ABSTRACT
tRNA-dependent addition of amino acids to lipids on the outer surface of the bacterial membrane results in decreased effectiveness of antimicrobials such as cationic antimicrobial peptides (CAMPs) that target the membrane, and increased virulence of several pathogenic species. After a brief introduction to CAMPs and the various bacterial resistance mechanisms used to counteract these compounds, this review focuses on recent advances in tRNA-dependent pathways for lipid modification in bacteria. Phenotypes associated with amino acid lipid modifications and regulation of their expression will also be discussed.
Introduction
The bacterial cell wall exhibits a high net negative charge due to the presence of phosphate as a building block in many of its molecular constituents. The phospholipids that form the membrane, as well as the teichoic acid chains spanning the cell wall in Gram-positive bacteria, contain phosphate groups that give these structures their negative charge. All forms of life- including bacteria, fungi, plants, and animals- have the ability to produce cationic antimicrobial peptides (CAMPs) to inhibit bacterial growth. CAMPs possess antimicrobial properties and exhibit a high affinity for the negatively charged components within the bacterial cell wall.
After a brief introduction on CAMPs and their distinctive modes of action, as well as various mechanisms used by bacteria to counteract CAMP activity, this review will focus on a particular mechanism of CAMP resistance that utilizes aminoacylated tRNAs (aa-tRNAs) to modify lipids in the bacterial envelope. Several reviews describing this mechanism have been published within the past few years.Citation1-Citation4 This manuscript sets out to review recent advances in the field and outline current insights regarding the structure-function relationship of tRNA-dependent lipid modification systems, and of the flipping mechanism of these modified lipids across the membrane. Examples of novel tRNA-dependent lipid modifications that have recently been discovered, and the enzymes responsible for their synthesis, will be discussed.
Cationic antimicrobial peptides (CAMPs) and bacterial resistance to CAMPs
CAMPs are found in all domains of life. They vary in sequence, size (10–50 amino acids (aa)), and secondary structure, but all share an amphipathic nature and exhibit a net positive charge (usually between +2 and +9) brought about by a lateral chain containing Arg and Lys residues. CAMPs fall into four distinct structural classes: α-helical, β-stranded, linear, and looped. α-helical, β-stranded, and linear CAMPs are mostly encoded in eukaryotic cells, whereas looped CAMPs (i.e., polymyxins) are primarily encoded in bacteria.Citation5 Some CAMPs are ribosomally synthesized, while others (e.g., polymyxinsCitation6) are produced by large multifunctional enzymes. The predominant mode of action used by these compounds involves pore formation and general disruption of the bacterial membrane upon binding of CAMPs to negatively charged phospholipids. Certain CAMPs, however, possess a targeted mode of action. For example, bacteriocins (CAMPs secreted by bacteria) such as nisin bind to Lipid II, which is the precursor for peptidoglycan synthesis. Another example, Polymyxin B, forms pores in the outer membrane of gram-negative bacteria after specific interaction with lipid A at the surface of the cell. It is worth mentioning, that at high concentrations, and in the absence of their high affinity molecular targets, both nisin and polymyxin B display general pore forming properties in the cytoplasmic membrane.Citation7 Beyond direct interaction with membrane components, certain CAMPs pass through the cytoplasmic membrane to inhibit intracellular targets. For instance, the bacterial peptide microcin J25 (MccJ25) inhibits RNA polymerase,Citation8 while apidaecins (in honeybees), oncocins (in insects), and Bactenecin-7 (in mammalian cells) inhibit bacterial translation by binding to ribosomal proteins.Citation9,Citation10
CAMPs secreted as part of the innate immune system in eukaryotes, as well as those produced by bacteria to inhibit growth of other microbial species, are the result of millions of years of co-evolution. It is proposed that the high diversity of structure and function among eukaryotic CAMPs has been shaped by a variety of crosstalk between host cells and bacterial species.Citation11,Citation12 The diversity of CAMPs in eukaryotes is illustrated in the Antimicrobial Peptide Database,Citation13 which lists more than 2,500 CAMPs of eukaryotic origin (122 in humanCitation14) and only 301 CAMPs of prokaryotic origin (only four were identified in archae). In mammals, CAMPs are produced and secreted by epithelial cells (i.e., skin, gastrointestinal, respiratory, and urogenital cells) and various cells of the immune system (e.g., monocytes, macrophages, neutrophils, and other granulocytes; for review see [Citation15], ). These systems use secreted CAMPs to kill bacteria outside the cell as well as intracellular pathogens enclosed inside the phagosome. Constitutive expression of CAMPs is developmentally regulated and influenced by age. Upregulation of CAMPs occurs during inflammation and infection. CAMPs produced in eukaryotic species not only play a direct role in the killing of bacterial pathogens, but also in the modulation of the immune response by, for example, decreasing inflammation and attracting various types of immune cells during infection (i.e., chemotaxis).Citation15 Additionally, certain CAMPs stimulate production of chemokines and cytokines from a variety of cell types, constituting an indirect mechanism for stimulating the immune response.Citation15
Figure 1. Multiple functions of CAMPs in host defense. CAMPs induce a variety of responses in host innate immune cells. They alter gene expression, induce production of chemokines and cytokines, promote recruitment of immune cells to the site of infection (chemotaxis), and inhibit growth of pathogenic species infecting the host. Selective control of inflammation in response to CAMP activity promotes wound healing and initiation of adaptive immune responses. LPS, lipopolysaccharide. Figure adapted from [Citation14,Citation15,Citation115].
![Figure 1. Multiple functions of CAMPs in host defense. CAMPs induce a variety of responses in host innate immune cells. They alter gene expression, induce production of chemokines and cytokines, promote recruitment of immune cells to the site of infection (chemotaxis), and inhibit growth of pathogenic species infecting the host. Selective control of inflammation in response to CAMP activity promotes wound healing and initiation of adaptive immune responses. LPS, lipopolysaccharide. Figure adapted from [Citation14,Citation15,Citation115].](/cms/asset/8c0d2175-2b35-4872-a895-63cca75141d5/krnb_a_1356980_f0001_b.gif)
When compared with the cell membranes of bacterial species, the cell membranes of multicellular organisms possess many fundamental differences in lipid composition that allow them to be more naturally resistant to CAMPs. The membranes of plant and animal cells are composed of zwitterionic lipids (e.g., phosphatidylethanolamine) that bear no net charge, and are therefore unable to interact efficiently with CAMPs. Eukaryotic membranes also contain lipids such as cholesterol, which stabilize the membrane and contribute to CAMP resistance.Citation16 In contrast, bacteria exhibit high levels of anionic components in their cell envelopes (phospholipids in the membrane, and teichoic acid chains in the cell walls of Gram-positive bacteria), which support efficient interaction with CAMPs. Because of this, bacteria have developed a battery of mechanisms to increase their resistance to CAMPs ( for review see [Citation17,Citation18]). Several of these mechanisms function by adding positively charged modifications to components of the cell wall to decrease binding to CAMPs.
Figure 2. Bacterial defenses against CAMPs and mechanisms for lowering the net negative charge of the cell envelope. CAMP resistance mechanisms include (1) efflux pumps, (2) CAMP-binding or hydrolyzing agents (3) formation of biofilms, and (4) addition of positively charged modifications to negatively charged substructures in the cell wall or lipid membrane. Addition of positively charged groups to the cell wall lowers the net negative charge of the envelope, decreasing binding of cationic compounds such as CAMPs and subsequently increasing bacterial resistance to these antimicrobials. In Gram-negative bacteria, lipid A in the outer membrane is the substrate for various positively charged modifications. Biosynthesis pathways for these modifications are not shown (For details see [Citation116-Citation118]). In certain species of both Gram-positive and Gram-negative bacteria, aminoacyl-phosphatidylglycerol synthases (aaPGSs) transfer aa from aa-tRNA (pre-formed by cytosolic aa-tRNA synthetases, aaRS) to phosphatidylglycerol (PG) in the membrane. In Gram positive bacteria, D-alanylation of lipoteichoic acid chains involves the enzymes encoded by the dltABCD operon. Positively and negatively charged molecules are represented in blue and red, respectively.
