and Vibrio cholerae have shown that lack of YbeY misregulates subsets of sRNAs and their predicted mRNA partners.Citation45,Citation50,Citation51 In particular, SmYbeY and SmHfq co-regulate a handful of sRNAs in a positive manner, thus suggesting that both proteins might share a chaperone function in sRNA pathways.Citation45 To test this hypothesis, the SmYbeY-dependent RNA network has been recently probed by genome-wide approaches (i.e., CoIP and transcriptomics) and experimental setups similar to those used previously for SmHfq.Citation26 SmYbeY CoIP-RNA was sparingly enriched with RNA species, contrary to what was observed for SmHfq, indicating either reduced competence of SmYbeY for RNA binding or transient contacts with its partner transcripts. Further, transcriptomics revealed scarce influence of SmYbeY on the expression of the ∼500 trans-sRNAs probed with the microarrays. Together, these findings argued against a major Hfq-like role of SmYbeY as RNA stabilizer and matchmaker in riboregulation.
The biochemical properties of YbeY were first reported for the E. coli homolog (EcoYbeY).Citation52 This protein behaved as a metallo endoribonuclease that specifically degraded in vitro synthetized single-stranded RNA substrates as well as several endogenous RNA molecules such as mRNAs and rRNA. This catalytic activity, which is largely conferred by the histidine triad H(X)3H(X)4DH characteristic of metallo-hydrolases, is also required for late-stage 70S ribosome quality control and 16S rRNA processing. Similarly, purified SmYbeY has metal-dependent endoribonuclease activity on synthetic and endogenous RNAs.Citation26 However, it is not required for 16S rRNA maturation and exhibits striking differences in substrate specificity with respect to what was proposed for its E. coli counterpart. SmYbeY is competent for cleaving both single-stranded (ssRNA) and double-stranded RNA (dsRNA) substrates, which is an unprecedented ability among bacterial endoribonucleases characterized to date. Nevertheless, in vitro assays evidenced certain preference of SmYbeY for degrading dsRNA. Specifically, reactivity patterns and catalytic efficiency of SmYbeY on the generic highly structured R1.1 RNA, which is typically used to assay RNase III activity, resembled cleavage of RNase III on the same substrate. Interestingly, SmYbeY-mediated catalysis showed strong metal co-factor dependence, with Mn2+ favoring overall cleavage efficiency on all substrates and Ca2+ specifically blocking reactivity on dsRNA and R1.1 RNA. This feature may have important implications for substrate selectivity and modulation of SmYbeY activity in vivo. In this regard, it is known that protection against the oxidative burst elicited in the host cells upon infection requires maintenance of Mn2+ and Ca2+ homeostasis in intracellular mammal pathogens and plant endosymbionts.Citation53,Citation54
This recent discovery extends the repertoire of RNases predicted to be encoded by the S. meliloti genome, which includes homologs of the known silencing endoribonucleases RNase E and RNase III, as well as the 3’-5’ exoribonuclease polynucleotide phosphorylase (PNPase), all of which have been shown to degrade the free pool of sRNAs in enterobacteria.Citation19,Citation55 It has been shown that S. meliloti RNase E promotes degradation of sinI and dnaA mRNAs upon base-pairing with EcpR1 and RcsR1 sRNAs in an Hfq-independent manner.Citation42,Citation43 RNase J is a ribonuclease functionally related to RNase E in the control of quorum sensing and S-adenosyl methionine (SAM) homeostasis in S. meliloti.Citation56 However, the contribution of these enzymes to RNA turnover and regulation in the group of plant symbiotic bacteria remains mostly unexplored.
SmYbeY influences decay of bulk and sRNA-regulated mRNAs
Transcriptomics uncovered a large SmYbeY-dependent RNA network integrated by more than 650 mRNAs (∼11% of the S. meliloti protein-coding genes) and hardly 35 trans-sRNAs (less than 6% of those identified in this bacterium).Citation26 Two-thirds of these transcripts, including most of trans-sRNAs, most likely are secondary molecular targets of SmYbeY, i.e., their steady-state levels are positively influenced by the endoribonuclease activity of SmYbeY. This set of mRNAs encodes fundamental cellular functions related to energy metabolism, RNA turnover and translation, whose downregulation would explain the pleiotropic phenotype associated to a SmYbeY loss-of-function. Comparison of the SmYbeY- and SmHfq-dependent gene sets revealed a rather discrete overlap, suggesting that despite of their commonalities, the free-living and symbiotic phenotypes of S. meliloti hfq and ybeY mutants rely on the alteration of largely different pathways.