![Figure 2. Bacterial defenses against CAMPs and mechanisms for lowering the net negative charge of the cell envelope. CAMP resistance mechanisms include (1) efflux pumps, (2) CAMP-binding or hydrolyzing agents (3) formation of biofilms, and (4) addition of positively charged modifications to negatively charged substructures in the cell wall or lipid membrane. Addition of positively charged groups to the cell wall lowers the net negative charge of the envelope, decreasing binding of cationic compounds such as CAMPs and subsequently increasing bacterial resistance to these antimicrobials. In Gram-negative bacteria, lipid A in the outer membrane is the substrate for various positively charged modifications. Biosynthesis pathways for these modifications are not shown (For details see [Citation116-Citation118]). In certain species of both Gram-positive and Gram-negative bacteria, aminoacyl-phosphatidylglycerol synthases (aaPGSs) transfer aa from aa-tRNA (pre-formed by cytosolic aa-tRNA synthetases, aaRS) to phosphatidylglycerol (PG) in the membrane. In Gram positive bacteria, D-alanylation of lipoteichoic acid chains involves the enzymes encoded by the dltABCD operon. Positively and negatively charged molecules are represented in blue and red, respectively.](/cms/asset/c19680ac-a207-4683-9428-acd6dd5f14e5/krnb_a_1356980_f0002_oc.jpg)
tRNA-dependent aminoacylation of membrane phosphatidylglycerol
Phosphatidylglycerol (PG) is a ubiquitous anionic phospholipid found in all domains of life and is particularly abundant in bacterial membranes.Citation19 PG contains a phosphate group, which confers a net negative charge (−1), and a terminal glycerol group characterized by hydroxyls that can be exploited to attach different types of modifications. These modifications include addition of a phosphate or sulfate group to form PG-phosphate or PG-sulfate in Archaea. Addition of a carbohydrate or an amino acid (aa) such as Lys, Ala, or Arg results in formation of the corresponding aminoacyl-PG (aa-PGs) in bacteria. Lastly, condensation of 2 PG molecules to form di-PG (also called cardiolipin, CL) is a process that occurs in all domains of life. Modified PGs provide the cell with a variety of altered components that can affect general properties of the membrane such as surface charge, fluidity, and morphology (for review see [Citation1,Citation2,Citation19]).
Lys-PG was discovered in the membrane of S. aureus in the mid 1960s by William Lennarz, who established that synthesis of this modified lipid requires lysyslated tRNA (Lys-tRNA) to serve as an amino acid donor.Citation20,Citation21 The bacterial enzyme responsible for synthesis of Lys-PG was not determined until 40 y later during a genetic screen aimed at identifying mutants of S. aureus that are sensitive to CAMPs.Citation22 Peschel and coauthors were the first to establish that lysyl-PG synthase (LysPGS), encoded by the gene mprF (multiple peptide resistance factor), enhances resistance of S. aureus to 10 CAMPs from various origins. The minimum inhibitory concentrations (MIC) for certain CAMPs was decreased by 30-fold in a strain lacking mprF.Citation23 In more recent years, it was shown that LysPGS is only one member of a large family of aminoacyl-PG synthase (aaPGS) homologs spanning nearly all known bacterial phyla. Some homologs are also present in archaeal methanogens,Citation24 but no examples have been identified in eukaryotic genomes.Citation1 Aminoacyl-phosphatidylglycerol synthases (aaPGSs), as these enzymes are collectively called, are bifunctional enzymes. They exhibit a cytosolic C-terminal domain bearing the tRNA-dependent aa-PG synthase activity, and an N-terminal integral membrane domain made up of a variable number (2 to 14) of predicted transmembrane helices (TMH). The membrane domain is responsible for flipping neosynthesized aa-PGs across the membrane from the cytoplasmic side where they are formed into the periplasm.Citation25-Citation28 Interestingly, some Actinobacteria such as Mycobacterium tuberculosis display a large aaPGS homolog called LysX. A bioinformatics analysis showed that LysX encompasses, like other aaPGSs, an integral membrane domain and an aaPGS synthase domain, but also includes a lysyl-tRNA synthetase (LysRS) domain at the C-terminus that exhibits a high level of similarity to the LysRS of the cytosolic translation machinery. It has been suggested that this protein is able to mediate each step of the lipid aminoacylation pathway, including tRNA aminoacylation, and by doing so increases the efficiency of lipid modification. The LysRS domain of LysX is required for synthesis of lysylated phosphatidylglycerol in M. tuberculosis, suggesting that free Lys-tRNALys produced by the translation machinery is not used by LysX for lipid modification.Citation29
Several bacterial species exhibit more than one aaPGS homolog (paralogs), suggesting a functional divergence exists among co-occurring proteins. Thus, in recent years the specificities of various aaPGS homologs have been investigated in different bacterial backgrounds. Several enzymes with altered aa-tRNA and lipid substrate specificities have been identified, and the recently solved crystal structures of two aaPGSs have shed light on the molecular mechanism of tRNA-dependent systems for lipid aminoacylation.Citation30
aaPGS mediated lipid remodeling uses several aa-tRNAs as substrates
Clostridium perfringens exhibits two aaPGSs: a LysPGS responsible for synthesis of Lys-PG and an AlaPGS responsible for synthesis of Ala-PG.Citation31 It has been established in vitro that some aaPGSs exhibit promiscuity for aa-tRNA recognition and can utilize multiple aa-tRNAs, while some are specific for a unique aa-tRNA.Citation32 For example, the LysPGS from Agrobacterium tumefaciens and the AlaPGS from C. perfringens specifically use Lys-tRNA and Ala-tRNA respectively, while the aaPGS from Bacillus subtilis utilizes both Lys- and Ala-tRNA as amino acid donors in vitro.Citation32 Interestingly both Lys-PG and Ala-PG were initially detected in the membrane of B. subtilis,Citation33 but a recent report suggests that D-Ala-PG and not L-Ala-PG is formed in B. subtilis by an aaPGS independent pathway.Citation34 One paralog in Enterococcus faecium (referred to as RakPGS or MprF2) displays a more relaxed specificity for its aa-tRNA and can utilize Ala-, Lys-, and Arg-tRNAs.Citation32,Citation35-Citation37 Organisms that harbor multi-specific aaPGSs produce an expanded repertoire of distinct aa-PGs in their bacterial membranes.
It was established early on that only the acceptor stem of the aa-tRNA is important for aaPGS activity. A minihelix encompassing just the acceptor stem and TYC stem loop of tRNAAla is as efficiently recognized as full length tRNA by AlaPGS.Citation31 Further investigations with the AlaPGS from P. aeruginosa revealed that a microhelix (a hairpin consisting of 7 base pairs) is the minimal substrate for AlaPGS activity. An in-depth study showed that the Ala moiety and the fifth base pair of the tRNA helix constitute the major elements for efficient recognition of Ala-tRNAAla.Citation38 Several studies have tried to determine the phenotypical changes correlated with synthesis of Lys-PG compared with Ala-PG in S. aureus and P. aeruginosa.Citation24,Citation30 These investigations showed that substitution of Lys-PG (which confers a net charge of +1) with Ala-PG (which bears a neutral net charge) did not affect bacterial susceptibility to CAMPs such as nisin and gallidermin, or to the CAMP-like antibiotic daptomycin. These findings suggest that the α amino group of the aa moiety of aa-PG alone is able to enhance antimicrobial resistance in bacteria.
The cytosolic domain of aaPGS exhibits a GCN5-like acetyltransferase fold
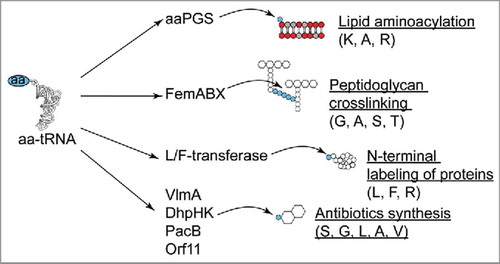
Until recently, no significant structural relationship was established between aaPGSs and other known proteins using conventional sequence search methodologies (i.e., BLAST). However, recent reports using more sensitive methods for homology detection (i.e., profile Hidden Markov Models, HMM) suggested that aaPGSs exhibited a domain resembling a GCN5-like acetyltransferase (GNAT) fold, which is a fold found in several other aa-tRNA transferases, (Citation39,Citation40 and see below). These observations were recently confirmed by the crystal structures of aaPGSs from Bacillus licheniformis and Pseudomonas aeruginosa.Citation30 In the Protein Family database (PfamCitation41), the GNAT fold is found in 39 families of transferases which altogether constitute the N-acyltransferases clan (CL0257). This clan includes many acyl-CoA and aa-tRNA transferases. Enzymes of the latter category (summarized in ) include the Fem proteins, which utilize aa-tRNAs to synthesize a connecting peptide to bridge peptidoglycan chains.Citation42 This bridging peptide exhibits up to 5 aa. Depending on the bacterial species, the connecting peptide can be made of Gly, Ala, Ser, or Thr. These aa are sequentially added to the cytoplasmic precursor of peptidoglycan (i.e., UDP-N-acetyl-muramyl-pentapeptide) by distinct Fem proteins (for review see [Citation3,Citation43]). This pathway is an alternate route to the direct bridging of peptidoglycan chains, and uses a set of penicillin binding proteins exhibiting a low affinity for β-lactam antibiotics (e.g., protein mecA in methicillin resistant S. aureus).Citation42 Another family of tRNA-dependent enzymes exhibiting a GNAT fold are Arg- and Leu/Phe-transferases, which transfer Arg, Leu, or Phe to the N-termini of proteins, thereby targeting them for degradation via the ClpS-ClpXP-mediated N-end rule pathway (see [Citation44] for review). The GNAT fold was recently identified in many more tRNA-dependent transferases in Streptomyces species, which utilize specific aa-tRNAs for synthesis of broad-spectrum antibiotics. For instance, the protein VlmA utilizes Ser-tRNASer for synthesis of the antibiotic valanimycinCitation45; DhpH and DhpK utilize Gly-tRNAGly and Leu-tRNALeu for biosynthesis of tripeptide phosphonateCitation46; the protein PacB utilizes Ala-tRNAAla in the biosynthetic pathway for pacidamycinCitation47; FzmI uses Val-tRNAVal for synthesis of fosfazinomycinCitation48; Orf11 uses Gly-tRNAGly for the synthesis of a streptothricin-related antibiotic.Citation49
Figure 3. Role of aa-tRNA transferases exhibiting a GNAT fold. aa-tRNA specificities are indicated in brackets. See text for details.