Notwithstanding this major divergence in their RNA networks, several putative SmYbeY substrates can be foreseen among the known SmHfq-binding mRNAs whose intracellular concentration increases in the absence of SmYbeY. The fix cluster (fixN1O1Q1P1GHI1S1) and the nifA mRNA, encoding cytochromes associated with the nitrogenase complex and a master regulator of nitrogen fixation, respectively, are all downregulated in S. meliloti hfq mutants and recovered in their entire lengths by CoIP with SmHfq (; upper and middle panels). These findings suggest that SmHfq interacts with these mRNAs in a protective mode, thus impairing their degradation by SmYbeY. Therefore, SmYbeY would fulfil a role in bulk mRNA turnover in rhizobia that in enterobacteria has been largely attributed to RNase E.Citation12 Nonetheless, silencing of the nifA mRNA could be driven by the yet uncharacterized asRNA265 (; middle panel). In this particular case, SmYbeY would serve an RNase III-like role promoting degradation of the nifA mRNA by a primary cleavage at the RNA duplex. Recent RNAseq surveys have shown pervasive antisense transcription in S. meliloti, which is strikingly biased toward nodulation and nitrogen fixation genes within the pSymA megaplasmid.Citation25,Citation57 Despite of S. meliloti chromosome almost triples pSymA in size and gene content, transcriptomics revealed that SmYbeY has similar relative impact on gene expression in both replicons.Citation26 It is therefore tempting to speculate on a major role of SmYbeY in the antisense control of mRNA levels from symbiotic genes clustered in pSymA.
Figure 1. SmYbeY influences turnover of bulk and sRNA-regulated mRNAs. Shown are enrichment patterns of putative SmYbeY mRNA substrates in SmHfq CoIP-RNA.Citation27 The vertical axis indicate fold enrichment with respect to a control CoIP-RNA. Relevant genomic information for each mRNA is provided in the schematics below the plots. Numbering denotes coordinates in the S. meliloti Rm1021 genome. SmYbeY-mediated silencing of nifA and prbA mRNAs is likely triggered by base-pairing with the trans-sRNA AbcR2 and the asRNA265, respectively. See text for further details.
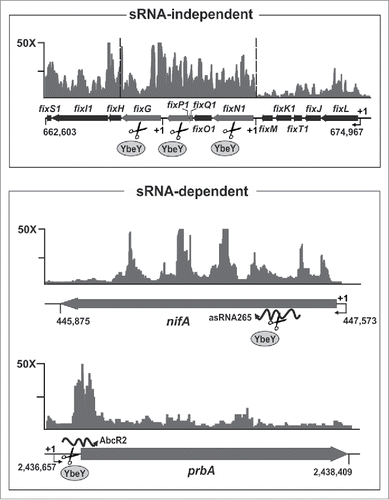
The set of SmYbeY and SmHfq co-regulated mRNAs includes several from amino acid transporter genes, whose expression is negatively influenced by both proteins.Citation26,Citation33 Interestingly, known targets of the SmHfq-dependent AbcR1/2 sRNAs were found among these mRNAs (e.g., livK or prbA).Citation27,Citation38 The enrichment patterns of these transcripts in SmHfq CoIP-RNA are compatible with their ribonucleolytic degradation upon SmHfq-assisted base pairing with AbcR1/2 at their 5’-UTR regions (; bottom panel). In vivo reporter and in vitro assays using the AbcR2-prbA interaction as a proof of principle further supported SmYbeY-mediated cleavage of this imperfect RNA duplex.Citation26 Existing in silico and experimental evidences suggest that the homologous AbcR1 and AbcR2 mRNAs govern a large regulon of transporter genes in S. meliloti, similar to that of GcvB in enterobacteria.Citation27,Citation38,Citation58 More recently, another S. meliloti trans-sRNA called NfeR1 has been shown to contribute to the Hfq-independent post-transcriptional silencing of ABC transporter mRNAs in response to external hyperosmolarity and during symbiosis.Citation59 Therefore, it is likely these 3 sRNAs and their suites of mRNA targets shape a dense overlapping regulon controlling nutrient uptake in response to different external cues, i.e., they show rather different expression profiles.Citation38,Citation59 Competition for shared regulatory sRNAs can promote mRNA cross-regulation via target mRNA-derived sponges that base-pair with and destabilize the sRNA, thereby relieving repression of the entire regulon.Citation60 In the GcvB network, RNase E catalyzes both biogenesis of the RNA sponge (i.e., SroC) and GcvB degradation.Citation37 It is therefore plausible that SmYbeY may be an active player in this layer of regulation of the particularly large network of S. meliloti ABC transporters.
Concluding remarks and future perspectives
As hubs in RNA networks, RNA-binding proteins and ribonucleases are global markers of sRNA function and activity mechanisms. In the α-rhizobia S. meliloti, the RNA chaperone SmHfq has a major role in riboregulation of nutrient uptake, but interacts with scarcely 14% of trans-sRNAs uncovered by RNAseq. This finding suggests that other yet unknown proteins may probably serve an Hfq-like function in S. meliloti. A recent screen of ribonucleoprotein complexes fractionated on a glycerol gradient (Grad-seq) identified ProQ as a new RNA-binding protein in S. enteria.Citation44 However, ProQ is not as ubiquitous as Hfq and, among α-rhizobia, only Rhizobium leguminosarum bv. viceae encodes a recognizable homolog of this protein.Citation61 Therefore, the search for new RNA-binding proteins assisting riboregulation in plant symbiotic bacteria remains opened.
Recent biochemical and genetic characterization of SmYbeY revealed that it behaves as a metal-dependent endoribonuclease rather than as an Hfq-like chaperone, as suggested by previous structural predictions and phenotypic evidences. SmYbeY indistinctly cleaves ssRNA and dsRNA, which is a unique feature among known bacterial endoribonucleases. The crystal structure, mutagenesis analysis and genome-wide mapping of SmYbeY cleavage sites will shed light about the molecular bases of this catalytic versatility and its regulation by different metal co-factors. SmYbeY influences turnover of bulk and sRNA-regulated mRNAs. The discrete contribution of SmYbeY to the different levels of RNA regulation, i.e., sRNA turnover and target mRNA decay, as well as the identity of its protein partners must be further explored in the near future. In particular, the role of SmYbeY in mRNA silencing driven by antisense transcription will be key to understand post-transcriptional regulation of genuine symbiotic genes.
Acknowledgments
We are grateful to Cecilia Arraiano for critical reading of the manuscript.
Funding
This work was funded by the Spanish Ministerio de Economía y Competitividad ERDF-cofinanced grant BFU2013–48282-C2–2-P. M.R. was funded by a contract of the program of “Formación Post-doctoral” from the same Ministry.
References
- Gottesman S, Storz G. Bacterial small RNA regulators: versatile roles and rapidly evolving variations. Cold Spring Harb Perspect Biol. 2011;3:a003798. doi:10.1101/cshperspect.a003798.
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi:10.1016/j.molcel.2011.08.022.
- Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr Opin Microbiol. 2004;7:140–4. doi:10.1016/j.mib.2004.02.015.
- Papenfort K, Vanderpool CK. Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev. 2015;39:362–78. doi:10.1093/femsre/fuv016.
- Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi:10.1038/nrmicro2615.
- Updegrove TB, Zhang A, Storz G. Hfq: the flexible RNA matchmaker. Curr Opin Microbiol. 2016;30:133–8. doi:10.1016/j.mib.2016.02.003.
- Sobrero P, Valverde C. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit Rev Microbiol. 2012;38:276–99. doi:10.3109/1040841X.2012.664540.
- Wagner EGH, Romby P. Small RNAs in bacteria and archaea: who they are, what they do, and how they do it. In: Theodore Friedmann JCD, Stephen FG, editors. Adv Genet. USA: Academic Press; 2015. p. 133–208.
- De Lay N, Schu DJ, Gottesman S. Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem. 2013;288:7996–8003. doi:10.1074/jbc.R112.441386.
- Morita T, Aiba H. RNase E action at a distance: degradation of target mRNAs mediated by an Hfq-binding small RNA in bacteria. Genes Dev. 2011;25:294–8. doi:10.1101/gad.2030311.
- Lalaouna D, Simoneau-Roy M, Lafontaine D, Massé E. Regulatory RNAs and target mRNA decay in prokaryotes. Biochimica et Biophysica Acta (BBA) – Gene Regulatory Mechanisms. 2013;1829:742–7. doi:10.1016/j.bbagrm.2013.02.013
- Arraiano CM, Andrade JM, Domingues S, Guinote IB, Malecki M, Matos RG, Moreira RN, Pobre V, Reis FP, Saramago M, et al. The critical role of RNA processing and degradation in the control of gene expression. FEMS Microbiol Rev. 2010;34:883–923. doi:10.1111/j.1574-6976.2010.00242.x.
- Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penadés JR, Valle J, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci USA. 2011;108:20172–7. doi:10.1073/pnas.1113521108.
- Saramago M, Bárria C, Arraiano CM, Domingues S. Ribonucleases, antisense RNAs and the control of bacterial plasmids. Plasmid. 2015;78:26–36. doi:10.1016/j.plasmid.2014.09.003.
- Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu Rev Genet. 2008;42:413–41. doi:10.1146/annurev.genet.42.110807.091427.
- Jiménez-Zurdo JI, Valverde C, Becker A. Insights into the noncoding RNome of nitrogen-fixing endosymbiotic α-proteobacteria. Mol Plant-Microbe Interact. 2013;26:160–7. doi:10.1094/MPMI-07-12-0186-CR.
- Jiménez-Zurdo JI, Robledo M. Unraveling the universe of small RNA regulators in the legume symbiont Sinorhizobium meliloti. Symbiosis. 2015;67:43–54. doi:10.1007/s13199-015-0345-z
- Becker A, Overlöper A, Schlüter J-P, Reinkensmeier J, Robledo M, Giegerich R, Narberhaus F, Evguenieva-Hackenberg E. Riboregulation in plant-associated α-proteobacteria. RNA Biol. 2014;11:550–62. doi:10.4161/rna.29625.
- Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, Barloy-Hubler F, Barnett MJ, Becker A, Boistard P, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–72. doi:10.1126/science.1060966.
- Sallet E, Roux B, Sauviac L, Jardinaud M-Fo, Carrère S, Faraut T, de Carvalho-Niebel F, Gouzy J, Gamas P, Capela D, et al. Next-generation annotation of prokaryotic genomes with EuGene-P: application to Sinorhizobium meliloti 2011. DNA Res. 2013;20:339–54. doi:10.1093/dnares/dst014.
- del Val C, Rivas E, Torres-Quesada O, Toro N, Jiménez-Zurdo JI. Identification of differentially expressed small non-coding RNAs in the legume endosymbiont Sinorhizobium meliloti by comparative genomics. Mol Microbiol. 2007;66:1080–91. doi:10.1111/j.1365-2958.2007.05978.x.
- Valverde C, Livny J, Schluter JP, Reinkensmeier J, Becker A, Parisi G. Prediction of Sinorhizobium meliloti sRNA genes and experimental detection in strain 2011. BMC Genomics. 2008;9:416. doi:10.1186/1471-2164-9-416.
- Ulve VM, Sevin EW, Cheron A, Barloy-Hubler F. Identification of chromosomal alpha-proteobacterial small RNAs by comparative genome analysis and detection in Sinorhizobium meliloti strain 1021. BMC Genomics. 2007;8:467. doi:10.1186/1471-2164-8-467.
- Schlüter JP, Reinkensmeier J, Daschkey S, Evguenieva-Hackenberg E, Janssen S, Janicke S, Becker JD, Giegerich R, Becker A. A genome-wide survey of sRNAs in the symbiotic nitrogen-fixing alpha-proteobacterium Sinorhizobium meliloti. BMC Genomics. 2010;11:245. doi:10.1186/1471-2164-11-245.
- Schlüter JP, Reinkensmeier J, Barnett MJ, Lang C, Krol E, Giegerich R, Long SR, Becker A. Global mapping of transcription start sites and promoter motifs in the symbiotic alpha-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics. 2013;14:156. doi:10.1186/1471-2164-14-156.
- Saramago M, Peregrina A, Robledo M, Matos RG, Hilker R, Serrania J, Becker A, Arraiano CM, Jiménez-Zurdo JI. Sinorhizobium meliloti YbeY is an endoribonuclease with unprecedented catalytic features, acting as silencing enzyme in riboregulation. Nucleic Acids Res. 2017;45:1371–91. doi:10.1093/nar/gkw1234.
- Torres-Quesada O, Reinkensmeier J, Schlüter J-P, Robledo M, Peregrina A, Giegerich R, Toro N, Becker A, Jiménez-Zurdo JI. Genome-wide profiling of Hfq-binding RNAs uncovers extensive post-transcriptional rewiring of major stress response and symbiotic regulons in Sinorhizobium meliloti. RNA Biol. 2014;11:563–79. doi:10.4161/rna.28239.
- Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet. 2008;4:e1000163. doi:10.1371/journal.pgen.1000163.