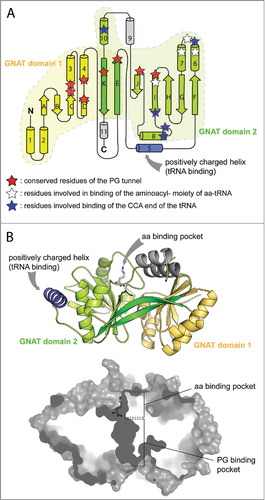
The structures of the transferase domains of the AlaPGS from P. aeruginosa and the LysPGS from B. licheniformis (in complex with the Lys analog lysinamide) revealed tandem repeated GNAT folds (). The strongest homologies with aaPGS structures were found within proteins of the Fem family. The active site of aaPGS is composed of two cavities located on opposite sides of the protein linked by a short bottlenecked tunnel.Citation30 One cavity, which is presumably facing the cytoplasm, is responsible for binding the aa born by the tRNA. The second cavity is more hydrophobic and, opening onto the membrane side, is responsible for binding to PG.Citation30 This structural work, in combination with biochemical experiments using protein variants and substrate analogs, showed that catalysis takes place at the bottleneck between the two substrate-binding pockets (). Regarding catalysis, an Arg residue, bound to the α carbonyl group of the co-crystalized amino acid analog, is proposed to increase the electrophilicity of the ester bond of the aa-tRNA mediating the nucleophilic episode of the 3′hydroxyl group of PG. A similar role in catalysis was proposed for a Lys residue in the structure of the alanyl-tRNA transferase FemX,Citation50 which is the strongest structural homolog of both crystallized aaPGSs.Citation30 These structural investigations also confirmed that the acceptor stem, the aminoacyl moiety of the aa-tRNA, and the terminal glycerol group of PG constitute the main determinants for substrate recognition. Comparison of the aaPGS structure to that of FemX in complex with an aa-tRNA analog showed that binding of the acceptor stem of the tRNA is mediated through interactions of a positively charged helix in the protein born by one GNAT fold, which represents an important structural feature common to all aa-tRNA transferases (i.e., Fem proteins, L/F-transferases, and aaPGS).Citation30
Figure 4. X-ray structures of the catalytic domains of AlaPGS from P. aeruginosa and LysPGS from B. licheniformis. A. Topology of the tandem GNAT fold repeat in aaPGSs. B. Structure of LysPGS with the Lys analog L-lysinamide bound to the aa binding pocket. Superposition of the FemX structure in complex with the tRNACitation50 reveal a possible binding mechanism of the tRNA, with helix 5 exhibiting basic residues. The cross-section of the structure shows the aa and PG binding pockets; the active site is located at the interface between the 2 pockets.
Flippase domain of aaPGS
It was only recently that the lipid flippase activity of aaPGS was directly demonstrated.Citation27 The flippase domain of aaPGS is an integral membrane domain consisting of a variable number (up to 14) of predicted TMHs. The aaPGS flippase activity has been investigated with the S. aureus LysPGS, whose membrane domain consists of 14 TMHs. In this organism, flipping of Lys-PG from the inner to the outer leaflet of the cytoplasmic membrane is necessary for optimal resistance to CAMPs.Citation27 Although the synthase domain of S. aureus LysPGS exhibits a strict specificity for its aa-tRNA, the flippase domain is able to flip Ala-PG or Lys-PG.Citation28 A recent study verified the membrane localization of the flippase domain, as well as the topology and oligomerization state of the protein inside the membrane.Citation25 This work showed that the region encompassing TMHs 1 through 6 supports the flippase activity of LysPGS, whereas TMHs 7 through 14 enhance LysPGS activity most likely by stabilizing the synthetic domain at the surface of the membrane, or by participating in selection and binding of PG. Mutagenesis of the protein revealed residues involved in the flippase mechanism. These residues, mostly hydrophilic, are thought to form a hydrophilic cleft within the flippase domain to facilitate the passage of the polar head of aa-PG through the membrane. Interestingly, using a bacterial 2-hybrid assay in vivo, one study showed that LysPGS forms dimers, and possibly higher oligomeric structures within the membrane. These oligomeric states are mediated through interactions between the membrane and cytoplasmic domains of each subunit.Citation25
Beyond PG modification: Aminoacylation of cardiolipin and diacylglycerol
Besides PG, three additional lipids (cardiolipin, lysophosphatidylglycerol, and diacylglycerol) have been identified as substrates participating in tRNA-dependent pathways for lipid modification by addition of amino acids ().
Aminoacylation of cardiolipin (CL)
Figure 5. Lipid and aa substrates of lipid aminoacylation pathways. Phosphatidylglycerol (PG), Cardiolipin (CL), and diacylglycerol (DAG). See text for details.
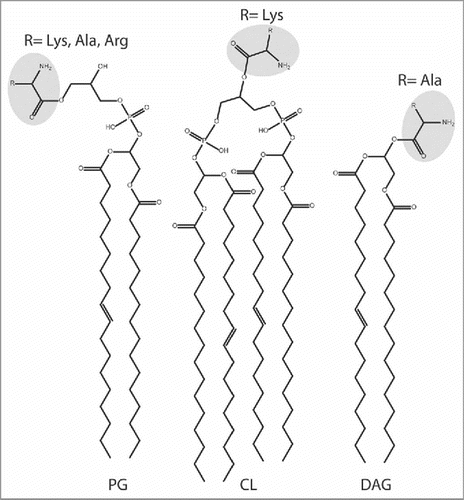
It was discovered early on that LysPGS from Listeria monocytogenes exhibits dual specificity for its lipid substrate. This enzyme is able to lysylate PG as well as the PG derivative CL, which exhibits a single hydroxyl group available for aminoacylationCitation51-Citation53(). The role of Lys-CL is not clear, but several studies showed that Lys-PG and Lys-CL increase resistance of L. monocytogenes to CAMPsCitation53 and osmolytic stress. Some evidence suggests that the two modified lipids may additionally influence expression of membrane proteins.Citation54
Aminoacylation of diacylglycerol (DAG)
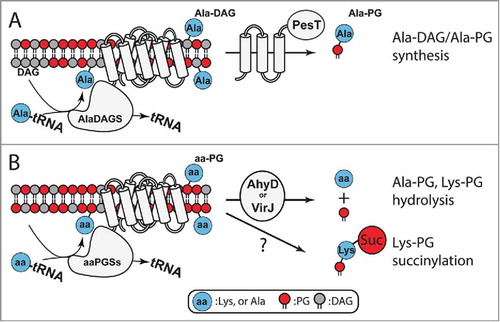
A recent phylogenetic analysis revealed 7 primary types of aaPGS homologs.Citation55 aaPGS homologs of types I and II are found in proteobacteria and firmicutes, respectively, and have been found to be specific for aminoacylation of PG and, more rarely, CL. Biochemical studies revealed the function of a novel type of aaPGS homolog found exclusively in Actinobacteria, a bacterial phylum comprising several important human pathogens. Specifically, various species of Corynebacterium exhibit an alanyl-diacylglycerol synthase (AlaDAGS) that uses Ala-tRNAAla and diacylglycerol (DAG, ) to form Ala-DAG. Interestingly, co-expression of AlaDAGS with a protein called PesT (a putative esterase encoded in operon with alaDAGS) induces formation of both Ala-DAG and Ala-PG in C. glutamicum. The role of PesT in this process is not clear. Since synthesis of Ala-PG requires co-expression of PesT and AlaDAGS, it has been proposed that PesT supports either i) an Ala-DAG/PG transferase activity (using Ala-DAG to form Ala-PG), or ii) an Ala-DAG hydrolase activity that allows formation of Ala-PG by AlaDAGSCitation55 ().
Figure 6. Homeostasis and utilization of aa-PG and Ala-DAG. A. In Corynebacterium, AlaDAGS utilizes Ala-tRNAAla and DAG to synthesize Ala-DAG. PesT, in concert with AlaDAGS, synthesizes Ala-PG. The role of PesT in this process is unknown.Citation55B. AhyD (in certain Firmicutes) and VirJ (in some proteobacteria) are aa-PG hydrolases. In B. subtilis, Lys-PG is N-succinylated by an unknown mechanism.
Homeostasis and utilization of aa-PG in the membrane
Three types of semi-conserved and structurally-unrelated hydrolases (AhyD, VirJ, and the putative esterase, PesT) have been identified.Citation35 Although the function of PesT remains to be clarified (see above), the functions of VirJ (found in various Gram-negative proteobacteria) and AhyD (found in several genera of Gram-positive bacteria) were recently revealed. Both of the latter enzymes are aa-PG hydrolases encoded in operon with aaPGS.Citation35,Citation56 These hydrolases are involved in controlling the levels of aminoacylated lipids in the membrane (), and both proteins were found to increase bacterial resistance to antimicrobial compounds.Citation35,Citation56,Citation57 While the cellular localization of AhyD remains undefined, VirJ is known to be anchored to the periplasmic surface in the cytoplasmic membrane of the gram-negative bacterium P. aeruginosa. VirJ exhibits broad substrate specificity and can hydrolyse the Ala-, Gly-, and Lys- moieties of artificial substrates.Citation56
In an interesting development, the modified lipid Lys-PG was shown to serve as a substrate for an additional membrane modification. A recent report showed that Lys-PG in B. subtilis is used to produce N-succinylated Lys-PG.Citation33 The biologic significance of and enzyme responsible for this modification are not known. However, it was proposed that a homolog of the enzyme responsible for protein succinylation, recently discovered in bacteria, may be accountable for Lys-PG modification. It is worth noting that attachment of succinyl to Lys reverses the net positive charge of Lys-PG (+1), yielding a net negative charge that is the same as that of unmodified PG (−1). This system may represent a means for bacteria lacking an aa-PG hydrolase (e.g., AhyD or VirJ are not found in B. subtilis) to reverse the net charge of the membrane without hydrolysing Lys-PG.
Increased virulence and antibiotic resistance linked to lipid aminoacylation
The effects of aa-PG on antibiotic resistance have been investigated in several important human pathogens such as S. aureus (for review see [Citation4]), L. monocytogenes,Citation53 B. anthracis,Citation58 P. aeruginosa,Citation24,Citation59 M. tuberculosis,Citation29,Citation60 Enterococcus species,Citation35,Citation36,Citation61 and B. subtilis Citation62,Citation63 (for review see [Citation1]). aa-PGs in these organisms primarily enhance bacterial resistance to positively charged compounds targeting the membrane (such as CAMPs) and last resort lipopetides like daptomycin.Citation4,Citation58 aa-PGs have been found to modestly enhance bacterial resistance (e.g., a 2-fold increase in the MIC) to other classes of antimicrobials such as β-lactams and glycopeptides. Lys-PG, for example, increases resistance of S. aureus to several β-lactams,Citation64,Citation65 and vancomycin.Citation66 Similarly, resistance phenotypes against β-lactams were observed in P. aeruginosa, which produces Ala-PG.Citation59 In M. tuberculosis, Lys-PG was shown to increase resistance to vancomycin.Citation29 β-lactams and vancomycin are antibiotics that do not interact with the cytoplasmic membrane; both compounds target distinct components of the peptidoglycan synthesis machinery inside the periplasm (i.e., penicillin binding proteins and peptidoglycan connecting pentapeptides, respectively). Increased resistance to these antibiotics may be due to indirect effects of the aaPGS lipid modification process on the homeostasis and general properties of the cell wall. For instance, Ala-PG found in the outer membrane of the Gram-negative bacterium P. aeruginosa may modulate the activity of porins in the outer membrane that are necessary for β-lactams to reach their periplasmic targets.Citation59 Lys-PG synthesis in S. aureus on the other hand might modulate the activity of proteins involved in peptidoglycan synthesis, which would indirectly affect resistance to β-lactams and vancomycin. These hypotheses are supported by several studies that demonstrated that the lack of aa-PG in the membranes of S. aureus and L. monocytogenes induces significant changes in the membrane proteome and its function.Citation54,Citation67,Citation68
aaPGSs have been recognized as virulence factors for prominent pathogens in several animal models. For instance, the lack of LysPGS in S. aureus decreases its capacity to proliferate in an endocarditis model in rabbits,Citation69 and in a sepsis model in mice.Citation23 The mortality rate of mice infected with a LysPGS deletion strain was significantly diminished.Citation23 Likewise, deletion of LysPGS in L. monocytogenes lowered the bacterial counts detected in the spleens and livers of infected mice as compared with those infected with the wild-type strain.Citation53 In M. tuberculosis, deletion of LysPGS (i.e., lysX) decreases bacterial growth in the lungs of guinea pigs relative to the wild-type strain.Citation29
Several studies demonstrated the importance of lipid aminoacylation in the interaction of pathogens with different types of cells in infected hosts. Lipid aminoacylation was also shown to increase the extra and intracellular survival of pathogens. For instance, LysPGS increases resistance of S. aureus to killing by neutrophils and leukocytes.Citation23,Citation70 In contrast, Lys-PG did not affect S. aureus survivability after phagocytosis by human neutrophils.Citation23 When deprived of Lys-PG, the obligate intracellular species L. monocytogenes poorly infects epithelial cells and macrophages, but is not compromised in cell-to-cell spreading ability.Citation53 Likewise, Lys-PG increases the survivability of M. tuberculosis inside phagocytic cells,Citation71 and is critical for maintaining intracellular pH levels while in activated macrophages.Citation60
Although each of the examples summarized above suggests that lipid aminoacylation increases resistance to CAMPs and virulence of pathogens, the associated phenotypes and their relative intensities are largely dependent on bacterial context. For example, lipid aminoacylation in Enterococcus faecalis does not affect virulence in a mouse bacteremia model, and only modestly affects the MICs of various CAMPs (up to a 4-fold increase). Mutants of this species defective for lipid aminoacylation do, however, exhibit increased resistance to opsonic killing, and a 42% decrease in biofilm formation compared with the wild-type strain.Citation36
Single nucleotide polymorphisms: Gain of function mutations in LysPGS of multi-resistant S. aureus
In recent years, as many multi-resistant strains of S. aureus have emerged, multiple independent studies have identified strains exhibiting single-nucleotide polymorphisms (SNPs) in mprF that are linked to resistance against daptomycin, a last resort CAMP-like pore forming lipopeptide used to combat acute microbial infections. Several studies identified SNPs in the mprF gene of S. aureus clinical isolates, or laboratory strains evolved in vitro under antibiotic pressure.Citation72-Citation83 Genome analysis revealed additional SNPs in genes such as rpoB (a polymerase subunit), capB (for capsular polysaccharide synthesis), and the promoter region of the dltABCD operon which is known to increase CAMP resistance (). However, SNPs in mprF were the most frequently encountered, and were accompanied by an increase in resistance to daptomycin and other CAMPs (nisin and human CAMPs e.g., [Citation75,Citation79]). Most mprF gain-of-function SNPs are located in the flippase domain, and they generally increase the total amount of Lys-PG in the membrane as well as the amount of Lys-PG translocated to the outer surface of the cytoplasmic membrane (e.g., [Citation77,Citation78]). Interestingly, SNPs in the lysX gene in M. tuberculosis were recently discovered; this finding further supports the hypothesis that this system plays a role in virulence.Citation84 The precise role of SNPs in the catalytic activities of aaPGSs (aa-PG synthesis and flipping) is not well understood, but they are predicted to increase both processes affecting aminoacylated lipids.
Physiological significance of lipid aminoacylation
Several studies showed that the lack of lipid aminoacylation in mutants of S. aureus and L. monocytogenes induces changes in the proteome of the membrane compared with that of wild-type strains. This indicates that aa-PGs not only modify the electrostatic properties of the membrane surface, but also affect expression and activity of membrane proteins.Citation54,Citation67 Interestingly, LysPGS in B. subtilis is localized at the septum of the bacterium, along with other components involved in membrane and cell wall biogenesis.Citation85 A recent study showed that LysPGS is also localized at the septum in E. faecalis, and demonstrated that human β-defensins (i.e., hBD2 and hBD3) interact with E. faecalis near this region to target the excretory system consisting of protein translocase (SecYEG) and sortase enzymes (SrtA). Altogether, these findings show that aa-PGs can provide positively charged lipid microdomains that protect the septum, along with important components involved in cell wall biosynthesis, from inhibition by CAMPs.Citation86
Most studies on lipid aminoacylation systems have emphasized their role in protecting bacteria from stresses located outside the cell. A single study reported the potential role of lipid aminoacylation in processes occurring inside the cell. Ichiahashi and coauthors demonstrated that the first step during genomic DNA replication (involving the protein DnaA) is inhibited by synthesis of Lys-PG in S. aureus.Citation87 These investigations suggest that stresses in the environment trigger expression of Lys-PG, not only to increase resistance to external challenges, but also to repress DNA replication and regulate cell cycling events.
Another important concept that has emerged in recent years, is the idea that low levels of aminoacylated lipids in the bacterial membrane may alter general membrane protein composition. In Firmicutes such as L. monocytogenes, S. aureus, and various Bacillus species, aminoacylated lipids represent a significant portion of the total membrane lipid content, ranging from 10–75% of the total.Citation52-Citation54,Citation88,Citation89 In M. tuberculosis, Lys-PG is a minor lipid constituent, representing less than 0.3% of the total lipids. Despite its low abundance, Lys-PG in this species is important for maintaining membrane potential, virulence, CAMP resistance, and survival in mononuclear phagocytes.Citation29,Citation71,Citation90 It was determined that the phenotypes observed in mutants deprived of Lys-PG may, in fact, be due to indirect effects of the modified lipids on phospholipid catabolism and cell division.Citation90 These findings indicate that PG-lysylation in M. tuberculosis plays a broader role outside of the typical electrostatic effects associated with aa-PGs, which might be relevant to many bacterial species.