- Berghoff BA, Glaeser J, Sharma CM, Zobawa M, Lottspeich F, Vogel J, Klug G. Contribution of Hfq to photooxidative stress resistance and global regulation in Rhodobacter sphaeroides. Mol Microbiol. 2011;80:1479–95. doi:10.1111/j.1365-2958.2011.07658.x.
- Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3' UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–19. doi:10.1038/emboj.2012.229.
- Sobrero P, Schluter JP, Lanner U, Schlosser A, Becker A, Valverde C. Quantitative proteomic analysis of the Hfq-regulon in Sinorhizobium meliloti 2011. PLoS One. 2012;7:e48494. doi:10.1371/journal.pone.0048494.
- Barra-Bily L, Pandey SP, Trautwetter A, Blanco C, Walker GC. The Sinorhizobium meliloti RNA chaperone Hfq mediates symbiosis of Sinorhizobium meliloti and alfalfa. J Bacteriol. 2010a;192:1710–8. doi:10.1128/JB.01427-09
- Torres-Quesada O, Oruezabal RI, Peregrina A, Jofre E, Lloret J, Rivilla R, Toro N, Jiménez-Zurdo JI. The Sinorhizobium meliloti RNA chaperone Hfq influences central carbon metabolism and the symbiotic interaction with alfalfa. BMC Microbiol. 2010;10:71. doi:10.1186/1471-2180-10-71.
- Gao M, Barnett MJ, Long SR, Teplitski M. Role of the Sinorhizobium meliloti global regulator Hfq in gene regulation and symbiosis. Mol Plant Microbe Interact. 2010;23:355–65. doi:10.1094/MPMI-23-4-0355.
- Barra-Bily L, Fontenelle C, Jan G, Flechard M, Trautwetter A, Pandey SP, et al. Proteomic alterations explain phenotypic changes in Sinorhizobium meliloti lacking the RNA chaperone Hfq. J Bacteriol. 2010b;192:1719–29. doi:10.1128/JB.01429-09
- Voss B, Holscher M, Baumgarth B, Kalbfleisch A, Kaya C, Hess WR, Becker A, Evguenieva-Hackenberg E. Expression of small RNAs in Rhizobiales and protection of a small RNA and its degradation products by Hfq in Sinorhizobium meliloti. Biochem Biophys Res Commun. 2009;390:331–6. doi:10.1016/j.bbrc.2009.09.125.
- Miyakoshi M, Chao Y, Vogel J. Cross talk between ABC transporter mRNAs via a target mRNA‐derived sponge of the GcvB small RNA. EMBO J. 2015;34:1478–92. doi:10.15252/embj.201490546.
- Torres-Quesada O, Millán V, Nisa-Martínez R, Bardou F, Crespi M, Toro N, Jiménez-Zurdo JI. Independent activity of the homologous small regulatory RNAs AbcR1 and AbcR2 in the legume symbiont Sinorhizobium meliloti. PLoS One. 2013;8:e68147. doi:10.1371/journal.pone.0068147.
- Overlöper A, Kraus A, Gurski R, Wright PR, Georg J, Hess WR, Narberhaus F. Two separate modules of the conserved regulatory RNA AbcR1 address multiple target mRNAs in and outside of the translation initiation region. RNA Biol. 2014;11:624–40. doi:10.4161/rna.29145.
- Caswell CC, Gaines JM, Ciborowski P, Smith D, Borchers CH, Roux CM, Sayood K, Dunman PM, Roop Ii RM. Identification of two small regulatory RNAs linked to virulence in Brucella abortus 2308. Mol Microbiol. 2012;85:345–60. doi:10.1111/j.1365-2958.2012.08117.x.
- MacLellan SR, Smallbone LA, Sibley CD, Finan TM. The expression of a novel antisense gene mediates incompatibility within the large repABC family of α-proteobacterial plasmids. Mol Microbiol. 2005;55:611–23. doi:10.1111/j.1365-2958.2004.04412.x.
- Robledo M, Frage B, Wright PR, Becker A. A stress-induced small RNA modulates alpha-rhizobial cell cycle progression. PLoS Genet. 2015;11:e1005153. doi:10.1371/journal.pgen.1005153.
- Baumgardt K, Šmídová K, Rahn H, Lochnit G, Robledo M, Evguenieva-Hackenberg E. The stress-related, rhizobial small RNA RcsR1 destabilizes the autoinducer synthase encoding mRNA sinI in Sinorhizobium meliloti. RNA Biol. 2015;13:486–99. doi:10.1080/15476286.2015.1110673.