Regulation of PG aminoacylation
Studies performed in various bacterial backgrounds have shown that lipid aminoacylation is triggered by environmental stimuli. For instance, in bacterial symbionts of plants such as Rhizobium tropici and Sinorhizobium medicae, Lys-PG formation is triggered by low pH and is required for acid tolerance and colonization of root nodules. Likewise, in species like B. subtilis,Citation91,Citation92 Bacillus megaterium,Citation89 S. aureus,Citation93,Citation94 and E. faecium,Citation35,Citation94,Citation95 acidic conditions increase the amount of aa-PG produced in the membrane.Citation55,Citation93,Citation94,Citation96 Because aa-PG does not provide any apparent tolerance to acidic conditions in E. faecium, it was recently proposed that lipid aminoacylation confers tolerance to inhibitors that are produced during exposure to acidic conditions, rather than to low pH levels themselves. For instance, aa-PGs enhance resistance of P. aeruginosa and E. faecium to lactic acid,Citation35,Citation59 a toxic osmolyte known to diffuse passively through the cytoplasmic membrane of bacteria.Citation97
The full regulatory network controlling aaPGS expression is not completely understood, but it has been studied in L. monocytogenes, R. tropici (seeCitation1 for review), and more extensively, in S. aureus. Various bacterial inhibitors targeting the cell envelope trigger expression of mprF in S. aureus. For instance, antibiotics such as methicillin, vancomycin, penicillin-G, D-cycloserine, and bacitracin, which all target the cellular envelope, induce expression of Lys-PG. Antibiotics such as chloramphenicol and puromycin, which target the translation machinery, do not induce Lys-PG synthesis.Citation98-Citation100 Recent studies showed that the 2-component system, LytSR, thought to be responsible for maintaining electrical potential and CAMP sensing at the surface of the cell membrane in S. aureus, does not control expression of mprF and other genes involved in maintenance of the cell surface charge.Citation101 Instead, the 2-component system GraSR (a.k.a. the antimicrobial peptide sensing system; aps) was shown to modulate expression of mprF, as well as the surface charge remodeling operon dltABCD involved in D-alanylation of teichoic acid chains inside the cell wall. This system also regulates expression of enzymes responsible for biosynthesis of Lys (a substrate of LysPGS), and VraFG, a putative ATP dependent CAMP efflux transporter.Citation102-Citation105 The cell surface sensor GraS binds to CAMPs using an extracellular loop at the surface of the membrane and activates the transcription factor GraR. The extracellular loop of GraS contains several anionic residues that interact with various CAMPs including polymyxin B, nisin, and histatin. A recent study shed light on the molecular mechanism used by GraS to sense certain CAMPs with more sensitivity than others.Citation106 Interestingly, the GraSR locus encodes a third subunit (GraX) that is predicted to form a part of the sensing mechanism.Citation102-Citation104 Together with previous observations showing that expression of mprF and dltABCD is dependent on a functional VraFG, new studies have shown that GraX is the central component of a “5-component system” for sensing of CAMPs (,Citation107). The GraSR sensing system requires both GraX and the VraGF transporter to function. In this complex signal transduction pathway VraGF does not play a role in detoxification of the cell as previously thought, but instead completes the GraSR sensing mechanism via its interaction with GraX to efficiently induce expression of dltABCD and mprF.Citation105
Figure 7. The 5-component signal transduction network controlling CAMP sensing and resistance pathways in S. aureus. CAMPs are sensed by VraFG and the signal is transduced to GraS through a mechanism that is likely to involve interaction between VraG and GraS. Activation of the GraSR system leads to increased transcription of the dltABCD operon and the mprF gene, leading to cell wall remodeling and CAMP resistance (figure adapted from Citation[107]).
![Figure 7. The 5-component signal transduction network controlling CAMP sensing and resistance pathways in S. aureus. CAMPs are sensed by VraFG and the signal is transduced to GraS through a mechanism that is likely to involve interaction between VraG and GraS. Activation of the GraSR system leads to increased transcription of the dltABCD operon and the mprF gene, leading to cell wall remodeling and CAMP resistance (figure adapted from Citation[107]).](/cms/asset/ab636924-e1e2-4909-bc75-b931a4cf83c6/krnb_a_1356980_f0007_oc.jpg)
Conclusion and outlook
Developments in recent years have shown that the tRNA-dependent system for lipid modification is more complex than originally hypothesized. Several novel aa-tRNA and lipid substrates were recently discovered, and new enzymes have been added to the repertoire of aaPGS homologs. Several hydrolases were found that are involved in maintaining the homeostasis of aminoacylated lipids. Lipid aminoacylation affects the biochemistry of the bacterial cell membrane and is involved in cellular adaptation to stress and other changes in the environment. In spite of recent progress, many open questions remain unanswered regarding the mechanism of aa-PG synthesis, flipping across the membrane, and the physiologic relevance of these modifications in bacteria. It is possible that, investigated through the prism of biomedical relevance, the fundamental physiologic role of lipid modifications has partially escaped previous investigations.
Another open question pertains to the relevance of aaPGS as a valid drug target. Sources in the literature state that lipid aminoacylation might represent an appealing drug target for which inhibitors would make pathogenic species more susceptible to existing drugs, or to natural defenses of the immune system of an infected host. Although several studies cited in this review have shown that aa-PG synthesis significantly enhances antimicrobial resistance and virulence in some pathogens, these effects are modest in the context of other bacterial species. Moreover, aa-PGs are not essential for bacterial growth, and one could argue that because the effects of aminoacylated lipids on antimicrobial resistance are weak, these modification systems might not represent practical targets for anti-infective strategies. With the rise of bacteria that are increasingly resistant to current drugs, new antimicrobials and/or drug combinations must be identified to find effective treatments of managing infections.Citation108,Citation109 In the last decade, many antibiotics that target bacterial systems that enhance antimicrobial resistance have been developed to potentiate the effect of existing antibiotics (e.g., [Citation110,Citation111]). For instance, multidrug efflux pumps, which have been shown to increase antibiotic resistance, have become a popular target. Indeed, recent studies suggest that multidrug transporters are a major determinant for the efficacy of both new and old antibiotics.Citation112 Efforts have also begun toward discovery of inhibitors of the lipid aminoacylation pathway. A high throughput assay was recently established to screen libraries of potential inhibitors of the aaPGS active site.Citation113 Also, a targeted antisense RNA strategy developed by Cubist Pharmaceuticals demonstrated that modulation of mprF expression in S. aureus can increase susceptibility to daptomycin.Citation114 More work is needed to identify lead compounds that might serve as inhibitors of lipid aminoacylation, and to determine whether inhibition of this pathway increases the efficacy of new and existing therapeutic strategies.
Acknowledgments
We would like to acknowledge A.M. Smith for critical reviewing the manuscript. The work performed in HR's laboratory was supported by National Institute of General Medical Sciences Grant R15–109404.
References
- Roy H. Tuning the properties of the bacterial membrane with aminoacylated phosphatidylglycerol. IUBMB Life. 2009;61:940–53. doi:10.1002/iub.240.
- Geiger O, Gonzalez-Silva N, Lopez-Lara IM, Sohlenkamp C. Amino acid-containing membrane lipids in bacteria. Prog Lipid Res. 2010;49:46–60. doi:10.1016/j.plipres.2009.08.002.
- Dare K, Ibba M. Roles of tRNA in cell wall biosynthesis. Wiley Interdiscip Rev RNA. 2011;3:247–64. doi:10.1002/wrna.1108
- Kuhn S, Slavetinsky CJ, Peschel A. Synthesis and function of phospholipids in Staphylococcus aureus. Int J Med Microbiol. 2015;305:196–202. doi:10.1016/j.ijmm.2014.12.016.
- Hancock RE. Peptide antibiotics. Lancet. 1997;349:418–22. doi:10.1016/S0140-6736(97)80051-7.
- Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004;58:453–88. doi:10.1146/annurev.micro.58.030603.123615.
- Hechard Y, Sahl HG. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie. 2002;84:545–57. doi:10.1016/S0300-9084(02)01417-7.
- Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol Cell. 2004;14:739–51. doi:10.1016/j.molcel.2004.06.010.
- Mardirossian M, Grzela R, Giglione C, Meinnel T, Gennaro R, Mergaert P, Scocchi M. The host antimicrobial peptide Bac71-35 binds to bacterial ribosomal proteins and inhibits protein synthesis. Chem Biol. 2014;21:1639–47. doi:10.1016/j.chembiol.2014.10.009.
- Krizsan A, Volke D, Weinert S, Strater N, Knappe D, Hoffmann R. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70S ribosome. Angew Chem Int Ed Engl. 2014;53:12236–9. doi:10.1002/anie.201407145.
- Hancock RE, Sahl HG. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol. 2006;24:1551–7. doi:10.1038/nbt1267.
- Peschel A, Sahl HG. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat Rev Microbiol. 2006;4:529–36. doi:10.1038/nrmicro1441.
- Wang G, Li X, Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016;44:D1087–93. doi:10.1093/nar/gkv1278.
- Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals (Basel). 2014;7:545–94. doi:10.3390/ph7050545.
- Lai Y, Gallo RL. AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol. 2009;30:131–41. doi:10.1016/j.it.2008.12.003.