- Smirnov A, Förstner KU, Holmqvist E, Otto A, Günster R, Becher D, Reinhardt R, Vogel J. Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci USA. 2016;113:11591–6. doi:10.1073/pnas.1609981113.
- Pandey SP, Minesinger BK, Kumar J, Walker GC. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 2011;39:4691–708. doi:10.1093/nar/gkr060.
- Gil R, Silva FJ, Peretó J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev. 2004;68:518–37. doi:10.1128/MMBR.68.3.518-537.2004.
- Davies BW, Walker GC. A highly conserved protein of unknown function is required by Sinorhizobium meliloti for symbiosis and environmental stress protection. J Bacteriol. 2008;190:1118–23. doi:10.1128/JB.01521-07.
- Davies BW, Köhrer C, Jacob AI, Simmons LA, Zhu J, Aleman LM, Rajbhandary UL, Walker GC. Role of Escherichia coli YbeY, a highly conserved protein, in rRNA processing. Mol Microbiol. 2010;78:506–18. doi:10.1111/j.1365-2958.2010.07351.x.
- Leskinen K, Varjosalo M, Skurnik M. Absence of YbeY RNase compromises the growth and enhances the virulence plasmid gene expression of Yersinia enterocolitica O:3. Microbiology. 2015;161:285–99. doi:10.1099/mic.0.083097-0.
- Vercruysse M, Köhrer C, Davies BW, Arnold MFF, Mekalanos JJ, RajBhandary UL, Walker GC. The highly conserved bacterial RNase YbeY is essential in Vibrio cholerae, playing a critical role in virulence, stress regulation, and RNA processing. PLoS Pathog. 2014;10:e1004175. doi:10.1371/journal.ppat.1004175.
- Pandey SP, Winkler JA, Li H, Camacho DM, Collins JJ, Walker GC. Central role for RNase YbeY in Hfq-dependent and Hfq-independent small-RNA regulation in bacteria. BMC Genomics. 2014;15:121. doi:10.1186/1471-2164-15-121.
- Jacob Asha I, Köhrer C, Davies Bryan W, RajBhandary Uttam L, Walker Graham C. Conserved bacterial RNase YbeY plays key roles in 70S ribosome quality control and 16S rRNA maturation. Mol Cell. 2013;49:427–38. doi:10.1016/j.molcel.2012.11.025.
- Davies BW, Walker GC. Disruption of sitA Compromises Sinorhizobium meliloti for manganese uptake required for protection against oxidative stress. J Bacteriol. 2007;189:2101–9. doi:10.1128/JB.01377-06.
- Papp-Wallace KM, Maguire ME. Manganese transport and the role of manganese in virulence. Ann Rev Microbiol. 2006;60:187–209. doi:10.1146/annurev.micro.60.080805.142149
- Saramago M, Bárria C, dos Santos RF, Silva IJ, Pobre V, Domingues S, et al. The role of RNases in the regulation of small RNAs. Curr Opin Microbiol. 2014;18:105–15. doi:10.1016/j.mib.2014.02.009.
- Baumgardt K, Melior H, Madhugiri R, Thalmann S, Schikora A, McIntosh M, Becker A, Evguenieva-Hackenberg E. RNase E and RNase J are needed for S-adenosylmethionine homeostasis in Sinorhizobium meliloti. Microbiology. 2017;163:570–83. doi:10.1099/mic.0.000442.
- Robledo M, Jiménez-Zurdo JI, Becker A. Antisense transcription of symbiotic genes in Sinorhizobium meliloti. Symbiosis. 2015;67:55–67. doi:10.1007/s13199-015-0358-7
- Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf H-J, Hinton JCD, Vogel J. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol. 2011;81:1144–65. doi:10.1111/j.1365-2958.2011.07751.x.
- Robledo M, Peregrina A, Millán V, García-Tomsig NI, Torres-Quesada O, Mateos PF, et al. A conserved α-proteobacterial small RNA contributes to osmoadaptation and symbiotic efficiency of rhizobia on legume roots. Environ Microbiol. 2017;19:2661–80. doi:10.1111/1462-2920.13757.
- Bossi L, Figueroa-Bossi N. Competing endogenous RNAs: a target-centric view of small RNA regulation in bacteria. Nat Rev Micro. 2016;14:775–84. doi:10.1038/nrmicro.2016.129
- Olejniczak M, Storz G. ProQ/FinO-domain proteins: Another ubiquitous family of RNA matchmakers? Mol Microbiol. 2017;104:905–15. doi:10.1111/mmi.13679.