- Matsuzaki K. Why and how are peptide-lipid interactions utilized for self-defense? Magainins and tachyplesins as archetypes. Biochim Biophys Acta. 1999;1462:1–10. doi:10.1016/S0005-2736(99)00197-2.
- Nawrocki KL, Crispell EK, McBride SM. Antimicrobial Peptide Resistance Mechanisms of Gram-Positive Bacteria. Antibiotics (Basel). 2014;3:461–92. doi:10.3390/antibiotics3040461.
- Band VI, Weiss DS. Mechanisms of Antimicrobial Peptide Resistance in Gram-Negative Bacteria. Antibiotics (Basel). 2015;4:18–41. doi:10.3390/antibiotics4010018.
- Luevano-Martinez LA, Kowaltowski AJ. Phosphatidylglycerol-derived phospholipids have a universal, domain-crossing role in stress responses. Arch Biochem Biophys. 2015;585:90–7. doi:10.1016/j.abb.2015.09.015.
- Lennarz WJ, Bonsen PP, van Deenen LL. Substrate specificity of O-L-lysylphosphatidylglycerol synthetase. Enzymatic studies on the structure of O-L-lysylphosphatidylglycerol. Biochemistry (Mosc). 1967;6:2307–12
- Lennarz WJ, Nesbitt JA, 3rd, Reiss J. The participation of sRNA in the enzymatic synthesis of O-L-lysyl phosphatidylgylcerol in Staphylococcus aureus. Proc Natl Acad Sci U S A. 1966;55:934–41. doi:10.1073/pnas.55.4.934.
- Peschel A, Collins LV. Staphylococcal resistance to antimicrobial peptides of mammalian and bacterial origin. Peptides. 2001;22:1651–9. doi:10.1016/S0196-9781(01)00500-9.
- Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J Exp Med. 2001;193:1067–76. doi:10.1084/jem.193.9.1067.
- Arendt W, Hebecker S, Jager S, Nimtz M, Moser J. Resistance phenotypes mediated by aminoacyl-phosphatidylglycerol synthases. J Bacteriol. 2012;194:1401–16. doi:10.1128/JB.06576-11.
- Ernst CM, Kuhn S, Slavetinsky CJ, Krismer B, Heilbronner S, Gekeler C, Kraus D, Wagner S, Peschel A. The lipid-modifying multiple Peptide resistance factor is an oligomer consisting of distinct interacting synthase and flippase subunits. MBio. 2015;6.
- Ernst CM, Peschel A. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol Microbiol. 2011;80:290–9. doi:10.1111/j.1365-2958.2011.07576.x.
- Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 2009;5:e1000660. doi:10.1371/journal.ppat.1000660.
- Slavetinsky CJ, Peschel A, Ernst CM. Alanyl-phosphatidylglycerol and lysyl-phosphatidylglycerol are translocated by the same MprF flippases and have similar capacities to protect against the antibiotic daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:3492–7. doi:10.1128/AAC.00370-12.
- Maloney E, Stankowska D, Zhang J, Fol M, Cheng QJ, Lun S, Bishai WR, Rajagopalan M, Chatterjee D, Madiraju MV. The two-domain LysX protein of Mycobacterium tuberculosis is required for production of lysinylated phosphatidylglycerol and resistance to cationic antimicrobial peptides. PLoS Pathog. 2009;5:e1000534. doi:10.1371/journal.ppat.1000534.
- Hebecker S, Krausze J, Hasenkampf T, Schneider J, Groenewold M, Reichelt J, Jahn D, Heinz DW, Moser J. Structures of two bacterial resistance factors mediating tRNA-dependent aminoacylation of phosphatidylglycerol with lysine or alanine. Proc Natl Acad Sci U S A. 2015;112:10691–6. doi:10.1073/pnas.1511167112.
- Roy H, Ibba M. RNA-dependent lipid remodeling by bacterial multiple peptide resistance factors. Proc Natl Acad Sci U S A. 2008;105:4667–72. doi:10.1073/pnas.0800006105.
- Roy H, Ibba M. Broad range amino acid specificity of RNA-dependent lipid remodeling by multiple peptide resistance factors. J Biol Chem. 2009;284:29677–83. doi:10.1074/jbc.M109.046367.
- Atila M, Katselis G, Chumala P, Luo Y. Characterization of N-Succinylation of L-Lysylphosphatidylglycerol in Bacillus subtilis Using Tandem Mass Spectrometry. J Am Soc Mass Spectrom. 2016;27:1606–13. doi:10.1007/s13361-016-1455-4.
- Atila M, Luo Y. Profiling and tandem mass spectrometry analysis of aminoacylated phospholipids in Bacillus subtilis. F1000Res. 2016;5:121. doi:10.12688/f1000research.7842.1.
- Smith AM, Harrison JS, Sprague KM, Roy H. A conserved hydrolase responsible for the cleavage of aminoacylphosphatidylglycerol in the membrane of Enterococcus faecium. J Biol Chem. 2013;288:22768–76. doi:10.1074/jbc.M113.484402.
- Bao Y, Sakinc T, Laverde D, Wobser D, Benachour A, Theilacker C, Hartke A, Huebner J. Role of mprF1 and mprF2 in the pathogenicity of Enterococcus faecalis. PLoS ONE. 2012;7:e38458. doi:10.1371/journal.pone.0038458.
- dos Santos Mota JM, den Kamp JA, Verheij HM, van Deenen LL. Phospholipids of Streptococcus faecalis. J Bacteriol. 1970;104:611–9.
- Hebecker S, Arendt W, Heinemann IU, Tiefenau JH, Nimtz M, Rohde M, et al. Alanyl-phosphatidylglycerol synthase: mechanism of substrate recognition during tRNA-dependent lipid modification in Pseudomonas aeruginosa. Mol Microbiol. 2011;80:935–50.
- Iyer LM, Abhiman S, Maxwell Burroughs A, Aravind L. Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol Biosyst. 2009;5:1636–60. doi:10.1039/b917682a.
- Aravind L, de Souza RF, Iyer LM. Predicted class-I aminoacyl tRNA synthetase-like proteins in non-ribosomal peptide synthesis. Biol Direct. 2010;5:48. doi:10.1186/1745-6150-5-48.
- Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi:10.1093/nar/gkr1065.
- Rohrer S, Berger-Bachi B. FemABX peptidyl transferases: a link between branched-chain cell wall peptide formation and beta-lactam resistance in gram-positive cocci. Antimicrob Agents Chemother. 2003;47:837–46. doi:10.1128/AAC.47.3.837-846.2003.
- Vollmer W, Blanot D, de Pedro MA. Peptidoglycan structure and architecture. FEMS Microbiol Rev. 2008;32:149–67. doi:10.1111/j.1574-6976.2007.00094.x.
- Tasaki T, Sriram SM, Park KS, Kwon YT. The N-end rule pathway. Annu Rev Biochem. 2012;81:261–89. doi:10.1146/annurev-biochem-051710-093308.
- Garg RP, Qian XL, Alemany LB, Moran S, Parry RJ. Investigations of valanimycin biosynthesis: elucidation of the role of seryl-tRNA. Proc Natl Acad Sci U S A. 2008;105:6543–7. doi:10.1073/pnas.0708957105.
- Bougioukou DJ, Mukherjee S, van der Donk WA. Revisiting the biosynthesis of dehydrophos reveals a tRNA-dependent pathway. Proc Natl Acad Sci U S A. 2013;110:10952–7. doi:10.1073/pnas.1303568110.
- Zhang W, Ntai I, Kelleher NL, Walsh CT. tRNA-dependent peptide bond formation by the transferase PacB in biosynthesis of the pacidamycin group of pentapeptidyl nucleoside antibiotics. Proc Natl Acad Sci U S A. 2011;108:12249–53. doi:10.1073/pnas.1109539108.
- Huang Z, Wang KA, van der Donk WA. New Insights into the Biosynthesis of Fosfazinomycin. Chem Sci. 2016;7:5219–23. doi:10.1039/C6SC01389A.
- Maruyama C, Niikura H, Izumikawa M, Hashimoto J, Shin-Ya K, Komatsu M, Ikeda H, Kuroda M, Sekizuka T, Ishikawa J, et al. tRNA-Dependent Aminoacylation of an Amino Sugar Intermediate in the Biosynthesis of a Streptothricin-Related Antibiotic. Appl Environ Microbiol. 2016;82:3640–8. doi:10.1128/AEM.00725-16.
- Fonvielle M, Li de La Sierra-Gallay I, El-Sagheer AH, Lecerf M, Patin D, Mellal D, Mayer C, Blanot D, Gale N, Brown T, et al. The structure of FemX(Wv) in complex with a peptidyl-RNA conjugate: mechanism of aminoacyl transfer from Ala-tRNA(Ala) to peptidoglycan precursors. Angew Chem Int Ed Engl. 2013;52:7278–81. doi:10.1002/anie.201301411.
- Gutberlet T, Dietrich U, Bradaczek H, Pohlentz G, Leopold K, Fischer W. Cardiolipin, alpha-D-glucopyranosyl, and L-lysylcardiolipin from gram-positive bacteria: FAB MS, monofilm and X-ray powder diffraction studies. Biochim Biophys Acta. 2000;1463:307–22. doi:10.1016/S0005-2736(99)00214-X.
- Fischer W, Leopold K. Polar lipids of four Listeria species containing L-lysylcardiolipin, a novel lipid structure, and other unique phospholipids. Int J Syst Bacteriol. 1999;2:653–62. doi:10.1099/00207713-49-2-653
- Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jänsch L. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol Microbiol. 2006;62:1325–39. doi:10.1111/j.1365-2958.2006.05452.x.
- Dare K, Shepherd J, Roy H, Seveau S, Ibba M. LysPGS formation in Listeria monocytogenes has broad roles in maintaining membrane integrity beyond antimicrobial peptide resistance. Virulence. 2014;5:534–46. doi:10.4161/viru.28359.
- Smith AM, Harrison JS, Grube CD, Sheppe AE, Sahara N, Ishii R, Nureki O, Roy H. tRNA-dependent alanylation of diacylglycerol and phosphatidylglycerol in Corynebacterium glutamicum. Mol Microbiol. 2015;98:681–93. doi:10.1111/mmi.13150.
- Arendt W, Groenewold MK, Hebecker S, Dickschat JS, Moser J. Identification and characterization of a periplasmic aminoacyl-phosphatidylglycerol hydrolase responsible for Pseudomonas aeruginosa lipid homeostasis. J Biol Chem. 2013;288:24717–30. doi:10.1074/jbc.M113.482935.
- Zhang X, Paganelli FL, Bierschenk D, Kuipers A, Bonten MJ, Willems RJ, van Schaik W. Genome-Wide Identification of Ampicillin Resistance Determinants in Enterococcus faecium. PLoS Genet. 2012;8:e1002804. doi:10.1371/journal.pgen.1002804.
- Samant S, Hsu FF, Neyfakh AA, Lee H. The Bacillus anthracis protein MprF is required for synthesis of lysylphosphatidylglycerols and for resistance to cationic antimicrobial peptides. J Bacteriol. 2009;191:1311–9. doi:10.1128/JB.01345-08.
- Klein S, Lorenzo C, Hoffmann S, Walther JM, Storbeck S, Piekarski T, Tindall BJ, Wray V, Nimtz M, Moser J. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol Microbiol. 2009;71:551–65. doi:10.1111/j.1365-2958.2008.06562.x.
- Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF, Ehrt S. Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. J Bacteriol. 2009;191:625–31. doi:10.1128/JB.00932-08.
- Kumariya R, Sood SK, Rajput YS, Saini N, Garsa AK. Increased membrane surface positive charge and altered membrane fluidity leads to cationic antimicrobial peptide resistance in Enterococcus faecalis. Biochim Biophys Acta. 2015;1848:1367–75. doi:10.1016/j.bbamem.2015.03.007.
- Hachmann AB, Angert ER, Helmann JD. Genetic analysis of factors affecting susceptibility of Bacillus subtilis to daptomycin. Antimicrob Agents Chemother. 2009;53:1598–609. doi:10.1128/AAC.01329-08.
- Salzberg LI, Helmann JD. Phenotypic and transcriptomic characterization of Bacillus subtilis mutants with grossly altered membrane composition. J Bacteriol. 2008;190:7797–807.
- Komatsuzawa H, Ohta K, Fujiwara T, Choi GH, Labischinski H, Sugai M. Cloning and sequencing of the gene, fmtC, which affects oxacillin resistance in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett. 2001;203:49–54. doi:10.1111/j.1574-6968.2001.tb10819.x.
- Nishi H, Komatsuzawa H, Fujiwara T, McCallum N, Sugai M. Reduced content of lysyl-phosphatidylglycerol in the cytoplasmic membrane affects susceptibility to moenomycin, as well as vancomycin, gentamicin, and antimicrobial peptides, in Staphylococcus aureus. Antimicrob Agents Chemother. 2004;48:4800–7. doi:10.1128/AAC.48.12.4800-4807.2004.
- Ruzin A, Severin A, Moghazeh SL, Etienne J, Bradford PA, Projan SJ, Shlaes DM. Inactivation of mprF affects vancomycin susceptibility in Staphylococcus aureus. Biochim Biophys Acta. 2003;1621:117–21. doi:10.1016/S0304-4165(03)00028-X.
- Sievers S, Ernst CM, Geiger T, Hecker M, Wolz C, Becher D, Peschel A. Changing the phospholipid composition of Staphylococcus aureus causes distinct changes in membrane proteome and membrane-sensory regulators. Proteomics. 2010;10:1685–93. doi:10.1002/pmic.200900772.
- Zemansky J, Kline BC, Woodward JJ, Leber JH, Marquis H, Portnoy DA. Development of a mariner-based transposon and identification of Listeria monocytogenes determinants, including the peptidyl-prolyl isomerase PrsA2, that contribute to its hemolytic phenotype. J Bacteriol. 2009;191:3950–64. doi:10.1128/JB.00016-09.
- Weidenmaier C, Peschel A, Kempf VA, Lucindo N, Yeaman MR, Bayer AS. DltABCD- and MprF-mediated cell envelope modifications of Staphylococcus aureus confer resistance to platelet microbicidal proteins and contribute to virulence in a rabbit endocarditis model. Infect Immun. 2005;73:8033–8. doi:10.1128/IAI.73.12.8033-8038.2005.
- Kristian SA, Durr M, Van Strijp JA, Neumeister B, Peschel A. MprF-mediated lysinylation of phospholipids in Staphylococcus aureus leads to protection against oxygen-independent neutrophil killing. Infect Immun. 2003;71:546–9. doi:10.1128/IAI.71.1.546-549.2003.
- Fol M, Globinska A, Staczek P, Kowalewicz-Kulbat M, Druszczynska M, Madiraju MV, Rudnicka W. The lack of L-PG production and the repercussions of it in regards to M. tuberculosis interactions with mononuclear phagocytes. Acta Microbiol Immunol Hung. 2013;60:127–44. doi:10.1556/AMicr.60.2013.2.4
- Yamaguchi T, Suzuki S, Okamura S, Miura Y, Tsukimori A, Nakamura I, Ito N, Masuya A, Shiina T, Matsumoto T. Evolution and single-nucleotide polymorphisms in methicillin-resistant Staphylococcus aureus strains with reduced susceptibility to vancomycin and daptomycin, based on determination of the complete genome. Antimicrob Agents Chemother. 2015;59:3585–7. doi:10.1128/AAC.05159-14.
- Chen CJ, Huang YC, Chiu CH. Multiple pathways of cross-resistance to glycopeptides and daptomycin in persistent MRSA bacteraemia. J Antimicrob Chemother. 2015;70:2965–72.
- Cameron DR, Jiang JH, Abbott IJ, Spelman DW, Peleg AY. Draft Genome Sequences of Clinical Daptomycin-Nonsusceptible Methicillin-Resistant Staphylococcus aureus Strain APS211 and Its Daptomycin-Susceptible Progenitor APS210. Genome Announc. 2015;3.
- Bayer AS, Mishra NN, Chen L, Kreiswirth BN, Rubio A, Yang SJ. Frequency and Distribution of Single-Nucleotide Polymorphisms within mprF in Methicillin-Resistant Staphylococcus aureus Clinical Isolates and Their Role in Cross-Resistance to Daptomycin and Host Defense Antimicrobial Peptides. Antimicrob Agents Chemother. 2015;59:4930–7. doi:10.1128/AAC.00970-15.
- Baek KT, Thogersen L, Mogenssen RG, Mellergaard M, Thomsen LE, Petersen A, et al. Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob Agents Chemother. 2015;59:6983–91.
- Yang SJ, Mishra NN, Rubio A, Bayer AS. Causal role of single nucleotide polymorphisms within the mprF gene of Staphylococcus aureus in daptomycin resistance. Antimicrob Agents Chemother. 2013;57:5658–64. doi:10.1128/AAC.01184-13.
- Mishra NN, Yang SJ, Chen L, Muller C, Saleh-Mghir A, Kuhn S, Peschel A, Yeaman MR, Nast CC, Kreiswirth BN, et al. Emergence of daptomycin resistance in daptomycin-naive rabbits with methicillin-resistant Staphylococcus aureus prosthetic joint infection is associated with resistance to host defense cationic peptides and mprF polymorphisms. PLoS ONE. 2013;8:e71151. doi:10.1371/journal.pone.0071151.
- Patel D, Husain M, Vidaillac C, Steed ME, Rybak MJ, Seo SM, Kaatz GW. Mechanisms of in-vitro-selected daptomycin-non-susceptibility in Staphylococcus aureus. Int J Antimicrob Agents. 2011;38:442–6. doi:10.1016/j.ijantimicag.2011.06.010.
- Friedman L, Alder JD, Silverman JA. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:2137–45. doi:10.1128/AAC.00039-06.
- Julian K, Kosowska-Shick K, Whitener C, Roos M, Labischinski H, Rubio A, Parent L, Ednie L, Koeth L, Bogdanovich T, et al. Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. Antimicrob Agents Chemother. 2007;51:3445–8. doi:10.1128/AAC.00559-07.
- Pillai SK, Gold HS, Sakoulas G, Wennersten C, Moellering RC, Jr., Eliopoulos GM. Daptomycin nonsusceptibility in Staphylococcus aureus with reduced vancomycin susceptibility is independent of alterations in MprF. Antimicrob Agents Chemother. 2007;51:2223–5. doi:10.1128/AAC.00202-07.
- Murthy MH, Olson ME, Wickert RW, Fey PD, Jalali Z. Daptomycin non-susceptible meticillin-resistant Staphylococcus aureus USA 300 isolate. J Med Microbiol. 2008;57:1036–8. doi:10.1099/jmm.0.2008/000588-0.
- Pardieu C, Casali N, Clark SO, Hooper R, Williams A, Velji P, Gonzalo X, Drobniewski F. Correlates between models of virulence for Mycobacterium tuberculosis among isolates of the Central Asian lineage: a case for lysozyme resistance testing? Infect Immun. 2015;83:2213–23. doi:10.1128/IAI.03080-14.
- Nishibori A, Kusaka J, Hara H, Umeda M, Matsumoto K. Phosphatidylethanolamine domains and localization of phospholipid synthases in Bacillus subtilis membranes. J Bacteriol. 2005;187:2163–74. doi:10.1128/JB.187.6.2163-2174.2005.
- Kandaswamy K, Liew TH, Wang CY, Huston-Warren E, Meyer-Hoffert U, Hultenby K, Schröder JM, Caparon MG, Normark S, Henriques-Normark B, et al. Focal targeting by human beta-defensin 2 disrupts localized virulence factor assembly sites in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2013;110:20230–5. doi:10.1073/pnas.1319066110.
- Ichihashi N, Kurokawa K, Matsuo M, Kaito C, Sekimizu K. Inhibitory effects of basic or neutral phospholipid on acidic phospholipid-mediated dissociation of adenine nucleotide bound to DnaA protein, the initiator of chromosomal DNA replication. J Biol Chem. 2003;278:28778–86. doi:10.1074/jbc.M212202200.
- Haest CW, de Gier J, den Kamp JO, Bartels P, van Deenen LL. Changes in permeability of Staphylococcus aureus and derived liposomes with varying lipid composition. Biochim Biophys Acta. 1972;255:720–33. doi:10.1016/0005-2736(72)90385-9.
- den Kamp JO, Houtsmuller UM, van Deenen LL. On the phospholipids of Bacillus megaterium. Biochim Biophys Acta. 1965;106:438–41. doi:10.1016/0005-2760(65)90059-7.
- Maloney E, Lun S, Stankowska D, Guo H, Rajagoapalan M, Bishai WR, Madiraju MV. Alterations in phospholipid catabolism in Mycobacterium tuberculosis lysX mutant. Front Microbiol. 2011;2:19. doi:10.3389/fmicb.2011.00019.
- den Kamp JA, Redai I, van Deenen LL. Phospholipid composition of Bacillus subtilis. J Bacteriol. 1969;99:298–303.
- van Iterson W, den Kamp JA. Bacteria-shaped gymnoplasts (protoplasts) of Bacillus subtilis. J Bacteriol. 1969;99:304–15.
- Houtsmuller UM, Van D. On the Accumulation of Amino Acid Derivatives of Phosphatidylglycerol in Bacteria. Biochim Biophys Acta. 1964;84:96–8.
- Houtsmuller UM, van Deenen LL. On the amino acid esters of phosphatidyl glycerol from bacteria. Biochim Biophys Acta. 1965;106:564–76. doi:10.1016/0005-2760(65)90072-X.
- Kocun FJ. Amino acid containing phospholipids as major components of the phospholipids of Streptococcus faecalis 10C1. Biochim Biophys Acta. 1970;202:277–82. doi:10.1016/0005-2760(70)90189-X.
- Gould RM, Lennarz WJ. Metabolism of phosphatidylglycerol and lysyl-phosphatidylglycerol in Staphylococcus aureus. J Bacteriol. 1970;104:1135–44.
- Rubin HE, Nerad T, Vaughan F. Lactate acid inhibition of Salmonella typhimurium in yogurt. J Dairy Sci. 1982;65:197–203. doi:10.3168/jds.S0022-0302(82)82177-2.
- Hebeler BH, Chatterjee AN, Young FE. Regulation of the bacterial cell wall: effect of antibiotics on lipid biosynthesis. Antimicrob Agents Chemother. 1973;4:346–53. doi:10.1128/AAC.4.3.346.
- Rozgonyi F, Kiss J, Jekel P, Vaczi L. Effect of methicillin on the phospholipid content of methicillin sensitive Staphylococcus aureus. Acta Microbiol Acad Sci Hung. 1980;27:31–40.
- Rozgonyi F, Biacs P, Szitha K, Kiss J. Effect of methicillin on the fatty acid composition of phospholipids in methicillin sensitive Staphylococcus aureus. Acta Microbiol Acad Sci Hung. 1981;28:97–110.
- Yang SJ, Xiong YQ, Yeaman MR, Bayles KW, Abdelhady W, Bayer AS. Role of the LytSR two-component regulatory system in adaptation to cationic antimicrobial peptides in Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57:3875–82. doi:10.1128/AAC.00412-13.
- Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol. 2007;66:1136–47. doi:10.1111/j.1365-2958.2007.05986.x.
- Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A. 2007;104:9469–74. doi:10.1073/pnas.0702159104.
- Yang SJ, Bayer AS, Mishra NN, Meehl M, Ledala N, Yeaman MR, Xiong YQ, Cheung AL. The Staphylococcus aureus Two-Component Regulatory System, GraRS, Senses and Confers Resistance to Selected Cationic Antimicrobial Peptides. Infect Immun. 2012;80:74–81. doi:10.1128/IAI.05669-11.
- Hiron A, Falord M, Valle J, Debarbouille M, Msadek T. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol Microbiol. 2011;81:602–22. doi:10.1111/j.1365-2958.2011.07735.x.
- Cheung AL, Bayer AS, Yeaman MR, Xiong YQ, Waring AJ, Memmi G, Donegan N, Chaili S, Yang SJ. Site-specific mutation of the sensor kinase GraS in Staphylococcus aureus alters the adaptive response to distinct cationic antimicrobial peptides. Infect Immun. 2014;82:5336–45. doi:10.1128/IAI.02480-14.
- Falord M, Karimova G, Hiron A, Msadek T. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2012;56:1047–58. doi:10.1128/AAC.05054-11.
- Werth BJ, Sakoulas G, Rose WE, Pogliano J, Tewhey R, Rybak MJ. Ceftaroline increases membrane binding and enhances the activity of daptomycin against daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus in a pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother. 2013;57:66–73. doi:10.1128/AAC.01586-12.
- Berti AD, Baines SL, Howden BP, Sakoulas G, Nizet V, Proctor RA, Rose WE. Heterogeneity of genetic pathways toward daptomycin nonsusceptibility in Staphylococcus aureus determined by adjunctive antibiotics. Antimicrob Agents Chemother. 2015;59:2799–806. doi:10.1128/AAC.04990-14.
- Stermitz FR, Lorenz P, Tawara JN, Zenewicz LA, Lewis K. Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5′-methoxyhydnocarpin, a multidrug pump inhibitor. Proc Natl Acad Sci U S A. 2000;97:1433–7. doi:10.1073/pnas.030540597.
- Kourtesi C, Ball AR, Huang YY, Jachak SM, Vera DM, Khondkar P, Gibbons S, Hamblin MR, Tegos GP. Microbial efflux systems and inhibitors: approaches to drug discovery and the challenge of clinical implementation. Open Microbiol J. 2013;7:34–52. doi:10.2174/1874285801307010034.
- Lomovskaya O, Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic–a vision for applied use. Biochem Pharmacol. 2006;71:910–8. doi:10.1016/j.bcp.2005.12.008.
- Grube CD, Roy H. A Quantitative Spectrophotometric Assay to Monitor the tRNA-Dependent Pathway for Lipid Aminoacylation In Vitro. J Biomol Screen. 2016. doi:10.1177/1087057116642987.
- Rubio A, Conrad M, Haselbeck R, Kedar GC, Driver V, Finn J, et al. Regulation of mprF by antisense restores daptomycin susceptibility to daptomycin-resistant isolates of Staphylococcus aureus. Antimicrob Agents Chemother. 2010.
- Mishra A, Apeksha B, Koppolu P, Lingam SA. Role of antimicrobial peptides in periodontal innate defense mechanism. J Oral Res Rev. 2015;7:74–6. doi:10.4103/2249-4987.172500
- Hankins JV, Madsen JA, Giles DK, Brodbelt JS, Trent MS. Amino acid addition to Vibrio cholerae LPS establishes a link between surface remodeling in gram-positive and gram-negative bacteria. Proc Natl Acad Sci U S A. 2012;109:8722–7. doi:10.1073/pnas.1201313109.
- Henderson JC, Fage CD, Cannon JR, Brodbelt JS, Keatinge-Clay AT, Trent MS. Antimicrobial peptide resistance of Vibrio cholerae results from an LPS modification pathway related to nonribosomal peptide synthetases. ACS Chem Biol. 2014;9:2382–92. doi:10.1021/cb500438x.
- Olaitan AO, Morand S, Rolain JM. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol. 2014;5:643. doi:10.3389/fmicb.2014.00643.
